Abstract
Gastric cancer (GC) is a common malignancy of the digestive system, characterized by high invasiveness and metastasis, making it a leading cause of cancer-related deaths. Long non-coding RNAs (lncRNAs) play a crucial role in various types of cancer. This study aimed to elucidate the function of LINC01615 and its potential regulatory mechanisms in GC. In tissue and serum samples from GC patients, LINC01615 expression was significantly elevated and was associated with recurrence, distant metastasis, and TNM stage. LINC01615 promoted the proliferation, migration, and invasion of GC cells, and accelerated the growth of subcutaneous tumors in nude mice. Mechanistically, LINC01615 activated the WNT2/β-Catenin pathway by preventing WNT2 mRNA degradation and enhancing the nuclear translocation of β-Catenin. Furthermore, we identified the transcription factor YY1 as a key regulator of LINC01615 expression, which directly binds to its promoter region. Analysis of clinical samples revealed a positive correlation between the YY1/LINC01615/WNT2 signaling axis and poor prognosis in GC patients. Taken together, this study uncovers that the YY1/LINC01615/WNT2 signaling axis plays a crucial role in GC progression and may serve as a novel diagnostic marker and therapeutic target.
Similar content being viewed by others
Introduction
GC ranks as the fifth most prevalent malignancy worldwide and the fourth leading cause of cancer-related mortality1. Although early-stage GC can be effectively treated with endoscopic intervention or surgical resection, most patients are diagnosed at an advanced stage, often with metastasis. Despite advances in multidisciplinary treatments, including surgery and chemotherapy, the management of metastatic GC remains a major clinical challenge. Consequently, gaining a deeper understanding of the molecular mechanisms driving GC progression and identifying novel therapeutic targets are key priorities in current research.
More than 70% of the human genome is transcribed into RNA, the majority of which comprises non-coding RNAs2. Long non-coding RNAs (lncRNAs) are traditionally defined as RNA molecules exceeding 200 nucleotides in length that lack significant protein-coding potential3. lncRNAs possess diverse gene regulatory functions, interacting with proteins, RNA, and DNA, thereby participating in nearly all biological processes and playing a crucial role in the development of various diseases. Nuclear-localized lncRNAs primarily regulate gene transcription and epigenetic modifications4, while cytoplasmic lncRNAs exert precise post-transcriptional control by interacting with proteins and microRNAs (miRNAs)5,6. lncRNAs have been recognized as critical regulatory factors in cancer initiation and progression, emerging as important biomarkers for various diseases. Studies have shown that lncRNA dysregulation may promote cancer cell proliferation, tumor progression, and metastasis7,8,9,10, making them potential targets for cancer diagnosis and therapy. However, although the mechanisms of lncRNAs in various cancers have been investigated, their specific regulatory roles in GC remain poorly understood.
LINC01615 is a newly identified long non-coding RNA. Although reports on its role in other diseases are limited, an increasing number of studies have demonstrated its oncogenic potential in various tumors. Ji Dong’s team identified LINC01615 as being closely associated with the extracellular matrix by screening differentially expressed lncRNAs in hepatocellular carcinoma, suggesting a possible role in liver cancer metastasis11. Pan-cancer analyses indicate that LINC01615 is a mesenchyme-specific lncRNA that is positively correlated with epithelial–mesenchymal transition (EMT) scores. It is highly expressed in most mesenchymal-type cancers and is associated with poor prognosis12. In colorectal cancer, LINC01615 promotes oncogenic activity in cancer cells by competitively binding to miR-3653-3p, thereby activating ZEB213. Yuan Xiang et al. also found that SIPA1 promotes metastasis in triple-negative breast cancer by upregulating LINC01615 expression14. Although several studies have confirmed the role of LINC01615 in various tumors, its specific function in GC has not yet been elucidated.
This study is the first to reveal that LINC01615 expression is significantly upregulated in GC tissues and serum samples compared to normal tissues and healthy individuals. Moreover, its high expression is closely associated with tumor metastasis, recurrence, advanced stage, and reduced patient survival. Further analysis demonstrated that LINC01615 is transcriptionally regulated by YY1 and stabilizes WNT2 mRNA in the cytoplasm, thereby activating the WNT/β-catenin signaling pathway and promoting GC cell proliferation, migration, and invasion. In conclusion, this study highlights the crucial role of LINC01615 in GC progression and suggests that targeting LINC01615 may provide a promising therapeutic strategy for GC treatment.
Materials and methods
Patients and specimens
A total of 78 GC tissue samples and matched adjacent non-tumor tissues, along with peripheral blood samples from 78 GC patients and 40 healthy controls (28 males and 12 females; age range: 36–83 years; median age: 58.5 years), were collected between April 2015 and April 2020 at the First Affiliated Hospital of Fujian Medical University. Clinical and pathological information of the GC patients is provided in Supplementary Table S1. None of the patients received chemotherapy or radiotherapy prior to surgery. Immediately after surgical resection, each tissue sample was divided into two portions: one portion was snap-frozen in liquid nitrogen and stored at − 80 °C for RNA extraction, and the other was fixed in 10% neutral buffered formalin within 30 min and paraffin-embedded for histological analysis. Peripheral blood samples were centrifuged to isolate serum, which was then stored at − 80 °C until further use.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University (Fuzhou, China). All procedures involving human participants were conducted in accordance with the ethical standards of the institutional and/or national research committee, as well as with the Declaration of Helsinki (1964) and its later amendments. Written informed consent was obtained from all GC patients and healthy donors prior to sample collection.
Cell culture
The GC cell lines (AGS and MKN45) and human gastric mucosal epithelial cells (GES-1) used in this study were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences, while the HEK293T cell line was obtained from the Key Laboratory of Digestive Cancer, Ministry of Education, Fujian Medical University. All cell lines were validated by short tandem repeat (STR) profiling and confirmed to be free of mycoplasma contamination. AGS cells were cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) supplemented with 10% fetal bovine serum (FBS), 100 µg/mL streptomycin, and 100 IU/mL penicillin. MKN45 cells were maintained in RPMI-1640 medium with the same supplements. GES-1 and HEK293T cells were cultured in RPMI-1640 and DMEM, respectively, with the same supplements. All cell cultures were maintained at 37 °C in a humidified atmosphere containing 5% CO₂.
Plasmid construction and cell transfection
The cDNAs of human LINC01615, ETS1, SP1, TFAP2A, and YY1 were cloned into the pcDNA3.1 vector using standard molecular cloning techniques. All plasmids used in this study were sequence-verified by DNA sequencing to confirm their accuracy. The primers used in the study are listed in Supplementary Table S2. Specific small interfering RNAs (siRNAs) targeting YY1, LINC01615, and WNT2 were provided by SunYa Biotechnology (Fuzhou, China). Transfection experiments were performed in HEK293T, AGS, and MKN45 cells using Lipofectamine™ 3000 reagent (Thermo Fisher Scientific, USA), following the manufacturer’s instructions. For lentiviral infection experiments, the cDNAs were cloned into the pCDH-GFP vector and co-transfected with the lentiviral packaging plasmids psPAX2 and pMD2.G into HEK293T cells. After 48 h, the cell culture supernatant was collected and used to infect the target cells. Stably infected cells were selected by adding puromycin 5 days post-infection. The same procedure was used for shRNA transfection and the subsequent selection of stable knockdown cell lines.
qRT-PCR
Total RNA was extracted from tissues, serum, and cells using TRIzol™ reagent (Life Technologies), followed by reverse transcription into cDNA using the Evo M-MLV RT Kit (AG11707, Accurate Biotechnology Co., Ltd., Hunan, China) for subsequent qRT-PCR analysis. The expression levels of target RNAs were determined using the SYBR® Green Premix Pro Taq HS qPCR Kit (AG11701, Accurate Biotechnology Co., Ltd., Hunan, China). GAPDH was used as an endogenous control for normalization, and fold changes in expression were calculated using the 2⁻ΔΔCt method. All primers used in this study are listed in Supplementary Table S3. For cytoplasmic and nuclear RNA fractionation, the Nuclear and Cytoplasmic RNA Purification Kit (Beyotime, China) was used to isolate nuclear and cytoplasmic fractions. RNA was subsequently extracted using TRIzol™ reagent and subjected to qRT-PCR analysis. GAPDH and U6 were used as positive controls for cytoplasmic and nuclear RNA, respectively. To ensure the accuracy of nuclear–cytoplasmic RNA separation, strict fractionation procedures and equal RNA input were applied prior to qRT-PCR. The nuclear-to-cytoplasmic ratio was calculated using the following formula:
Cell proliferation, edu incorporation, and colony formation assays
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Japan). Cells were seeded at a density of 1,000 cells per well in 96-well plates and incubated at 37 °C with 5% CO₂ for 24, 48, 72, or 96 h. At the indicated time points, serum-free DMEM containing 10% CCK-8 solution was added to each well, followed by an additional 2-hour incubation. The absorbance at 450 nm was measured using a microplate reader to evaluate cell proliferation.
