Abstract
Hematoxylin and eosin (H&E) staining is time-consuming, costly, hazardous, and prone to technician-dependent quality variations. This calls for fast, low-cost, and standardized computational alternatives. Lately, generative adversarial networks (GANs) have shown promising results by generating virtual stains from unstained tissue sections. However, no prior study has systematically benchmarked GANs for optimizing skin histology. Moreover, prior evaluations have focused mostly on the perceptual quality of virtual stains rather than their diagnostic utility. In this paper, we introduce VISGAB, a virtual staining-driven GAN benchmark. To our knowledge, it is the first to systematically evaluate and compare common GAN architectures for skin histology. We have also introduced a novel histology-specific fidelity index (HSFI), which focuses on diagnostic accuracy. VISGAB has been systematically applied to Cycle Consistent GAN (CycleGAN), Contrastive Unpaired Translation GAN (CUTGAN), and Dual Contrastive Learning GAN (DCLGAN) using the E-Staining DermaRepo skin histology dataset. The dataset contains 87 whole-slide images (WSIs) of normal, carcinoma, and inflammatory dermatoses tissues. VISGAB findings identify CycleGAN with superior structural fidelity (SSIM: 0.93, HSFI: 0.81), diagnostic sufficiency (75% nuclear atypia detection), and Turing test success (81%), despite higher mean inference time (~ 1.96 min) and mode collapse risk (~ 25%). Although CUTGAN and DCLGAN offer faster training, artifacts (blurring, overstaining, hallucinations) limit their diagnostic utility. Qualitative evaluations by experts and statistical rigor further substantiate our findings in favor of CycleGAN. This work supports AI-driven histopathology by addressing critical gaps in the literature.
Similar content being viewed by others
Introduction
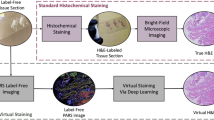
Histological staining is pivotal in clinical diagnostics, enabling visualization of tissue architecture and cellular morphology for diseases such as cancer1. Conventional hematoxylin and eosin (H&E) staining, considered the gold standard, achieves clarity in the delineation of nuclear and cytoplasmic structures and contrast, which enables pathologists to discriminate subtle morphological differences between normal and pathological tissues. However, conventional chemical staining methods are not only time-consuming and labor-intensive, as highlighted in Fig. 1, but also prone to variability in staining quality and consistency2. Such drawbacks have triggered strong interest in creating alternative computational means to virtually mimic H&E staining, thus providing fast, low-cost, and standardized means for histopathological examination. The emergence of generative adversarial networks (GANs) has transformed image synthesis and translation processes.
Two categories of staining frameworks are shown in a top-to-bottom order. The top image (a) shows a conventional chemical staining process (H&E staining) from tissue extraction till microscopic histological image. The bottom image (b) demonstrates a virtual/digital staining process, highlighting a chemical free environment.
Goodfellow3 et al., proposed that GANs can produce high-quality images and set the stage for many image-to-image translation techniques. On the basis of this idea, Isola4 et al. introduced Pix2Pix, a conditional GAN (c-GAN), and improved paired image-to-image translation tasks. Zhu5 et al. applied these concepts to an unpaired scenario with a cycle consistent GAN (CycleGAN). The use of GANs in virtual staining has since been a viable line of research. Various studies6,7,8 have demonstrated that deep learning methods can successfully produce virtually stained histopathological images from unstained tissue sections. These methods focus on maintaining fine-grained details, structural fidelity, and local texture while preserving crucial histological features.
Despite such improvements, no prior study systematically benchmarks GANs for skin histology. This gap is critical, as melanin-rich tissues (e.g., melanomas) pose unique challenges for stain fidelity and structural preservation. Melanoma misdiagnosis rates increase (4–19%) due to variability in histopathological diagnosis9,10, underscoring the need for standardized histological evaluation parameters. The core contributions of this paper include the following: -.
-
VISGAB: To the best of our knowledge, this is the first virtual-staining-driven GAN benchmark for optimizing skin histology using the E-Staining DermaRepo dataset11.
-
Building on this, we introduced and implemented a novel histology-specific fidelity index (HSFI) to quantify the diagnostic utility of staining models alongside perceptual quality.
-
To further evaluate our approach, we performed clinical validation via pathologist ratings and Turing tests.
