Abstract
Endometriosis (EMS) is a chronic gynecological condition affecting 6–10% of reproductive-age women. These lesions, albeit of benign nature, present cancer-like features, and may progress to epithelial ovarian cancer (EOC) through a multistep process. The coexistence of EMS and EOC in the presence of transitional lesions (TL) - i.e. atypical EMS/borderline lesions - is termed as endometriosis-correlated ovarian cancer (ECOC). Given the regulatory role of miRNAs in gene expression and biological pathways, we aimed to identify miRNAs that may drive or modulate the malignant transformation of ovarian EMS. Global miRNA profiling was evaluated on 81 samples (EMS, TL and ECOC) from 44 patients. Principal component analysis and unsupervised clustering revealed two distinct clusters formed by ECOC and ovarian EMS samples, whereas TL were dispersed among the other two. Differential expression analysis (DEA) of TL vs. ovarian EMS, revealed 30 significantly upregulated miRNAs. Comparison of ECOC vs. TL samples yielded 118 significantly upregulated and 77 downregulated miRNAs. Finally, DEA of ECOC vs. ovarian EMS lesions yielded 200 upregulated and 128 downregulated miRNAs. Evaluation of miRNAs commonly deregulated between the three groups showed 14 shared upregulated miRNAs (miR-429, miR-425-5p, miR-200c-3p, miR-200c-5p, miR-200b-3p, miR-200a-3p, miR-183-5p, miR-182-5p, miR-141-5p, miR-141-3p, miR-96-5p, miR-93-5p, miR-10a-5p, miR-10a-3p) with a progressive increase in expression levels, from ovarian EMS to TL and ultimately to ECOC. The identified miRNA expression profiles associated with the progression from ovarian EMS, TL and ECOC provide valuable insights into the molecular progression from benign to malignant lesions and could represent potential biomarkers for the early detection of ECOC.
Similar content being viewed by others
Introduction
Endometriosis (EMS) is a chronic, invaliding, inflammatory, and hormonal dependent gynecological condition affecting approximately 6–10% of reproductive-age women 1. Despite their benign nature, these lesions exhibit cancer-like features such as increased ability to spread and infiltrate in other tissues. In some patients, EMS can develop through a multistep process into epithelial ovarian cancer (EOC), more frequently in the peri- and post-menopausal years 2,3. Endometriosis-correlated ovarian cancer (ECOC) is defined by the Scott criteria as the coexistence of EMS and cancer within the same ovary, with the presence of histological evidence of a transitional/intermediate state between the two conditions 4,5. These transitional lesions (TL) are histologically described as atypical EMS or borderline ovarian tumors. The most frequent EOC histotypes associated with EMS are endometrioid ovarian cancer (ENOC) and clear cell ovarian cancer (CCC) 3.
A few genetic alterations potentially contributing to the malignant transformation of ovarian EMS have been identified 6,7. For instance, driver mutations in the PI3K/AKT/mTOR pathway and its upstream effectors have been frequently found in these lesions 7. Moreover, early occurrence of mTOR pathway alterations in EMS implies that sustained disruption of this pathway may increase the carcinogenic potential of endometriotic cells. Additionally, mutations in other genes such as PIK3CA, PTEN, and ARID1A have been described in ECOC and EMS lesions, but without a clear understanding of the direct contribution to the pathogenesis of ECOC 7. Further, late-stage transformation in ENOC and CCC is associated with mutations in the KRAS gene which activate the RAS/ERK signaling cascade 6,7. Nevertheless, the complete molecular mechanisms of this malignant transformation and progression are yet to be fully understood 6.
It is well known that miRNAs may contribute to the multi-step process of oncogenesis by deregulating expression levels of specific target genes 8,9,10. Some of the altered miRNAs within EMS lesions have been tied to several processes including epithelial to mesenchymal transition (EMT), angiogenesis, cell proliferation, cell adhesion, and invasion 8,11,12. Moreover, miRNAs have an important role in EMS by promoting aberrant activation of the mTOR and PTEN signaling pathways, thus contributing to the pathogenesis of EMS, such as progesterone resistance, altered cellular behaviors, and fostering the transition to cancer 8,9.
The study of miRNA dysregulation has highlighted the deep molecular connections between EMS and OC. In both diseases, miRNAs act as critical modulators of signaling pathways that govern inflammation, cell survival, EMT, angiogenesis, and hormone responsiveness. In EMS, persistent inflammatory and pro-survival miRNA profiles (e.g., miR-21 upregulation, miR-200 family downregulation) allow ectopic endometrial cells to survive in hostile environments, resist apoptosis, and acquire invasive properties. Similarly, in OC, these same miRNAs contribute to oncogenic transformation, chemoresistance, and metastasis by converging on central axes such as PI3K/AKT/mTOR, NF-κB, and TGF-β/EMT signaling. A particularly striking overlap is the shared dysregulation of miR-21, miR-200 family, and miR-451, which appear across both conditions. While the directionality of change may vary depending on tissue type and disease stage, their recurrent involvement strongly suggests that these miRNAs represent a molecular continuum linking benign and malignant states. This is especially relevant in the pathogenesis of ECOC where long-standing endometriotic lesions are thought to accumulate additional genetic and epigenetic “hits,” facilitated in part by aberrant miRNA landscapes, that drive malignant transformation.
