Abstract
Olanzapine, a second-generation antipsychotic, is commonly used to manage agitation in patients with schizophrenia and bipolar disorder, though its underlying mechanism of action remains unclear. In this study, network pharmacology and molecular docking were applied to explore the potential molecular mechanisms of olanzapine. Targets related to olanzapine were retrieved from GeneCards, SwissTargetPrediction, and the Comparative Toxicogenomics Database (CTD), while disease-associated targets were collected from DisGeNET and GeneCards. A protein–protein interaction (PPI) network was constructed to identify key targets. Functional enrichment analyses using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and Bioinformatics platforms indicated involvement in cell proliferation, apoptosis, and inflammation. The core targets identified included brain-derived neurotrophic factor (BDNF), insulin (INS), AKT serine/threonine kinase 1 (AKT1), tumor protein p53 (TP53), insulin-like growth factor 1 (IGF1), nerve growth factor (NGF), nuclear factor kappa B subunit 1 (NFKB1), and fibroblast growth factor 2 (FGF2). These targets are primarily involved in the phosphoinositide 3-kinase (PI3K)-AKT and mitogen-activated protein kinase (MAPK) signaling pathways. Molecular docking demonstrated strong binding affinities between olanzapine and these targets, with docking scores ranging from − 5.3 to − 8.8 kilocalories per mole (kcal/mol). These findings suggest that olanzapine, as an antipsychotic, may alleviate acute agitation symptoms by modulating signaling pathways associated with neuroinflammation and neuroplasticity, providing a basis for further research into its mechanism of action in neuropsychiatric disorders.
Similar content being viewed by others
Introduction
Schizophrenia and bipolar disorder are major psychiatric conditions, typically characterized by core features such as psychotic manifestations (hallucinations, delusions, disorganized thought processes) and recurrent mood disturbances including mania, hypomania, and depression1,2,3. These fundamental symptoms lead to marked impairments in social and occupational functioning, heavy burdens for caregivers, and reduced quality of life4,5. In addition to these defining domains, patients often present with other clinically significant manifestations, among which agitation is common during acute episodes and poses considerable challenges for clinical management.
Agitation is a multifactorial syndrome characterized by restlessness, irritability, uncooperativeness, anxiety, and excessive movement or speech6,7. Agitation is a common and urgent issue in psychiatry, occurring not only in emergency situations but also in inpatient and outpatient psychiatric care settings. Symptoms of agitation range from mild (such as restlessness and anxiety) to severe (including violence and aggression) with rapid fluctuations8,9,10. Acute agitation is relatively common in patients with schizophrenia6,11 and bipolar disorder6,12. In a European study, the prevalence of agitation in psychiatric emergencies was 4.6%; among the affected patients, half were with schizophrenia and a quarter with bipolar disorder13. In an investigation of 1000 violent incidents, approximately 29% were associated with patients experiencing agitation14. It is crucial to promptly manage agitation and prevent aggression to reduce the risk of injuries to patients, caregivers, and medical staff9,15. A recent expert consensus on the assessment and management of agitation in psychiatry recommended verbal de-escalation and environmental changes as first-line options, followed by pharmacological interventions, with physical restraint reserved as a last resort16.
Olanzapine is a second-generation antipsychotic classified as a thienobenzodiazepine. Olanzapine has an affinity for dopamine receptors D1, D2, D3, and D4, as well as for serotonin receptors such as 5-hydroxytryptamine receptors 2 A (HTR2A), 2 C (HTR2C), 3 (HTR3), and 6 (HTR6); histamine H1; α1-adrenergic; and multiple muscarinic receptors17,18,19. Compared with first-generation antipsychotics, olanzapine is associated with a decreased risk of acute extrapyramidal side effects and cardiovascular events, but an increased risk of metabolic side effects (such as increased weight, hyperglycemia, and hyperlipemia)20,21. Olanzapine was first approved by the Food and Drug Administration (FDA) for schizophrenia in 1996. The FDA-approved indications have since expanded to include the treatment of bipolar disorder22, acute agitation of schizophrenia and bipolar disorder20,23, and in combination with fluoxetine, the treatment of bipolar disorder type 1 depression and treatment-resistant depression24,25. Olanzapine, the best-studied second-generation antipsychotic for psychiatric agitation16,17,22,23,26, is currently accessible in the form of an oral tablet, orally disintegrating tablet, and intramuscular and intravenous formulations20,26. Numerous studies have shown that intramuscular olanzapine is a prompt, efficient, and safe therapy for acute agitation in schizophrenia and bipolar disorder17,27,28,29. Nevertheless, the mechanism of olanzapine’s effects on agitation in schizophrenia and bipolar disorder is unclear.