-
Additionally, we analyzed the correlation between HSFI and both quantitative and qualitative evaluations, establishing the diagnostic potential, accuracy, and alignment of HSFI with these evaluations.
-
Finally, we assessed the risk of mode collapse; a typical issue in GANs, via feature-space entropy.
The VISGAB results identify CycleGAN as the optimal choice among evaluated GANs for virtual H&E staining, citing its superior structural fidelity and diagnostic sufficiency, despite computational, mode collapse trade-offs, and single institutional dataset. Building on these findings, this work advances AI-driven histopathology by establishing reproducible GAN benchmarks and prioritizing clinical relevance. The remainder of this paper unfolds in six sections. Section II summarizes the related work on tissue staining and GANs, while Section III describes the materials and methods for GAN frameworks. Section IV then presents the experimental results, which are comprehensively discussed in Section V. Following this, Section VI addresses limitations and future work, and finally, Section VII offers the conclusion.
Related work
Since the 19th century, histopathological staining has aided disease diagnosis and tissue assessment. Staining techniques have evolved, improving contrast and enabling clearer visualization of cellular structures. H&E staining remains the gold standard in histopathology, but its labor-intensive process, hazardous chemicals, batch variability, and need for expertise drive research into alternatives. Approaches like immunohistochemistry (IHC) and special stains aim to improve diagnostic precision. Histopathological staining highlights H&E-driven nuclear-cytoplasmic differentiation: hematoxylin stains nuclei blue by binding to nucleic acids, while eosin colors cytoplasm pink. Despite improved protocols, batch variability and manual steps still affect reproducibility and diagnostic consistency12,13,14,15, fueling interest in computational solutions.
Studies in the early 2000 s revealed that variable staining intensities and inter-batch differences could impact diagnostic outcomes, especially in cancer pathology. By the late 2000s, researchers investigated automation in H&E processing, but differences in tissue fixation, thickness, and reagent freshness still affect consistency16,17. Early digital pathology efforts focused on color transfer, stain normalization, classification, segmentation, and detection in medical images18,19,20. Concurrent with these methodological breakthroughs, the integration of artificial intelligence (AI) in digital pathology also started reengineering diagnostic processes21, thereby paving the way for novel virtual staining strategies. GANs transform virtual staining by learning domain mappings. Pix2Pix4 pioneered paired translation but required aligned datasets, a limitation addressed by CycleGAN5 via cyclic consistency losses. Accordingly, a U-Net-based virtual staining framework22 was introduced, but it struggled with stain color variability across datasets. StainGAN23, a new stain style transfer method, accurately mimics staining patterns by highlighting the maintenance of tissue morphology throughout the staining process. Subsequent models, such as contrastive unpaired translation GAN (CUTGAN)7, used contrastive learning to preserve local details, whereas dual contrastive learning GAN (DCLGAN)8 combines dual contrastive losses with cyclic consistency for enhanced feature retention.
Despite deep learning advances in digital pathology tasks such as segmentation and classification, critical gaps in virtual staining persist24. These include including benchmarking, diagnostic fidelity, and domain-specific knowledge. All previous studies evaluated GANs in isolation and did not include systematic, standardized comparisons. Traditional evaluation metrics such as structure similarity index (SSIM), peak signal-to-noise ratio (PSNR), Fréchet inception distance (FID), and Kernel inception distance (KID) focus on perceptual and distributional details instead of diagnostic fidelity. This limitation is addressed by HSFI, which is correlated with pathologist concordance (see Section V). Additionally, melanin interference in melanocytic lesions is understudied36. As detailed in Table 1, the literature lacks virtual-staining-driven GAN benchmarking for skin histology optimization. Previous works focused on stain normalization or on demonstrating GAN feasibility for image synthesis. This work bridges these gaps with benchmarking via VISGAB on a dermatopathology-specific dataset, using pathologist-driven evaluations (HSFI, visual Turing tests). It also quantifies computational overhead and mode collapse risks through feature-space entropy to align synthetic outputs with diagnostic needs.