To the best of our knowledge, this is the first study to perform global miRNome profiling on the full spectrum of ovarian EMS, TL, and ECOC in a prospectively enrolled and pathologically validated cohort. Although previous studies examined miRNA alterations in EMS or ovarian cancer separately, they often lacked TL that represents the histological and molecular bridge between benign and malignant tissue. Our study fills this critical gap by identifying miRNA signatures that change progressively across these three disease states. By providing an integrated view of miRNA dysregulation, including their targeted pathways, during this stepwise transformation of EMS into ECOC, our work advances the understanding of disease pathogenesis and opens avenues for early biomarker development and targeted therapeutic strategies.
Materials and methods
Study design and patient population
This was a single-center study which enrolled patients prospectively. The study was funded by the Italian Ministry of Health (ENDO-2021–12371926). Patient enrollment took place between November 1, 2021, and December 30, 2023, at the Division of Oncologic Gynecology, IRCCS, Azienda Ospedaliero-Universitaria di Bologna, Italy. Ethical approval was obtained from the Ethics Committee of Area Vasta Emilia Centro (AVEC) under reference number CE 923/2021/Oss/AOUBo. Informed consent was obtained from the patients involved in the study. All methods were performed in accordance with the relevant guidelines and regulations.
Inclusion criteria were: (i) histologically proven ovarian EMS; (ii) histologically proven ECOC and/or borderline or atypical EMS both arising from ovarian EMS according to the Scott criteria 4; (iii) surgical resection performed at our center; (iv) available material for RNA extraction; (v) written informed consent; (vi) women above 18 years old.
Exclusion criteria were: (i) a documented history of cancer within the previous 5 years; (ii) prior pelvic radiotherapy, or (iii) systemic chemotherapy administered in the 5 years preceding the study.
Demographic, clinical, surgical, pathological and follow-up data were collected from each patient’s medical records.
MiRNA isolation from FFPE samples
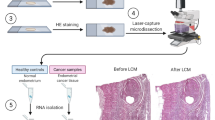
Tissue specimens were collected during the patient’s surgical procedure and processed using standard hospital laboratory procedures to obtain formalin-fixed paraffin-embedded (FFPE) tissues. Three types of tissues were collected from patients. Namely, EMS lesions, TL – comprising atypical endometriosis and borderline tumors, and ECOC (Supplementary Methods).
Serial sections of 3 μm and 10 μm thickness were sliced from the FFPE blocks. The 3 μm sections were stained with hematoxylin and eosin (H&E) and reviewed by a dedicated pathologist, who identified and marked the regions of interest for dissection. Using microscopic guidance and sterile surgical blades, the marked areas were dissected to ensure the selection of regions containing ovarian EMS, TL (atypical and borderline lesions), or ECOC. This methodology also ensured the sample purity for downstream analyses. Atypical EMS and borderline ovarian tumors were grouped together and designated as ‘transitional lesions’ (TL).
RNA was isolated separately from each sample, including those obtained from the same patient, to ensure independent processing and analysis. Specifically, total RNA was isolated from FFPE sections using the RecoverAll Total NucleicAcid Isolation Kit (Thermo Scientific), in accordance with manufacturer’s instructions. RNA quantification was performed using Qubit fluorometer (Thermo Scientific). Only samples yielding a minimum of 100 ng of RNA were deemed suitable and thus selected for subsequent analyses.
MiRNome profiling and annotation
Global miRNome profiling was performed for all samples by high-throughput sequencing. QIAseq miRNA Library Kit (Qiagen) was used to prepare the cDNA libraries following manufacturer’s instructions. Specifically, RNA libraries were prepared from 50 ng of RNA. Library quality and quantification were assessed using High Sensitivity DNA Kit on the TapeStation system (Agilent). Barcoded cDNA libraries were pooled to achieve a final concentration of 4 nM, with the final concentration being verified on the same Agilent system. Sequencing was conducted on the NextSeq 500 platform (Illumina, San Diego, CA) using 75-bp single-end reads. Data analysis was carried out using Qiagen GeneGlobe miRNA sequencing pipeline, which included sample demultiplexing, adapter and low-quality read trimming, alignment to the human transcriptome (GRCh38) for miRNA identification, and quantification of Unique Molecular Identifiers (UMIs) to determine raw expression values for each sample. The resulting data matrix containing the raw counts for each miRNA was used for further analyses.
Annotation of miRNAs association with cancer and their tumor suppressor or oncomiR role was done using the Cancer miRNA Census (CMC) classification 13.
Differential expression and functional enrichment analysis
Downstream analyses were conducted in R version 4.4.0 (2024-04-24 ucrt) - “Puppy Cup" 14. Batch effects across different runs were adjusted using the ComBat_seq function from the Bioconductor sva (version 3.52.0) package 15. Normalized expression values obtained through DESeq2’s variance-stabilizing transformation (VST) were used to assess samples variation and pattern distribution through Principal Component Analysis (PCA). Heatmaps 16,17 were used to visualize overall expression profiles and perform sample clustering through unsupervised learning methods. Differential expression analysis (DEA) was performed using DESeq2 (version 1.44.0) R package 18, that models count-based RNA-seq data with a negative binomial distribution to account for biological variability and overdispersion. This approach also uses shrinkage estimators for variance and fold change, improving stability and interpretability, while accounting for differences in sequencing depth between samples and is specifically recommended for count-based RNA-seq data. All p-values were adjusted following the Benjamini-Hochberg (FDR) procedure 19. Differentially expressed miRNAs were presented using Vulcano plots generated with the EnhancedVolcano (version 1.22.0) package 20. Shared miRNAs between comparisons were illustrated using Venn diagrams 21. MiRNA annotation, gene target 22 and pathways enrichment analyses 23 were carried out using the miEAA tool 24. Targeted transcription factors were identified using the JASPAR database 25. Relationships and gene interactions were evaluated using STRING 26.
