Abstract
Pancreatic cancer is a highly aggressive malignancy with a poor prognosis, often diagnosed at advanced stages. Current treatments are limited, underscoring the need for effective therapies. Recent studies suggest that luteolin exhibits significant anti-tumor activity, yet its mechanisms in pancreatic cancer remain poorly understood. This study explored the anti-tumor mechanism of luteolin in pancreatic cancer (PC), focusing on its regulation of cyclin B1 (CCNB1)-mediated cell cycle progression. PANC-1 and SW1990 cell lines were used for assays of cell proliferation, migration, and invasion, and flow cytometry was used to assess cell cycle distribution and apoptosis. The efficacy of luteolin was further evaluated in patient-derived organoids (PDOs) and a mouse xenograft model. To identify molecular targets, we employed network pharmacology and transcriptomic sequencing, followed by experimental validation using Western blot, molecular docking, and surface plasmon resonance (SPR) assays to confirm luteolin’s interaction with CCNB1 and its effects on downstream signaling pathways. Luteolin exhibited a dose-dependent inhibitory effect on pancreatic cancer cell proliferation, migration, and invasion. It induced G2/M cell cycle arrest and apoptosis, with significant suppression of PDO growth. In vivo, luteolin effectively inhibited subcutaneous tumor growth in a xenograft mouse model without causing systemic toxicity or organ damage. Network pharmacology and transcriptomic analyses identified CCNB1 as a pivotal target, and experimental validation confirmed luteolin’s direct binding to CCNB1. This interaction disrupted the CCNB1/cyclin-dependent kinase 1 (CDK1) complex, leading to cell cycle arrest and reduced tumor progression. Luteolin suppresses pancreatic cancer growth by targeting CCNB1-mediated cell cycle regulation, inducing G2/M arrest, and promoting apoptosis. This highlights its potential as a therapeutic agent in pancreatic cancer treatment.
Similar content being viewed by others
Introduction
Pancreatic cancer (PC) is one of the most lethal malignancies, characterized by rapid progression and a dismal prognosis1. It ranks as the fourth leading cause of cancer-related deaths globally, with most patients diagnosed at an advanced stage, making effective treatment challenging2. Although treatments for PC — including surgery, chemotherapy, radiotherapy, immunotherapy, and targeted therapy — have advanced in recent years3,4, while the therapeutic effect is limited, underscoring the urgent need for novel and effective treatment strategies.
Natural products have attracted increasing research interest in recent years because they can enhance efficacy and reduce toxicity during drug discovery and development5,6. Luteolin (Lut), 3′,4′,5,7-tetrahydroxyflavone, is a member of the flavone subclass of flavonoids. It exists abundantly in fruits, vegetables, and herbs such as apples, beets, broccoli, celery, peppers, parsley, and thyme7,8. It has been reported that luteolin produces strong anti-proliferation, anti-inflammatory and antioxidant activities in different human cancer cell lines9,10,11,12,13. However, its specific mechanisms of action in pancreatic cancer remain largely unexplored.
In this study, the anti-cancer effects of luteolin were evaluated, and its underlying mechanisms were comprehensively investigated. Luteolin significantly inhibited the migration and proliferation of pancreatic cancer (PC) cells. Furthermore, we demonstrated that luteolin targets cyclin B1 (CCNB1), leading to cell cycle arrest at the G2/M phase and induction of apoptosis in PC cells. In vivo, a subcutaneous tumor model in nude mice confirmed that luteolin effectively suppressed solid tumor growth without causing significant organ toxicity. These findings suggest that luteolin may be a promising therapeutic agent for the treatment of pancreatic cancer.
Materials and methods
Compounds and reagents
CCNB1 protein (Cat# ab128445) were bought from Abcam (Cambridge, UK). Luteolin (Cat# HY-N0162) was purchased from MedChemexpress CO., Ltd (NJ, USA). Antibodies for CCNB1 (Cat#28603-1-AP), P53 (Cat#60283-2-Ig), Bcl-2 (Cat#68103-1-Ig), Bax (Cat#50599-2-Ig), Caspase3 (Cat#19677-1-AP), PI3K (Cat# 20584-1-AP), Akt (Cat# 60203-2-Ig), p-Akt (Cat# 66444-1-Ig), mTOR (Cat#66888-1-Ig), p-mTOR (Cat#67778-1-Ig) and β-actin (Cat# 81115-1-RR) were all purchased from Proteintch (USA). p-PI3K (Cat# AP0153) were purchased from Bioworld Technology (Nanjing, China).
Cell culture
Human PC cell lines PANC-1 and SW1990 were purchased from American Type Culture Collection (ATCC, USA). PANC-1 and SW1990 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) and supplemented with 10% fetal bovine serum (FBS) (Gibco, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco, USA). All cells were incubated at 37 °C in a humidified atmosphere containing 5% CO₂.
3D culture
Human PC samples were cut into 3 mm × 3 mm pieces, minced and digested in Collagenase/Hyaluronidase solution (Stemcell Technologies, #07912) for 1–2 h at 37 °C. Digested fragments were embedded in Matrigel (Corning, #356231) and cultured in complete Human Pancreatic Organoid Medium [advanced DMEM/F12 supplemented with 1x B27, 1.25 mM N-Acetylcysteine, 10 nM Gastrin, 50 ng/ml EGF, 100 ng/ml FGF10, 10 mM Nicotinamide, 5 µM A83-01, 10 µM SB202190, 1x Primocin]. For drug sensitivity assays, dissociated organoids were seeded in 96-well plates at a density of 1000 cells per well in Matrigel. After 4 days of culture, organoids were treated with different concentrations of luteolin for 48 h; morphological changes were observed using an Olympus microscope. Cell viability was assessed using the CellTiter-Glo 3D assay (Promega, #G9681) according to the manufacturer’s instructions. Luminescence was measured using a SpectraMax microplate reader. The half-maximal inhibitory concentration (IC₅₀) was calculated by nonlinear regression (dose–response curve fitting) using GraphPad Prism 9.0. The study is in accordance with the Declaration of Helsinki and received ethical approval from the Fudan University Shanghai Cancer Center (approval number 2212267-7).
Cell viability assay
Cell viability was assessed using the enhanced Cell Counting Kit-8 (Beyotime, Shanghai, China). Briefly, 100 µL of cell suspension (3 × 104 cells/mL) was seeded into each well of 96-well plates. After incubation overnight, different concentrations of luteolin were added, followed by incubation for 24, 48, and 72 h. Then, 10 µL of CCK-8 solution was added to cultured cells and incubated at 37 ◦C for 30 min. Next, Then, the absorbance at 450 nm was measured using a microplate reader (SpectraMax ABS plus, Molecular Devices, USA).
