Abstract
Gastric cancer (GC) is a leading cause of cancer-related death, with poor prognosis due to metastasis. Despite improvements in early detection and treatment, effective therapies for metastatic GC are limited. Studies suggest that microRNAs play a key role in GC progression and metastasis. The expression of miR-5095, CEACAM5 and EMT-related proteins were determined by RT-qPCR and western blotting. The effects of miR-5095 on GC cell proliferation were assessed using CCK-8 and EdU assays, while the impact of miR-5095 on GC cell migration and invasion was evaluated using transwell assays. Bioinformatic tools were used to predict potential target genes of miR-5095, and the interaction between miR-5095 and CEACAM5 was confirmed through dual-luciferase reporter assays. Additionally, the expression of CEACAM5 was reversed by overexpressing plasmids to verify whether miR-5095 exerts its effects through CEACAM5. The in vivo effects of miR-5095 on tumor growth and metastasis were studied using a xenograft mouse model. miR-5095 expression was significantly reduced, and CEACAM5 expression was elevated in GC tissues and cell lines compared to adjacent normal tissues and cells. In vitro, miR-5095 overexpression notably inhibited CEACAM5 expression and suppressed GC cell proliferation, migration, invasion, and EMT in AGS and HGC-27 cells. Moreover, luciferase reporter assays confirmed that miR-5095 directly targets CEACAM5. The inhibitory effects of miR-5095 on GC cells were reversed by CEACAM5 overexpression. In vivo, miR-5095 significantly inhibits the proliferation and metastasis of gastric cancer.
Similar content being viewed by others
Introduction
Gastric cancer is a major global public health concern, with particularly high incidence and mortality rates in regions like East Asia. In China, it ranks among the most common malignancies, with 358,700 new cases reported in 2022, accounting for 7.44% of all new cancer cases. That year, 260,400 deaths from gastric cancer were recorded, representing 10.11% of cancer-related deaths1. Globally, there were 596,835 new cases, making up 4.9% of all new cancers, with 509,853 deaths, or 6.8% of cancer-related fatalities, positioning it as the fourth leading cause of cancer mortality2. Surgical treatment is the primary approach for early-stage gastric cancer and significantly improves survival rates. However, many patients are diagnosed at advanced stages, limiting the effectiveness of surgery. Therefore, other therapeutic modalities, including chemotherapy, radiotherapy, targeted therapy, and immunotherapy, are crucial for managing advanced cases3. Despite their importance, the use of these treatments is hindered by side effects, the heterogeneity of gastric cancer, and unclear metastasis mechanisms4,5,6. The evolution of cancer treatment—from broad cytotoxic strategies to molecularly targeted therapies—highlights the necessity of understanding tumor biology to develop more effective and specific interventions7. A better understanding of the molecular processes involved in gastric cancer initiation, progression, and metastasis, along with the development of novel therapeutic strategies, is vital to improving patient outcomes and advancing clinical diagnosis and treatment.
MicroRNAs (miRNAs) are fundamental post-transcriptional regulators, integral to the control of gene expression networks governing cell proliferation, differentiation, and apoptosis. Their dysregulation is a hallmark of cancer, where they can function as potent oncogenes or tumor suppressors8. In gastric cancer, the aberrant expression of specific miRNAs has been extensively linked to tumorigenesis, metastasis, and chemo-resistance. For instance, the oncogenic role of CEACAM5, a well-established biomarker in GC, is further underscored by its regulation via miRNA networks; it was recently demonstrated that miR-498 acts as a tumor suppressor by directly targeting and downregulating CEACAM5, thereby inhibiting GC cell proliferation, migration, and EMT in vitro and in vivo9. Conversely, miR-96-5p is excessively expressed in GC and promotes cancer cell proliferation by directly targeting and downregulating the tumor suppressor FOXO310. Emerging within this complex landscape is miR-5095, which has recently been implicated as a putative tumor suppressor in several malignancies. In non-small cell lung cancer (NSCLC), its tumor-suppressive function is antagonized by the oncogenic lncRNA LINC01296, which acts as a molecular sponge to sequester miR-5095, thereby facilitating proliferation and migration11. A parallel mechanism is observed in esophageal cancer, where circRAD23B upregulates oncogenes PARP2 and AKT2 by sponging miR-509512. This recurring theme of miR-5095 inactivation through ceRNA networks underscores its significant role in constraining oncogenic pathways; its loss appears to be a critical event in cancer progression across diverse tissue types.
Carcinoembryonic Antigen-Related Cell Adhesion Molecule 5 (CEACAM5) is a glycosylated cell surface protein and a member of the carcinoembryonic antigen (CEA) family, primarily involved in intercellular adhesion, signal transduction, and immune regulation13. CEACAM5 is highly expressed in various malignancies and plays a significant role in the initiation and progression of gastric cancer, making it a potential tumor marker and therapeutic target14,15. Clinically, the detection of CEACAM5 serves as a potential biomarker for the early diagnosis and monitoring of gastric cancer progression. Furthermore, the high expression of CEACAM5 is considered an independent prognostic factor for poor outcomes in gastric cancer patients, highlighting its importance in tumor biology16. Studies have shown that CEACAM5 expression in gastric cancer tissues is significantly higher than in normal gastric tissues, and its overexpression is closely associated with tumor invasiveness and metastatic potential9.
Epithelial-mesenchymal transition (EMT) is a crucial biological process associated with tumor invasiveness and metastasis, playing a key role in the development and progression of gastric cancer. Research indicates that the occurrence of EMT is closely linked to the invasiveness and metastatic ability of gastric cancer cells, directly influencing patient prognosis. During EMT, there is a decrease in the expression of the epithelial marker E-cadherin, while the expression of mesenchymal markers such as Vimentin increases. This transition enables cancer cells to acquire enhanced migratory capacity, thereby promoting tumor metastasis and recurrence17. Additionally, EMT is closely associated with immune cell infiltration in the tumor microenvironment18. Studies have found that EMT-related cytokines can promote the infiltration of tumor-associated macrophages and lymphocytes, thereby affecting tumor growth and metastasis19.