Real-time PCR validation analyses
To check the goodness of the statistical analysis, two significantly dysregulated miRNAs - identified from the NGS analysis - were randomly selected for validation by quantitative real-time PCR (qRT-PCR). A set of 20 samples used for sequencing (n = 10 EMS and n = 10 ECOC) was analyzed. RNA was retrotranscribed using the TaqMan™ Advanced miRNA cDNA Synthesis Kit. (Thermo Fisher Scientific), following the manufacturer’s protocol. and qRT-PCR was carried out with TaqMan probes (miR-10a-5p (ID: 479241), miR-200c-3p (ID: 478351)); miR-16-5p (ID:477860) was used as internal reference. Each reaction was run in triplicate on a QuantStudio 7 Flex system (Applied Biosystems). The data were then analyzed using the Thermofisher Cloud App, using the method of ΔCt.
Statistical analysis
MiRNA expression levels were plotted using boxplots, with the whiskers’ length set to 1.5*interquartile range (IQR). Individual data points were also presented. A p-value of 0.05 was considered statistically significant for single comparisons, while the Benjamini-Hochberg (FDR) 19 adjusted p-value was used to control multiple comparisons and reduce the likelihood of false positives.
Results
Patients’ characteristics
A total of 81 samples obtained from 44 patients were analyzed in this study. The median age at diagnosis was 56 years, with more than half of the patients having been through menopause (59%). The most frequent stage at diagnosis was FIGO stage I found in 48% of patients followed by stage II in 37% and stage III in 14% of cases. Multiple sample types were obtained from individual patients due to the co-presence of different tissue states, i.e. EMS, TL and ECOC tissues (Fig. 1A). This resulted in a total of n = 30 (37.0%) ovarian EMS samples, n = 22 (27.2%) TL, and n = 29 (35.8%) ECOC samples. General and clinical details of the cases included in the study can be found in Supplementary Table 1.
A) Presentation of study workflow and main analyses. B) Unsupervised clustering heatmap showing distinct grouping of ECOC and ovarian EMS samples while transitional lesions samples lacked a clear separation. miRNAs commonly upregulated (from Fig. 1E) in the ECOC and transitional lesions samples are marked with a star. C) PCA with PC1 density plot based on the miRNAs expression of 81 samples. D) Volcano plots showing the differentially expressed miRNAs between ovarian EMS, transitional lesions and ECOC tissues. The threshold for the FDR adjusted p-value was set at 0.05 while the threshold for log2FC was set at 1.5. Blue dots represent downregulated miRNAs, and red dots represent upregulated miRNAs. E) Venn diagrams showing the distribution of significantly upregulated and downregulated miRNAs across the differentially expressed comparative analyses. PCA: principal component analysis. EMS: ovarian endometriosis; TL: transitional lesions; ECOC: endometriosis-correlated ovarian cancer.
MiRNA profiles in ovarian EMS, transitional and ECOC lesions
The RNA sequencing yielded an average of 9.07 × 106 reads per sample with a range of 4.66 × 106 – 1.25 × 107. A total of 1181 miRNAs were detected; miRNAs with low counts defined as less than 5 reads per sample (on average) were removed for the subsequent analyses.
First, we sought to evaluate the overall trends and expression patterns of the miRNome by performing a PCA and hierarchical clustering on the samples. PCA of the entire cohort of samples revealed two distinct clusters formed by ovarian EMS and ECOC samples whereas TL samples were not grouped in one independent cluster but rather dispersed among the other two. Principal component (PC) 1, which explains 22.5% of the total variance, showed a good separation of ECOC from ovarian EMS as illustrated in the density plots represented above the PCA graph (Fig. 1B). Unsupervised clustering heatmap of samples and miRNAs highlighted a good grouping of ECOC and ovarian EMS samples. Similarly to the PCA, TL samples did not form their own group and were scattered throughout the heatmap (Fig. 1C).
Next, we carried out a DEA to compare miRNA expressions between the three types of samples (EMS, TL and ECOC). Volcano plots in Fig. 1D highlight the deregulated miRNAs observed in each comparison. In particular, when comparing TL with ovarian EMS samples, we identified 30 significantly upregulated miRNAs out of which 23 had a logFC > 1.5. However, no miRNAs were significantly downregulated in this analysis. Comparison of ECOC vs. TL samples, revealed a total of 118 significantly upregulated miRNAs out of which 40 had a logFC > 1.5 and 77 downregulated, with 15 passing the logFC>|1.5| threshold. Among the significantly upregulated miRNAs, two were classified as cancer associated (miR-375, miR-107). Among the significantly downregulated miRNAs, three (miR-320a, miR-133b and miR-451a) were cancer associated with the latter two being tumor suppressors. Finally, DEA of ECOC vs. ovarian EMS lesions yielded the greatest differences with 200 upregulated and 128 downregulated miRNAs, out of which 98 and 26, respectively, with a logFC>|1.5|. Out of the significantly upregulated miRNAs, three (miR-375, miR-449a, miR-107) were cancer associated. Interestingly, the same three miRNAs (miR-320a, miR-133b and miR-451a) were also significantly downregulated in ECOC.