The EdU incorporation assay was performed using the EdU Detection Kit (C0075S, Beyotime Biotechnology, Shanghai, China). Cells from each group were seeded in 96-well plates and treated with EdU at a final concentration of 10 µM, followed by incubation at 37 °C for 2 h. Cells were then fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.3% Triton X-100 in PBS for 10–15 min, and washed three times with PBS. The click reaction solution was added, and the cells were incubated in the dark at room temperature for 30 min, followed by another three washes. Nuclei were stained with Hoechst 33,342 for 10 min, and cell proliferation was visualized using a fluorescence microscope.
For the colony formation assay, 1,000 AGS or MKN45 cells were seeded into cell culture dishes and incubated for 10 days. Cells were then washed 2–3 times with PBS, fixed with 4% paraformaldehyde, and stained with 1% crystal violet solution (Beyotime Biotechnology) to visualize colony formation. Excess dye was removed by rinsing with running water. The number of colonies was quantified using ImageJ software (National Institutes of Health, USA).
Wound healing, transwell migration, and invasion assays
For the wound healing assay, 1 × 10⁶ cells were seeded into six-well plates and cultured until reaching 90% confluence. A linear scratch was made across the cell monolayer using a sterile 200 µL pipette tip (~ 1 mm wide). Images of the wound area were captured at 0 h and 48 h, and the scratch width was measured at each time point to assess cell migration ability.
The cell migration assay was performed using Transwell chambers (BD Biosciences, San Jose, CA, USA). The lower chamber was filled with DMEM supplemented with 20% fetal bovine serum (FBS), while the upper chamber was seeded with 2 × 10⁵ cells in 0.6 mL of serum-free DMEM. Cells were incubated at 37 °C for 48 h. After incubation, the membranes were fixed with methanol and stained with crystal violet. Non-migrated cells on the upper surface of the membrane were removed using a cotton swab. Cells that had migrated to the lower surface of the membrane were then counted under an inverted phase-contrast microscope.
The cell invasion assay was conducted following the same procedure as the migration assay, except that Matrigel-coated Transwell chambers (BD Biosciences) were used. The subsequent steps were identical to those described in the migration assay.
Animal experiments
All animal experiments were conducted in accordance with the ARRIVE guidelines and the ethical standards for animal research. This study was approved by the Ethics Committee of Fujian Medical University. Two independent subcutaneous tumor xenograft experiments were performed using BALB/c nude mice, which were obtained from the Model Animal Research Center of Nanjing University (Jiangsu, China).
In the first experiment, twenty male BALB/c nude mice (6 weeks old, 18–20 g) were randomly assigned to four groups (n = 5 per group): LINC01615 overexpression, LINC01615 knockdown, and their respective negative controls. Each mouse was subcutaneously injected with 1 × 10⁶ cells suspended in 200 µL PBS into the right axillary region. Mice were monitored for food intake, general activity, and tumor growth. Tumor size was measured every three days using calipers, and tumor volume (mm³) was calculated using the formula: Volume = 0.5 × W² × L (W = width, L = length). On day 24, mice were anesthetized with isoflurane and euthanized by cervical dislocation in accordance with the AVMA Guidelines for the Euthanasia of Animals (2020). Tumors were then excised, photographed, weighed, and fixed in 10% neutral-buffered formalin for further analysis.
The second xenograft experiment was conducted using an additional cohort of twenty BALB/c nude mice, randomly divided into four groups (n = 5 per group): negative control, LINC01615 knockdown, SKL2001 treatment, and LINC01615 knockdown combined with SKL2001. The experimental procedures and analyses were identical to those described in the first experiment.
Western blot
For cell samples, proteins were extracted using RIPA lysis buffer (Beyotime Biotechnology) supplemented with 1% PMSF (Sangon Biotech). Cell lysates were incubated on ice for 10 min, sonicated, and centrifuged at 12,000 rpm for 10 min to collect the supernatant. For tissue samples, proteins were extracted by homogenizing the tissues on ice in RIPA buffer containing PMSF, followed by centrifugation at 12,000 rpm for 10 min to obtain the supernatant. Protein concentration was determined using the BCA assay, and 30 µg of protein per lane was loaded onto 8% or 10% SDS-PAGE gels. Electrophoresis was performed at 90 V for 30 min, then increased to 110 V until the dye front reached the bottom of the gel. After electrophoresis, proteins were transferred to PVDF membranes (Sigma-Aldrich) using the wet transfer method at 110 V for 1.5 h at 4 °C. PVDF membranes were pre-activated in methanol and blocked with 5% BSA at room temperature for 2 h, then incubated overnight at 4 °C with primary antibodies (details provided in Supplementary Table S4). On the following day, membranes were washed three times with TBST (10 min each), incubated with HRP-conjugated anti-rabbit or anti-mouse IgG secondary antibodies at room temperature for 2 h, and washed again three times with TBST. Protein bands were visualized using ECL detection and quantified using ImageJ software, with β-actin or GAPDH serving as loading controls.
For nuclear and cytoplasmic protein fractionation, the Nuclear and Cytoplasmic Protein Extraction Kit (P0027, Beyotime Biotechnology) was used according to the manufacturer’s instructions. GAPDH was used as a cytoplasmic marker, and PCNA as a nuclear marker. Equal amounts of cytoplasmic and nuclear proteins were then subjected to SDS-PAGE and Western blot analysis.
Fluorescence in situ hybridization (FISH)
The experiment was conducted using the RNA Fluorescence In Situ Hybridization (FISH) Detection Kit (R0306S, Beyotime Biotechnology) to localize and quantify target non-coding RNAs in tissues and cells. For paraffin-embedded tissue sections, samples were fixed in 4% paraformaldehyde for 1–2 h, and section thickness was kept within 4 μm. The sections were deparaffinized with xylene and ethanol, rehydrated, and permeabilized. Proteinase K (5–20 µg/mL in PBS) was added, and samples were digested at room temperature for 2–10 min. For cell samples, cells were cultured in 24-well plates to reach a density of 70–90%, and washed 2–3 times with DNase/RNase-free PBS. Cells were then fixed with 4% paraformaldehyde at room temperature for 10 min, followed by three washes with DNase/RNase-free PBS (5 min each). Next, samples were incubated in prehybridization solution containing 1× yeast RNA at 37 °C in the dark for 20 min. The probe was diluted to a final concentration of 0.5–1 µg/mL, and hybridization was carried out at the same temperature for 2–12 h. After hybridization, samples were sequentially washed with preheated Washing Buffers I, II, and III. Nuclei were stained with 0.5–1 µg/mL DAPI solution in the dark for 3–5 min, followed by imaging using a fluorescence microscope. To prevent fluorescence quenching, samples were mounted with antifade mounting medium (P0126-25 mL, Beyotime Biotechnology) and stored in the dark at 4 °C. The probes were FITC-labeled at the 5′ end as follows: LINC01615: GGACTCAGACAGTGGAGGCCCCTCGCTGAC; U6: AGTATATGTGCTGCCGAAGCGAGCACT.