Colony formation assay
Colony formation assays were performed by seeding PANC-1 and SW1990 cells at 1 × 103 cells per well in 6-well plates. Cells were then treated with different concentrations of luteolin. After ten days of cultivation, cells that formed visible colonies were washed with phosphate-buffered saline (PBS) three times, fixed with 4% paraformaldehyde for 15 min, and further stained with 0.1% crystal violet for 30 min. Cell colonies containing more than 50 cells were counted using a microscope.
Transwell migration and invasion assay
Cell migration and invasion was measured in 24-well transwell cell culture chambers (Corning, NY, USA). For migration assay, PANC-1 and SW1990 cells were seeded into upper chambers at a density of 1 × 105 cells per well in serum-free DMEM. The lower chambers were filled with DMEM containing 10% FBS. While for invasion assay, cells in 200 µL of serum-free DMEM were seeded to the upper chamber which was pre-treated by Matrigel (BD Biosciences, USA) for 2 h at 37 ◦C. Next the cells were treated with different concentration of luteolin for 48 h. Then non-migrating and invading cells were removed from the upper surface using a cotton swab, and chambers were stained with 1% crystal violet for 30 min. Migrating and invading cells adhered to underside of chambers were counted under a light microscope (Olympus, Japan), and mean values of five fields were determined.
Cell apoptosis and cell cycle assay
Apoptosis was assessed using the Annexin-V/FITC Apoptosis Detection Kit (BD, USA) according to the manufacturer’s instructions. Briefly, cells were seeded in 6-well plates, treated with luteolin, and analyzed by flow cytometry (BD Biosciences, USA).
Cell cycle assay was performed in accordance with the Cell Cycle and Apoptosis Analysis Kit (Solarbio, China). In brief, the collected cells were treated with different concentration of luteolin, then the cells were fixed with ice-cold 70% ethanol for 24 h. Then, propidium iodide staining solution was added to the cell samples, the cell precipitation was slowly and adequately resuspended, and the cells were incubated at 37 ◦C for 30 min without light. Cell cycle was detected by flow cytometer and analyzed by the NovoExpress software14.
Network Pharmacology and bioinformatics analysis
Targets of luteolin were retrieved from the TCMSP database15 and SwissTargetPrediction (http://swisstargetprediction.ch/)16. After removing duplicates, unique targets were retained. Gene expression data from GSE101448 and GSE62165 were downloaded and analyzed for differentially expressed genes (DEGs) using the limma package in R (v4.0.3). DEGs were defined by |log2 fold-change| > 1 and P < 0.05. Finally, the targets of luteolin in the treatment of pancreatic cancer were obtained.
RNA sequencing and data analysis
PANC-1 cells, either untreated (NC) or treated with 20 µM luteolin for 48 h, were harvested for total RNA extraction. RNA concentration and purity were determined by NanoPhotometer® (IMPLEN, CA, USA), and integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Three biological replicates per condition (six libraries total) were prepared with 1 µg of RNA each, employing the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer’s protocol; unique dual indices were incorporated to multiplex samples. Raw sequencing reads were processed to remove adapter contamination, poly-N stretches, and low-quality bases, yielding high-confidence clean reads. The reference genome index was generated and reads were aligned using HISAT2 v2.0.5. Gene-level quantification and normalization were performed, and differential expression analysis was conducted with DESeq2 (v1.16.1), applying an adjusted p-value cutoff of < 0.05 and absolute log₂ fold-change > 1.
Cellular thermal shift assay (CETSA)
Cultured PANC-1 and SW1990 cells were harvested and lysed to obtain cell lysates. Cell lysates were incubated with 20 µM luteolin for 48 h at room temperature to allow binding. After incubation, the mixture was divided into 100 µL aliquots in individual tubes. Aliquots were heated for 4 min at the indicated temperatures and then cooled for 4 min at room temperature. SDS-PAGE loading buffer was added and samples were boiled prior to analysis by Western blot. The prepared samples were analyzed by Western blot to determine the interaction between luteolin and target proteins.
Surface plasmon resonance (SPR) and molecular docking
SPR was conducted with a Biacore T200 instrument (Cytiva, MA, USA). CCNB1 (20 µg/mL) was covalently immobilized on a NTA chip (Cat# BR100532, Cytiva) across ligand flow channels. Then, the chip was equilibrated with PBS. A concentration series of luteolin were added into the flow system to test the binding affinity between luteolin and CCNB1. Luteolin was dissolved in PBS with 0.1% Dimethyl sulfoxide (DMSO), 10 µl/min flow rate, 120 s contact time, and 240 s dissociation time were set. The software of T200 evaluation state model was utilized to analyze the binding affinity data and calculated the compound’s dissociation constant (KD) value.
3D structures of active ligand components were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The 3D structure format of the target was found in the Protein Data Bank (PDB) database (https://www1.rcsb.org/). The software PyMOL 2.4.1 (Schrodinger, USA) was used for protein dehydrating and ligand removal. AutoDock 4.2.6 (Scripps Research, USA) was used for target protein hydrogenation and charge calculation, and AutoDock Vina 1.1.2 (Scripps Research, USA) was used for docking. The docking results were visualized by PyMOL 2.4.1 software (Schrodinger, USA).
Immunofluorescence staining
PANC-1 and SW1990 cells were grown in 24-well plates and exposed to luteolin for 48 h. After treatment, cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with Beyotime Saponin for 15 min, and rinsed three times with ice-cold PBS before being blocked with 5% bovine serum albumin for 20 min. Primary antibodies were then applied overnight at 4 °C, followed by three PBS washes and a 30-min incubation at room temperature with a 1:200 dilution of FITC-conjugated goat anti-rabbit IgG (Affinity, China). After another three washes in ice-cold PBS, nuclei were counterstained with 200 µl 4’,6-diamidino-2-phenylindole (DAPI) solution (Beyotime, China) for 5 min at room temperature. Images were captured using an Olympus confocal laser scanning microscope17.