In this study, we demonstrated that miR-5095 significantly inhibits gastric cancer (GC) cell proliferation, migration, and invasion by directly targeting CEACAM5. These results were further validated in in vivo experiments. Additionally, our findings suggest that miR-5095 may serve as a potential prognostic biomarker and therapeutic target for GC, offering new insights into its role in GC progression and metastasis. This research lays the groundwork for future studies exploring the therapeutic potential of miR-5095 in GC treatment.
Materials and methods
Clinical samples
A total of 9 gastric cancer (GC) tissues and paired adjacent normal tissues were collected from GC patients who underwent surgical resection at the General Hospital of Ningxia Medical University. This study was approved by the Research Ethics Committee of the General Hospital of Ningxia Medical University and was performed in accordance with the ethical standards of the Declaration of Helsinki and relevant national and international guidelines. All patients provided written informed consent prior to inclusion in the study. No patient had received radiotherapy or chemotherapy before surgery.
Cell Lines, cell culture, and cell transfection
Human gastric cancer cell lines AGS, HGC-27, and the normal gastric cell line GES-1 were purchased from Procell (Wuhan, China). All cell lines were maintained in DMEM (Procell) supplemented with 10% fetal bovine serum (Procell), 1× GlutaMAX™ (Thermo Fisher Scientific), and 50 µg/ml gentamicin (Procell), in a humidified incubator at 37 °C with 5% CO₂. When cells reached approximately 90% confluence, they were passaged using Recombinant Trypsin EDTA Solution (Procell). For transfection, miR-5095 mimics, mimic-NC, CEACAM5 overexpression plasmid, and empty vector were synthesized by Hanbio Tech (Shanghai, China). Transfection was performed using LipoFiter 3.0 and RNAFit (Hanbio Tech) to introduce vectors or mimics into the cells for subsequent experiments.
CCK-8 assay
AGS and HGC-27 cells, transfected with either vector or mimics, were seeded in 96-well plates (1000 cells per well) with triplicates. After 72 h of incubation, 10 µL of CCK-8 solution (AR1160, Boster, China) was added to each well and incubated for 1–4 h. Absorbance was measured at 450 nm using a microplate reader to assess cell viability.
RNA isolation and RT-qPCR
Total RNA and miRNA were extracted using the Cell/Animal/Plant Tissue Total RNA Extraction Kit (Cat#: G3640-50T, Servicebis, China) and miRcute miRNA Isolation Kit (Cat#: DP501, TIANGEN, China), respectively. Reverse transcription was performed using the TransScript® II One-Step gDNA Removal and cDNA Synthesis SuperMix (Cat#: AH311, TRAN, China) or miRcute Plus miRNA First-Strand cDNA Kit (Cat#: KR211, TIANGEN, China). Quantitative PCR (qPCR) was conducted using the DyNAmo* Capillary SYBR* Green 2-Step qRT-PCR Kit (Thermo Fisher Scientific, USA). Relative expression levels of mRNA or miRNA were normalized to GAPDH or U6, and fold differences were calculated using the ΔΔCt method with respect to control groups. Primer sequences used in the study are as follows: miR-5095,forward 5’-TTACAGGCGTGAACCACCGCG-3’; CEACAM5,forward 5’-GCCTCAATAGGACCACAGTCAC-3’ and reverse 5’-CAGGTTAAGGCTACAGCATCCTC-3’.
Migration and invasion assays
For migration and invasion assays, 1 × 10⁵ cells in serum-free medium were seeded in the top chamber of a Transwell (Corning, New York, USA), with or without Matrigel coating, and complete medium was added to the bottom chamber. After 24 h of incubation, cells on the lower membrane were fixed with 4% formaldehyde and stained with 0.1% crystal violet. The number of migrated and invaded cells was counted under a microscope (Olympus, Japan) at 100× magnification in five random fields.
Western blot analysis
Total cellular protein was extracted using lysis buffer (Keygen BioTech) containing protease inhibitor cocktail (Thermo Fisher Scientific, USA), while nuclear protein was isolated using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China). Protein samples were separated by SDS-PAGE and transferred to PVDF membranes (Merck Millipore, USA). Membranes were incubated overnight at 4 °C with primary antibodies against CEACAM5 (1:1000, Proteintech), E-cadherin (1:1000, Proteintech), N-cadherin (1:1000, Proteintech), and Vimentin (1:1000, Proteintech). After washing, membranes were incubated with corresponding horseradish peroxidase-conjugated secondary antibodies (1:5000, Proteintech) for 1 h at room temperature. Protein bands were detected using enhanced chemiluminescence (ECL, 4 A BIOTECH).
Online database analysis
Differentially expressed genes (DEGs) in stomach adenocarcinoma (STAD) were obtained from the GEPIA2 database (http://gepia2.cancer-pku.cn/), accessed in December 2024. Gastric cancer-related genes were downloaded from GeneCards (https://www.genecards.org/), accessed in December 2024. Potential target genes regulated by miR-5095 were predicted using TargetScan (https://www.targetscan.org/), accessed in December 2024. Protein-protein interactions involving CEACAM5 were identified using STRING (https://cn.string-db.org/), and enrichment analysis was subsequently performed, with access in December 2024.
Luciferase reporter assay
To construct the luciferase reporter vector, the full-length 3’UTR of CEACAM5, including both wild-type and mutant variants, was synthesized by GenePharma (Shanghai, China) and cloned into the pGL3-basic vector (Genecreate, Wuhan, China). 293T cells were co-transfected with the plasmids and either hsa-miR-5095 mimics or miR-NC using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, luciferase activities of firefly and Renilla were measured using the Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA).