RNA in situ hybridization (RNA-ISH)
To detect the expression of LINC01615 in tissue samples, a 5′-biotin-labeled antisense RNA probe (same sequence as used in the fluorescence in situ hybridization assay) was employed. Paraffin-embedded tissue sections were deparaffinized and rehydrated in PBS, followed by fixation with 4% paraformaldehyde (PFA) for 10 min. The sections were then permeabilized with proteinase K (20 µg/mL, Roche) for 15 min, followed by an additional 5-minute fixation in 4% PFA. Prior to hybridization, the sections were treated with 0.25% acetic anhydride for 10 min. Hybridization was carried out overnight at 65 °C using the biotin-labeled probe diluted 1:100 in hybridization buffer (Amresco). After hybridization, the sections were washed twice at 65 °C in a buffer containing 1× SSC, 50% formamide, and 0.1% Tween-20 (30 min each). Unbound probes were removed by incubation with RNase A (20 µg/mL, Roche) at 37 °C for 30 min in a humid chamber. The sections were then incubated overnight at 4 °C with horseradish peroxidase (HRP)-conjugated streptavidin (A0305, Beyotime Biotechnology). Signal detection was performed using a DAB substrate, with color development for 3–10 min until a brown precipitate was observed. The sections were counterstained with hematoxylin, dehydrated through a graded ethanol series (70%, 80%, 90%, 100%), cleared in xylene, and mounted with neutral balsam. The localization and expression levels of LINC01615 were examined under a light microscope.
Immunohistochemistry (IHC)
Tissue sections were first deparaffinized in xylene and then rehydrated through a graded ethanol series, followed by antigen retrieval using citrate buffer. To reduce nonspecific staining, the sections were incubated in 3% hydrogen peroxide for 30 min to block endogenous peroxidase activity, and then incubated in 10% normal goat serum for 10 min to block nonspecific binding sites. Primary antibodies, including anti-Ki67 (1:1000), anti-Vimentin (1:4000), anti-YY1 (1:5000), and anti-WNT2 (1:500), were added and incubated at room temperature for 1 h to ensure specific binding to their targets. After primary antibody incubation, the sections were treated with a biotinylated secondary antibody for 10 min, followed by incubation with a streptavidin–peroxidase complex to amplify the signal. DAB (3,3′-diaminobenzidine, Promega) was used as the chromogenic substrate for color development, and the sections were counterstained with hematoxylin to visualize cell nuclei morphology and localization.
RNA-ISH and IHC evaluation methods
Positive staining is primarily distributed in the nucleus or cytoplasm, and a semi-quantitative scoring method is used for evaluation. In five randomly selected high-power fields (×400), staining intensity and the percentage of positive cells were assessed. The staining intensity was scored as follows: 0 = no staining, 1 = weak staining, 2 = moderate staining, 3 = strong staining. The percentage of positive cells was scored as: 0 = < 5%, 1 = 5%−25%, 2 = 26%−50%, 3 = 51%−75%, 4 = > 75%. The final comprehensive score was calculated by multiplying staining intensity by the percentage of positive cells, with a score range of 0 to 12. All sections were independently scored by two pathologists, who were blinded to the experimental groupings, and the average score was taken as the final score.
Fluorescence staining
AGS and MKN45 cells were seeded in 3.5 cm culture dishes and transiently co-transfected with plasmids for 48 h. Cells were then fixed with pre-cooled 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 at room temperature for 10 min. After fixation, cells were blocked with 5% bovine serum albumin (BSA) at room temperature for 30 min to reduce nonspecific binding. Subsequently, cells were incubated overnight at 4 °C with an anti-β-catenin antibody (1:200, Cell Signaling Technology). On the following day, cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:200, Abcam) for 1 h in the dark. Nuclei were stained with DAPI, and β-catenin localization in the nucleus and cytoplasm was observed using a laser scanning confocal microscope (TCS SP8, Leica Microsystems, Germany).
TOP/FOPflash and dual-luciferase reporter assay
To investigate the regulatory effect of LINC01615 on the Wnt/β-catenin signaling pathway, a TOP/FOPflash luciferase reporter assay was performed. Cells were transfected with either the TOPflash plasmid (containing TCF/LEF binding sites, D2501-1 µg, Beyotime Biotechnology) or the FOPflash plasmid (containing mutated TCF/LEF binding sites, D2503-1 µg, Beyotime Biotechnology), along with the pRL-TK Renilla luciferase plasmid as an internal control. Forty-eight hours post-transfection, cells were lysed using the Dual-Luciferase Reporter Assay Kit (Vazyme), and luminescence was measured using an Orion II microplate luminometer (Berthold Detection Systems). The TOP/FOP ratio (TOPflash luminescence/FOPflash luminescence) was calculated to evaluate Wnt/β-catenin pathway activity.
To analyze the transcriptional activity of the LINC01615 promoter, cells were transfected with recombinant pGL4.10 firefly luciferase reporter plasmids containing either the full-length or truncated promoter sequences of LINC01615, along with the pRL-TK plasmid as an internal control. Forty-eight hours after transfection, luciferase activity was measured following the same procedure as the TOP/FOPflash assay. Primers were synthesized by Boshang Biotechnology Co., Ltd. (Fuzhou, China), and the specific sequences are listed in Supplementary Table S5.
Chromatin Immunoprecipitation assay (ChIP)
Following the manufacturer’s instructions, the Enzymatic Chromatin IP Kit (9002 S, Cell Signaling Technology) was used to perform the chromatin immunoprecipitation (ChIP) assay. Briefly, GC cells were treated with 1% formaldehyde for 10 min to crosslink DNA and proteins, followed by the addition of 125 mM glycine to terminate the crosslinking reaction. Cells were then washed with cold PBS and lysed using cell lysis buffer to release chromatin, which was subsequently sheared into 200–500 bp fragments by sonication. At 4 °C, 5 µg of YY1 antibody (#46395, Cell Signaling Technology) was added and incubated on a rotator for 4–6 h for immunoprecipitation. Rabbit IgG was used as a negative control. To capture the antibody–antigen complexes, Protein A/G magnetic beads were added, followed by an additional 1–2 h of incubation. Several washes were performed to remove unbound chromatin. To reverse the crosslinks, immunocomplexes were incubated at 65 °C for several hours, followed by treatment with proteinase K to ensure complete protein degradation, thereby facilitating the extraction of high-quality DNA. After DNA extraction, concentration and purity were assessed to confirm suitability for PCR amplification targeting the − 400 to − 200 bp region of the LINC01615 promoter. PCR products were loaded onto a 1.8% agarose gel for electrophoretic separation, stained with a nucleic acid dye, and visualized under UV light. To avoid signal oversaturation in the input group, only 10% of the input sample was used. PCR primer sequences: Forward: AGGGTCAGGACTTTGGCCC; Reverse: CTTTCCAAAAGAGAAAACTACATGGA.