Network pharmacology is an emerging subject based on the theories of system biology, pharmacology, network biology, and computer analysis technology to explore the complex relationships between drugs, diseases, and targets, as well as to guide the discovery of new drugs30,31,32. The basic idea of network pharmacology is to effectively use information from various databases, based on “drug-pathway-target-disease” networks, to scientifically explore the mechanisms of action of drugs in treating diseases. Molecular docking is mainly used to predict binding modes and affinities between ligands and receptors33. Network pharmacology and molecular docking can complement each other in investigating the molecular mechanisms of drugs in disease therapy.
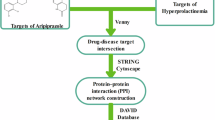
In the present study, we used network pharmacology to predict potential targets of olanzapine, schizophrenia, bipolar disorder, and agitation, and signaling pathways of olanzapine against agitation in schizophrenia and bipolar disorder. The mechanism of olanzapine against agitation in schizophrenia and bipolar disorder was explored. The workflow is shown in Fig. 1.
Methods
Target identification of olanzapine
The chemical structure of olanzapine was obtained from the DrugBank database34 (https://go.drugbank.com). The corresponding targets of olanzapine were predicted and identified using several target prediction databases, including GeneCards35 (https://www.genecards.org/), SwissTargetPrediction36 (http://www.swisstargetprediction.ch/), and the Comparative Toxicogenomics Database (CTD)37 (http://ctdbase.org/). Known validated pharmacological targets of olanzapine were retrieved from the DrugBank database. GeneCards offers information on all annotated and predicted human genes. The SwissTargetPrediction database provides target predictions based on a combination of two-dimensional (2D) and three-dimensional (3D) chemical structure similarities. The CTD provides manually curated information about the relationships among chemicals, genes, and diseases to help characterize disease mechanisms.
Target genes for schizophrenia, bipolar disorder, and agitation
Disease-related target genes were identified from the DisGeNET38 (https://www.disgenet.org/) and GeneCards databases using the keywords “schizophrenia” and “bipolar disorder.” Symptom-related target genes were obtained from the same databases using the keyword “agitation.” DisGeNET contains a large number of genes and variants associated with human diseases. The median value was set as the cut-off value. Disease target genes were obtained after eliminating repeated targets.
Drug-disease target interactions
The overlapping targets among the olanzapine-related targets and the predicted schizophrenia targets, predicted bipolar disorder targets, and predicted agitation targets were obtained and visualized using Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny).
Protein-protein interaction (PPI) network construction and core target identification
The interaction targets of the four datasets were imported into the String database39 (https://string-db.org). The species was limited to “homo sapiens,” the minimum interaction confidence score was set to ≥ 0.4, and the disconnected nodes were hidden. Cytoscape 3.10.0 software (https://www.cytoscape.org/) was used to construct and perform a systematic analysis of the PPI network40. The CytoHubba plug-in was used to screen the top 20 hub genes ranked by the maximal clique centrality (MCC) method41.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of core targets
The Database for Annotation, Visualization, and Integrated Discovery (DAVID)42 (https://david.ncifcrf.gov/), a functional enrichment analysis tool, was used to perform GO analysis and KEGG43,44 pathway analysis on the core targets screened from the PPI network. The top 10 enriched KEGG pathways that met the criteria of P < 0.05 and FDR < 0.05 were selected for presentation. The GO enrichment analysis contains three terms: biological process (BP), cellular component (CC), and molecular function (MF). The bioinformatics platform (http://www.bioinformatics.com.cn/) was used to visualize the results.
Drug-pathway-target-disease network construction
To visualize and illustrate the complex relationships between olanzapine, pathways, targets, and diseases, a drug-pathway-target-disease network was constructed and analyzed in Cytoscape 3.10.0.
Molecular Docking
Olanzapine was docked with eight proteins selected from the core targets using AutoDockTools 1.5.6 and AutoDock Vina45. Briefly, docking was performed as follows. First, ligand preparation was conducted; the olanzapine structure file (in. sdf format) was downloaded from the PubChem database46 (https://pubchem.ncbi.nlm.nih.gov/), saved as a. pdb file using ChemBio3D Ultra 14.0, and then calculated with AutoDockTools 1.5.6 software, and saved as a pdbqt file. Second, the proteins were prepared; the crystal structures of core targets were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB)47 (http://www.rcsb.org) and imported into PyMOL 2.4.1 to remove water and original ligands. The receptor proteins were prepared by adding polar hydrogens and appropriate charges, as well as by assigning atom types using AutoDockTools 1.5.6. The third step was grid preparation, in which the grid size and coordinates were adjusted to contain the binding sites predicted by DoGSiteScorer (https://proteins.plus/). The fourth step was molecular docking, which was performed using AutoDock Vina. In the final step, PyMOL 2.4.1 and Discovery Studio 2021 were used to visualize the interactions between olanzapine and core proteins.