Materials and methods
Dataset
The E-Staining DermaRepo11 comprises 87 pairs of unstained whole-slide images (WSIs) and their stably de-identified corresponding H&E-stained counterparts. All procedures involving human participants, including skin biopsy collection, followed the Declaration of Helsinki. Relevant institutional and national guidelines were also observed. The dataset was curated under an Advarra institutional review board (Protocol No. Pro00035376). Written informed consent was obtained from all participants. All samples were de-identified before analysis. Patient privacy and data handling complied with Health insurance portability and accountability act (HIPAA) standards, where applicable. Skin biopsy samples are from 22 participants (15 males, 7 females), ages 34–83 (median: 67.7). Approximately 20% of WSIs contain more than one tissue section, resulting in 104 paired tissues. The dataset includes tissue from both normal skin and various pathological conditions: basal cell carcinoma (BCC), squamous cell carcinoma (SCC), intra-epidermal carcinoma (IEC), and inflammatory dermatoses. Tissue morphology consists of 47 normal samples, 40 carcinomas, and 17 inflammatory dermatoses. All samples were imaged using a Leica Aperio microscope at 20× magnification (0.50 μm/pixel resolution), under controlled illumination to minimize interslide variation. High-resolution scans ranged from 0.22 to 1.7 gigapixels. To mitigate batch effects, slides were processed within 6 months using identical H&E staining protocols.
Each WSI was resized to 512 × 512-pixel patches with a 256-pixel overlap (50% overlap in both horizontal and vertical directions) between consecutive patches, using a sliding window and bicubic interpolation. For patch-level embedding, we subdivided each 512 × 512 patch into non-overlapping 16 × 16-pixel tiles. The 50% overlap ensures each interior pixel is covered by up to four times, which reduces seam artifacts at reconstruction by providing controlled redundancy. Overlapping regions were fused during WSI reconstruction by seam-aware blending across overlapping patches. To prevent data leakages, all image patches from the same slides were restricted to a single split. For diagnostic consistency, a board-certified pathologist verified concordance between unstained and H&E-stained WSIs, rejecting 5% of the slides due to folding or staining artifacts. Subsequently, as a quality control step, plain background regions (intensity variance < 10%) were discarded with adaptive thresholding25. After quality filtering, 12,450 usable patches (6,225 paired unstained H&E images) remained, of which 80% (9,960) were set aside for training and 20% (2,490) were set aside for testing.
Methodology
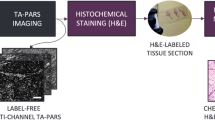
The proposed methodology consists of four main stages i.e., preprocessing, training frameworks as shown in Fig. 2, followed by optimization and convergence, and patch merger at inference.
The top image (a) features preprocessing pipeline, highlighting various steps involved from acquisition of an unstained tissue slide to yielding of training patches. The bottom image (b) features a typical GAN architecture, highlighting its generator(s), discriminator(s), and associated loss function(s).
Preprocessing
A comprehensive preprocessing pipeline was implemented to ensure consistency and reduce potential bias. To produce consistent inputs and improve model robustness, we employed a three-phase preprocessing pipeline of stain normalization, image resizing, and data augmentation. The combined protocol, along with normalizing the dataset, also increases its diversity, providing a rich source of training data. By reducing the inherent variability of histopathology images, the pipeline allows the GANs to learn robust, clinically relevant features.
Each patch undergoes z-score intensity normalization, i.e., scaling pixel values to zero mean and unit variance (µ = 0, σ = 1). This strict transformation mitigates scanner-induced intensity variations while preserving diagnostically critical texture patterns26, an empirically proven method for reproducibility in computational histopathology. Resizing ensures that each patch adheres to the predefined spatial resolution, which is crucial for maintaining the homogeneity required by deep learning algorithms. The images were resized to 512 × 512 pixel patches with a 256-pixel overlap between consecutive patches, using a sliding window and bicubic interpolation, which balances computational efficiency with the retention of cellular-level details. For patch-level embedding, each patch was subdivided into non-overlapping 16 × 16-pixel tiles. This sampling approach provided maximum tissue coverage without redundant feature extraction27,28. The unstained and stained pairs were spatially registered using an affine transformation based on feature matching. During training, we augment the data through random rotations of up to ± 15° and both horizontal and vertical flipping29. This strategy simulates diverse histopathological orientations and spatial arrangements, thereby enhancing the model’s robustness and avoiding overfitting to specific tissue orientations.