Gene set enrichment analysis (GSEA)
Gene Set Enrichment Analysis (GSEA) was conducted to explore the potential mechanisms by which LINC01615 promotes malignant biological behaviors in GC. Transcriptomic data for stomach adenocarcinoma (STAD) were obtained from The Cancer Genome Atlas (TCGA) database, and samples were categorized into high- and low-expression groups based on the median expression level of LINC01615. GSEA was performed using GSEA software (version 4.0.3) with the Hallmark and KEGG gene sets from the Molecular Signatures Database (MSigDB) as reference datasets. A continuous-type CLS file was generated to associate phenotype labels with gene expression profiles. The ranking metric was set to “Signal2Noise”, and samples were permuted 1,000 times to assess statistical significance, with all other parameters set to default. Pathways with a normalized enrichment score (NES) > 1 and a false discovery rate (FDR) q-value < 0.25 were considered significantly enriched.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9.0 and SPSS 22.0 software. Data are presented as mean ± standard deviation (SD) from at least three independent experiments. Student’s t-test was used to compare two groups of independent data, and the Pearson χ² test was applied to evaluate the association between LINC01615 expression and the clinicopathological parameters of GC patients. Survival curves were generated using Kaplan–Meier analysis and further evaluated with the log-rank test. Statistical significance was defined as follows: ns = not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Results
High expression of LINC01615 correlates with poor prognosis in GC
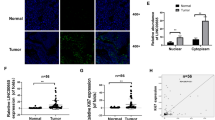
To identify lncRNAs associated with the malignant progression of GC, we retrieved gene expression profiles and corresponding clinical information from the TCGA database. Differential expression analysis revealed that 475 lncRNAs were significantly upregulated and 83 lncRNAs were significantly downregulated in GC tissues compared with normal tissues (Fig. 1A; Supplementary Table S6). The criteria for differential expression were an absolute log₂FC > 2.5 and an adjusted p-value (padj) < 0.001. In subsequent studies, we prioritized upregulated lncRNAs in GC tissues as candidate molecules for further investigation, since they are more easily detectable and more likely to contribute to tumor progression. By integrating patient survival data (selection criteria: hazard ratio (HR) > 1.5 and p-value < 0.05), we identified eight lncRNAs significantly associated with the prognosis of GC patients, namely LINC01235, DIRC1, LNCOG, LINC01615, HMGA, LINC02588, KIF26B, and POU6F2-AS2 (Fig. 1B–C). Guided by these high-throughput findings, we further validated the expression of these eight lncRNAs in GC tissues. qRT-PCR analysis using three paired GC and adjacent normal tissues (n = 3 pairs) confirmed that all eight lncRNAs were significantly upregulated in GC, with LINC01615 showing the most pronounced increase (Fig. 1D). Therefore, we focused on investigating the biological role of LINC01615 in GC in subsequent studies. FISH analysis revealed markedly higher fluorescence intensity of LINC01615 in GC tissues compared with adjacent normal tissues (Fig. 1E), consistent with the qRT-PCR results and further supporting its potential role in GC progression. We next examined LINC01615 expression in clinical tissue samples from 78 GC patients and analyzed correlations with clinicopathological parameters. High LINC01615 expression was significantly associated with cancer recurrence (Fig. 1F), distant metastasis (Fig. 1G), and TNM stage (Fig. 1H), whereas no significant associations were observed with other parameters (Supplementary Fig. S1A–S1F).
High expression of LINC01615 correlates with poor prognosis in GC. (A) Volcano plot of differentially expressed non-coding RNAs in GC versus normal tissues (TCGA database). Green and red dots represent downregulated and upregulated genes, respectively. (B) Forest plot of prognostic non-coding RNAs associated with overall survival (OS) in GC patients. (C) Venn diagram showing the overlap between overexpressed and prognostic genes. (D) qRT-PCR analysis of eight non-coding RNAs in paired GC and adjacent normal tissues (n = 3 pairs). (E) FISH images showing LINC01615 expression in GC tissues (tumor) and adjacent noncancerous tissues (peritumor). Scale bar = 100 μm. (F–H) Correlation of LINC01615 expression with recurrence (F), distant metastasis (G), and TNM stage (H) in 78 GC tissue samples. (I) Serum LINC01615 levels in GC patients (n = 78) versus healthy controls (n = 40). ROC curve shows discriminatory ability (AUC = 0.7662). (J–L) ROC analyses of serum LINC01615 levels for recurrence (J, AUC = 0.7616), distant metastasis (K, AUC = 0.8150), and TNM stage (L, AUC = 0.8080) in GC patients (n = 78). ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Furthermore, we evaluated LINC01615 expression in the serum of GC patients to assess its potential as a non-invasive biomarker. LINC01615 levels were significantly elevated in the serum of GC patients (Fig. 1I) and strongly correlated with cancer recurrence (Fig. 1J), distant metastasis (Fig. 1K), and TNM stage (Fig. 1L). ROC curve analysis based on serum samples from 78 GC patients and 40 healthy controls revealed an AUC value of 0.7662, indicating a significant difference between the two groups.
LINC01615 enhances GC cell proliferation, migration, and invasion in vitro and in vivo
We analyzed the expression of LINC01615 in normal human gastric epithelial cells (GES-1) and GC cell lines (AGS and MKN45). The results showed that LINC01615 expression was significantly higher in AGS and MKN45 cells compared with GES-1 cells (Fig. 2A). To further investigate its function, we established stable LINC01615 knockdown in AGS and MKN45 cells using shRNA lentiviral vectors (Fig. 2B; Supplementary Fig. S2A). CCK-8 assay revealed that knockdown of LINC01615 significantly reduced GC cell viability (Fig. 2C; Supplementary Fig. S2B). Similarly, colony formation and EdU incorporation assays confirmed that silencing LINC01615 markedly suppressed GC cell proliferation compared with control cells (Fig. 2D–E; Supplementary Fig. S2C–D). In terms of cell motility, wound healing and Transwell assays demonstrated that LINC01615 knockdown significantly impaired GC cell migration and invasion (Fig. 2F–G; Supplementary Fig. S2E–F). Western blot analysis showed that stable knockdown of LINC01615 led to upregulation of the tumor suppressor proteins p53 and p21, as well as the epithelial marker E-cadherin, while the mesenchymal markers N-cadherin and Vimentin were significantly downregulated (Fig. 2H; Supplementary Fig. S2G). In contrast, stable overexpression of LINC01615 was achieved by lentiviral transfection (Fig. 2I; Supplementary Fig. S2H). These cells exhibited markedly enhanced proliferative, migratory, and invasive capacities (Fig. 2J–N; Supplementary Fig. S2I–M). Consistently, LINC01615 overexpression caused downregulation of p53, p21, and E-cadherin, along with notable upregulation of N-cadherin and Vimentin (Fig. 2O; Supplementary Fig. S2N).
LINC01615 promotes proliferation, migration, and invasion of GC cells in vitro and in vivo. (A) qRT-PCR analysis showing differential LINC01615 expression in normal gastric epithelial cells (GES-1) versus GC cells (AGS and MKN45). (B) LINC01615 knockdown (sh01615#1, sh01615#2) significantly reduces its expression compared with control (shControl). (C–E) CCK-8 (C), colony formation (D), and EdU incorporation assays (E) showing that LINC01615 knockdown suppresses AGS cell proliferation. (F) Wound healing assay showing impaired migration of LINC01615-silenced AGS cells at 48 h. (G) Transwell migration and invasion assays demonstrating reduced motility of AGS cells after LINC01615 knockdown. (H) Western blot showing that LINC01615 knockdown increases p53 and p21, decreases EMT markers (Vimentin, N-cadherin), and elevates E-cadherin. (I) qRT-PCR confirming LINC01615 overexpression in AGS cells. (J–L) CCK-8 (J), colony formation (K), and EdU incorporation assays (L) showing enhanced proliferation of AGS cells upon LINC01615 overexpression. (M–N) Wound healing (M) and Transwell (N) assays showing promoted migration and invasion after LINC01615 overexpression. (O) Western blot showing reduced p53 and p21, increased EMT markers (Vimentin, N-cadherin), and reduced E-cadherin in LINC01615-overexpressing AGS cells. (P–Q) Representative images (P) and tumor growth curves (Q) of xenografts derived from AGS cells with LINC01615 knockdown or overexpression. (R–S) Tumor volume (R) and weight (S) at day 24 of the in vivo experiment. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To evaluate the role of LINC01615 in GC progression in vivo, we established a xenograft nude mouse model. LINC01615 silencing significantly inhibited tumor growth, whereas its overexpression promoted tumor growth, as reflected by changes in tumor volume and weight compared with controls (Fig. 2P–S). Western blot analysis of xenograft tissues confirmed molecular alterations consistent with the in vitro findings, showing opposite trends following knockdown and overexpression (Supplementary Fig. S3A). Furthermore, RNA-ISH and IHC analyses demonstrated that expression of Ki67 and Vimentin correlated with LINC01615 levels, with knockdown reducing their expression and overexpression increasing it (Supplementary Fig. S3B).
LINC01615 activates the WNT pathway and promotes β-catenin nuclear translocation
To investigate the mechanism by which LINC01615 promotes malignant biological behaviors in GC, we performed KEGG pathway enrichment analysis using R software (version 3.4.3) and GSEA (version 4.0.3). The results suggested that LINC01615 may promote GC progression through activation of the WNT signaling pathway (Fig. 3A). Consistently, Western blot analysis confirmed that LINC01615 overexpression upregulated the protein levels of downstream WNT pathway effectors, including TCF, LEF, MET, and c-MYC (Fig. 3B), whereas silencing LINC01615 significantly suppressed their expression (Fig. 3C). The qRT-PCR results were consistent with the Western blot findings (Fig. 3D–E).