Xenograft tumor model in nude mice
The mice used in this study were strictly in accordance with the animal ethics and protection regulations of the Laboratory Animal Center of Ningxia Medical University. And all animal experiments were conducted in accordance with the ARRIVE guidelines and the NIH Guide for the Care and Use of Laboratory Animals. The euthanasia method was performed in compliance with the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Animals (2020). Four-week-old male BALB/c nude mice weighing 14–16 g were purchased from the Laboratory Animal Center of Ningxia Medical University and housed in SPF-class barrier facilities. After the cell transfection steps, logarithmic-phase HGC-27 cells were selected. The old medium was discarded, and the cells were washed once with PBS. Then, trypsin was added for 1–2 min to digest the cells, and the digestion was terminated with complete culture medium after the cells detached. The cell suspension was collected and centrifuged at 1000 rpm for 5 min. The supernatant was discarded, and the cells were washed twice with PBS to remove any residual culture medium. The cells were then resuspended in serum-free medium and the cell concentration was adjusted to 2 × 10⁶ cells/ml. The HGC-27 cell suspension was mixed with Matrigel at a 1:1 ratio and injected into the axillary fat pads of the mice (200 µL per mouse). Mice were housed in standard SPF-class animal rooms. After tumor formation, the activity of the mice was observed daily. On day 21, the mice were deeply anesthetized and then euthanized by cervical dislocation, after which the tumors were excised, weighed, and processed for subsequent experiments.
Gastric cancer lung metastasis model in nude mice
After the cell transfection steps, logarithmic-phase HGC-27 cells were selected. The original medium was discarded, and the cells were washed once with PBS. Trypsin was added for 1–2 min to digest the cells, and digestion was terminated with complete culture medium after the cells detached. The cell suspension was collected and centrifuged at 1000 rpm for 5 min. The supernatant was discarded, and the cells were washed twice with PBS to ensure the removal of any residual culture medium. The cells were then resuspended in serum-free medium, and the cell concentration was adjusted to 1 × 10⁶ cells/ml. The HGC-27 cell suspension (200 µL per mouse) was injected into the tail vein of nude mice. After the model was established, the activity of the mice was observed daily. On day 21, the mice were deeply anesthetized and euthanized by cervical dislocation, after which the lungs were excised for subsequent experimental processing.
H&E staining for tumor histopathology
Mouse tumor samples were fixed in 4% paraformaldehyde and embedded in paraffin. Tissue Sects. (4–6 μm) were deparaffinized, rehydrated, and stained with hematoxylin for 5–10 min, followed by eosin for 1–2 min. After dehydration and clearing, the sections were mounted and observed under a microscope.
Immunohistochemistry for Ki67 detection
Tumor sections were deparaffinized and rehydrated. After antigen retrieval and blocking with 5% BSA, the sections were incubated with Ki-67 primary antibody overnight at 4 °C. The next day, secondary antibodies were applied, and the sections were developed with DAB chromogen. Images were captured using a microscope.
Statistical analysis
All experiments were repeated at least three times. Data are presented as the mean ± standard error (SE) and were analyzed using GraphPad Prism 8.0.1 (GraphPad, La Jolla, CA, USA). Group comparisons were performed using t-tests and one-way ANOVA with post hoc correction. Statistical significance was defined as *P < 0.05, **P < 0.01.
Result
Expression of miR-5095 in gastric cancer tissues and cells
To assess the expression level of miR-5095 in gastric cancer tissues and cells, we performed RT-qPCR on nine pairs of gastric cancer tissues and their corresponding normal tissues. Additionally, we compared miR-5095 expression in the normal gastric cell line GES-1 and gastric cancer cell lines AGS and HGC-27. The results showed that miR-5095 was significantly downregulated in gastric cancer tissues compared to adjacent normal tissues, suggesting that miR-5095 may play an important role in gastric cancer initiation and progression (Fig. 1A). Similarly, miR-5095 expression was significantly lower in gastric cancer cell lines than in normal gastric cells, further supporting its potential suppressive role in gastric cancer (Fig. 1B). These findings suggest that the downregulation of miR-5095 might be associated with the malignant characteristics of gastric cancer, indicating its potential regulatory role in the biological behavior of the cancer.
Expression of miR-5095 in gastric cancer tissues and cells. (A) miR-5095 expression in gastric cancer tissues and matched adjacent normal tissues (n = 9 pairs) was quantified by RT-qPCR. Statistical analysis was performed using a two-tailed paired t-test (P = 0.0003). (B) miR-5095 expression in GES-1, AGS, and HGC-27 cells (n = 3 per group) was measured by RT-qPCR. Statistical analysis was performed using a two-tailed unpaired t-test: AGS vs. GES-1 (P = 0.0015), HGC-27 vs. GES-1 (P = 0.0005). *P < 0.05, **P < 0.01.
miR-5095 transfection efficiency and its effect on gastric cancer cell proliferation
To evaluate the transfection efficiency of miR-5095, we transfected AGS and HGC-27 gastric cancer cells with miR-5095 mimics and assessed miR-5095 expression using RT-qPCR. The results showed that miR-5095 expression was significantly increased in the miR-5095 mimic transfection group compared to the negative control (NC) mimic and control groups, confirming successful transfection (Fig. 2A). This validated the efficiency of the transfection method, providing a reliable foundation for subsequent functional experiments.
Next, we investigated the impact of miR-5095 overexpression on gastric cancer cell proliferation using a CCK-8 assay. Cells transfected with miR-5095 mimics exhibited a significant reduction in proliferation compared to the NC mimic group (Fig. 2B), suggesting that miR-5095 overexpression effectively inhibits gastric cancer cell proliferation. This was further supported by EdU assays, which showed a significant decrease in DNA synthesis in miR-5095-transfected cells (Fig. 2C).
miR-5095 transfection efficiency and its effect on gastric cancer cell proliferation. (A) Transfection efficiency of miR-5095 mimics in AGS and HGC-27 cells was evaluated by RT-qPCR (n = 3). Statistical analysis was performed using a two-tailed unpaired t-test: mimic NC vs. CON (AGS: P = 0.8518, HGC-27: P = 0.0764), mimics vs. mimic NC (AGS: P = 0.0003, HGC-27: P = 0.0002). (B) CCK-8 assay showing the change in cell viability in transfected AGS and HGC-27 cells (n = 3). Statistical analysis was performed using a two-tailed unpaired t-test: miR-5095 mimic vs. mimic NC (P < 0.0001 for both AGS and HGC-27). (C) EdU assay demonstrating the change in DNA synthesis capacity of gastric cancer cells (n = 3, scale bar = 100 μm). *P < 0.05, **P < 0.01.