Training frameworks – GANs
GANs were trained with a composite loss, balancing adversarial, cyclicconsistency, reconstruction, and contrastive components. Training used floating point16 (FP16) mixed-precision30 on NVIDIA A100 GPU (80 GB VRAM), lasting ~ 48 hours per model. The AdamW optimizer, with learnable affine normalization (β₁, β₂) and cosine annealing, was used. Learning rate and optimizer hyperparameters were searched within (1 × 10− 3, 2 × 10− 3, 1 × 10− 4, 2 × 10− 4), and varied with β1 ∈ (0.5,0.9) and β2 ∈ (0.999,0.9995). The best configuration (2 × 10− 3, β1 = 0.5–0.9, β2 = 0.999) was then manually refined by early stopping on validation SSIM. The learning rate decayed smoothly to zero over 200 epochs. This warm-restart schedule helps the model escape sharp minima. A batch size of 8 was maintained. During training, we alternate gradient updates: Discriminator updates maximized classification loss, while generator updates minimized the combined loss. Progress was monitored on a held-out validation set to prevent overfitting. Networks learned realistic staining patterns in ~ 190 epochs. Training details and composite loss functions are provided in Table 2 and Eq. (1) to (12), respectively.
CycleGAN extends the GANs to unpaired image-to-image translation by learning two-sided mappings. CycleGAN enforces cyclic consistency so that the translated image recovers the original image during two-sided mappings. It employs two generators (GC1, GC2) and two discriminators (DC1, DC2) each. Its dual adversarial approach is augmented with cyclic consistency loss (Lcycle) to obtain unpaired image-to-image translation. Its least-squares adversarial losses (LGAN) stabilize the training31, whereas the cyclic consistency loss (Lcycle) preserves original structures while translating an image into the target domain and vice versa, as depicted from Eq. (1) to (4).
where λcycle penalizes deviations in both forward and backward mapping.
CUTGAN is a contrastive learning architecture for unpaired image-to-image translation, focusing on maintaining fine-grained local details. It has a single generator (GCUT) and a discriminator (DCUT). Its standard adversarial loss (LGAN) ensures that the translated image lies in the target distribution, as expressed from Eq. (5) to (7). CUTGAN uses a patchwise contrastive loss (LpatchNCE) that helps the generated image patch resemble its input and vice versa in a learned feature space. Typically, features are extracted by an encoder (). In addition to the adversarial loss (LGAN), the LpatchNCE is calculated between the source and target domain encoder outputs, allowing the model to learn fine-grained structural information.
where λpatchNCE maximizes mutual information between corresponding unstained and H&E patches. The parameter τ scales the logits as a temperature parameter. The similarity in the feature space is measured by the dot product.
The DCLGAN uses a dual contrastive framework for learning two-sided embedding across domains, instead of using cyclic consistency for training stabilization. DCLGAN consists of generators (G1, G2), discriminators (D1, D2), and a multi-layered perceptron (H1, H2), each with two layers, for learning feature embedding and loss functions to guide model training. The loss functions shown in Eq. (8) to (12) include adversarial loss (LGAN), patch noise contrastive estimation loss (LpatchNCE), and identity loss (Lidentity). The LGAN motivates the generators to produce virtual H&E images that are indistinguishable from actual stained samples. LpatchNCE helps the generated image patch resemble its input and vice versa. The Lidentity restricts generators from yielding unreasonable variations while creating image patches, retaining color, and learning model embeddings. This dual contrastive loss helps retain global and local histological structures.
Optimization and convergence
Periodic checking tracks convergence and avoids overfitting on a holdout subset of patches for all GANs. Effective learning of intricate mappings for virtual staining was facilitated by using composite loss functions, robust mixed-precision optimization, and systematic grid-search hyperparameter tuning. Additionally, incorporating contrastive loss into DCLGAN and CUTGAN, drawing inspiration from advances in contrastive learning32, was key for maintaining both context and tissue information, as shown in Figs. 3 and 4.
Virtually stained H&E-equivalent image patches generated by the respective staining frameworks are represented in three rows from top to bottom. The top row (a) shows the virtually stained patches generated by CycleGAN, with strong structural coherence, high distributional details, the lowest hallucinations, and excellent stain specificity. Middle row (b) depicts CUTGAN patches, demonstrating weak distributional details. The bottom row (c) depicts DCLGAN output patches with slightly lower artifacts and hallucinations than CUTGAN.