LINC01615 promotes activation of the WNT/β-catenin signaling pathway in GC cells. (A) GSEA of TCGA stomach adenocarcinoma dataset showing enrichment of the WNT/β-catenin signaling pathway in the high-LINC01615 group. (B–C) Western blot analysis of WNT target proteins (TCF4, LEF, Met, c-Myc) in AGS and MKN45 cells after LINC01615 overexpression (B) or knockdown (C), normalized to β-actin. Relative gray value analysis indicates protein levels. (D–E) qRT-PCR analysis of TCF4, LEF, Met, c-Myc mRNA in AGS and MKN45 cells following LINC01615 overexpression (D) or knockdown (E), normalized to GAPDH. (F–H) TOP/FOP luciferase reporter assays showing WNT/β-catenin activity in AGS and MKN45 cells with LINC01615 overexpression (F) or knockdown (G–H). (I–J) Western blot analysis of cytoplasmic and nuclear β-catenin in AGS and MKN45 cells with LINC01615 overexpression (I) or knockdown (J). β-actin and PCNA served as cytoplasmic and nuclear controls, respectively. (K–L) Western blot analysis of β-catenin, p-β-catenin (Ser33/37/Thr41), GSK3β, and p-GSK3β (Ser9) in AGS and MKN45 cells after LINC01615 overexpression (K) or knockdown (L). (M–N) Immunofluorescence staining showing β-catenin localization in AGS (M) and MKN45 (N) cells with LINC01615 overexpression or knockdown. Nuclei were counterstained with DAPI. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
β-catenin is a key mediator of the WNT pathway15. Activation of this pathway is characterized by β-catenin nuclear translocation, where it exerts transcriptional regulatory functions16. To test this, we performed a TOP/FOPflash luciferase reporter assay, which demonstrated that β-catenin–driven transcriptional activity was markedly enhanced in AGS and MKN45 cells overexpressing LINC01615 (Fig. 3F), but significantly reduced in LINC01615 knockdown cells (Fig. 3G–H). To further validate these findings, cytoplasmic and nuclear proteins were fractionated, and Western blot analysis showed increased nuclear β-catenin in LINC01615-overexpressing cells (Fig. 3I), whereas nuclear β-catenin levels were reduced upon LINC01615 knockdown (Fig. 3J). Moreover, in LINC01615-overexpressing GC cells, total β-catenin levels increased, phosphorylated β-catenin (Ser33/37/Thr41) levels decreased, and phosphorylated GSK3β (Ser9) levels increased, while total GSK3β levels remained unchanged. Conversely, opposite trends were observed in LINC01615 knockdown cells (Fig. 3K–L).
Finally, immunofluorescence staining revealed that LINC01615 promoted β-catenin translocation into the nucleus in AGS and MKN45 cells, whereas LINC01615 knockdown produced the opposite effect (Fig. 3M–N).
LINC01615 promotes WNT2 mRNA stability
To further elucidate the mechanism by which LINC01615 regulates the WNT signaling pathway, we first examined its subcellular localization using FISH analysis. The results showed that LINC01615 was primarily localized in the cytoplasm (Fig. 4A). This finding was further validated by qRT-PCR analysis of RNA isolated from nuclear and cytoplasmic fractions, which revealed significantly higher levels of LINC01615 in the cytoplasm compared with the nucleus (Fig. 4B). These results suggest that LINC01615 may exert its regulatory functions at the post-transcriptional level. Given that β-catenin exerts its transcriptional regulatory activity predominantly in the nucleus, we hypothesized that LINC01615 does not directly bind to β-catenin but instead regulates upstream molecules influencing β-catenin activity. Correlation analysis between LINC01615 and core WNT pathway components revealed that WNT2 showed the strongest association (Supplementary Table S7). Further analysis demonstrated that high WNT2 expression was significantly associated with poor prognosis in GC (Fig. 4C). In addition, TCGA database analysis showed a significant positive correlation between LINC01615 and WNT2 expression (Fig. 4D), suggesting that LINC01615 may activate the WNT signaling pathway by regulating WNT2.
LINC01615 stabilizes WNT2 mRNA and enhances its expression in GC cells. (A) FISH analysis showing cytoplasmic localization of LINC01615 and nuclear localization of U6 in AGS cells. (B) Nuclear and cytoplasmic fractionation confirming predominant cytoplasmic localization of LINC01615 in AGS and MKN45 cells. U6 and GAPDH served as nuclear and cytoplasmic controls, respectively. (C) Cox regression analysis of overall survival in TCGA GC patients stratified by WNT2 expression (low vs. high). (D) Correlation between LINC01615 and WNT2 expression in the TCGA database. (E–G) qRT-PCR showing that LINC01615 overexpression (E) increases WNT2 mRNA levels, whereas knockdown (F–G) decreases them in AGS and MKN45 cells. (H–J) Western blot showing changes in WNT2 protein levels in AGS and MKN45 cells with LINC01615 overexpression (H) or knockdown (I–J). β-actin served as the loading control. (K–N) qRT-PCR analysis of WNT2 mRNA stability in AGS and MKN45 cells treated with Actinomycin D, following LINC01615 overexpression (K, M) or knockdown (L, N). (O–P) DNA gel imaging showing WNT2 mRNA levels in AGS cells treated with Actinomycin D (5 µg/ml) after LINC01615 overexpression or knockdown. GAPDH served as the loading control. Time points (0, 4, 8, 12 h) indicate the duration post-treatment. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Based on this hypothesis, we first measured the levels of mature WNT2 mRNA in GC cells with LINC01615 overexpression or knockdown. The results showed that upregulation or downregulation of LINC01615 led to a significant increase or decrease in the levels of mature WNT2 mRNA, including the 3′-UTR, CDS, and 5′-UTR regions (Fig. 4E–G; Supplementary Fig. S4A). In contrast, the levels of pre-WNT2 mRNA (containing four intronic regions) remained unchanged (Supplementary Fig. S5A–C). These findings suggest that LINC01615 does not affect WNT2 transcription but instead regulates mRNA stability through a post-transcriptional mechanism. Consistently, Western blot analysis confirmed that WNT2 protein levels increased upon LINC01615 overexpression and decreased following its knockdown (Fig. 4H–J; Supplementary Fig. S4B).
Subsequently, we used actinomycin D (ActD) to inhibit DNA transcription and evaluated the degradation rate of WNT2 mRNA. The results showed that LINC01615 overexpression significantly prolonged the half-life of WNT2 mRNA (Fig. 4K and M), whereas LINC01615 knockdown accelerated its degradation (Fig. 4L and N). DNA gel electrophoresis further supported these findings (Fig. 4O–P; Supplementary Fig. S6).
LINC0615 drives malignant behaviors of GC through the WNT2/β-catenin signaling axis
SKL2001 has been reported to inhibit the interaction between β-catenin and its degradation complex, thereby preventing β-catenin degradation, promoting its nuclear accumulation, and activating the WNT signaling pathway. To further investigate the role of LINC01615 in this process, we treated LINC01615-knockdown cells with SKL2001. The results showed that SKL2001 rescued the downregulation of WNT pathway proteins induced by LINC01615 silencing (Fig. 5A). In addition, SKL2001 restored GC cell proliferation (Fig. 5B–C), as well as migration and invasion abilities (Fig. 5D–E). Furthermore, SKL2001 reversed the expression changes of tumor suppressors (p53, p21) and EMT-related markers (E-cadherin, N-cadherin, Vimentin) caused by LINC01615 knockdown (Fig. 5F).