Western blot analysis
Western blotting was performed using standard methods. Briefly, cells were lysed on ice in radioimmunoprecipitation assay buffer (RIPA) buffer (Biosharp, China), protease inhibitors (Biosharp, China) and PMSF (Biosharp, China). Protein concentrations were measured using a bicinchoninic acid (BCA) assay kit (Cat# P0010; Beyotime, Shanghai, China). Proteins were separated by electrophoresis in an SDS-PAGE gel and blotted onto a polyvinylidene fluoride (PVDF) membrane (Millipore, CA). Membranes were incubated with specific primary antibodies overnight at 4 ◦C as following: Bcl-2, Bax, Caspase3, P53, CCNB1, CDK1, CDC40, TPX2 and β-actin. After the membrane was washed three times with TBST, it was incubated with horseradish peroxidaseconjugated secondary antibody (Zs-BIO; diluted 1/10000). β-actin was used as an internal control. Then, protein bands were detected by using the Western Lightning Plus-ECL Kit (Thermo, USA). The intensity of protein fragments was scanned and quantified by Quantity One imaging software (Bio-Rad, USA).
Establishment of CCNB1-knockdown and overexpression cell lines
We utilized a lentiviral vector system to perform CCNB1 knockdown in PANC-1 cells. First, lentiviral vectors containing shRNA sequences targeting CCNB1 were designed and constructed. These vectors were co-transfected with packaging plasmids into 293 T cells to produce lentiviral particles. Next, the generated lentiviral particles were used to infect PANC-1 cells, followed by selection with appropriate antibiotics post-infection to obtain stable cell lines with CCNB1 knockdown. Knockdown efficiency was validated by Western blotting and qPCR. The specific shRNA sequences were as follows (Table 1).
For the rescue experiments, a full-length human CCNB1 cDNA sequence was cloned into a lentiviral overexpression vector (pLVX-EF1α, Clontech). The constructed plasmid or an empty vector control was co-transfected with packaging plasmids into 293 T cells to produce lentiviral particles. PANC-1 cells were infected with the viruses and selected with antibiotics to generate stable CCNB1-overexpressing (OE-CCNB1) and vector control cell lines. Overexpression efficiency was confirmed by Western blot analysis.
Immunoprecipitation-mass spectrometry (IP-MS)
IP-MS To identify luteolin’s potential target proteins, immunoprecipitation (IP) followed by mass spectrometry (MS) was performed. Briefly, PANC-1 cells were treated with 20 µM luteolin for 48 h, and cell lysates were prepared using RIPA buffer containing protease inhibitors. Protein A/G beads were used to capture luteolin-bound proteins using an anti-CCNB1 antibody. After washing, the immunoprecipitates were eluted, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The resulting data were analyzed using Proteome Discoverer software (Thermo Fisher) to identify luteolin-binding proteins.
Animal experiment
Ten 5-week-old female BALB/c nude mice were purchased. After one week of acclimatization, 6-week-old mice were used for the establishment of the xenograft model. For the PANC-1 xenograft mouse model, PANC-1 cells were injected subcutaneously into 6-week-old female nude mice at a rate of 1 × 107 cells. When the tumor volume reaches about 50-70mm3, all mice were distributed into control and treatment groups, with 5 mice each group. The drug doses for the control and treatment groups are shown below: control group, by gavage with PBS every day; and treatment group by gavage with luteolin at 180 mg/kg every day. Mouse body weight and tumor volume were recorded every 3 days. Survival rate was recorded daily. After 21 days of continuous administration, the mice were euthanized, tumors were harvested and tumor weight was recorded. Tumor volume was calculated using the following formula: tumor volume (mm3) = 0.5 × Length × Width2. The tumors, hearts, livers and kidneys were excised immediately, fixed with 4% paraformaldehyde. And the tumors, hearts, livers and kidneys were cut into sections. All the tissue was sectioned, dewaxed, and evaluated for structural changes with hematoxylin and eosin (H&E) staining. The tumor tissue sections were stored in an oven at 60℃ for 24 h, then dehydrated with xylene and ethanol gradient (100%−70%) in sequence. After continuous incubation with antigen solution and 3% H2O2 for 30 min, the slides were rinsed with water and incubated with primary antibody at 4℃ overnight. The slides were rinsed again, incubated with the corresponding secondary antibody for 30 min, and then stained with hematoxylin. The expressions of CCNB1 and Ki67 were then assessed using fluorescence microscopy. Euthanasia was performed by gradual CO₂ inhalation at a flow rate displacing 20–30% of the chamber volume per minute for at least 5 min. Death was confirmed by cessation of breathing and absence of heartbeat. This protocol ensured rapid unconsciousness and minimized suffering. All experiments and procedures were carried out according to the principles of the Committee on Ethics of Animal Experiments of Fudan University Shanghai Cancer Center. And the experiment was approved by the Institutional Animal Care and Use Committee (permission number: FUSCC-IACUC-2023241) of Fudan University Shanghai Cancer Center.
Statistical analysis
Statistical analysis was conducted by using GraphPad Prism 9.0 software (La Jolla, CA, USA). Data are presented as mean ± standard deviation (SD) from at least three independent experiments. Statistical analyses were performed using two-tailed Student’s t-test or one-way ANOVA with Tukey’s multiple comparisons test. P < 0.05 was considered significant. Values of P < 0.05 were considered significant.
Results
Luteolin exhibited anti-cancer activity in vitro
To assess the effect of luteolin on cell viability, PANC-1 and SW1990 cells were treated with different concentrations of luteolin for 24 h, 48 h, 72 h and detected by CCK-8 assay. Luteolin demonstrated a concentration-dependent inhibitory effect on the activity of PANC-1 and SW1990 cells, with IC50 values at 48 h being 46.98 µM for PANC-1 cells and 36.12 µM for SW1990 cells (Fig. 1A-B). Based on the IC50, 10, 20 µM were selected as the working concentration for the subsequent experiments. Luteolin reduced the viability of PANC-1 and SW1990 cells in a dose- and time-dependent manner at concentrations of 10 µM and 20 µM (Fig. 1C-D).
We then utilized patient-derived tumor organoids (PDOs) from freshly resected tumor tissues to evaluate the anti-cancer activity of luteolin in PC. Consistent with results in 2D cell culture, the PDOs exhibited varying degrees of growth inhibition (Fig. 1E-F). We treated three independent patient-derived organoid (PDO) lines with a concentration gradient of luteolin. Dose-response curves were generated (Fig. 1G), with IC₅₀ values of 18.8 µM (PDO-01), 13.7 µM (PDO-02), and 30.1 µM (PDO-03).
Effects of the luteolin on pancreatic cancer growth. (A) The viability of the PANC-1 cells by CCK‑8 assay. (B) The viability of the SW1990 cells by CCK‑8 assay. (C-D) CCK8 assay of PANC-1 and SW1990 cell proliferation at 24, 48, and 72 h after treated with luteolin. (E-F) Effect of luteolin on pancreatic cancer organoids. (G) Dose-response curves of three independent PDO lines treated with a concentration gradient of luteolin.