Patch merger at inference
WSIs are broken down into smaller, manageable tiles, or patches, for GANs inference. However, a particular concern is that recomposition related to visual consistency, especially color variation and contrast disparity across adjacent virtual patches, could lead to boundary artifacts and compromise diagnostic fidelity. To alleviate these issues, we implemented a 50% patch-overlap policy between neighboring patches in both the horizontal and vertical directions. The overlapping areas are fused further through alpha blending to smoothly transition pixel intensities across overlapping regions33. This spatially aware blending eliminates edge discontinuities and reduces tiling artifacts. The final stitched image, therefore, has seamless transitions across patch boundaries, with a more natural and diagnostically correct look, as depicted in Fig. 5.
Results
Quantitative evaluations
We compared VISGAB with each GAN model using two key image pairs (unstained vs. virtually stained and H&E vs. virtually stained). The image pairs contained both normal and pathological lesions (BCC, SCC, and IEC). We used quantitative metrics such as SSIM, PSNR, FID, KID, LPIPS, and a novel HSFI to capture diagnostic details. Results for SSIM, PSNR, FID, and KID are in Fig. 6; LPIPS and HSFI appear in Fig. 7. All values are reported as mean ± standard deviation with 95% confidence intervals. We used paired t-tests (p ≤ 0.05, n = 100 paired patches).
The SSIM measures perceptual image similarity between two images by specifically identifying failures in melanin-rich areas, especially granularity34,35,36. SSIM values range from 0 to 1, with 1 representing ideal structural correspondence. The consistently lower mean SSIM values of 0.3604–0.5916 across the utilized GANs between unstained and virtually stained images reaffirm the inherent domain differences, which underscore the necessity for GAN-based translation. When comparing H&E-stained to virtually stained images, CycleGAN achieves an SSIM of 0.93 ± 0.005, indicating that its synthetic images are practically indistinguishable from actual stains in nuclear/cytoplasmic structure. In contrast, both CUTGAN (0.22 ± 0.013) and DCLGAN (0.20 ± 0.010) lagged behind with respect to epidermal stratification.
PSNR measures how closely images match at the pixel level through exact color and intensity comparison. A PSNR ≥ 30 dB defines adequate color fidelity in synthetic H&E images37,38,39. All the GANs yield lower mean PSNR values (10.40–11.74 dB) between unstained and virtually stained images, revealing a domain gap and affirming the application of GANs as a bridge between unstained and stained images. For H&E-stained versus virtually stained images, CycleGAN achieves a mean PSNR of 29.06 ± 0.43 dB, maintaining adequate color registration essential for melanin differentiation. In contrast, CUTGAN (13.12 ± 0.29 dB) and DCLGAN (12.61 ± 0.26 dB) fail at this mapping and do not achieve sufficient stain separation.
The FID measures distribution similarity and coherence of high-level features, making it sensitive to textural and structural fidelity. Specifically, the FID computes histopathological realism based on features40,41,42 from a pretrained Inception-v3 network. An FID ≤ 25 is equated with perceived realism in computational histopathology. In contrast, higher mean FID values of 341.1–352.8 between unstained and virtually stained images signify a domain shift between these two types of histology. Comparing H&E-stained to virtually stained images, CycleGAN’s lowest mean FID (27.7 ± 0.64) indicates close correspondence with actual H&E-stained feature distributions. Conversely, the higher mean FID values of CUTGAN (233.6 ± 5.43) and DCLGAN (228.3 ± 5.31) between H&E-stained and virtually stained images indicate degraded distribution similarity, especially in melanin-rich areas.
KID quantifies the distributional dissimilarity between images using maximum mean discrepancy (MMD) of features from a pretrained inception-v3 network43. Lower KID values indicate greater similarity between image sets, while higher values reflect increased dissimilarity. Notably, KID is less sensitive to small sample sizes and feature outliers. For example, higher mean KID values of 0.19–0.25 between unstained and virtually stained images highlight an inherent domain gap, emphasizing the need for sophisticated translation frameworks. In contrast, for H&E-stained versus virtually stained images, CycleGAN’s mean KID (0.010 ± 0.001) suggests synthetic textures are statistically indistinguishable from actual stains. Meanwhile, CUTGAN (0.036 ± 0.008) and DCLGAN (0.017 ± 0.004) maintain global contrast but fall behind in melanin-dense textures.