LINC01615 promotes malignant behaviors of GC through the WNT2/β-catenin signaling pathway. (A) Western blot of key WNT/β-catenin pathway proteins (β-catenin, p-β-catenin [Ser33/37/Thr41], GSK3β, p-GSK3β [Ser9]) in AGS cells with LINC01615 knockdown (sh01615#1, sh01615#2) or control (shControl), treated with SKL2001 (+) or vehicle (DMSO, –). β-actin served as the loading control. (B–C) CCK-8 assay (B) and colony formation assay (C) assessing proliferation and colony-forming ability of AGS cells. (D–E) Wound healing (D) and Transwell assays (E) evaluating migration and invasion. (F) Western blot of p53, p21, E-cadherin, Vimentin, N-cadherin in AGS cells. β-actin served as the loading control. (G) Western blot of WNT/β-catenin pathway proteins in AGS cells overexpressing LINC01615, with or without WNT2 knockdown. β-actin served as the loading control. (H) Immunofluorescence showing β-catenin localization in AGS cells. β-catenin (green); nuclei counterstained with DAPI (blue). (I) Western blot of p53, p21, E-cadherin, Vimentin, N-cadherin in AGS cells. β-actin served as the loading control. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Conversely, in cells overexpressing LINC01615, SKL2001 further enhanced the activation of WNT pathway proteins (Supplementary Fig. S7A), accelerated cell proliferation (Supplementary Fig. S7B–C), and promoted migration and invasion (Supplementary Fig. S7D–E). In addition, SKL2001 augmented the upregulation of EMT markers and further suppressed tumor suppressor proteins, thereby reinforcing the effects mediated by LINC01615 (Supplementary Fig. S7F).
To further confirm that LINC01615 promotes GC progression through the WNT2/β-catenin axis, we transiently transfected si-WNT2 into AGS cells stably overexpressing LINC01615. Western blot analysis showed that WNT2 knockdown abrogated the protein-level activation of WNT pathway components induced by LINC01615 overexpression (Fig. 5G). In addition, immunofluorescence staining demonstrated that the nuclear accumulation of β-catenin, driven by LINC01615 overexpression, was reversed by WNT2 silencing (Fig. 5H). Consistently, WNT2 knockdown also counteracted the upregulation of EMT markers and the downregulation of tumor suppressor proteins induced by LINC01615 overexpression (Fig. 5I).
Xenograft experiments in nude mice further demonstrated that the tumor-suppressive effect induced by Linc01615 knockdown could be compensated by SKL2001 treatment (Supplementary Fig. S8).
YY1 transcriptionally regulates the expression of LINC01615
Given that transcriptional regulation is a crucial step in controlling gene expression17,18, we next investigated the regulatory mechanisms underlying LINC01615 transcription. We cloned the full-length LINC01615 promoter and a series of 5′-deletion fragments into the pGL4.10 vector, designated as pGL4.10–2000 bp, pGL4.10–1600 bp, pGL4.10–1200 bp, pGL4.10–800 bp, and pGL4.10–400 bp (Fig. 6A). These plasmids were transfected into AGS cells, and transcriptional activity was measured by dual-luciferase reporter assay. The results showed that the pGL4.10–400 bp construct exhibited transcriptional activity comparable to the full-length promoter (− 200 to 2000 bp) (Fig. 6B), suggesting that the core regulatory elements reside within the − 200 to 400 bp region.
YY1 transcriptionally regulates LINC01615 expression in GC cells. (A) Gel electrophoresis of PCR-amplified LINC01615 promoter fragments (pGL4.10–2000 bp, −1600 bp, −1200 bp, −800 bp, −400 bp) cloned into the pGL4.10 vector. (B) Luciferase reporter assay of truncated promoter constructs in AGS and MKN45 cells. pGL4.10-WT served as control. (C) Venn diagram of predicted transcription factors binding the LINC01615 promoter (databases: JASPAR, PROMO, Gene-regulation, GTRD). (D) Luciferase activity of the pGL4.10–400 bp construct in AGS cells co-transfected with ETS1, SP1, TFAP2A, or YY1. (E) YY1 expression levels (log2 TPM + 1) in normal vs. tumor samples from TCGA-STAD. (F) Cox regression analysis of overall survival in TCGA patients stratified by YY1 expression. (G) Correlation between LINC01615 and YY1 expression in TCGA. (H) Western blot showing YY1 protein levels in AGS and MKN45 cells transfected with increasing doses of pcDNA3.1-YY1. (I) Relative luciferase activity of LINC01615 promoter constructs in AGS and MKN45 cells with increasing YY1 expression. (J) qRT-PCR showing LINC01615 upregulation after YY1 overexpression in AGS and MKN45 cells. (K) Western blot confirming YY1 knockdown by siRNAs in AGS and MKN45 cells. (L) Luciferase activity of promoter constructs (pGL4.10 and pGL4.10–400 bp) showing reduced activity after YY1 knockdown. (M) qRT-PCR showing decreased LINC01615 expression after YY1 knockdown in AGS and MKN45 cells. (N) ChIP assay detecting YY1 binding to the LINC01615 promoter. PCR detection and quantification confirm YY1 enrichment at the promoter region. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Using the JASPAR, PROMO, Gene-regulation, and GTRD databases, we predicted potential transcription factors that may regulate LINC01615. Four candidates were identified: ETS1, SP1, TFAP2A, and YY1 (Fig. 6C; Supplementary Table S8). Subsequent dual-luciferase assays revealed that YY1 exerted the strongest transcriptional regulatory effect on LINC01615 (Fig. 6D). Furthermore, YY1 expression was significantly upregulated in GC tissues compared with normal tissues (Fig. 6E), associated with poor prognosis (Fig. 6F), and positively correlated with LINC01615 levels (Fig. 6G), making it a suitable candidate for further study.
To explore the relationship between YY1 expression and the pGL4.10–400 bp promoter, we co-transfected varying doses of a YY1 expression plasmid with pGL4.10–400 bp. Overexpression of YY1 significantly enhanced pGL4.10–400 bp promoter activity in a dose-dependent manner (Fig. 6H–I). Consistently, qRT-PCR analysis confirmed that YY1 overexpression increased LINC01615 expression (Fig. 6J), whereas siRNA-mediated YY1 knockdown suppressed pGL4.10–400 bp promoter activity and decreased LINC01615 expression (Fig. 6K–M).
Finally, chromatin immunoprecipitation (ChIP) assays validated the direct association of YY1 with the LINC01615 promoter. PCR amplification confirmed that YY1 specifically bound to the 400 bp promoter fragment (Fig. 6N).
YY1/LINC01615 axis regulates Proliferation, Migration, and invasion of GC cells
Previous studies have confirmed that YY1 can transcriptionally activate LINC01615 by directly binding to its promoter, thereby upregulating its expression. We also established that LINC01615 functions as an oncogene in GC. Notably, although YY1 has been reported to promote GC progression through various mechanisms, it remains unclear whether this effect is mediated by regulating LINC01615.
To address this, we established an AGS GC cell line with stable YY1 overexpression using lentiviral transfection. qRT-PCR (Fig. 7A) and Western blot analysis (Fig. 7B) confirmed robust YY1 overexpression, validating the cell model. CCK-8 assays showed that YY1 overexpression markedly enhanced AGS cell proliferation, whereas LINC01615 knockdown in YY1-overexpressing cells significantly reversed this proliferative effect (Fig. 7C). Similarly, colony formation assays (Fig. 7D) and EdU incorporation assays (Fig. 7E) demonstrated that silencing LINC01615 attenuated YY1-induced proliferation.
YY1 promotes proliferation, migration, and invasion of GC cells by upregulating LINC01615. (A–B) qRT-PCR (A) and Western blot (B) confirming YY1 overexpression in AGS cells. (C) CCK-8 assay showing that YY1 overexpression enhances AGS cell proliferation, whereas LINC01615 knockdown reverses this effect. (D–E) Colony formation (D) and EdU incorporation (E) assays further validating that LINC01615 knockdown inhibits YY1-induced proliferation. (F) Wound healing assay showing increased migration with YY1 overexpression, reversed by LINC01615 knockdown. (G) Transwell assays confirming that LINC01615 knockdown suppresses the migration and invasion enhanced by YY1. (H) Western blot showing that YY1 overexpression downregulates p53, p21, E-cadherin and upregulates Vimentin, N-cadherin; these effects are reversed by LINC01615 knockdown. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Regarding cell motility, the wound healing assay revealed that YY1 overexpression promoted AGS cell migration, while LINC01615 knockdown markedly reversed this effect (Fig. 7F). Consistently, Transwell assays further confirmed that LINC01615 knockdown abrogated the enhanced migration and invasion induced by YY1 overexpression (Fig. 7G).