Effect of miR-5095 on EMT, migration, and invasion of gastric cancer cells
To explore the effect of miR-5095 on the migration and invasion of gastric cancer cells, we assessed the expression of EMT-related proteins by Western blot. miR-5095 transfection led to an upregulation of E-cadherin and downregulation of N-cadherin and Vimentin expression, indicating that miR-5095 might inhibit the EMT process and thus reduce the migratory and invasive abilities of gastric cancer cells (Fig. 3A). Additionally, Transwell assays showed that miR-5095 significantly inhibited both migration (Fig. 3B) and invasion (Fig. 3C) of gastric cancer cells compared to the control groups. These results indicate that miR-5095 plays a significant role in suppressing migration and invasion in gastric cancer, and can regulate the expression of EMT-related proteins.
Effect of miR-5095 on EMT, migration, and Invasion of gastric cancer cells. (A) Western blot analysis of EMT-related proteins in AGS and HGC-27 gastric cancer cells transfected with miR-5095 mimics or mimic NC (n = 3). Statistical analysis was performed using multiple unpaired two-tailed t-tests. In AGS cells, E-cadherin (P = 0.000020), N-cadherin (P < 0.0001), and Vimentin (P < 0.0001); in HGC-27 cells, E-cadherin (P < 0.0001), N-cadherin (P < 0.0001), and Vimentin (P = 0.0001). Representative blots are shown from three independent experiments. (B) Transwell migration assay assessing the change in migration ability of gastric cancer cells (n = 3). (C) Transwell invasion assay assessing the change in invasion ability of gastric cancer cells (n = 3, scale bar = 200 μm). *P < 0.05, **P < 0.01.
Target gene screening for miR-5095
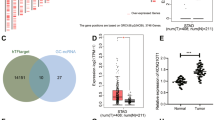
Bioinformatics analysis identified 24 potential target genes of miR-5095. From a dataset of 5021 genes predicted by TargetScan, 1660 candidate genes were selected based on a threshold score ≤ -0.2. Further filtering using GEPIA2 and GeneCards narrowed down the list to 1461 and 1651 genes, respectively. The intersection of these three datasets revealed 24 candidate genes (Fig. 4A), with the top five genes exhibiting the highest fold change being CEACAM5, FUT3, CDKN3, KLK10, and RAD51 (Fig. 4B). STRING database analysis revealed 26 proteins with high confidence interactions with CEACAM5, which were involved in key biological processes such as cell adhesion and negative regulation of cell death (Fig. 4C and D).
Target gene screening for miR-5095. (A) Venn diagram showing the intersection of genes from the GEPIA2, GeneCards, and TargetScan databases. (B) The 24 genes regulated by miR-5095 identified through screening. (C) Protein-protein interaction network for CEACAM5 from STRING database. (D) Enrichment analysis for the top 26 proteins interacting with CEACAM5.
Validation of CEACAM5 as a target gene of miR-5095
The GEPIA2 analysis confirmed that the expression of CEACAM5 in gastric cancer tissues was significantly higher than that in normal tissues (Fig. 5A). Dual-luciferase reporter assays demonstrated that the fluorescence ratio in the CEACAM5-WT + miR-5095 mimic group was significantly lower than that in the CEACAM5-WT + NC mimic group, while no significant change was observed in the CEACAM5-Mut + miR-5095 mimic group. This indicates that miR-5095 specifically binds to wild-type CEACAM5 (Fig. 5B). Western blot analysis revealed that CEACAM5 protein levels were significantly reduced in AGS and HGC-27 cells transfected with miR-5095 mimics (Fig. 5C), and RT-qPCR further confirmed this reduction at the mRNA level (Fig. 5D). These findings provide compelling evidence confirming that CEACAM5 is a direct target of miR-5095 and suggest that miR-5095 may suppress the proliferation, migration, and invasion of gastric cancer cells by targeting CEACAM5.
Validation of CEACAM5 as a target gene of miR-5095. (A) mRNA expression of CEACAM5 in tumor tissues (n = 408) and normal tissues (n = 211) from the GEPIA2 database. (B) Dual-luciferase reporter assay showing the binding sites and fluorescence ratio changes (ns: no significance). Statistical comparisons between groups were performed using unpaired two-tailed t-tests. CEACAM5-WT + miR-5095 mimic vs. CEACAM5-WT + NC mimic group (P < 0.0001). CEACAM5-Mut + miR-5095 mimic vs. CEACAM5-Mut + NC mimic group (P = 0.9598). (C) Western blotting assay detecting the expression of CEACAM5 in gastric cancer cells (n = 3). Statistical analysis was performed using unpaired two-tailed t-tests comparing miR-5095 mimics to mimic NC: AGS cells (P < 0.0001); HGC-27 cells (P = 0.0002). Representative image shown from three independent experiments. (D) RT-qPCR measurement of CEACAM5 expression in gastric cancer cells. Statistical analysis was performed using unpaired two-tailed t-tests comparing miR-5095 mimics to mimic NC: AGS cells (P < 0.0001); HGC-27 cells (P = 0.0001). *P < 0.05, **P < 0.01.
miR-5095 targets CEACAM5 to inhibit gastric cancer cell proliferation, migration, and invasion
To confirm that miR-5095 inhibits gastric cancer cell proliferation, migration, and invasion through CEACAM5 targeting, we overexpressed CEACAM5 in gastric cancer cells. Western blot analysis confirmed successful overexpression (Fig. 6A). The overexpression of CEACAM5 reversed the inhibitory effects of miR-5095 on cell proliferation (Fig. 6B), the EMT process (Fig. 6C), migration (Fig. 6D), and invasion (Fig. 6E). These results suggest that CEACAM5 promotes gastric cancer cell biological behaviors, while miR-5095 suppresses these behaviors by downregulating CEACAM5 expression, supporting miR-5095 as a potential therapeutic target.