Table 1 summarizes the tools and databases utilized in our study, providing a comprehensive overview of the methods and resources applied.
Results
Targets of olanzapine against agitation in schizophrenia and bipolar disorder
Screening of GeneCards, SwissTargetPrediction, and the CTD yielded 348, 109, and 148 olanzapine targets, respectively. Known validated pharmacological targets of olanzapine are listed in Supplementary Table S1. Screening of the DisGeNET and GeneCards databases yielded 1622 and 1891 schizophrenia target genes, respectively; 1183 and 2106 bipolar disorder target genes, respectively; and 109 and 1984 agitation target genes, respectively. After eliminating duplicate targets, our sample identified a total of 517 targets for olanzapine, 2582 targets for schizophrenia, 2796 targets for bipolar disorder, and 1996 targets for agitation. The interaction targets of olanzapine, schizophrenia, bipolar disorder, and agitation included 185 common targets. These targets were screened as potential targets of olanzapine against agitation in schizophrenia and bipolar disorder (Fig. 2a).
Protein-protein interaction (PPI) network of common target proteins of olanzapine, schizophrenia, bipolar disorder, and agitation. (a) Venn diagram of intersecting targets. (b) The original PPI network was obtained from the STRING database, consisting of 181 nodes and 3004 edges. (c) The PPI network was visualized using Cytoscape software. Node colors range from yellow to red, representing increasing node degree (i.e., the number of connections). (d) The top 20 hub targets in the PPI network were identified using the maximal clique centrality (MCC) method via the CytoHubba plugin in Cytoscape. Node colors represent MCC scores, ranging from yellow (low) to red (high).
Construction and analysis of a PPI network to screen core targets
The 185 common targets of olanzapine against agitation in schizophrenia and bipolar disorder were imported into the String database to construct the original PPI network (Fig. 2b). To further analyze and visualize target interactions, the initial PPI data were imported into Cytoscape 3.10.0 to construct a new PPI network (Fig. 2c). The PPI network included 181 nodes and 3004 edges (Fig. 2c). In the network, node colors from yellow to red are proportional to the target’s degree. The top 20 hub targets ranked by the MCC method were screened by the CytoHubba plug-in (Fig. 2d).
GO enrichment analysis
To further explore the mechanism of olanzapine against agitation in schizophrenia and bipolar disorder, we performed a GO enrichment analysis of the top 20 hub targets. A total of 264 GO enriched items with P < 0.05 were identified, including 215 BP, 17 CC, and 32 MF. The top 10 items of BP, CC, and MF were screened as biological functions of potential hub targets (Fig. 3a–c). The four most highly enriched BP terms mainly involved positive regulation of gene expression; positive regulation of transcription, DNA-templated; negative regulation of apoptotic process; and positive regulation of transcription from RNA polymerase II promoter. Highly enriched CC terms were extracellular region, extracellular space, and endoplasmic reticulum lumen. Moreover, the MF terms included protease binding, identical protein binding, and growth factor activity.
KEGG pathway analysis and drug-pathway-target-disease network construction
To explore potential pathways of olanzapine against agitation in schizophrenia and bipolar disorder, we performed a KEGG pathway enrichment analysis of the top 20 hub targets. A total of 114 KEGG pathways were identified based on P < 0.05. The top 10 highly enriched pathways were processed for further analyses (Table 2; Fig. 3d). Furthermore, the pathways with the top three highest gene numbers were the PI3K-Akt signaling pathway, pathways in cancer, and the MAPK signaling pathway. Among them, the PI3K-Akt and MAPK signaling pathways were enriched with the top three highest number of core genes, which could be the most important potential pathways (Fig. 3d). The common core targets in the PI3K-Akt and MAPK signaling pathways were brain-derived neurotrophic factor (BDNF), insulin (INS), AKT serine/threonine kinase 1 (AKT1), tumor protein p53 (TP53), insulin-like growth factor 1 (IGF1), nerve growth factor (NGF), nuclear factor kappa B subunit 1 (NFKB1), and fibroblast growth factor 2 (FGF2).