Luteolin inhibited PC cells proliferation, migration and invasion
The colony formation assay was conducted to assess the colony-forming capability of PANC-1 and SW1990 cells. Prolonged exposure of luteolin at 10 and 20µM for 14 days suppressed colony formation in both cell lines (Fig. 2A-B).
As shown in Fig. 2C-D, luteolin markedly inhibited the migration of both PANC-1 and SW1990 cells. Additionally, the invasion capacity of both cell lines was also reduced in a concentration-dependent manner following luteolin treatment (Fig. 2E-F).
Luteolin induced apoptosis and the effect on cell cycle in PC cells
After 48 h incubation of luteolin, Annexin V/PI double staining was conducted to evaluate cell apoptosis in PC cells treated with luteolin. The results indicated that luteolin effectively promoted apoptosis in a dose-dependent manner (Fig. 3A-B). Western blot analysis revealed increased levels of caspase 3, P53, and Bax in PANC-1 and SW1990 cells, while Bcl-2 levels decreased with increasing luteolin concentrations (Fig. 3C).
Flow cytometry showed that luteolin treatment increased the proportion of cells in G1 and decreased the proportion in G2 (Fig. 4A–D). Although this profile may seem inconsistent with a G2/M arrest, subsequent molecular data (downregulation of CCNB1/CDK1 and genetic knockdown experiments) support a primary G2/M blockade leading to secondary accumulation in G1. Flow cytometry analysis of cell cycle distribution revealed a complex phenotype. While luteolin treatment significantly increased the proportion of cells in the G1 phase, it concomitantly reduced the proportion in the G2 phase (Fig. 4A-D). This apparent discrepancy—an observation of G1 accumulation alongside a proposed G2/M arrest—is not uncommon in cell cycle studies and can arise from a potent primary G2/M arrest. Cells stalled in G2/M are unable to divide, which can lead to a subsequent accumulation of daughter cells in the G1 phase in the following cell cycle, or reflect population heterogeneity. Critically, the molecular evidence strongly supports a G2/M arrest mechanism: as detailed in subsequent sections, we observed profound downregulation of the core G2/M regulator CCNB1 and its complex partner CDK1 (Fig. 7), and genetic knockdown of CCNB1 alone was sufficient to recapitulate the G2/M arrest phenotype (Fig. 8). Therefore, we interpret the flow cytometry profile as a secondary G1 accumulation resulting from a primary and potent blockade at the G2/M checkpoint mediated by CCNB1 inhibition. The concomitant increase in G1 phase cells, as detected by flow cytometry, is consistent with a model where a primary arrest at G2/M disrupts normal cell cycle progression, leading to an accumulation of cells in the subsequent G1 phase.
Luteolin induced pancreatic cancer cell apoptosis in vitro. (A, B) Flow cytometric analysis of PANC-1 and SW1990 cells. (C) The bands of apoptosis‑related proteins Bax, Bcl-2, caspase‑3, p53 and beta-actin in PANC-1 and SW1990 cells following treatment of luteolin by western blot analysis. All experiment were run in triplicate. The statistical significance is indicated as ***P < 0.0001.
The effect of luteolin on the cell cycle. (A-D) Luteolin increased the proportion of cells in the G1 phase and reduced the proportion of cells in G2 phase in PANC-1 and SW1990 cells. The statistical significance is indicated as ***P < 0.0001. Note: The observed increase in G1 population alongside a decrease in G2 may represent a secondary consequence of a primary G2/M arrest, as supported by the molecular data in Figs. 7 and 8.
CCNB1 as a potential target of Luteolin in PC
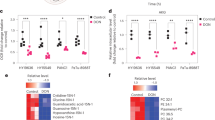
Using the TCMSP and SwissTargetPrediction databases, we identified 56 luteolin-related targets (Supplementary Table 1). Additionally, we obtained 1,102 and 1,745 differentially expressed genes (DEGs) from the GSE101448 and GSE62165 datasets, respectively (Supplementary Tables 2–3). A Venn diagram revealed nine overlapping genes (Fig. 5A).
To further investigate gene expression changes in untreated (NC) versus luteolin-treated PANC-1 cells, we conducted RNA sequencing analysis on triplicate samples. The resulting RNA sequencing dataset was utilized for DEG screening, generating a volcano plot that illustrated the overall distribution of significantly expressed genes. A total of 8,904 DEGs were identified, with 4,138 genes upregulated and 4,766 downregulated (Fig. 5B). The top 30 DEGs are displayed in Fig. 5C.
By integrating the findings from network pharmacology and RNA sequencing, we identified six key genes through Venn diagram (Fig. 5D). Comparison of expression levels of these genes in the TCGA database revealed significant differences between normal and tumor tissues (Fig. 5E). Furthermore, survival analysis based on the TCGA database indicated that CCNB1, NFU2, TOP2A (Figs. 5F-I) and MMP1 were associated with overall survival (OS) in PC patients, while MMP9 and PTGS1 showed no significant differences. Among the four genes assessed in the RNA sequencing data, CCNB1 exhibited the most pronounced differences between the control and treatment groups.
Collectively, these results indicate that CCNB1 is a direct target of luteolin in PC.
CCNB1 may be the target of luteolin on pancreatic cancer. (A) The veen diagram showed the results of network pharmacology and bioinformatics. (B) The volcano map of DEGs. (C) The heatmap of top 30 DEGs. (D) The veen diagram of network pharmacology and RNA-sequencing. (E) The expression difference of key genes between normal and tumor tissue in TCGA database. (F-I) The KM-plot of CCNB1, MMP1, NUF2 and TOP2A. The statistical significance is indicated as ***P<0.001 and **** P < 0.0001.
The intersection between Luteolin and CCNB1
Given that luteolin positively affects PC growth and may target CCNB1, we sought to further explore the interplay between luteolin and CCNB1. Hence we performed SPR and molecular docking. Molecular docking predicted a binding energy of − 7.9 kcal/mol between luteolin and CCNB1 (Fig. 6A–B), suggesting favorable binding. Consistent with this, SPR assays showed a concentration-dependent increase in binding response (Fig. 6D), with an equilibrium dissociation constant (K_D) of approximately 36.7 µM (Fig. 6E).
Next, to confirm the direct binding of luteolin to CCNB1, we employed a cellular thermal shift assay (CETSA). Treatment of PANC-1 and SW1990 cells with luteolin led to significant thermal stabilization of CCNB1 protein (Fig. 6C), demonstrating the intracellular binding between luteolin and CCNB1.