Quantitative results for perceptual and distributional metrics across two image distributions are shown from left to right. The left image (a) displays quantitative results for unstained versus virtually stained patches, while the right image (b) highlights the same comparison for H&E versus virtually stained patches using the respective staining frameworks. The color insertion highlights the individual quantitative metrics.
LPIPS estimates the similarity between two images by comparing deep feature representations from a pretrained neural network, such as the visual geometry group (VGG) or AlexNet. By design, LPIPS emphasizes semantic and textural differences. Importantly, perceptual realism increases with decreasing LPIPS values44,45,46,47. For context, an LPIPS ≤ 0.15 aligns with pathologists’ acceptability of synthetic histology. In contrast, higher mean LPIPS values in the range of 0.71–0.78 between unstained and virtually stained images emphasize the perceptual gap and indicate a need for strong translation models. Regarding H&E-stained vs. virtually stained images, CycleGAN achieves a mean LPIPS of 0.083 ± 0.002, ensuring that the synthetic images are perceptually indistinguishable from actual stains. On the other hand, the LPIPS of both CUTGAN (0.49 ± 0.01) and DCLGAN (0.48 ± 0.01) maintained global contrast but fell short in semantic and textural differences.
The HSFI is a novel diagnostic fidelity index conceived, derived, and formalized with expert guidance. It targets morphological details and domain knowledge using a weighted scoring system. HSFI mirrors pathologists’ diagnostic pipelines, emphasizing nuclear morphology (form, size, chromatin structure), tissue architecture (epidermal stratification, stromal texture), and stain consistency48,49,50. The HSFI score ranges from 0 to 1, with 1 indicating ideal fidelity. The specific calculation is shown in Eq. (13): -
where α, β, and γ are the optimized weighting coefficients. These are determined through grid search (0.2, 0.3, 0.4, 0.5) to maximize agreement with expert pathologist ratings. After cross-validation, the final weights were set to α = 0.3, β = 0.4, and γ = 0.3. Meanwhile, NMS, TAS, and SCS represent respective scores from nuclear morphology, tissue architecture, and stain consistency, respectively.
The lower mean HSFI values of 0.19–0.23, measured between unstained and virtually stained images, indicate that GAN-based translation is necessary to bridge the gap between these two types of images. Comparing H&E-stained and virtually stained images, the mean HSFI of CycleGAN (0.81 ± 0.02) suggests that synthetic images usually meet diagnostic criteria. CUTGAN (0.58 ± 0.01) and DCLGAN (0.59 ± 0.01) preserve the stromal architecture but do not achieve strong nuclear–cytoplasmic contrast.
Quantitative results for perceptual error and diagnostic fidelity across two image distributions, i.e., unstained versus virtually stained patches, and H&E versus virtually stained patches, are shown as box & whisker plots from left to right. The plots on the left (a) signify perceptual error (LPIPS) while the plots on the right (b) compare diagnostic fidelity (HSFI) of respective staining frameworks.
To demonstrate the accuracy of HSFI and its alignment with perceptual and structural metrics, we computed the Pearson correlation (r) using quantitative evaluations of all the virtual stains generated by the considered GANs and real H&E images. These evaluations are highlighted in Table 3. We then compared these results with HSFI. All correlations are significant at p < 0.05, across four (n = 4) aggregated observations (H&E, CycleGAN, CUTGAN, DCLGAN), and 95% CI were computed using Fisher’s z-transformation. The corresponding correlations are listed in Table 4. HSFI shows consistent and interpretable correlations with established quantitative indices. Its near-perfect positive correlation with SSIM (r = 0.982) and PSNR (r = 0.967) confirms that HSFI captures structural and pixel-level reconstruction fidelity. This aligns HSFI with conventional metrics used in virtual staining and medical image synthesis34,37. The strong negative correlations with FID (r = −0.956), KID (r = −0.931), and LPIPS (r = −0.942) indicate that HSFI inversely tracks perceptual distance and realism metrics. These metrics are widely accepted in generative image evaluation43,51,52. This suggests that HSFI not only aligns with pixel-level quality but also captures perceptual and distributional fidelity across the generative domain.