To further explore the complicated mechanism of olanzapine’s effect against agitation in schizophrenia and bipolar disorder, a drug-pathway-target-disease network was constructed using Cytoscape 3.10.0. The drug–pathway–target–disease network diagram included 67 nodes (one drug, 10 pathways, 53 targets, and three disease nodes) and 269 edges (Fig. 4a). To illustrate the positions of core targets in the key pathways, we constructed PI3K-Akt and MAPK signaling pathway diagrams (Fig. 4b-c).
Drug-pathway-target-disease network diagram and the distribution of core targets in the most related pathways. (a) Drug-pathway-target-disease network diagram illustrating the relationships among olanzapine, pathways, targets, and diseases to explain olanzapine’s effects on agitation in schizophrenia and bipolar disorder. Orange inverted triangles represent the drug olanzapine; diamonds represent the top 10 signaling pathways; blue circles indicate common targets enriched in these pathways; red diamonds and triangles denote core pathways and core targets, respectively; pink hexagons represent diseases; and yellow rounded rectangles represent the agitation symptoms. (b–c) Distribution of the eight common core targets in the PI3K/AKT and MAPK signaling pathways. The eight common core targets are BDNF, INS, AKT1, TP53, IGF1, NGF, NFKB1, and FGF2. Red symbols indicate enriched common targets; red ovals and arrows show the positions of the core targets within the pathways. (b) PI3K/AKT signaling pathway. (c) MAPK signaling pathway.
Molecular Docking verification
Since the drug targets of olanzapine were mainly enriched in the PI3K-Akt and MAPK signaling pathways, a network pharmacological analysis showed that eight core proteins were common in these two pathways, namely BDNF (PDB ID: 5UGG), INS (PDB ID: 4AJX), AKT1 (PDB ID: 3CQU), TP53 (PDB ID: 5O1D), IGF1 (PDB ID: 1IMX), NGF (PDB ID: 5JFW), NFKB1 (PDB ID: 7LFC), and FGF2 (PDB ID: 1BFF). We selected these eight targets as potential core targets. Details of the core targets are shown in Table 3. Discovery Studio 2021 and PyMOL 2.4.1 were employed to illustrate the 2D and 3D binding interactions between olanzapine and potential core proteins (Fig. 5). Docking scores ranged from − 5.3 to − 8.8 kcal/mol between olanzapine and core targets (Table 4), suggesting stable binding. Olanzapine had the highest binding affinity with NGF (Fig. 5g). The binding interactions involved a hydrogen bond with GLU-518 and hydrophobic interactions with LEU-516, VAL-524, PHE-589, PHE-669, LEU-657, and ALA-542 (Fig. 5g). Binding to AKT1 involved a hydrogen bond with ARG-243 and hydrophobic interactions with ARG-2, ARG-243, ARG-346, and TYR-350 (Fig. 5a). For BDNF, the binding was mediated by hydrogen bonds with HIS-603 and CYS-604, an electrostatic interaction with LYS-607, and hydrophobic interactions with LYS-607 and PHE-587 (Fig. 5b). Binding to FGF2 included a hydrogen bond with TYR-123, electrostatic interactions with ARG-105 and GLU-107, and a hydrophobic interaction with PRO-104 (Fig. 5c). In the case of IGF1, olanzapine formed hydrogen bonds with ASP-53 and GLU-46, electrostatic interactions with ARG-56 and GLU-46, and a hydrophobic interaction with ARG-56 (Fig. 5d). Binding to INS involved a hydrogen bond with ASN-3, hydrophobic interactions with ASN-3, VAL-2, and LEU-6, and an additional interaction with CYS-7 (Fig. 5e). For NFKB1, olanzapine engaged in hydrophobic interactions with ALA-363, LEU-328, and ALA-367 (Fig. 5f). Binding to TP53 was characterized by hydrogen bonds with ASP-186 and SER-166, an electrostatic interaction with GLU-198, a hydrophobic interaction with LYS-101, and an additional interaction with MET-237 (Fig. 5h).
Discussion
The present study integrated the information on olanzapine, targets, pathways, and diseases from numerous databases and constructed a drug–pathway–target–disease network. Based on network pharmacology and molecular docking analyses, the potential mechanism of reducing agitation in schizophrenia and bipolar disorder by olanzapine was explored.
We screened 185 common targets among olanzapine, schizophrenia, bipolar disorder, and agitation. These targets were considered potential targets of olanzapine in treating agitation in schizophrenia and bipolar disorder. To screen core targets, a PPI network was constructed, and the top 20 hub targets were screened by the MCC method. Based on the results of the PPI network and KEGG pathway analyses, eight core targets were obtained, namely, BDNF, INS, AKT1, TP53, IGF1, NGF, NFKB1, and FGF2.