miR-5095 targets CEACAM5 to inhibit gastric cancer cell proliferation, migration, and invasion. (A) Western blotting assay detecting CEACAM5 protein expression (n = 3). OE-NC + miR-5095 mimics vs. OE-NC + NC mimics group (AGS: P = 0.0009, HGC-27: P < 0.0001, unpaired two-tailed t-test). OE-CEACAM5 + miR-5095 mimics group vs. OE-NC + miR-5095 mimics group (AGS: P < 0.0001, HGC-27: P < 0.0001, unpaired two-tailed t-test). Representative image shown from three independent experiments. (B) CCK-8 assay showing the change in cell viability (n = 3). OE-NC + miR-5095 mimics vs. OE-NC + NC mimics group (AGS: P < 0.0001, HGC-27: P < 0.0001, unpaired two-tailed t-test). OE-CEACAM5 + miR-5095 mimics group vs. OE-NC + miR-5095 mimics group (AGS: P < 0.0013, HGC-27: P = 0.0008, unpaired two-tailed t-test). (C) Western blotting assay detecting the expression of EMT-related proteins (n = 3). Statistical analysis was performed using multiple unpaired two-tailed t-tests. OE-NC + miR-5095 mimics vs. OE-NC + NC mimics group: AGS: E-cadherin (P = 0.0003), N-cadherin (P < 0.0001), Vimentin (P = 0.0002); HGC-27: E-cadherin (P = 0.0006), N-cadherin (P = 0.0060), Vimentin (P = 0.0017). OE-CEACAM5 + miR-5095 mimics group vs. OE-NC + miR-5095 mimics group: AGS: E-cadherin (P = 0.0028), N-cadherin (P < 0.0001), Vimentin (P < 0.0001); HGC-27: E-cadherin (P = 0.0313), N-cadherin (P < 0.0091), Vimentin (P = 0.0046). Representative image shown from three independent experiments. Transwell migration (D) and invasion (E) assay assessing the migration and invasion ability of gastric cancer cells (n = 3, scale bar = 200 μm). *P < 0.05, **P < 0.01.
Effect of miR-5095 on tumor growth and metastasis in mouse models
To assess the effect of miR-5095 on tumor growth, we established a xenograft model by injecting NC mimic- and miR-5095 mimic-transfected HGC-27 cells into immunodeficient mice. Tumor growth was significantly inhibited in the miR-5095 mimic group, with reduced tumor size (Fig. 7A), volume (Fig. 7B), and weight (Fig. 7C) compared to the NC mimic group. Histological analysis using H&E staining revealed that tumors in the miR-5095 mimic group exhibited a more dispersed cell structure and enlarged intercellular spaces (Fig. 7D). Furthermore, immunohistochemistry showed a significant reduction in Ki-67-positive cells in the miR-5095 group (Fig. 7E), confirming the inhibitory effect of miR-5095 on tumor cell proliferation.
Additionally, to evaluate the impact of miR-5095 on metastasis, we established a tail vein lung metastasis model. H&E staining of lung tissue showed that the miR-5095 mimic group exhibited significantly fewer metastatic foci, suggesting that miR-5095 effectively inhibits the metastatic ability of gastric cancer cells (Fig. 7F). These findings provide further evidence for the potential of miR-5095 as an anti-tumor therapeutic target in clinical applications.
Effect of miR-5095 on tumor growth and metastasis in mouse models. (A) Representative images of tumors from xenograft mice (n = 5). (B) Changes in tumor volume.Statistical analysis was performed using a two-tailed paired t-test (P = 0.0006). (C) Changes in tumor weight. Statistical analysis was performed using a two-tailed paired t-test (P < 0.0001). (D) H&E staining to observe tissue morphology (scale bar = 100 μm). (E) Immunohistochemistry for Ki-67 expression (scale bar = 50 μm). Statistical analysis was performed using a two-tailed paired t-test (P = 0.0026). (F) H&E staining showing metastatic lesions in the lungs of mice (n = 3, scale bar = 200 μm). Statistical analysis was performed using a two-tailed paired t-test (P = 0.0008). *P < 0.05, **P < 0.01.
Discussion
Gastric cancer is a leading gastrointestinal malignancy with high incidence and mortality rates worldwide, and its metastatic characteristics significantly impact patient prognosis, making it a critical factor in clinical management20. Consequently, increasing attention is being focused on understanding cancer initiation and progression, with the goal of identifying new therapeutic strategies to reduce metastasis and discovering early diagnostic biomarkers for gastric cancer. In gastric cancer, mounting evidence indicates frequent alterations in epigenetic regulatory factors, which play crucial roles in its development21. MicroRNAs (miRNAs) are key epigenetic regulators that can induce target mRNA degradation or suppress translation based on the degree of complementarity with their target mRNAs. During tumorigenesis and progression, miRNAs exhibit dual roles: some miRNAs promote cancer development22, while others inhibit tumor progression23. Our study found that miR-5095 expression was significantly lower in gastric cancer tissues compared to adjacent normal tissues, and also differed markedly between gastric cancer cells and human gastric mucosal epithelial cells. These results suggest that miR-5095 could function as a potential tumor suppressor, regulating gastric cancer progression. Additionally, miR-5095 inhibited the proliferation and migration of gastric cancer cells, aligning with findings from other studies that suggest miR-5095 may play a suppressive role in gastric cancer. Previous studies have shown that miR-5095 is generally downregulated in cancers, and its upregulation has been found to inhibit tumor progression. For instance, overexpression of miR-5095 has been shown to effectively suppress the proliferation and migration of non-small cell lung cancer (NSCLC) cells11. Similarly, circRAD23B in esophageal cancer promotes aggressive tumor cell growth by acting as a sponge for miR-5095, enhancing the expression of PARP2 and AKT212. In a mouse model, we further validated the impact of miR-5095 on tumor growth and metastasis. The results demonstrated that overexpression of miR-5095 significantly inhibited tumor growth in xenograft models and reduced the occurrence of lung metastasis.