Qualitative evaluations
Three board-certified dermatopathologists with a combined 35 years of experience assessed both virtual and real H&E images. The experts were unaware of the image source (whether real H&E or virtual stain), patient identifiers, or any case metadata. The images were presented in pairs. Fifty full WSIs and fifty cropped patches at 20× magnification were shown to each of the dermatopathologist. Experts used a standardized rubric53 to rate staining specificity on a 5-point Likert scale. Diagnostic trustworthiness was rated on a binary scale (Yes/No), and artifact detection was graded on a severity scale. To set a baseline for clinical agreement, the same dermatopathologists evaluated the real H&E slides and patches under the same blinded conditions. We calculated pairwise Fleiss’ κ for inter-rater agreement. Diagnostic concordance54 for each image type is the percentage of cases where a dermatopathologist’s diagnosis matches the reference diagnosis, defined as the majority vote of the H&E consensus panel. For each metric, we report mean ± standard deviation with a 95% CI and a p-value ≤ 0.05. The average values of the qualitative evaluations by the dermatopathologists are shown in Table 5.
CycleGAN achieved good H&E consistency (4.4 ± 0.2) and melanin differentiation (4.2 ± 0.3), both critical for melanoma diagnosis. It has demonstrated significant diagnostic trustworthiness in tissue architecture (90 ± 3%) and nuclear atypia (75 ± 5%), but struggled with the accuracy of mitotic figures (60 ± 8%). The minimal blurring, overstaining, and hallucinations (10 ± 2%) each, falling well within clinically acceptable thresholds. Its Turing test success rate (81 ± 3%) is validated by κ = 0.85 ± 0.08, achieving very good inter-rater agreement. CUTGAN struggled with both stain specificity and diagnostic trustworthiness. Pathologists observed severe artifacts in its virtually stained patches: severe blurring (25%), over-staining (20 ± 5%), and hallucinations (45 ± 15%). The Turing test success rate (43 ± 7%) of CUTGAN was mainly restricted by hallucinations, resulting in fair inter-rater agreement (κ = 0.58 ± 0.15). DCLGAN preserved tissue architecture (70 ± 5%), but struggled with nuclear atypia (50 ± 15%), mitotic figure (30 ± 15%), melanin differentiation (3.9 ± 0.4), and over-staining (20 ± 5%). Cytoplasmic blurring in 20% of cases limited its Turing test success rate (57 ± 6%), as validated by κ = 0.68 ± 0.14, reflecting moderate expert agreement.
To show that HSFI matches expert evaluations, we computed Pearson correlations (r) between HSFI and each qualitative metric over four model variants. We considered stain specificity, tissue architecture, artifacts, and hallucinations. For this, we used the qualitative data from Table 5, which includes all virtual stains from the GANs, real H&E images, and the HSFI values. All correlations are significant at p<0.05, based on four aggregated observations (H&E, CycleGAN, CUTGAN, DCLGAN). We calculated 95% CI using Fisher’s z-transformation. Table 6 lists the correlations, their directions, and inferences. Because this analysis uses four model-level observations (one per model), it provides evidence for face validity at the model level and not patch-level clinical validation. Key findings are as follows: HSFI was strongly correlated (r ≥ 0.97) with H&E consistency, melanin differentiation, nuclear atypia sufficiency, mitotic figure accuracy, Fleiss’ κ, and Turing-test success. Artifact metrics, such as over-staining and hallucinations, were both strongly and negatively correlated with HSFI (r = -0.99). The correlation between tissue architecture and HSFI was comparatively lower (r = 0.88), suggesting that the weighting coefficient (β) of tissue architecture in TAS may need refinement. These results suggest HSFI aligns well with expert-rated diagnostic quality at the model level, but additional patch-level validation is required.