Among the eight core targets, BDNF, INS, IGF1, NGF, and FGF2, belong to neurotrophic/growth factor systems, which may play a key role in neuroplasticity. Increased levels of BDNF and NGF in the cerebrospinal fluid of children with Epstein-Barr virus meningoencephalitis were significantly associated with psychomotor agitation48. In an olfactory bulbectomy mouse model, high-intensity repetitive transcranial magnetic stimulation reduced psychomotor agitation and increased BDNF and neurogenesis levels49. Olanzapine can have a direct effect on pancreatic tissue and induce insulin resistance50. Compared with no-stressed mice, the hippocampal levels of trophic factors, including BDNF, glial cell line-derived neurotrophic factor (GDNF), and IGF1, were increased in mice with chronic mild stress51. In addition, the serum levels of IGF1, NGF, and FGF2 were significantly increased in manic episodes of bipolar disorder52. Olanzapine increased NGF in the hippocampus, occipital cortex, and hypothalamus of Wistar rats53. Aggressive behavior in mice caused by 6–8 weeks of isolation led to a rise in NGF levels in the hypothalamus54. These studies demonstrated increased levels of neurotrophic/growth factors in the hippocampus and hypothalamus under conditions of psychological and environmental stress. A literature review found that damage to the frontal lobe, hypothalamus, and hippocampus can result in reduced self-control, impulsiveness, inability to regulate emotions, and an unregulated release of hormones, leading to an increased sympathetic response55.
Dopamine-induced activation of the D2 receptors inhibited AKT1 signaling via dephosphorylation mediated by the β-arrestin 2/phosphatase PP2A complex56. Conversely, the interaction of trophic factors (such as IGF1 and BDNF) with their cognate receptors activating AKT was essential to the survival of neuronal cells57. AKT1 genetic variants have been found in both schizophrenia and bipolar disorder58. The p53 protein, encoded by the TP53 gene, plays a key role in the control of apoptosis and is involved in the development of oligodendrocytes59. TP53 may affect myelin and white matter integrity in the frontal lobe of patients with schizophrenia59. NFKB stimulates the expression of numerous proinflammatory cytokines in the immune system60. Elevated NF-kB activity correlates with increased immune activation in the cortex61. Based on previous studies, the mechanism of olanzapine in treating agitation in schizophrenia and bipolar disorder may involve apoptosis, cell survival, and immune activity.
GO enrichment analyses revealed many biological processes, which were related to the treatment of olanzapine in agitation, schizophrenia, and bipolar disorder, including positive regulation of gene expression; positive regulation of transcription, DNA-templated; and negative regulation of apoptotic process. The core targets in our study were enriched in immune-related and inflammatory pathways, including the PI3K-Akt and MAPK signaling pathways. Immune activation is a component of the stress response, and psychosocial stress has been linked to the development of psychiatric disorders62. The PI3K/AKT/GSK3 pathway may act as a central intracellular network in psychiatric illnesses, regulating synaptic neuroplasticity, cell proliferation, migration, and apoptosis63. Patients with type 1 bipolar disorder were unable to reduce NF-κB and MAPK signaling after stress, potentially resulting from immune imbalance64. Olanzapine may have neuroprotective effects on neuronal cells via the PKA, PI3K, PKC, and CaMKII signaling pathways65. Astrocytes activate cellular protection, leading to chemotaxis and release of pro-inflammatory cytokines via NF-κB and p38 MAPK signaling66. The PI3K-Akt and MAPK signaling pathways were key pathways in the drug-pathway-target-disease network, which could be associated with immune responses, cell proliferation, apoptosis, and inflammation.
Based on the network pharmacology results, we systematically explored the potential mechanisms by which olanzapine may exert therapeutic effects on agitation, a common symptom in both schizophrenia and bipolar disorder. While these disorders differ in their overall pathogenesis, our analysis focused on shared molecular mechanisms related to agitation rather than disease-specific features. Therefore, the shared pathways identified in our study may contribute to agitation in both disorders, but not necessarily explain the distinct clinical manifestations of schizophrenia and bipolar disorder. Further research focusing on other symptom domains (e.g., mania, depression, hallucinations) is warranted to clarify how olanzapine may affect broader symptomatology via distinct or overlapping molecular mechanisms.
Finally, molecular docking was used to evaluate the binding affinities between olanzapine and eight core targets (including BDNF, INS, AKT1, TP53, IGF1, NGF, NFKB1, and FGF2). It is generally accepted that a docking score below − 5 kcal/mol indicates a good binding capacity between ligands and receptors67,68,69. The docking scores in our study, ranged from − 5.3 to − 8.8 kcal/mol and indicated good binding abilities between olanzapine and core targets. Hence, olanzapine may exert therapeutic effects on agitation in schizophrenia and bipolar disorder via all these eight core targets.