Epithelial-to-mesenchymal transition (EMT) is a key step in tumor metastasis, as it regulates cell adhesion molecules and reconfigures the cytoskeleton, enabling tumor cells to acquire invasive and migratory capabilities. EMT is characterized by the downregulation of epithelial markers (such as E-cadherin) and upregulation of mesenchymal markers (such as vimentin and fibronectin). This process is driven by core transcription factors, such as SNAIL, TWIST, and ZEB1/ZEB2, which directly suppress E-cadherin expression and activate mesenchymal genes, facilitating the detachment of tumor cells from the primary site and their entry into the circulation24. Our study, through Western blot analysis, found that overexpression of miR-5095 significantly reduced the expression of mesenchymal markers, N-cadherin and Vimentin, while upregulating the expression of the epithelial marker E-cadherin, indicating that miR-5095 may regulate the EMT process. This modulation of key EMT markers could be a significant mechanism through which miR-5095 inhibits tumor metastasis. Additionally, the observed changes in the expression of related EMT proteins further support the potential role of miR-5095 in inhibiting cell migration and invasion. By regulating the EMT process, miR-5095 may influence the metastatic potential of tumor cells, which is consistent with the mechanisms of other miRNAs in gastric cancer. For example, miR-223 regulates the EMT process in gastric cancer cells by targeting Sp1, thereby inhibiting cell migration and invasion25. Similarly, the miR-200 family maintains the epithelial phenotype by directly inhibiting ZEB1/ZEB2, thus suppressing EMT and metastasis26. Furthermore, changes in the exosomal miRNA profile during EMT, such as the upregulation of miR-23a, can transmit pro-metastatic signals to adjacent epithelial cells, creating a malignant feedback loop27. This miRNA-mediated intercellular communication suggests that miRNAs could serve as biomarkers and therapeutic targets for EMT-associated metastasis.
Our study employed a systematic bioinformatics approach to identify potential targets of miR-5095, a strategy that aligns with the network-based paradigm increasingly used to decipher complex oncogenic processes28,29,30,31. This integrated bioinformatics analysis identified 24 target genes of miR-5095, with CEACAM5 showing the highest fold change. We confirmed the direct interaction between miR-5095 and CEACAM5 through dual-luciferase reporter assays. The high expression of CEACAM5 in gastric cancer tissues is associated with tumor malignancy, and miR-5095 significantly downregulates CEACAM5 expression, providing theoretical support for its potential as a therapeutic target32. Functional enrichment analysis revealed that the candidate target gene CEACAM5, along with its interacting proteins, is involved in biological processes such as the negative regulation of cell death and cell adhesion, further supporting the promotive role of CEACAM5 in cancer and the potential application of miR-5095 in gastric cancer therapy. This finding is consistent with the role of CEACAM5 in tumor cells, where its overexpression is believed to promote the EMT process and enhance tumor cell invasiveness33. For example, in esophageal squamous cell carcinoma (ESCC), CEACAM5 is highly expressed in cases with lymph node metastasis and cooperates with the FOXA1/LOXL2 pathway to promote EMT, suggesting its potential as a poor prognostic marker34. However, in head and neck squamous cell carcinoma (HNSCC), high CEACAM5 expression is significantly associated with the absence of lymph node metastasis and better clinical prognosis. Mechanistically, CEACAM5 inhibits MDM2-mediated EMT progression, reducing tumor invasiveness and improving prognosis. These studies indicate that the biological function of CEACAM5 is tissue-specific, and its value as a prognostic marker or therapeutic target needs to be assessed in the context of specific cancer types. In HNSCC, CEACAM5 may act as a tumor suppressor, whereas in ESCC and other tumors, it may promote tumor progression. This duality may arise from the complexity of its regulatory network, including interactions with epigenetic regulators and the cross-talk between different signaling pathways. Future research should further investigate the molecular partners and downstream effectors of CEACAM5 in different tumor microenvironments.
Furthermore, the multi-step research strategy employed by the Jackson laboratory to investigate voltage-gated sodium channels (VGSCs) provides a valuable roadmap that could be applied to CEACAM5. Their approach began with a comprehensive pan-cancer analysis of VGSC expression and function35, which identified their broad potential in oncology. This was followed by focused studies on specific cancer types, such as glioma, where the β3 subunit (SCN3B) was validated as a biomarker36. Finally, deep mechanistic in vitro studies elucidated SCN3B’s channel-independent role in modulating cytoskeletal dynamics and cell motility37. Given that CEACAM5, like VGSCs, exhibits pan-cancer expression and context-dependent functions, a similar strategy is highly applicable. Future research should first execute a systematic pan-cancer analysis of CEACAM5 using platforms like TCGA and GTEx to prioritize cancer types where its role is most clinically significant. Subsequently, the biological functions and molecular mechanisms of CEACAM5—particularly its influence on immune regulation and metastasis through pathways such as EMT—should be rigorously investigated in these top-priority cancers using in vitro and in vivo models.
This structured strategy, moving from broad bioinformatic discovery to focused functional validation, would efficiently illuminate CEACAM5’s diverse roles across cancers. More importantly, the identification of such key molecular drivers robustly informs the development of targeted therapies. In this context, it is worthwhile to explore the translational potential of CEACAM5 and its regulators, like miR-5095, from the perspective of traditional medicine. Modern network pharmacology approaches, as demonstrated in the mapping of Hypericum perforatum’s active compounds to targets for major depressive disorder, provide a powerful blueprint for systematically linking herbal medicine components to oncological targets38. Compelling pre-clinical and clinical reports further substantiate that phytochemicals and complex herbal formulations can modulate tumor growth and therapy response39. Therefore, future efforts could leverage this bioactivity-guided framework to screen natural compounds capable of targeting the miR-5095/CEACAM5 axis, thereby harnessing the multi-target synergy of traditional medicine to develop novel therapeutic strategies for cancers dependent on this pathway.