To identify the early sign of deregulations potentially contributing to ECOC development and progression, we evaluated deregulated miRNAs in common between TL vs. ovarian EMS and ECOC vs. TL. The results showed a total of 14 shared upregulated miRNAs and 68 downregulated miRNAs (Fig. 1E). The 14 miRNAs (miR-429, miR-425-5p, miR-200c-3p, miR-200c-5p, miR-200b-3p, miR-200a-3p, miR-183-5p, miR-182-5p, miR-141-5p, miR-141-3p, miR-96-5p, miR-93-5p, miR-10a-5p, miR-10a-3p) showed a progressing increase in expression levels, from ovarian EMS to TL and ultimately to ECOC (Fig. 2A, Supplementary Table 2). Conversely, no significant downregulation in the early stages of progression from EMS to TL was observed. However, a marked reduction in miRNA expression was observed in ECOC tissues compared to both TL and EMS tissues (Supplementary Table 3). Figure 2B highlights the progressive decrease in expression of the most significantly downregulated miRNAs in ECOC samples. Validation of NGS results for miR-10a-5p and miR-200c-3p was done for a subset of samples and confirmed their significant upregulation in ECOC tissues (Supplementary Fig. 1).
MiRNAs commonly upregulated and downregulated in both TL and ECOC samples (from Fig. 1E). A) The upregulated miRNAs show a gradual increase in expression levels, with ECOC samples having the highest levels. B) A similar trend was seen in downregulated miRNAs (subset of the topmost significant), albeit EMS and TL samples had comparable expression levels. EMS: ovarian endometriosis; TL: transitional lesions; ECOC: endometriosis-correlated ovarian cancer.
MiRNA targeted genes and enriched deregulated pathways
MiRNA targets prediction through miRTarBase showed multiple significant (adjusted p-value < 0.05) genes in all comparisons, some of which were shared among groups (Fig. 3, Supplementary Table 4). In particular, progression from EMS to TL was associated with the deregulation of miRNAs targeting 199 genes, some of which being transcription factors involved in EMT, indicating plasticity and microenvironmental adaptation begin before the complete malignant transformation. In ECOC vs. TL, additional pathway alterations related to growth/survival and metabolism (nucleotide and protein metabolism, vitamins and drug metabolism), with SMAD3 transcription factor, involved in transforming growth factor β (TGFβ) signaling, resulting as the most significantly deregulated. downregulation of several tumor-suppressive miRNAs appears predominantly at this step, consistent with the jump in cell-cycle and apoptosis pathway enrichment. Comparison of ECOC with EMS revealed a total of 2365 deregulated genes. The topmost significant pathways were related to drug metabolism and Metabolism of xenobiotics by cytochrome P450. Glycolysis/gluconeogenesis, TCA cycle, oxidative phosphorylation, and pyruvate metabolism are consistently enriched, indicating a broad rewiring of energy production.
A) Top targeted genes by the significantly deregulated miRNAs. Transcription factors are marked with red bars. B) Venn diagrams showing the commonly targeted genes by significantly upregulated and downregulated miRNAs. Transcription factors are marked in red. C) String analysis of the commonly targeted genes. EMS: ovarian endometriosis; ECOC: endometriosis-correlated ovarian cancer.
A total of three genes were commonly deregulated throughout all the stages of the oncogenic process, namely BCL2, IGF1R and GRHL1 (Fig. 3A).
Functional enrichment analysis was performed on the significantly deregulated miRNAs to identify the most important pathways potentially affected by their dysregulation (Fig. 4, Supplementary Table 4). The topmost significant results revealed multiple pathways involved in signal transduction, metabolism and DNA repair that were altered during EMS progression to TL. Further progression towards ECOC was associated with significant alterations in pathways responsible for nucleotide metabolism, protein processing and drug metabolism. Interestingly, 14 KEGG pathways were significantly deregulated from the early stages of transformation to TL, and showed an even more pronounced deregulation in the ECOC tissues. This involved multiple signaling pathways such as PI3K/AKT signaling, Wnt, Hippo and FoxO signaling. Moreover, processes well-known to play a role in tumorigenesis such as apoptosis, cell cycle and cellular adhesion and communication, were significantly enriched (Fig. 4B).
A) Topmost significant KEGG 23 targeted pathways by the deregulated miRNAs. B) Venn diagram showing the commonly targeted pathways by significantly upregulated and downregulated miRNAs. EMS: ovarian endometriosis; ECOC: endometriosis-correlated ovarian cancer.
Discussion
Summary of the main findings
Our study identified specific miRNA signatures associated with benign ovarian EMS, TL and malignant stages of the disease (Fig. 5). The DEA of ECOC vs. ovarian EMS samples showed a high number of deregulated miRNAs with 98 upregulated and 26 downregulated miRNAs with a logFC>|1.5|. Moreover, our study revealed 14 miRNAs potentially involved in modulating the shift from ovarian EMS to ECOC, as their expression gradually increased from EMS to TL and to ECOC. These dysregulated miRNAs in ECOC, could serve as potential new liquid biopsy biomarkers for early and non-invasive detection of malignant transformation in patients with EMS, and may carry a prognostic value as has been demonstrated for other solid tumors 27. Moreover, pathway enrichment of the significantly deregulated miRNAs showed marked alterations of critical pathways early in the malignant process which are involved in cell cycle, apoptosis, metabolism and immune signaling.
Results in the context of literature
Most of these miRNAs mainly belong to the miR-200 family (miR-141-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-200c-5p, miR-429), which is a crucial regulator of multiple cancer hallmarks 28. For instance, several studies highlighted that they have a critical role in regulating EMT, self-renewal and differentiation of cancer stem cells and apoptosis 28,29. In this context, it is important to highlight that key triggers for EMT include TGFβ family members, which are actively produced by different cell types within a specific tumor and metastatic environment 30. In our cohort, all these miRNAs were upregulated in ECOC samples and the TGFβ signaling cascade turned out to be among the top targeted pathways by the deregulated miRNAs. These results are in agreement with multiple reports by independent research groups who characterized the expression of miR-200 family in EOC patients compared to healthy controls 31,32,33,34,35. Besides the mir-200 family, miR-183 has been shown to play a tumor-promoting role in EOC, potentially through Smad4 regulation via the TGF-β/Smad4 signaling pathway 36 as well as modulate apoptosis and drug resistance 37.