This study has several limitations. First, although network pharmacology and molecular docking provide useful exploratory tools, the results are inherently predictive, and binding affinities observed in silico do not necessarily reflect in vivo pharmacological or clinical effects. Second, the accuracy and timeliness of database-derived information remain limited, and some unrecorded or unconfirmed targets may have been missed in our analysis. Third, our focus on agitation, a clinically important but secondary symptom domain, may narrow the interpretability of the findings in the broader context of schizophrenia and bipolar disorder. Finally, this study relied solely on data mining approaches without experimental or clinical validation, and further in vitro, in vivo, and clinical studies are needed to confirm the predicted targets and pathways.
Conclusions
In summary, this study used network pharmacology and molecular docking technology to explore the molecular mechanisms of olanzapine against agitation in schizophrenia and bipolar disorder. Olanzapine may influence common core targets (including BDNF, INS, AKT1, TP53, IGF1, NGF, NFKB1, and FGF2) in the PI3K-Akt and MAPK signaling pathways to affect agitation in schizophrenia and bipolar disorder.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Tasic, L. et al. Peripheral biomarkers allow differential diagnosis between schizophrenia and bipolar disorder. J. Psychiatr Res. 119, 67–75 (2019).
McIntyre, R. S. et al. Bipolar Disorders. Lancet 396, 1841–1856 (2020).
Song, J. et al. Differences in Gray matter volume corresponding to delusion and hallucination in patients with schizophrenia compared with patients who have bipolar disorder. Neuropsychiatr Dis. Treat. 11, 1211–1219 (2015).
Global and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry 9, 137–150 (2022).
Díaz Zuluaga, A. M. et al. Evaluation and Socio-occupational intervention in bipolar and schizophrenic patients within a multimodal intervention Program- PRISMA. Rev. Colomb Psiquiatr. (Engl Ed). 47, 4–12 (2018).
Perrin, E. et al. A prospective, observational study of the safety and effectiveness of intramuscular psychotropic treatment in acutely agitated patients with schizophrenia and bipolar mania. Eur. Psychiatry. 27, 234–239 (2012).
Lesem, M. D. et al. Rapid acute treatment of agitation in individuals with schizophrenia: multicentre, randomised, placebo-controlled study of inhaled loxapine. Br. J. Psychiatry. 198, 51–58 (2011).
Zeller, S. L. & Citrome, L. Managing agitation associated with schizophrenia and bipolar disorder in the emergency setting. West. J. Emerg. Med. 17, 165–172 (2016).
Pompili, M. et al. The management of psychomotor agitation associated with schizophrenia or bipolar disorder: A brief review. Int J. Environ. Res. Public. Health 18 (2021).
Roberts, J., Gracia Canales, A., Blanthorn-Hazell, S., Craciun Boldeanu, A. & Judge, D. Characterizing the experience of agitation in patients with bipolar disorder and schizophrenia. BMC Psychiatry. 18, 104 (2018).
Allen, M. H. et al. Effect of nicotine replacement therapy on agitation in smokers with schizophrenia: a double-blind, randomized, placebo-controlled study. Am. J. Psychiatry. 168, 395–399 (2011).
Preskorn, S. H. et al. Effect of Sublingual Dexmedetomidine vs placebo on acute agitation associated with bipolar disorder: A randomized clinical trial. Jama 327, 727–736 (2022).
San, L. et al. State of acute agitation at psychiatric emergencies in europe: the STAGE study. Clin. Pract. Epidemiol. Ment Health. 12, 75–86 (2016).
Powell, G., Caan, W. & Crowe, M. What events precede violent incidents in psychiatric hospitals? Br. J. Psychiatry. 165, 107–112 (1994).
Holloman, G. H. Jr. & Zeller, S. L. Overview of project BETA: best practices in evaluation and treatment of agitation. West. J. Emerg. Med. 13, 1–2 (2012).
Garriga, M. et al. Assessment and management of agitation in psychiatry: expert consensus. World J. Biol. Psychiatry. 17, 86–128 (2016).
Wagstaff, A. J., Easton, J. & Scott, L. J. Intramuscular olanzapine: a review of its use in the management of acute agitation. CNS Drugs. 19, 147–164 (2005).
Bymaster, F. P. et al. Antagonism by olanzapine of dopamine D1, serotonin2, muscarinic, Histamine H1 and alpha 1-adrenergic receptors in vitro. Schizophr Res. 37, 107–122 (1999).