At the molecular level, Western blot analysis showed that YY1 overexpression decreased the expression of tumor suppressors (p53, p21) and the epithelial marker E-cadherin, while upregulating mesenchymal markers (N-cadherin, Vimentin). Importantly, LINC01615 knockdown significantly reversed these molecular alterations in YY1-overexpressing cells (Fig. 7H).
The YY1/LINC01615/WNT2 signaling axis as a prognostic biomarker in GC
To further validate the regulatory mechanism identified in our study, we analyzed the expression of YY1, LINC01615, and WNT2 in 78 paired GC tissues and adjacent normal tissues using IHC and RNA-ISH. The results showed that all three molecules were significantly upregulated in GC tissues compared with adjacent normal tissues (Fig. 8A–D). In addition, total RNA and protein extracted from 10 paired samples further confirmed the elevated expression of YY1, LINC01615, and WNT2 in GC tissues by RT-PCR and Western blot analysis (Supplementary Fig. S9).
Expression and prognostic role of YY1, LINC01615, and WNT2 in GC. (A) Representative IHC and RNA-ISH images of YY1, LINC01615, and WNT2 in 78 paired GC and adjacent normal tissues at 100× and 400× magnifications. Hematoxylin and eosin (H&E) staining is shown at the top. (B–D) Quantification of IHC or RNA-ISH scores showing significantly higher levels of LINC01615 (B), YY1 (C), and WNT2 (D) in GC tissues compared with normal tissues. (E–G) Correlation analyses showing positive associations between YY1 protein and LINC01615 RNA (E), WNT2 protein and LINC01615 RNA (F), and WNT2 protein and YY1 protein (G), indicating a YY1/LINC01615/WNT2 regulatory axis in GC. (H–J) Kaplan–Meier survival curves showing that high expression of YY1 (H), LINC01615 (I), and WNT2 (J) predicts poor overall survival in GC patients. (K) Schematic model of the YY1/LINC01615/WNT2 signaling axis in GC progression: YY1 activates LINC01615 transcription, which stabilizes WNT2 mRNA and upregulates its expression, thereby activating the WNT/β-catenin pathway and promoting proliferation and metastasis in GC cells. ns = not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Correlation analysis revealed that the expression levels of YY1, LINC01615, and WNT2 were positively correlated with each other (Fig. 8E–G), suggesting a potential linear regulatory relationship within the YY1/LINC01615/WNT2 axis during GC progression. Prognostic analysis further demonstrated that higher expression levels of YY1, LINC01615, and WNT2 were significantly associated with poor overall survival (OS) in GC patients (Fig. 8H–J).
Discussion
Previous studies have shown that lncRNAs, a novel class of non-coding RNAs, play critical roles in the development and progression of malignancies19,20,21,22, including GC23,24,25. Mechanistically, lncRNAs regulate cancer progression through multiple pathways, such as acting as miRNA sponges or directly interacting with proteins or mRNAs. In this study, we identified LINC01615 as a novel oncogenic lncRNA involved in GC progression. Our findings demonstrated that LINC01615 was significantly upregulated in both GC tissues and patient serum. High LINC01615 expression was strongly correlated with metastasis, recurrence, advanced tumor stage, and poor prognosis. Functionally, LINC01615 knockdown inhibited GC cell proliferation, migration, and invasion in vitro and suppressed tumor growth in vivo, whereas LINC01615 overexpression exerted the opposite effects. Collectively, these results highlight the oncogenic role of LINC01615 in GC and provide new insights into its clinical significance.
The canonical WNT pathway is highly conserved and is activated through autocrine or paracrine mechanisms by the binding of extracellular WNT ligands to their membrane receptors. Once activated, the pathway induces the stabilization of β-catenin and its nuclear translocation, ultimately promoting the expression of genes involved in cell proliferation, survival, differentiation, and migration26. Previous studies have shown that lncRNAs exert strong regulatory effects on the WNT/β-catenin pathway. Several lncRNAs, such as JPX, LALR1, and NEAT1, directly or indirectly enhance β-catenin stability, thereby activating the canonical WNT pathway. For example, in metastatic lung cancer, lncRNA-JPX suppresses GSK-3β expression and promotes β-catenin nuclear translocation27. In hepatocytes, lncRNA-LALR1 reduces AXIN1 expression by recruiting CTCF to the AXIN1 promoter, thereby inhibiting β-catenin phosphorylation and enhancing the transcription of c-Myc and Cyclin D128. Similarly, lncRNA-NEAT1 interacts with DDX5, facilitating the formation of a DDX5/β-catenin complex and promoting downstream gene transcription29. In line with these findings, our study demonstrates that LINC01615 overexpression significantly upregulates the expression of key WNT pathway downstream molecules, including TCF, LEF, Met, and c-Myc, whereas LINC01615 knockdown exerts the opposite effect. Moreover, LINC01615 overexpression enhances β-catenin stability and promotes its nuclear accumulation, thereby activating the WNT/β-catenin pathway and ultimately driving GC cell proliferation, migration, and invasion.
WNT proteins are secreted glycoproteins that can become aberrantly activated when extracellular signaling is disrupted, leading to developmental abnormalities and even tumorigenesis30. Overactivation of the WNT signaling pathway contributes to the proliferation, growth, and metastasis of various malignancies, with WNT2 being a critical component of this pathway. Previous studies have demonstrated that excessive WNT2 secretion activates the downstream β-catenin signaling cascade, ultimately promoting cancer cell growth31. Furthermore, WNT2 expression has been positively correlated with invasive potential, tumor stage, and pathological grade in multiple malignancies32. In this study, we identified a novel regulatory mechanism whereby LINC01615 activates the WNT2/β-catenin signaling axis by stabilizing WNT2 mRNA, thereby promoting the malignant phenotype of GC cells. This finding reveals a previously unrecognized mechanism of WNT2 regulation by a lncRNA and provides new insights into non-coding RNA–mediated modulation of the WNT pathway.
YY1 is a zinc finger protein belonging to the GLI-Krüppel family, initially identified as a repressor of the P5 promoter of adeno-associated virus33. Depending on cellular context, YY1 can function as either a transcriptional activator or a tumor suppressor34. Numerous studies have demonstrated that YY1 transcriptionally activates lncRNAs, thereby contributing to tumor progression35,36,37. In GC, YY1 has been widely recognized as a crucial oncogenic factor38,39. For instance, Lu et al. reported that YY1 promotes GC malignancy by upregulating LINC0085840. In our study, dual-luciferase reporter assays confirmed that YY1 transcriptionally activates LINC01615, while ChIP assays validated its direct binding to the LINC01615 promoter. As a transcription factor, YY1 may therefore promote GC progression through the regulation of multiple lncRNAs. In addition, previous studies have shown that YY1 activates various signaling pathways, including the WNT pathway, to drive GC development41,42, which is consistent with our findings. Collectively, these observations suggest that the role of YY1 in GC is complex and multifaceted, involving multiple signaling networks and potential cross-regulation between pathways. Thus, YY1 represents a promising target for future mechanistic studies and therapeutic interventions in GC.
Despite uncovering the critical role of LINC01615 in GC, this study has several limitations. First, although we confirmed the role of LINC01615 in stabilizing WNT2 mRNA, the underlying molecular mechanisms remain to be clarified, such as potential interactions with RNA-binding proteins or other non-coding RNAs. Second, the sample size of this study was relatively small and primarily derived from a single center, which may introduce selection bias and limit the generalizability of our conclusions. Finally, the role of LINC01615 in other cancer types has not been systematically investigated, and its generalizability and specificity across different malignancies warrant further study.
Conclusion
In conclusion, we elucidated the mechanism by which YY1 transcriptionally regulates LINC01615, thereby promoting the malignant progression of GC. Upregulation of LINC01615 stabilizes WNT2 mRNA, resulting in elevated WNT2 expression and subsequent activation of the WNT/β-catenin pathway. This activation facilitates β-catenin nuclear translocation, enabling its transcriptional regulatory functions (Fig. 8K). Collectively, our study provides novel insights into the oncogenic mechanism of the YY1/LINC01615/WNT2 axis in GC.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
H, S. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 71, 209–249 (2021).