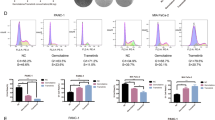
Additionally, we performed immunofluorescence assay to explore the location of CCNB1 in PANC-1 and SW1990 cells. As shown in Fig. 6F–I, luteolin-treated cells exhibited slightly decreased CCNB1 immunofluorescence compared with controls.
The intersection between luteolin and CCNB1. (A) Molecular docking model between luteolin and CCNB1. (B) The detailed binding site of luteolin on CCNB1. (C) The Western blot band of CCNB1 for cellular thermal shift assay. (D) The change of affinity response intensity with the passage of time. (E) The variation of response intensity with the increase of luteolin concentration. (F-I) Immunofluorescence of CCNB1(green) and DAPI (blue). The statistical significance is indicated as *** P < 0.0001.
Luteolin downregulated CCNB1 to regulate cell cycle
CCNB1, a crucial protein for the G2/M phase transition, forms a complex with CDK1 to regulate mitosis entry. Gene Set Enrichment Analysis (GSEA) revealed that luteolin treatment significantly downregulated cell cycle-related pathways, especially those associated with the G2/M phase (Fig. 7A). This suggested that luteolin might exert its anti-cancer effects by modulating cell cycle-related signaling pathways. Further analysis using immunoprecipitation-mass spectrometry (IP-MS) combined with differential gene analysis identified CDK1, CDC40, and TPX2 as significantly downregulated genes in luteolin-treated cells (Fig. 7B-C). Western blotting confirmed these findings, showing that luteolin downregulated CCNB1 expression, which in turn reduced the expression of CDK1, CDC40, and TPX2 (Fig. 7D-H). These results suggest that luteolin inhibits cell cycle progression by downregulating CCNB1 and affecting key G2/M regulatory proteins.
To confirm the role of CCNB1 in luteolin’s mechanism of action, we performed CCNB1 knockdown experiments. In the absence of CCNB1, the expression of CDK1, CDC40, and TPX2 was significantly reduced, indicating the importance of CCNB1 in regulating these proteins during the G2/M phase (Fig. 8). When CCNB1 was knocked down, luteolin’s impact on these proteins was attenuated, with their expression remaining low or unchanged (Fig. 8). This suggests that luteolin’s effects on G2/M phase proteins are dependent on CCNB1, further highlighting the crucial role of CCNB1 in mediating luteolin’s anti-cancer action.
To determine the specificity of luteolin’s interaction, we analyzed our IP-MS data for the presence of other cyclins and core pathway components. Notably, CCNB1 was the only cyclin family member significantly enriched in the luteolin-bound protein complexes, as cyclins A, D, and E were not detected. This indicates a highly specific binding interaction between luteolin and CCNB1(Supplementary Fig. 1 A). Furthermore, interrogation of our transcriptomic data (Supplementary Fig. 1B) revealed that luteolin treatment selectively downregulated CCNB1 mRNA expression without exerting a consistent or significant effect on the mRNA levels of CCNA2, CCND1, CCNE1. Together, these findings demonstrate that luteolin exerts its anti-tumor effects primarily through the specific targeting of CCNB1 at both the protein and transcriptional levels.
Luteolin downregulated CCNB1 and regulated cell cycle-related proteins. (A) The results of GSEA functional analysis. (B) The intersection genes between IP-MS and DEGs. (C) The heatmap of the cell cycle-related genes. (D-H) The cell cycle-related proteins and CCNB1 in PANC-1 and SW1990 cells following treatment of luteolin by western blot analysis. All experiment were run in triplicate. The statistical significance is indicated as *** P < 0.0001.
Overexpression of CCNB1 rescues the molecular effects of Luteolin
To provide direct functional evidence that the effects of luteolin are specifically dependent on CCNB1, we performed a rescue experiment by establishing a stable CCNB1-overexpressing PANC-1 cell line (OE-CCNB1) (Supplementary Fig. 2 A). Western blot analysis confirmed that the expression levels of key G2/M regulatory proteins (CDK1, CDC40, TPX2) were significantly downregulated upon luteolin treatment in vector control cells, recapitulating our previous findings.
Crucially, in OE-CCNB1 cells, the suppressive effects of luteolin on these downstream proteins were markedly attenuated (Supplementary Fig. 2B-C). The expression levels of CDK1, CDC40, and TPX2 remained significantly higher in luteolin-treated OE-CCNB1 cells compared to treated vector controls.
These results demonstrate that maintaining CCNB1 expression levels can reverse the molecular consequences of luteolin treatment, confirming that the disruption of the G2/M regulatory complex is a direct downstream effect of luteolin’s interaction with CCNB1, rather than an off-target effect. This provides compelling functional validation that luteolin’s anti-tumor effects are CCNB1-dependent.
Luteolin suppressed the growth of xenograft tumors
To investigate the anti-tumor activity of luteolin in vivo, we established mouse xenograft models by subcutaneously injecting PANC-1 cells into nude mice, as outlined in Fig. 9A. The nude mice were equally divided into two groups. Results from the tumorigenesis assay indicated that the tumor formation rate, weight, and volume in the luteolin-treated group were significantly lower than those in the saline control group (Fig. 9B-D). Of note, no significant differences in body weights during the administration of luteolin, implying good tolerance of luteolin (Fig. 9E). As demonstrated in Fig. 9F, the results of the H&E staining of organs supported the above results. Additionally, immunohistochemical (IHC) staining of tumor tissues revealed a reduced intensity of Ki67 in the luteolin group compared to the control. Furthermore, the expression level of the target protein CCNB1 was significantly lower in the tumors of mice treated with luteolin (Fig. 9G). These results confirm that luteolin effectively inhibits tumor growth in PANC-1 xenograft models by targeting CCNB1.
Luteolin inhibits the tumor growth of PANC-1 xenografts in nude mice in vivo. (A) The management of PANC-1 xenografts nude mice. (B) Images of tumors at the end of experiment. The mice were administered normal saline (vehicle control), luteolin (180 mg/kg/day, treatment control). Luteolin and normal saline were orally administered by gavage once a day for 14 days. (C) The weight of tumor between luteolin and control group. (D) The tumor volume between the luteolin and control group. (E) The bodyweight of mice between the luteolin and control group. (F) Histopathological examination of organs from PANC-1 xenograft mice after the administration of luteolin or saline. Formalin-fixed organs (heart, liver and kidney) were stained by hematoxylin and eosin. Scale bars represent 100 μm. (G) Luteolin significantly decreased the mitotic event index of the tumor and the expression level of CCNB1. A total of 5 mice were used in each group. The data are expressed as the mean ± SD. ***P<0.001.