As described for other epithelial malignancies, the increased circulating levels of miR-200 in EOC patients seems to be in contrast with the downregulation observed in EOC-infiltrated abdominal lymph nodes 38. However, this can be explained considering that miR-200 expression could be downregulated during EMT and invasion in the ascitic fluid, while reorganization into metastatic colonies and reactivation of an epithelial differentiation transition (MET) might be associated with increased miR-200 level and possibly their release into the bloodstream of patients with metastatic EOC. Considering the progressive upregulation observed in our cohort of ECOC, this could indicate that the miR-200 family acts from the initial phases of malignant transformation up to the advanced phase of tumor spreading with MET activation. Interestingly, no significant downregulation of miRNAs was observed during the transition from EMS to TL. Such changes in expression became evident only in the later stage of progression to ECOC. This suggests that the pronounced downregulation of specific tumor suppressor miRNAs may play a role in facilitating the transition from TL to ECOC.
Deregulated pathways in the tumorigenic transformation from EMS to TL and ECOC
In our study, regulation of PTEN tumor suppressor activity appeared to be significantly influenced by miRNA upregulation in TL and ECOC samples. PTEN is one of the most frequently inactivated tumor suppressor genes in different tumor types and functions as dual specificity phosphatase. According to a recent systematic review on miRNAs involved in the transformation of EMS into ECOC, the miR-200 family is involved in targeting PTEN 39 as confirmed by our results, showing a progressive increase in expression levels from ovarian EMS to TL and ultimately to ECOC. Within a complex network of target genes and pathways, as highlighted in the aforementioned systematic review, PTEN turned out as one of the primary genes targeted by dysregulated miRNAs (including miR-200, miR-21, miR-214, miR-362, miR-3613, miR-451 and miR-205) that are involved in the transformation of ovarian EMS into ECOC. Additionally, PTEN is targeted by miR-183 cluster members (miR-96, miR-182 and miR-183), that in our study resulted to be present during ECOC onset. miR-182 and miR-183 consistently behave as oncomiRs, showing elevated levels in several cancers; indeed, this cluster has been found to control the expression of the PTEN gene in non-small cell lung cancer, breast cancer and head and neck squamous cell carcinoma 40,41,42. In the study by Braicu et al. 43 miR-182 was overexpressed in ECOC compared to EMS tissue and has been described to play a role in malignant transformation in high-grade serous EOC by sustaining cell proliferation, invasion and migration processes, as was emphasized by Liu et al. 44.
Overall, our research showed that multiple pathways were deregulated during the tumorigenic transformation from EMS to TL and to ECOC. As expected, the PI3K-Akt signaling pathway emerged as a critical driver, promoting cell growth and proliferation through mTOR activation and facilitating immune evasion by interacting with tumor-infiltrating immune cells 45. The Wnt and Hippo signaling pathways were also targeted by the deregulated miRNAs in TL and ECOC, which might lead to excessive cell proliferation and resistance to apoptosis 46,47. The Wnt pathway maintains the balance between proliferation, differentiation, and apoptosis 47, while the Hippo pathway typically acts as a tumor suppressor, by preventing uncontrolled cell growth 46. FoxO signaling, which promotes stress resistance and metabolic efficiency, was also enriched, which might result in increased proliferation and reduced apoptosis 48. Deregulation of HIF-1α signaling could ensure the shift of cellular metabolism toward anaerobic glycolysis to sustain energy production under hypoxic conditions and thus promote tumor invasion and metastasis 49. Finally, deregulation of VEGF signaling could promote angiogenesis, contributing to progression, metastasis, and poor prognosis 50. Together, these deregulated pathways highlight the complex molecular mechanisms underlying this transformation, interestingly beginning as early as the TL stage.
Similarities and differences with the existing literature
Our results build upon and significantly extend current understanding in the field of gynecological oncology. A central finding of our study - the progressive upregulation of the miR-200 family resonates strongly with existing literature that identifies these miRNAs as crucial regulators of EMT, cancer stem cell dynamics, and apoptosis 28. Their established role in OC, particularly in regulating the TGF-β signaling cascade, provides a robust context for our observations 35. Similarly, our identification of miR-183 as an oncomiR in ECOC, possibly through the TGF-β/Smad4 pathway, is supported by prior investigations 51. The deregulated pathways highlighted in our study, including PI3K-Akt, Wnt, Hippo, FoxO, HIF-1α, and VEGF signaling, are universally recognized as important drivers of tumor progression and are consistently described in various cancer types 45,52.
Despite these significant areas of convergence with existing knowledge, our work provides a novel and critical contributions that address several gaps in the literature. A key strength of our work is the identification of a specific panel of 14 miRNAs that exhibit a gradual and consistent increase in expression levels from EMS to TL, and ultimately to ECOC. This progressive upregulation offers unique insights into the molecular drivers of malignant progression, distinguishing early from late-stage alterations.
Interestingly, our study revealed no significant downregulation of miRNAs during the early transition from EMS to TL. Significant reductions in miRNA expression became evident only in the later stage of progression to ECOC. This specific temporal pattern suggests that pronounced downregulation of certain tumor suppressor miRNAs may play a more critical role in facilitating the transition from TL to ECOC, rather than initiating the benign-to-transitional shift. However, as previously mentioned, given the lack of other studies investigating TL lesions, we are unable to compare this result with independent works. Nevertheless, while our data should be considered as exploratory, it is worth of interest in light of the potential scientific impact.