Bymaster, F. P. et al. Potent antagonism of 5-HT(3) and 5-HT(6) receptors by olanzapine. Eur. J. Pharmacol. 430, 341–349 (2001).
Ward, K. & Citrome, L. The treatment of acute agitation associated with schizophrenia or bipolar disorder: investigational drugs in early stages of their clinical development, and their clinical context and potential place in therapy. Expert Opin. Investig. Drugs. 29, 245–257 (2020).
Correll, C. U. et al. Effects of olanzapine combined with Samidorphan on weight gain in schizophrenia: A 24-Week phase 3 study. Am. J. Psychiatry. 177, 1168–1178 (2020).
Schulz, C. & Haight, R. J. Safety of olanzapine use in adolescents. Expert Opin. Drug Saf. 12, 777–782 (2013).
Battaglia, J. Pharmacological management of acute agitation. Drugs 65, 1207–1222 (2005).
DeBattista, C. & Hawkins, J. Utility of atypical antipsychotics in the treatment of resistant unipolar depression. CNS Drugs. 23, 369–377 (2009).
McIntyre, R. S., Cha, D. S., Kim, R. D. & Mansur, R. B. A review of FDA-approved treatment options in bipolar depression. CNS Spectr. 18 (Suppl 1), 4–20 (2013). quiz 21.
Cole, J. B. et al. A prospective observational study of patients receiving intravenous and intramuscular olanzapine in the emergency department. Ann. Emerg. Med. 69, 327–336e322 (2017).
Wright, P. et al. Double-blind, placebo-controlled comparison of intramuscular olanzapine and intramuscular haloperidol in the treatment of acute agitation in schizophrenia. Am. J. Psychiatry. 158, 1149–1151 (2001).
Tollefson, G. D. et al. Olanzapine versus haloperidol in the treatment of schizophrenia and schizoaffective and schizophreniform disorders: results of an international collaborative trial. Am. J. Psychiatry. 154, 457–465 (1997).
San, L. et al. A naturalistic multicenter study of intramuscular olanzapine in the treatment of acutely agitated manic or schizophrenic patients. Eur. Psychiatry. 21, 539–543 (2006).
Zhou, Z. et al. Applications of Network Pharmacology in Traditional Chinese Medicine Research. Evid Based Complement Alternat Med. 1646905 (2020). (2020).
Hopkins, A. L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690 (2008).
Onisiforou, A. & Zanos, P. One path, two solutions: Network-based analysis identifies targetable pathways for the treatment of comorbid type II diabetes and neuropsychiatric disorders. Comput. Struct. Biotechnol. J. 23, 3610–3624 (2024).
Pinzi, L. & Rastelli, G. Molecular docking: shifting paradigms in drug discovery. Int J. Mol. Sci 20 (2019).
Wishart, D. S. et al. DrugBank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 46, D1074–d1082 (2018).
Stelzer, G. et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinf. 54, 13031–313033 (2016).
Daina, A., Michielin, O. & Zoete, V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–w364 (2019).
Davis, A. P. et al. CTD tetramers: a new online tool that computationally links curated chemicals, genes, phenotypes, and diseases to inform molecular mechanisms for environmental health. Toxicol Sci. (2023).
Piñero, J., Saüch, J., Sanz, F. & Furlong, L. I. The disgenet cytoscape app: exploring and visualizing disease genomics data. Comput. Struct. Biotechnol. J. 19, 2960–2967 (2021).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–d612 (2021).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Chin, C. H. et al. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 (Suppl 4), S11 (2014).
Dennis, G. Jr. et al. Database for Annotation, Visualization, and integrated discovery. Genome Biol. 4, P3 (2003).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Trott, O. & Olson, A. J. AutoDock vina: improving the speed and accuracy of Docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Kim, S. et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 49, D1388–d1395 (2021).
Burley, S. K. et al. Protein data bank (PDB): the single global macromolecular structure archive. Methods Mol. Biol. 1607, 627–641 (2017).
Chiaretti, A. et al. Increased levels of neurotrophins in the cerebrospinal fluid of children with Epstein-Barr virus meningoencephalitis. Int. J. Infect. Dis. 20, 52–57 (2014).
Heath, A. et al. Medium- and high-intensity rTMS reduces psychomotor agitation with distinct neurobiologic mechanisms. Transl Psychiatry. 8, 126 (2018).
Suzuki, Y. et al. Quetiapine-induced insulin resistance after switching from Blonanserin despite a loss in both bodyweight and waist circumference. Psychiatry Clin. Neurosci. 66, 534–535 (2012).