Discussion
Pancreatic cancer (PC) remains one of the most aggressive and difficult-to-treat malignancies, with a notably high mortality rate despite advances in surgical techniques and chemotherapeutic strategies, such as gemcitabine-based adjuvant therapy3,18,19. The poor prognosis of PC is largely due to its late diagnosis, aggressive metastasis, and limited efficacy of available treatments, underscoring the need for the development of novel therapeutic strategies. Current therapeutic options for pancreatic cancer primarily include chemotherapy and surgical resection, but the survival rates remain dismal due to the aggressive nature of the disease and the inherent resistance to chemotherapeutic agents20,21,22. Therefore, identifying effective agents and therapeutic targets remains a critical challenge for improving patient outcomes.
Luteolin has garnered increasing attention due to its widespread presence in nature, low toxicity, and significant anti-tumor effects. Previous studies have demonstrated that luteolin can inhibit the growth of various malignant tumors through anti-proliferative and pro-apoptotic mechanisms23,24,25,26. Notably, in the context of pancreatic cancer, several studies have revealed specific mechanisms underlying luteolin’s anti-tumor effects. For instance, luteolin was reported to induce apoptosis and cell cycle arrest in pancreatic cancer cells through modulation of STAT3 signaling27,28 and to inhibit epithelial-mesenchymal transition (EMT) and metastasis via suppression of the TGF-β pathway29. Furthermore, other work has shown that luteolin30. While these studies highlight the multi-faceted anti-cancer properties of luteolin in PC, the specific target and complete mechanism, particularly concerning cell cycle regulation, have remained incompletely elucidated. In this study, we evaluated the inhibitory effects of luteolin on PC. In vitro, luteolin significantly suppressed PC cells growth and inhibited colony formation in a time- and dose-dependent manner, receded PC cells migration and invasion activity, and induced apoptosis. Importantly, luteolin also significantly inhibited the growth of patient-derived organoids (PDOs), providing further evidence of its potent anti-cancer effects in a more physiologically relevant model. In vivo, luteolin administration suppressed tumor growth in a xenograft mouse model without causing significant body weight loss or signs of organ toxicity, suggesting its favorable safety profile.
Building on the in vitro and in vivo effects of luteolin, we explored its mechanism of action in PC. Through network pharmacology and RNA-Sequencing analysis, CCNB1 was recognized as the direct target of luteolin against PC. CCNB1, as the regulatory subunit of maturation-promoting factor (MPF), binds to CDK1 and plays a critical role at the G2/M phase junction, driving cell cycle progression and initiating mitosis. The cell cycle is a tightly regulated process essential for cellular proliferation and tissue homeostasis, and its dysregulation is a hallmark of cancer31. Among the critical phases of the cell cycle, the G2/M transition represents a pivotal checkpoint that ensures genomic integrity before mitotic entry32. This transition is primarily regulated by the maturation-promoting factor (MPF), a complex formed by cyclin B1 (CCNB1) and cyclin-dependent kinase 1 (CDK1)33. In malignancies such as pancreatic cancer (PC), aberrant cell cycle regulation contributes to unchecked proliferation, increased genomic instability, and resistance to therapy34. Dysregulated expression of CCNB1 has been observed in various tumors, including liver35, breast36,37, prostate38 and other cancers39, where it contributes to cell growth, differentiation, apoptosis, and metastasis. In liver cancer, for instance, overexpression of CCNB1 is associated with poor prognosis and aggressive tumor behavior40,41,42, while in breast and prostate cancers, CCNB1 is also frequently upregulated, promoting tumor growth and progression38,43,44,45. However, research on the specific role of CCNB1 in pancreatic cancer remains limited, making this finding particularly noteworthy.
Through a combination of network pharmacology, RNA sequencing, and experimental validation, we demonstrated that luteolin directly targets CCNB1, leading to its downregulation. Western blotting, surface plasmon resonance (SPR), and cellular thermal shift assay (CETSA) confirmed the direct interaction between luteolin and CCNB1, further supported by molecular docking studies that revealed a strong binding affinity between the two. These findings suggest that luteolin’s anti-cancer effects are mediated, at least in part, through its ability to disrupt the CCNB1/CDK1 complex, leading to cell cycle arrest and inhibition of mitosis. This is consistent with previous studies showing that targeting CCNB1 can effectively suppress tumor growth and enhance the sensitivity of cancer cells to chemotherapy. For instance, in a study on liver cancer, targeting CCNB1 with RNA interference led to cell cycle arrest and apoptosis in tumor cells, demonstrating the therapeutic potential of inhibiting CCNB138.
Moreover, our findings are supported by the work of other researchers who have reported the involvement of cyclins, including CCNB1, in the progression of pancreatic cancer. Overexpression of CCNB1 has been linked to the promotion of tumorigenesis and metastasis in pancreatic ductal adenocarcinoma (PDAC), the most common form of PC. Inhibition of CCNB1 has been shown to reduce the proliferative capacity of PDAC cells and impair tumor growth, suggesting that targeting CCNB1 could be a viable therapeutic strategy for treating this malignancy. Furthermore, our study provides additional evidence that luteolin may offer a new approach to treating PC by modulating cell cycle regulatory pathways, specifically through the downregulation of CCNB1 and its downstream targets. The term ‘CCNB1-mediated signalling’ in our title reflects the core finding that luteolin directly targets CCNB1. However, our work extends beyond mere target identification to demonstrate the functional disruption of its key downstream pathway, evidenced by the downregulation of CDK1, CDC40, and TPX2.
An interesting observation in our study was the concurrent increase in G1 population and decrease in G2 population upon luteolin treatment, as detected by flow cytometry. While this may seem at odds with a G2/M arrest mechanism at first glance, such complex cell cycle profiles can be explained by the dynamics of cell cycle perturbation. A strong and sustained block at the G2/M phase prevents mitotic entry and cell division. This can result in an overall increase in the number of cells that, upon eventual adaptation or escape from arrest, enter the next interphase and accumulate in G1 Furthermore, the robust downregulation of the CCNB1/CDK1 complex and its downstream effectors (CDC40, TPX2), the induction of G2/M arrest in CCNB1-knockdown cells, and most importantly, the reversal of the cell cycle phenotype upon CCNB1 overexpression provide compelling molecular evidence that the primary and direct target of luteolin is the G2/M transition via CCNB1. The flow cytometry data thus likely captures a mixed population response, with the molecular data unequivocally pointing to CCNB1-mediated G2/M arrest as the initiating event.