A critical environmental factor in gastric cancer is Helicobacter pylori (HP) infection. Recent studies have confirmed that HP infection significantly enhances the migratory ability of gastric cancer cells and alters the expression of a gene network involved in migration, immunity, and drug sensitivity40. Although our study focuses on the miR-5095/CEACAM5 axis, it is intriguing to consider its potential interaction with HP infection. Future research could explore whether HP infection affects the expression of miR-5095 and CEACAM5 in gastric tumors. It is plausible that HP-induced inflammation and the resulting genetic alterations may modulate the levels of miR-5095 and CEACAM5, which in turn could influence immune cell infiltration in the tumor microenvironment and ultimately promote metastatic progression. Investigating the potential link between this key bacterial carcinogen and the miR-5095/CEACAM5 pathway could contribute to a more comprehensive understanding of the pathogenesis of gastric cancer and uncover novel therapeutic strategies targeting both host and microbial factors.
Despite the promising in vitro and in vivo findings supporting the role of miR-5095 in targeting CEACAM5 in gastric cancer, this study has several limitations. First, the in vitro experiments were limited to only two gastric cancer cell lines, which may not fully represent the heterogeneity of the disease. In vivo, we employed two common models: a subcutaneous xenograft model to assess tumor growth and a tail vein injection model to evaluate hematogenous lung metastasis. While these models were selected for their operational simplicity and high success rates, they represent a simplification of the complex metastatic process in gastric cancer. Our study does not encompass other critical metastatic routes, such as lymphatic dissemination or peritoneal carcinomatosis, which are highly prevalent in gastric cancer patients. Furthermore, all animal studies were conducted in immunodeficient mice, lacking validation in immunocompetent or genetically engineered/chemically induced models that better recapitulate the tumor microenvironment and immune interactions. Second, the function of CEACAM5 appears to be tissue-specific, as previously reported, and the current results derived from gastric cancer models may not be generalizable to other cancer types or biological contexts. Moreover, the study did not explore the upstream regulators of miR-5095 or construct a complete lncRNA–miRNA–mRNA regulatory axis, nor did it investigate other potential targets and downstream pathways of miR-5095 in detail. Clinical relevance is also limited by the absence of patient survival data, serum miR-5095 levels, or treatment response correlations. Lastly, the relatively small sample size may not fully capture inter-individual heterogeneity. Future studies with larger cohorts, more comprehensive models (including orthotopic, lymphatic metastasis, and immunocompetent models), and clinical data are needed to validate these findings and better evaluate the therapeutic potential of miR-5095 in gastric cancer.
Conclusion
In conclusion, CEACAM5, as a target gene of miR-5095, may play a key role in the onset and progression of gastric cancer by regulating the proliferation, migration, and metastatic potential of tumor cells. This study demonstrates the targeting relationship between miR-5095 and CEACAM5 in vitro and in vivo, suggesting that miR-5095 could be a promising candidate for further investigation. However, clinical data, including patient survival, serum miR levels, and treatment responses, are lacking, and future studies are needed to validate its potential as a prognostic biomarker and therapeutic target in gastric cancer.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Han, B. et al. Cancer incidence and mortality in China, 2022. J. Natl. Cancer Cent. 4, 47–53 (2024).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Wang, F. H. et al. The Chinese society of clinical oncology (CSCO): clinical guidelines for the diagnosis and treatment of gastric cancer. Cancer Commun. (Lond.) 44 127–172 (2024).
Pihlak, R., Fong, C. & Starling, N. Targeted therapies and developing precision medicine in gastric cancer. Cancers (Basel), 15 (2023).
Ratti, M. et al. Emerging therapeutic targets and future directions in advanced gastric cancer: A comprehensive review. Cancers (Basel), 16 (2024).
Guan, W. L., He, Y. & Xu, R. H. Gastric cancer treatment: recent progress and future perspectives. J. Hematol. Oncol. 16, 57 (2023).
Sonkin, D., Thomas, A. & Teicher, B. A. Cancer treatments: Past, present, and future. Cancer Genet. 286-287, 18–24 (2024).
Smolarz, B., Durczynski, A., Romanowicz, H. & Szyllo, K. P. Hogendorf, MiRNAs in cancer (Review of Literature). Int. J. Mol. Sci. 23 (2022).
Zhang, L., Zhang, C. & Liu, N. CEACAM5 targeted by miR-498 promotes cell proliferation, migration and epithelial to mesenchymal transition in gastric cancer. Transl Oncol. 24, 101491 (2022).
He, X. & Zou, K. MiRNA-96-5p contributed to the proliferation of gastric cancer cells by targeting FOXO3. J. Biochem. 167, 101–108 (2020).
Hu, X., Duan, L., Liu, H. & Zhang, L. Long noncoding RNA LINC01296 induces non-small cell lung cancer growth and progression through sponging miR-5095. Am. J. Transl Res. 11, 895–903 (2019).
Lan, X., Liu, X., Sun, J., Yuan, Q. & Li, J. CircRAD23B facilitates proliferation and invasion of esophageal cancer cells by sponging miR-5095. Biochem. Biophys. Res. Commun. 516, 357–364 (2019).
Azari, F. et al. Singhal, carcinoembryonic Antigen-Related cell adhesion molecule type 5 Receptor-Targeted fluorescent intraoperative molecular imaging tracer for lung cancer: A nonrandomized controlled trial. JAMA Netw. Open. 6, e2252885 (2023).
Kawakami, H. New therapeutic target molecules for gastric and gastroesophageal junction cancer. Int. J. Clin. Oncol. 29, 1228–1236 (2024).
Zhu, X. Y. et al. A novel human single-domain antibody-drug conjugate targeting CEACAM5 exhibits potent in vitro and in vivo antitumor activity. Acta Pharmacol. Sin. 45, 609–618 (2024).