Boulle, F. et al. Hippocampal and behavioral dysfunctions in a mouse model of environmental stress: normalization by agomelatine. Transl Psychiatry. 4, e485 (2014).
Liu, X. et al. Elevated serum levels of FGF-2, NGF and IGF-1 in patients with manic episode of bipolar disorder. Psychiatry Res. 218, 54–60 (2014).
Angelucci, F., Aloe, L., Iannitelli, A., Gruber, S. H. & Mathé, A. A. Effect of chronic olanzapine treatment on nerve growth factor and brain-derived neurotrophic factor in the rat brain. Eur. Neuropsychopharmacol. 15, 311–317 (2005).
Spillantini, M. G. et al. Nerve growth factor mRNA and protein increase in hypothalamus in a mouse model of aggression. Proc. Natl. Acad. Sci. U S A. 86, 8555–8559 (1989).
Woon, C. A. & Literature Review Violence and aggression in neuroscience nursing. J. Neurosci. Nurs. 55, 60–64 (2023).
Beaulieu, J. M. et al. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell 122, 261–273 (2005).
Patapoutian, A. & Reichardt, L. F. Trk receptors: mediators of neurotrophin action. Curr. Opin. Neurobiol. 11, 272–280 (2001).
Karege, F. et al. Genetic overlap between schizophrenia and bipolar disorder: a study with AKT1 gene variants and clinical phenotypes. Schizophr Res. 135, 8–14 (2012).
Molina, V. et al. Convergent evidence of the contribution of TP53 genetic variation (Pro72Arg) to metabolic activity and white matter volume in the frontal lobe in schizophrenia patients. Neuroimage 56, 45–51 (2011).
Cartwright, T., Perkins, N. D. & C, L. W. NFKB1: a suppressor of inflammation, ageing and cancer. Febs J. 283, 1812–1822 (2016).
Volk, D. W., Moroco, A. E., Roman, K. M., Edelson, J. R. & Lewis, D. A. The role of the nuclear Factor-κB transcriptional complex in cortical immune activation in schizophrenia. Biol. Psychiatry. 85, 25–34 (2019).
Daniels, T. E., Olsen, E. M. & Tyrka, A. R. Stress and psychiatric disorders: the role of mitochondria. Annu. Rev. Clin. Psychol. 16, 165–186 (2020).
Matsuda, S. et al. Roles of PI3K/AKT/GSK3 pathway involved in psychiatric illnesses. Diseases 7 (2019).
Wieck, A. et al. Differential neuroendocrine and immune responses to acute psychosocial stress in women with type 1 bipolar disorder. Brain Behav. Immun. 34, 47–55 (2013).
Lee, J. G., Cho, H. Y., Park, S. W., Seo, M. K. & Kim, Y. H. Effects of olanzapine on brain-derived neurotrophic factor gene promoter activity in SH-SY5Y neuroblastoma cells. Prog. Neuropsychopharmacol. Biol. Psychiatry. 34, 1001–1006 (2010).
Hol, E. M. & Pekny, M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr. Opin. Cell. Biol. 32, 121–130 (2015).
Li, X. et al. Network Pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu Decoction against sepsis. Comput. Biol. Med. 144, 105389 (2022).
Li, X., Tang, H., Tang, Q. & Chen, W. Decoding the mechanism of Huanglian Jiedu Decoction in treating pneumonia based on network Pharmacology and molecular Docking. Front. Cell. Dev. Biol. 9, 638366 (2021).
Zeng, Z. et al. Network Pharmacology and Molecular Docking-Based Prediction of the Mechanism of Qianghuo Shengshi Decoction against Rheumatoid Arthritis. Biomed Res Int 6623912 (2021). (2021).
Acknowledgements
We would like to express heartfelt thanks to the developers of all the databases used in this manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 82171053 and 81871052) to Chuanjun Zhuo.
Author information
Authors and Affiliations
Contributions
Chuanjun Zhuo, Xuemin Shi, Xingying Chen, Xiao Yan performed research and drafted the manuscript. Xuemin Shi, Xingying Chen, Xiao Yan, Ranli Li, Jiangshun Yang, Chunmian Chen, Langlang Chen contributed significantly to data analysis and interpretation. Guangdong Chen, Chao Li, Chuanjun Zhuo contributed to conception, research design, manuscript preparation and editing. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
The current analysis did not require ethical approval because it only involved collecting uploaded data information from public database searches.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, X., Chen, X., Ma, X. et al. Molecular mechanisms of olanzapine against agitation in schizophrenia and bipolar disorder based on network pharmacology and molecular docking. Sci Rep 15, 40279 (2025). https://doi.org/10.1038/s41598-025-24139-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24139-9