Study strengths, limitations, and future directions
Our study has several strengths such as the prospective enrolment which limited potential biases associated with patient selection and data collection. Moreover, the monocentric nature assured the homogeneity of our cohort from a molecular point of view allowing us to limit potential biases associated with tissue sampling and data analysis. Nevertheless, the small sample size and single center design may limit the generalizability of the findings. Other limitations are related to the samples themselves and the technical aspects of the analyses. Specifically, given the small size of the lesions and the use of FFPE tissues, we have to mention that in some cases we could not collect biological material, and in other cases, even with the available material, RNA isolation was suboptimal, leading to the exclusion of a few samples from our analysis. Moreover, unfortunately, we could not perform any functional validation of our results due to the lack of commercial cell lines for EMS or ECOC. While our analysis identified several candidate miRNAs and pathways of interest, the study design did not allow for longitudinal monitoring of miRNA dynamics during disease progression in the same patients.
Our findings have several potential clinical applications. The progressive upregulation of specific miRNAs, particularly members of the miR-200 family, from EMS to TL and finally to ECOC suggests that they may serve as early biomarkers for malignant transformation. If validated in larger cohorts and in liquid biopsy settings, such circulating miRNAs could enable the non-invasive identification of patients with EMS with an increased risk for ECOC. In addition, because many of the deregulated miRNAs converge on key oncogenic pathways such as PI3K/AKT, TGFβ, and PTEN signaling, they also represent potential therapeutic targets by antagomiRs or miRNA mimics so to limit tumor progression, or improve treatment response. While functional studies and clinical trials are still required, our work lays the foundation for developing miRNA-based diagnostic and therapeutic strategies for these patients.
Conclusions
Our study highlights the crucial role of the miR-200 family in the malignant transformation of ovarian EMS into ECOC. The progressive upregulation of these miRNAs, from ovarian EMS to TL and ultimately to ECOC, suggests their involvement in key tumorigenic pathways, including EMT, PTEN suppression, and activation of TGFβ signaling. These molecular alterations may drive the initiation and progression of malignancy as well as contribute to reshaping the tumor microenvironment, promoting proliferation and metastasis.
The clinical relevance of these findings is twofold: on one hand, miRNAs from the miR-200 family could serve as early biomarkers for identifying patients at risk of malignant progression of endometriosis; on the other, targeting these miRNAs may open new therapeutic avenues to modulate the molecular mechanisms underlying neoplastic transformation. Further functional studies will be essential to validate the precise role of these miRNAs and explore their potential application in personalized treatment strategies.
Data availability
The datasets used and/or analyzed during the current study have been submitted to GEO accession code: GSE292134.
Abbreviations
- CCC:
-
clear cell ovarian cancer
- DEA:
-
differential expression analysis
- EMS:
-
endometriosis
- EMT:
-
epithelial to mesenchymal transition
- ENOC:
-
endometrioid ovarian cancer
- EOC:
-
epithelial ovarian cancer
- ECOC:
-
endometriosis-correlated ovarian cancer
- FFPE:
-
formalin-fixed paraffin-embedded
- PCA:
-
principal component analysis
- TL:
-
transitional lesions
References
Burney, R. O. & Giudice, L. C. Pathogenesis and pathophysiology of endometriosis. Fertil. Steril. 98 (3), 511–519. https://doi.org/10.1016/j.fertnstert.2012.06.029 (2012).
Vercellini, P. et al. Perimenopausal management of ovarian endometriosis and associated cancer risk: when is medical or surgical treatment indicated? Best Pract. Res. Clin. Obstet. Gynaecol. 51, 151–168. https://doi.org/10.1016/j.bpobgyn.2018.01.017 (2018).
Livori, K. & Calleja-Agius, J. Rare but still there: A scoping review on Endometriosis-Associated ovarian cancer. Discov Med. 36 (182), 467–481. https://doi.org/10.24976/Discov.Med.202436182.44 (2024).
Scott, R. B. Malignant changes in endometriosis. Obstet. Gynecol. 2 (3), 283–289 (1953).
Mezzapesa, F. et al. Two possible entities of endometriosis-associated ovarian cancer: correlated or incidental? Int J. Gynecol. Cancer Published Online January. 10, 101634. https://doi.org/10.1016/j.ijgc.2025.101634 (2025).
Herreros-Villanueva, M., Chen, C. C., Tsai, E. M. & Er, T. K. Endometriosis-associated ovarian cancer: what have we learned so far? Clin. Chim. Acta Int. J. Clin. Chem. 493, 63–72. https://doi.org/10.1016/j.cca.2019.02.016 (2019).
Kajiyama, H. et al. Endometriosis and cancer. Free Radic Biol. Med. 133, 186–192. https://doi.org/10.1016/j.freeradbiomed.2018.12.015 (2019).
Zubrzycka, A., Migdalska-Sęk, M., Jędrzejczyk, S. & Brzeziańska-Lasota, E. Circulating MiRNAs related to Epithelial-Mesenchymal transitions (EMT) as the new molecular markers in endometriosis. Curr. Issues Mol. Biol. 43 (2), 900–916. https://doi.org/10.3390/cimb43020064 (2021).
Begum, M. I. A., Chuan, L., Hong, S. T. & Chae, H. S. The pathological role of MiRNAs in endometriosis. Biomedicines 11 (11), 3087. https://doi.org/10.3390/biomedicines11113087 (2023).