While luteolin’s effects on CCNB1 and cell cycle progression are compelling, further studies are needed to fully elucidate its broader molecular mechanisms in pancreatic cancer. It will be important to explore whether luteolin’s effects extend beyond CCNB1 to other critical cell cycle regulators, such as CDK2, cyclin E, and p21, which also play essential roles in the progression of pancreatic cancer. Additionally, the potential synergy between luteolin and conventional chemotherapeutic agents, such as gemcitabine, should be investigated to determine whether luteolin could enhance the therapeutic efficacy of existing treatments and overcome drug resistance in pancreatic cancer. While this study identifies CCNB1-mediated cell cycle arrest as a primary mechanism, luteolin is known to modulate other signaling pathways (e.g., PI3K/AKT, STAT3) in other contexts. The potential contribution of or crosstalk with these pathways in pancreatic cancer remains an interesting question for future investigation.
Conclusions
Luteolin effectively suppresses PC by targeting CCNB1 and disrupting cell cycle progression. These findings highlight luteolin’s potential as a therapeutic agent, either alone or in combination with other treatments.
Data availability
The RNA sequencing data will be available in NCBI following a year embargo from the date of publication. The data that support the findings of this study are available on request from the corresponding author upon reasonable request. All raw and processed data are publicly available in the NCBI Sequence Read Archive under accession PRJNA1010518.
Abbreviations
- PC:
-
Pancreatic cancer
- RNA:
-
Ribonucleic acid CCNB1cyclin B1
- CDK1:
-
Cyclin-dependent kinases-1
- CDC40:
-
Cell division cycle 40
- CDX:
-
Cell line-derived xenograft
- DMEM:
-
Dulbecco’s modified eagle’s medium
- FBS:
-
Fetal bovine serum
- CCK-8:
-
Cell counting Kit-8
- PBS:
-
phosphate-buffered saline
- TCMSP:
-
Traditional Chinese medicine systems pharmacology database and analysis platform
- DEGs:
-
Differentially expressed genes
- CETSA:
-
Cellular thermal shift assay
- SPR:
-
Surface plasmon resonance
- DMSO:
-
Dimethyl sulfoxide
- KD:
-
Dissociation constant
- RIPA:
-
Radio Immunoprecipitation assay
- PMSF:
-
Phenylmethanesulfonyl fluoride
- BCA:
-
Bicinchoninic acid
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PVDF:
-
Polyvinylidene fluoride
- GSEA:
-
Gene set enrichment analysis
References
Grossberg, A. J. et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma [J]. Cancer J. Clin. 70 (5), 375–403 (2020).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries [J]. CA Cancer J. Clin. 71 (3), 209–249 (2021).
Mizrahi, J. D. et al. Pancreatic cancer [J]. Lancet (London England). 395 (10242), 2008–2020 (2020).
Canto, W. O. O. D. L. D. et al. E M,. Pancreatic Cancer: Pathogenesis, Screening, Diagnosis and Treatment [J]. Gastroenterology, (2022).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs from 1981 to 2014 [J]. J. Nat. Prod. 79 (3), 629–661 (2016).
Arena, A. C. et al. Natural Products as Sources of New Analgesic Drugs [J]. Evidence-based complementary and alternative medicine: eCAM, 2022: 9767292. (2022).
Ahmadi, S. M. et al. Structure-Antioxidant activity relationships of Luteolin and Catechin [J]. J. Food Sci. 85 (2), 298–305 (2020).
Singh Tuli, H. et al. Luteolin, a potent anticancer compound: from chemistry to cellular interactions and synergetic perspectives [J]. Cancers, 14(21). https://doi.org/10.3390/cancers14215373 (2022).
Cho, H. J. et al. Luteolin acts as a radiosensitizer in non–small cell lung cancer cells by enhancing apoptotic cell death through activation of a p38/ROS/caspase cascade [J]. Int. J. Oncol. 46 (3), 1149–1158 (2015).
Im, E., Yeo, C. & Lee, E. O. Luteolin induces caspase-dependent apoptosis via inhibiting the AKT/osteopontin pathway in human hepatocellular carcinoma SK-Hep-1 cells [J]. Life Sci. 209, 259–266 (2018).
Juszczak, A. M. et al. Skin cancer, including related pathways and therapy and the role of Luteolin derivatives as potential therapeutics [J]. Med. Res. Rev. 42 (4), 1423–1462 (2022).
Chen, Y. H. et al. Synergistic combination of Luteolin and Asiatic acid on cervical cancer in vitro and in vivo [J]. Cancers, 15(2). https://doi.org/10.3390/cancers15020548 (2023).
Lin, H. W. et al. Luteolin reduces aqueous extract PM2.5-induced metastatic activity in H460 lung cancer cells [J]. Int. J. Med. Sci. 19 (10), 1502–1509 (2022).
Li, X. Z. et al. Flavopiridol induces cell cycle arrest and apoptosis by interfering with CDK1 signaling pathway in human ovarian granulosa cells [J]. Sci. Rep. 14 (1), 26239 (2024).
Ru, J. et al. TCMSP: a database of systems Pharmacology for drug discovery from herbal medicines [J]. J. Cheminform. 6, 13 (2014).
Daina, A., Michielin, O. & Zoete, V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules [J]. Nucleic Acids Res. 47 (W1), W357–w64 (2019).
Yang, S. et al. Disulfiram downregulates ferredoxin 1 to maintain copper homeostasis and inhibit inflammation in cerebral ischemia/reperfusion injury [J]. Sci. Rep. 14 (1), 15175 (2024).
Siegel, R. L. et al. Cancer statistics, 2023 [J]. CA Cancer J. Clin. 73 (1), 17–48 (2023).
Singhi, A. D. et al. Early detection of pancreatic cancer: opportunities and challenges [J]. Gastroenterology 156 (7), 2024–2040 (2019).
De Dosso, S. et al. Treatment landscape of metastatic pancreatic cancer [J]. Cancer Treat. Rev. 96, 102180 (2021).
Neoptolemos, J. P. et al. Therapeutic developments in pancreatic cancer: current and future perspectives [J]. Nat. Rev. Gastroenterol. Hepatol. 15 (6), 333–348 (2018).
Ko, A. H. Progress in the treatment of metastatic pancreatic cancer and the search for next opportunities [J]. J. Clin. Oncology: Official J. Am. Soc. Clin. Oncol. 33 (16), 1779–1786 (2015).