Zhou, J. et al. Identification of CEACAM5 as a biomarker for prewarning and prognosis in gastric cancer. J. Histochem. Cytochem. 63, 922–930 (2015).
Jamal Eddin, T. M., Nasr, S. M. O., Gupta, I. & Zayed, H. Al Moustafa, Helicobacter pylori and epithelial mesenchymal transition in human gastric cancers: an update of the literature. Heliyon 9, e18945 (2023).
Li, H. et al. High expression of vinculin predicts poor prognosis and distant metastasis and associates with influencing tumor-associated NK cell infiltration and epithelial-mesenchymal transition in gastric cancer. Aging (Albany NY). 13, 5197–5225 (2021).
Guo, R. & Yang, B. Hypoxia-Induced LXRalpha contributes to the migration and invasion of gastric cancer cells. Folia Biol. (Praha). 67, 91–101 (2021).
Lin, Z. et al. Prediction of distant metastasis and survival prediction of gastric cancer patients with metastasis to the liver, lung, bone, and brain: research based on the SEER database. Ann. Transl Med. 10, 16 (2022).
Fattahi, S., Amjadi-Moheb, F., Tabaripour, R., Ashrafi, G. H. & Akhavan-Niaki, H. PI3K/AKT/mTOR signaling in gastric cancer: epigenetics and beyond. Life Sci. 262, 118513 (2020).
Benacka, R., Szaboova, D., Gulasova, Z., Hertelyova, Z. & Radonak, J. Classic and new markers in diagnostics and classification of breast cancer. Cancers (Basel) 14 (2022).
Pan, W., Chai, B., Li, L. & Lu, Z. Ma, p53/MicroRNA-34 axis in cancer and beyond. Heliyon 9, e15155 (2023).
Wang, S., Sun, Z., Lei, Z. & Zhang, H. T. RNA-binding proteins and cancer metastasis. Semin Cancer Biol. 86, 748–768 (2022).
Hu, J. et al. miRNA-223 inhibits epithelial-mesenchymal transition in gastric carcinoma cells via Sp1. Int. J. Oncol. 49, 325–335 (2016).
Korpal, M. & Kang, Y. The emerging role of miR-200 family of MicroRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 5, 115–119 (2008).
Tang, Y. T. et al. Alterations in Exosomal MiRNA profile upon epithelial-mesenchymal transition in human lung cancer cell lines. BMC Genom. 19, 802 (2018).
Liu, H. Pan-cancer profiles of the Cuproptosis gene set. Am. J. Cancer Res. 12, 4074–4081 (2022).
Liu, H. & Tang, T. Pan-cancer genetic analysis of Cuproptosis and copper metabolism-related gene set. Front. Oncol. 12, 952290 (2022).
Liu, H. & Tang, T. Pan-cancer genetic analysis of disulfidptosis-related gene set. Cancer Genet. 278-279, 91–103 (2023).
Rasteh, A. M., Liu, H. & Wang, P. Pan-cancer genetic profiles of mitotic DNA integrity checkpoint protein kinases. Cancer Biomark. 41, CBM240119 (2024).
Beauchemin, N. & Arabzadeh, A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 32, 643–671 (2013).
Wu, Z., Zeng, X., Wang, H. & Wang, X. LncRNA ARAP1-AS1 contributes to lung adenocarcinoma development by targeting miR-8068 to upregulate CEACAM5. Cancer Biomark. 38, 177–189 (2023).
Sano, M. et al. Forkhead box A1 transcriptional pathway in KRT7-expressing esophageal squamous cell carcinomas with extensive lymph node metastasis. Int. J. Oncol. 36, 321–330 (2010).
Liu, H., Weng, J., Huang, C. L. & Jackson, A. P. Voltage-gated sodium channels in cancers. Biomark. Res. 12, 70 (2024).
Liu, H., Weng, J., Huang, C. L. & Jackson, A. P. Is the voltage-gated sodium channel beta3 subunit (SCN3B) a biomarker for glioma? Funct. Integr. Genomics. 24, 162 (2024).
Liu, H. et al. The voltage-gated sodium channel beta3 subunit modulates C6 glioma cell motility independently of channel activity. Biochim. Biophys. Acta Mol. Basis Dis. 1871, 167844 (2025).
Xu, Z., Rasteh, A. M., Dong, A., Wang, P. & Liu, H. Identification of molecular targets of hypericum perforatum in blood for major depressive disorder: a machine-learning Pharmacological study. Chin. Med. 19, 141 (2024).
Liu, H. R. Harnessing traditional medicine and biomarker-driven approaches to counteract trichostatin A-induced esophageal cancer progression. World J. Gastroenterol. 31, 106443 (2025).
Ou, L. et al. Helicobacter pylori infection facilitates cell migration and potentially impact clinical outcomes in gastric cancer. Heliyon 10, e37046 (2024).
Funding
This work was supported by grants from the Ningxia Natural Science Foundation Project (2022AAC03569, 2022AAC03573).
Author information
Authors and Affiliations
Contributions
W.L. and C.Z. contributed the original draft preparation. H.S. and Y.Z. provides the conceptualization. W.L., C.Z., W.F. and F.H. contributed to the interpretation of the data. H.S. and Y.Z. reviewed and edited the article. X.Z., Y.M. and H.Q. provided the methodology. W.L. and W.F. provided funding for the project. C.Z. supervised work and wrote manuscripts. All the authors have contributed to the paper and have reviewed the manuscript and agree with its contents.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethic statements
The animal study protocol conducted in accordance with the ARRIVE guidelines and approved by the Laboratory Animal Ethics and Welfare Committee of the Laboratory Animal Center of Ningxia Medical University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, W., Cai, R., Fan, W. et al. MiR-5095 inhibits proliferation, migration, and invasion of gastric cancer cells by targeting CEACAM5. Sci Rep 15, 39950 (2025). https://doi.org/10.1038/s41598-025-23669-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23669-6