Dey Bhowmik, A., Shaw, P., Gopinatha Pillai, M. S., Rao, G. & Dwivedi, S. K. D. Evolving landscape of detection and targeting miRNA/epigenetics for therapeutic strategies in ovarian cancer. Cancer Lett. 611, 217357. https://doi.org/10.1016/j.canlet.2024.217357 (2025).
Azam, I. N. A., Wahab, N. A., Mokhtar, M. H., Shafiee, M. N. & Mokhtar, N. M. Roles of MicroRNAs in regulating apoptosis in the pathogenesis of endometriosis. Life Basel Switz. 12 (9), 1321. https://doi.org/10.3390/life12091321 (2022).
Marí-Alexandre, J. et al. MicroRNAs and angiogenesis in endometriosis. Thromb. Res. 135 (Suppl 1), S38–40. https://doi.org/10.1016/S0049-3848(15)50439-8 (2015).
Suszynska, M. et al. CMC: cancer MiRNA Census – a list of cancer-related MiRNA genes. Nucleic Acids Res. 52 (4), 1628–1644. https://doi.org/10.1093/nar/gkae017 (2024).
R Core Team. — European Environment Agency. Accessed May 1, 2024. https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (2020).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The Sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28 (6), 882–883. https://doi.org/10.1093/bioinformatics/bts034 (2012).
Gu, Z. Complex heatmap visualization. iMeta 1 (3), e43. https://doi.org/10.1002/imt2.43 (2022).
Neuwirth, E. & RColorBrewer ColorBrewer Palettes. Published online April 3, 2022. Accessed February 4, https://cran.r-project.org/web/packages/RColorBrewer/index.html (2024).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Korthauer, K. et al. A practical guide to methods controlling false discoveries in computational biology. Genome Biol. 20 (1), 118. https://doi.org/10.1186/s13059-019-1716-1 (2019).
Blighe, K., Rana, S., Lewis, M. & EnhancedVolcano Publication-ready volcano plots with enhanced colouring and labeling. Bioconductor. 2023;R package version 1.20.0. https://doi.org/10.18129/B9.bioc.EnhancedVolcano
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P. & Minghim, R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 16 (1), 169. https://doi.org/10.1186/s12859-015-0611-3 (2015).
Cui, S. et al. MiRTarBase 2025: updates to the collection of experimentally validated microRNA–target interactions. Nucleic Acids Res. 53 (D1), D147–D156. https://doi.org/10.1093/nar/gkae1072 (2025).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53 (D1), D672–D677. https://doi.org/10.1093/nar/gkae909 (2025).
Kern, F. et al. MiEAA 2.0: integrating multi-species MicroRNA enrichment analysis and workflow management systems. Nucleic Acids Res. 48 (W1), W521–W528. https://doi.org/10.1093/nar/gkaa309 (2020).
Castro-Mondragon, J. A. et al. JASPAR. : the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50(D1):D165-D173. (2022). https://doi.org/10.1093/nar/gkab1113
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51 (D1), D638–D646. https://doi.org/10.1093/nar/gkac1000 (2023).
S, E. et al. Prognostic stratification of metastatic prostate cancer patients treated with abiraterone and enzalutamide through an integrated analysis of Circulating free MicroRNAs and clinical parameters. Front. Oncol. 11 https://doi.org/10.3389/fonc.2021.626104 (2021).
Cavallari, I. et al. The miR-200 family of micrornas: fine tuners of Epithelial-Mesenchymal transition and Circulating cancer biomarkers. Cancers 13 (23), 5874. https://doi.org/10.3390/cancers13235874 (2021).
Feng, X., Wang, Z., Fillmore, R. & Xi, Y. MiR-200, a new star MiRNA in human cancer. Cancer Lett. 344 (2), 166–173. https://doi.org/10.1016/j.canlet.2013.11.004 (2014).
Kumari, A. et al. TGFβ signaling networks in ovarian cancer progression and plasticity. Clin. Exp. Metastasis. 38 (2), 139–161. https://doi.org/10.1007/s10585-021-10077-z (2021).
Meng, X. et al. Diagnostic and prognostic relevance of Circulating Exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 7 (13), 16923–16935. https://doi.org/10.18632/oncotarget.7850 (2016).
Pan, C. et al. Exosomal MicroRNAs as tumor markers in epithelial ovarian cancer. Mol. Oncol. 12 (11), 1935–1948. https://doi.org/10.1002/1878-0261.12371 (2018).
Choi, P. W. et al. Characterization of miR-200 family members as blood biomarkers for human and laying Hen ovarian cancer. Sci. Rep. 10 (1), 20071. https://doi.org/10.1038/s41598-020-77068-0 (2020).
Halvorsen, A. R. et al. Evaluation of prognostic and predictive significance of Circulating MicroRNAs in ovarian cancer patients. Dis. Markers. 2017, 3098542. https://doi.org/10.1155/2017/3098542 (2017).
Ravegnini, G. et al. MicroRNA profiling reveals potential biomarkers for the early transformation of endometriosis towards endometriosis-correlated ovarian cancer. Transl Oncol. 55, 102367. https://doi.org/10.1016/j.tranon.2025.102367 (2025).
Zhou, J., Zhang, C., Zhou, B. & Jiang, D. miR-183 modulated cell proliferation and apoptosis in ovarian cancer through the TGF-β/Smad4 signaling pathway. Int. J. Mol. Med. 43 (4), 1734–1746. https://doi.org/10.3892/ijmm.2019.4082 (2019).