Wang, Q. et al. Luteolin induces apoptosis by ROS/ER stress and mitochondrial dysfunction in gliomablastoma [J]. Cancer Chemother. Pharmacol. 79 (5), 1031–1041 (2017).
Zang, M. D. et al. Luteolin suppresses gastric cancer progression by reversing epithelial-mesenchymal transition via suppression of the Notch signaling pathway [J]. J. Transl Med. 15 (1), 52 (2017).
Zhang, H. W. et al. Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kγ mediated PI3K/AKT/mTOR/p70S6K/ULK signaling pathway in human breast cancer cells [J]. Sci. Rep. 8 (1), 11255 (2018).
Li, Y. et al. Luteolin directly binds to KDM4C and attenuates ovarian cancer stemness via epigenetic suppression of PPP2CA/YAP axis [J]. Biomed. Pharmacother. 160, 114350 (2023).
Cai, X. et al. The molecular mechanism of luteolin-induced apoptosis is potentially related to Inhibition of angiogenesis in human pancreatic carcinoma cells [J]. Oncol. Rep. 28 (4), 1353–1361 (2012).
Kato, H. et al. DPYD, down-regulated by the potentially chemopreventive agent luteolin, interacts with STAT3 in pancreatic cancer [J]. Carcinogenesis 42 (7), 940–950 (2021).
Huang, X. et al. Luteolin decreases invasiveness, deactivates STAT3 signaling, and reverses interleukin-6 induced epithelial-mesenchymal transition and matrix metalloproteinase secretion of pancreatic cancer cells [J]. Onco Targets Ther. 8, 2989–3001 (2015).
Li, Z. et al. The dietary compound Luteolin inhibits pancreatic cancer growth by targeting BCL-2 [J]. Food Funct. 9 (5), 3018–3027 (2018).
Matthews, H. K., Bertoli, C. & DE Bruin R A M. Cell cycle control in cancer [J]. Nat. Rev. Mol. Cell. Biol. 23 (1), 74–88 (2022).
Fischer, M. et al. Coordinating gene expression during the cell cycle [J]. Trends Biochem. Sci. 47 (12), 1009–1022 (2022).
Pan, Z. N. et al. Propylparaben exposure impairs G2/M and metaphase-anaphase transition during mouse oocyte maturation [J]. Ecotoxicol. Environ. Saf. 283, 116798 (2024).
Qu, M. et al. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle [J]. Proc. Natl. Acad. Sci. U S A. 120 (2), e2214829120 (2023).
Zou, Y. et al. CDK1, CCNB1, and CCNB2 are prognostic biomarkers and correlated with immune infiltration in hepatocellular carcinoma [J]. Med. Sci. Monit. 26, e925289 (2020).
Fang, L. et al. Bioinformatics analysis highlight differentially expressed CCNB1 and PLK1 genes as potential Anti-Breast cancer drug targets and prognostic markers [J]. Genes (Basel), 13(4). https://doi.org/10.3390/genes13040654 (2022).
Fu, H. et al. High expression of CCNB1 driven by NcRNAs is associated with a poor prognosis and tumor immune infiltration in breast cancer [J]. Aging (Albany NY). 14 (16), 6780–6795 (2022).
Chen, X. et al. CCNB1 and AURKA are critical genes for prostate cancer progression and castration-resistant prostate cancer resistant to vinblastine [J]. Front. Endocrinol. (Lausanne). 13, 1106175 (2022).
Chen, E. B. et al. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis [J]. Aging (Albany NY). 11 (18), 7473–7491 (2019).
Hao, L. et al. Study on the mechanism of Quercetin in Sini Decoction plus ginseng soup to inhibit liver cancer and HBV virus replication through CDK1 [J]. Chem. Biol. Drug Des. 103 (6), e14567 (2024).
Gao, Q. et al. Identification of the hub and prognostic genes in liver hepatocellular carcinoma via bioinformatics analysis [J]. Front. Mol. Biosci. 9, 1000847 (2022).
Rong, M. H. et al. CCNB1 promotes the development of hepatocellular carcinoma by mediating DNA replication in the cell cycle [J]. Exp. Biol. Med. (Maywood). 247 (5), 395–408 (2022).
Alwadi, D. et al. Endocrine disrupting chemicals influence hub genes associated with aggressive prostate cancer [J]. Int. J. Mol. Sci., 24(4). https://doi.org/10.3390/ijms24043191 (2023).
Wu, Y. et al. Z-Guggulsterone induces cell cycle arrest and apoptosis by targeting the p53/CCNB1/PLK1 pathway in Triple-Negative breast cancer [J]. ACS Omega. 8 (2), 2780–2792 (2023).
Aljohani, A. I. et al. The clinical significance of Cyclin B1 (CCNB1) in invasive breast cancer with emphasis on its contribution to lymphovascular invasion development [J]. Breast Cancer Res. Treat. 198 (3), 423–435 (2023).
Funding
This study was supported by the National Natural Science Foundation of China (81973616), Project of International Cooperation and Exchanges of the National Natural Science Foundation of China (No. 82020108017), Innovation Group Project of National Natural Science Foundation of China (No.81921002), National Key Research and Development Program of China (No. 2017FYA0205301).
Author information
Authors and Affiliations
Contributions
Linjia Peng Concepted and performed the methodology, investigation, formal analysis and writing. Xiaonan Guo performed the methodology, validation, investigation, data Curation. Xinxin Kong performed the investigation, resources, software.Haiting Zhang performed the investigation, validation. Yanfeng Liang performed the resources, project administration.Qiuli Zhang and Zhiguang Ren performed the methodology, validation, writing – Review & Editing. Daxiang Cui performed the conceptualization, funding acquisition, supervision, writing – Review & Editing, project administration. Linjia Peng, Haiting Zhang and Xinxin Kong were responsible for the work related to the two rounds of revision. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declaration
All procedures involving human tissues were conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Fudan University Shanghai Cancer Center (Approval Number: 2212267-7). Patient-derived organoids (PDOs) were established from freshly resected pancreatic cancer tissues with informed consent. All animal studies were performed in compliance with the Guide for the Care and Use of Laboratory Animals, in accordance with the ARRIVE guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) of Fudan University Shanghai Cancer Center (Permission Number: FUSCC-IACUC-2023241).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, L., Guo, X., Kong, X. et al. The mechanism of luteolin suppressing pancreatic cancer (PC) via cyclin B1 (CCNB1)-mediated signalling. Sci Rep 15, 42044 (2025). https://doi.org/10.1038/s41598-025-25973-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25973-7