Geng, Y., Jiang, J. & Wu, C. Function and clinical significance of circrnas in solid tumors. J. Hematol. Oncol. J. Hematol. Oncol. 11, 98 (2018).
Djebali, S. et al. Landscape of transcription in human cells. Nature. 489, 101–108 (2012).
Herman, A. B., Tsitsipatis, D. & Gorospe, M. Integrated LncRNA function upon genomic and epigenomic regulation. Mol. Cell. 82, 2252–2266 (2022).
Tay, Y., Rinn, J. & Pandolfi, P. P. The multilayered complexity of CeRNA crosstalk and competition. Nature. 505, 344–352 (2014).
Statello, L., Guo, C. J., Chen, L. L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118 (2021).
Hashemi, M. et al. Long non-coding RNA (lncRNA) H19 in human cancer: from proliferation and metastasis to therapy. Pharmacol. Res. 184, 106418 (2022).
Liang, H. et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer. 17, 119 (2018).
Wu, D. D. et al. Role of the LncRNA ABHD11-AS1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol. Cancer. 16, 138 (2017).
Yang, B. et al. Overexpression of LncRNA IGFBP4–1 reprograms energy metabolism to promote lung cancer progression. Mol. Cancer. 16, 154 (2017).
Ji, D. et al. Identification of LINC01615 as potential metastasis-related long noncoding RNA in hepatocellular carcinoma. J. Cell. Physiol. 234, 12964–12970 (2019).
Kumar, P., Khadirnaikar, S., Bhandari, N., Chatterjee, A. & Shukla, S. An epithelial-mesenchymal plasticity signature identifies two novel LncRNAs with the opposite regulation. Front Cell. Dev. Biol. 10, 885785 (2022).
Hu, Z., Yang, C., Guo, S., Li, Y. & Li, Y. LINC01615 activates ZEB2 through competitively binding with miR-3653-3p to promote the carcinogenesis of colon cancer cells. Cell. Cycle. 21, 228–246 (2022).
Xiang, Y. et al. SIPA1 regulates LINC01615 to promote metastasis in Triple-Negative breast cancer. Cancers. 14, 4815 (2022).
Moon, R. T., Bowerman, B., Boutros, M. & Perrimon, N. The promise and perils of Wnt signaling through β-Catenin. Science. 296, 1644–1646 (2002).
Gumbiner, B. M. Signal transduction of beta-catenin. Curr. Opin. Cell. Biol. 7, 634–640 (1995).
Lambert, S. A. et al. The hum transcription factors. Cell. 172, 650–665 (2018).
Haberle, V. & Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell. Biol. 19, 621–637 (2018).
Liu, M. et al. The interplay between non-coding RNAs and alternative splicing: from regulatory mechanism to therapeutic implications in cancer. Theranostics. 13, 2616–2631 (2023).
Singh, D., Assaraf, Y. G. & Gacche, R. N. Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist. Updat Rev. Comment Antimicrob. Anticancer Chemother. 63, 100851 (2022).
Huang, Z., Zhou, J. K., Peng, Y., He, W. & Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer. 19, 77 (2020).
Nemeth, K., Bayraktar, R., Ferracin, M. & Calin, G. A. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat. Rev. Genet. 25, 211–232 (2024).
Luo, Y. et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy. 17, 4083–4101 (2021).
Yang, H. et al. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J. Adv. Res. 37, 91–106 (2022).
Wei, L. et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol. Cancer. 19, 62 (2020).
MacDonald, B. T., Tamai, K. & He, X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 17, 9–26 (2009).
Pan, J. et al. LncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol. Cancer. 19, 9 (2020).
Xu, D. et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/β-catenin signaling. Hepatol. Baltim. Md. 58, 739–751 (2013).
Zhang, M. et al. The LncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. J. Hematol. Oncol. 11, 113 (2018).
Zhang, Q. et al. Wnt/β-catenin signaling mediates the suppressive effects of Diallyl trisulfide on colorectal cancer stem cells. Cancer Chemother. Pharmacol. 81, 969–977 (2018).
Zhang, H., Qin, D., Jiang, Z. & Zhang, J. SNHG9/miR-199a-5p/Wnt2 axis regulates cell growth and aerobic Glycolysis in glioblastoma. J. Neuropathol. Exp. Neurol. 78, 939–948 (2019).
Kimura, M., Nakajima-Koyama, M., Lee, J. & Nishida, E. Transient expression of WNT2 promotes somatic cell reprogramming by inducing β-Catenin nuclear accumulation. Stem Cell. Rep. 6, 834–843 (2016).
Shi, Y., Seto, E., Chang, L. S. & Shenk, T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 67, 377–388 (1991).
Meliala, I. T. S., Hosea, R., Kasim, V. & Wu, S. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics. 10, 4183–4200 (2020).
Yan, Z. et al. YY1-induced LncRNA ZFPM2-AS1 facilitates cell proliferation and invasion in small cell lung cancer via upregulating of TRAF4. Cancer Cell. Int. 20, 108 (2020).
Zhou, W. et al. Yin Yang 1-Induced long noncoding RNA DUXAP9 drives the progression of oral squamous cell carcinoma by blocking CDK1-Mediated EZH2 degradation. Adv. Sci. Weinh Baden-Wurtt Ger. 10, e2207549 (2023).
Yang, Z. et al. YY1-inudced activation of LncRNA DUXAP8 promotes proliferation and suppresses apoptosis of triple negative breast cancer cells through upregulating SAPCD2. Cancer Biol. Ther. 22, 216–224 (2021).
Wang, J. et al. The CCDC43-ADRM1 axis regulated by YY1, promotes proliferation and metastasis of gastric cancer. Cancer Lett. 482, 90–101 (2020).
Zhang, L., Zou, L. & Sun, P. Relationship between miR-378c and YY1 expression in patients with gastric cancer and the clinicopathological features. Cell. Mol. Biol. Lett. 26, 12 (2021).
Lu, Y. et al. LINC00858 stabilizes RAN expression and promotes metastasis of gastric cancer. Biol. Direct. 17, 41 (2022).
Kang, W. et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J. Transl Med. 12, 80 (2014).
Bhaskar Rao, D., Panneerpandian, P., Balakrishnan, K. & Ganesan, K. YY1 regulated transcription-based stratification of gastric tumors and identification of potential therapeutic candidates. J. Cell. Commun. Signal. 15, 251–267 (2021).
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Grant No. 82372958), the Natural Science Foundation of Fujian Province (Grant No. 2022J02032), the Fujian Provincial Health Technology Project (Grant No. 2020CXA032), and the Fujian Province Eagle Plan Youth Top Talent Project (Grant No. SCYJHJRC-ZGW2024).
Author information
Authors and Affiliations
Contributions
X. Lin: Conceptualization, formal analysis, supervision, validation, investigation, writing the original draft. S. Liu: Investigation, methodology. H. Chen: Investigation, methodology. R. He: Investigation, software. P. Lin: Methodology. Y. Lin: Methodology. X. Pan: Methodology. J. Ye: Conceptualization, resources, data curation, supervision, funding acquisition, project administration, writing–review and editing. G. Zhu: Conceptualization, data curation, supervision, funding acquisition, validation, investigation, project administration, writing–review and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All animal experiments were conducted at the Animal Experimental Center of Fujian Medical University and approved by the university’s ethics committee (IACUC FJMU 2023-Y-0735). This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Fujian Medical University (Nos. 2018 [030]).
Consent for publication
The authors confirm that they have obtained consent for publication from all individuals whose personal data or images are included in this manuscript, and that these individuals are aware that their data will be made publicly available. The authors declare that there are no known financial or personal conflicts of interest that could have influenced the research reported in this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, X., Liu, S., Chen, H. et al. YY1-mediated transcriptional regulation of LINC01615 inhibits WNT2 mRNA degradation to promote gastric cancer progression. Sci Rep 15, 41067 (2025). https://doi.org/10.1038/s41598-025-24956-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24956-y