Kandettu, A. et al. Deregulated MiRNA clusters in ovarian cancer: imperative implications in personalized medicine. Genes Dis. 9 (6), 1443–1465. https://doi.org/10.1016/j.gendis.2021.12.026 (2022).
Xu, S. et al. The biphasic expression pattern of miR-200a and E-cadherin in epithelial ovarian cancer and its correlation with clinicopathological features. Curr Pharm. Des 20(11):1888–1895. https://doi.org/10.2174/13816128113199990523
Hamberg, M. et al. MiRTargetLink–miRNAs, genes and interaction networks. Int. J. Mol. Sci. 17 (4), 564. https://doi.org/10.3390/ijms17040564 (2016).
Wang, H. et al. MiR-183-5p is required for non-small cell lung cancer progression by repressing PTEN. Biomed. Pharmacother Biomedecine Pharmacother. 111, 1103–1111. https://doi.org/10.1016/j.biopha.2018.12.115 (2019).
Vahabi, M. et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J. Exp. Clin. Cancer Res. CR. 38 (1), 141. https://doi.org/10.1186/s13046-019-1119-x (2019).
Zhao, Y. S., Yang, W. C., Xin, H. W., Han, J. X. & Ma, S. G. MiR-182-5p knockdown targeting PTEN inhibits cell proliferation and invasion of breast cancer cells. Yonsei Med. J. 60 (2), 148–157. https://doi.org/10.3349/ymj.2019.60.2.148 (2019).
Braicu, O. L. et al. MiRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. OncoTargets Ther. 10, 4225–4238. https://doi.org/10.2147/OTT.S137107 (2017).
Liu, Z. et al. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J. Pathol. 228 (2), 204–215. https://doi.org/10.1002/path.4000 (2012).
He, Y. et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal. Transduct. Target. Ther. 6 (1), 1–17. https://doi.org/10.1038/s41392-021-00828-5 (2021).
Wang, D., He, J., Dong, J., Meyer, T. F. & Xu, T. The HIPPO pathway in gynecological malignancies. Am. J. Cancer Res. 10 (2), 610–629 (2020).
Liu, J. et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal. Transduct. Target. Ther. 7 (1), 1–23. https://doi.org/10.1038/s41392-021-00762-6 (2022).
Tan, L. et al. FoxO1 promotes ovarian cancer by increasing transcription and METTL14-mediated m6A modification of SMC4. Cancer Sci. 115 (4), 1224–1240. https://doi.org/10.1111/cas.16120 (2024).
Qin, J. et al. Hypoxia-inducible factor 1 alpha promotes cancer stem cells-like properties in human ovarian cancer cells by upregulating SIRT1 expression. Sci. Rep. 7 (1), 10592. https://doi.org/10.1038/s41598-017-09244-8 (2017).
Sopo, M. et al. Expression profiles of VEGF-A, VEGF-D and VEGFR1 are higher in distant metastases than in matched primary high grade epithelial ovarian cancer. BMC Cancer. 19 (1), 584. https://doi.org/10.1186/s12885-019-5757-3 (2019).
Naser Al Deen, N. et al. Over-expression of miR-183-5p or miR-492 triggers invasion and proliferation and loss of Polarity in non-neoplastic breast epithelium. Sci. Rep. 12 (1), 21974. https://doi.org/10.1038/s41598-022-25663-8 (2022).
Fu, M. et al. The Hippo signalling pathway and its implications in human health and diseases. Signal. Transduct. Target. Ther. 7 (1), 376. https://doi.org/10.1038/s41392-022-01191-9 (2022).
Acknowledgements
We thank Stefano Friso for the most valuable support in data collection and management and LOTO ONLUS Association for its support of cancer research in women. This work was funded by the Italian Ministry of Health grant code ENDO-2021-12371926 and grant code RC-2022-0505/22; by ALMA IDEA22 “NextGenerationEU” CUP J45F21002000001. Prof. Giorgi was supported by the Italian Ministry of University and Research, with the following grants: PON “Ricerca e Innovazione” 2014–2020; PRIN project 2022CEHEX8; PNRR program for HPC, Big Data, and the Quantum Computing; and by AIRC under MFAG 2021—ID 26319 project—P.I. De Leo Antonio.
Funding
The publication fee of this article was funded by the Italian Ministry of Health, RC-2025-2797531.
Author information
Authors and Affiliations
Contributions
All authors provided substantial contributions to the study and had the opportunity to review and approve the final manuscript. Conceptualization: GR; AMP; Methodology: GR; FGo; Software: CAC; GR; FMG; Validation: FGo; GM; Formal analysis: CAC; FMG; Investigation: GR; FGo; ADL; DDB; SDC; Resources: CAC; ADL; SDC; AA; FMG; Data Curation: SDC; FGo; FM; Writing - Original Draft: CAC; GR; Writing - Review & Editing: FMG; GM; Visualization: CAC; FMG; Supervision: AMP; PDI; FMG; SA; Project administration: PDI; LS; GT; Funding acquisition: AMP; LS; PDI; FMG;
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Ethical approval was obtained from the Ethics Committee of Area Vasta Emilia Centro (AVEC) under reference number CE 923/2021/Oss/AOUBo.
Consent to participate and for publication
Informed consent to participate and for publication was obtained from the patients involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Coada, C.A., Perrone, A.M., Gorini, F. et al. MiRNome alterations drive the malignant transformation of endometriosis into endometriosis-correlated ovarian cancer. Sci Rep 15, 41434 (2025). https://doi.org/10.1038/s41598-025-26466-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26466-3