To further establish HSFI’s validity, we analyzed qualitative monotonic trends between two model distributions: Model I compares H&E images against CycleGAN, while Model II compares H&E images against the aggregated means of CUTGAN and DCLGAN. With only two data points per metric (n = 2), Pearson correlation coefficients are mathematically degenerate. Therefore, rather than treating these as true correlations, we interpreted them as indicators of perfect monotonic trends. This directionally consistent proportional relationship between HSFI and all qualitative indices confirms HSFI’s clinical interpretability, its alignment with expert evaluations, and its role as a perceptually valid quality index in virtual staining. For this analysis, we used qualitative evaluation data from Table 5, which includes all virtual stains produced by the evaluated GANs and real H&E images, along with the respective HSFI values. Table 7 details the corresponding trend, direction, relative difference magnitude, and diagnostic interpretations. Figures 8 and 9 display two-point comparative radar charts for the model distributions. When comparing H&E to CycleGAN, HSFI shows consistent directional, monotonic alignment with all qualitative evaluation indices. The largest performance gaps are observed in mitotic figure accuracy (~29%) and nuclear atypia (~17%), confirming HSFI’s sensitivity to diagnostic-level fidelity and expert consensus. This demonstrates that as the visual and diagnostic quality of virtual stains diverges from real H&E, HSFI decreases proportionally. Across ten qualitative axes, the average relative reduction from H&E to CUTGAN+DCLGAN is approximately 35–45%. The most significant drops occur in mitotic figure accuracy (~65%), tissue architecture (~42%), and nuclear atypia (~50%). These changes match the greatest HSFI declines (~42%), demonstrating close directional coupling.
The two-point radar plot compares H&E and CycleGAN’s performance across ten qualitative evaluation dimensions, all scaled from 0 to 1. The blue shape (H&E) sets the upper limit across each axis. The orange shape (CycleGAN) has the same form but is smaller, which results in lower scores on each criterion. CycleGAN maintains structure and appearance close to H&E but loses detail in areas where exact shapes matter the most. For fair comparison, metrics where lower scores are better (overstaining, hallucinations) are inverted, so higher numbers always mean better results on the chart.
Computational cost
The computational cost for training and inference is estimated for all the utilized GANs under identical conditions: NVIDIA A100 GPU hardware, FP16 mixed-precision, batch size, optimizer setup, and epoch training schedule. With this standardized setup on NVIDIA A100 (80GB VRAM), the training time varied across all the GANs, as illustrated in Fig. 10. Specifically, CycleGAN is the most resource-intensive (~ 48.33 h, 2.01 days), mainly due to its dual generator-discriminator and cyclic consistency overhead. CUTGAN is the most computationally efficient method (~ 36.67 h, 1.53 days) because of its lightweight architecture and contrastive loss optimization. In contrast, DCLGAN incurs a moderate training cost (~ 43.33 h, 1.80 days) due to the dual contrastive loss and overlap ratios. Despite these differences in training time, all considered GANs require approximately 1.72–1.96 min for inference, as shown in Fig. 11. This inference efficiency results from using fixed pretrained generators and eliminating adversarial or contrastive losses during testing.
Ablation experiment
To rigorously quantify the contribution of each architectural component in CycleGAN, we performed an ablation experiment on the held-out test set. All hyperparameters were held constant (512 × 512 input patches, 16 × 16 tiles, two generators, two discriminators, encoder-decoder, composite loss, learning rate, β₁/β2 ratios, etc.). We evaluated five model variants as depicted in Table 8. We began by assessing training parameters, FLOPs, training time, inference latency, peak GPU memory, SSIM, PSNR, FID, and HSFI. To systematically compare each modification, we first applied 30% pruning with fine-tuning and then performed knowledge distillation to report feature losses and training time. Next, we applied quantized model compression (FP16 to INT8) via calibration for quant-aware training. Following this, we adapted the model with coarser tiles (32 × 32) and then performed runtime engine conversion from ONNX (FP16) to TensorRT (INT8). Timing measurements were averaged across n = 30 representative images. Structured pruning decreased the mean per-image latency by ~ 28%, from 117.6 s (1.96 min) to ~ 84 s (1.40 min), with a minor decrease in HSFI (−0.01). Knowledge distillation nearly halved the runtime (~ 57 s per image, HSFI drop − 0.02). Quantization further reduced per-image latency to ~ 46 s, and runtime engine conversion reduced it again to ~ 29 s, both with minimal performance loss (HSFI ≤ 0.005, SSIM ≤ 0.002). Notably, adapting the model with a 32 × 32 tiles size yielded the best HSFI improvement (+ 0.005) and reduced artifact-related boundary effects, while also providing per-image speedups (~ 88 s, 1.47 min) due to fewer overlapping tiles. Overall, the ablation experiment demonstrates the unique efficiency and fidelity effects of each modification, providing a unified comparison across all model variants and optimization methods. This approach also identifies promising avenues for efficiency optimizations with corresponding performance gains or losses.
The Two-point comparative radar plot: H&E vs. CUTGAN + DCLGAN. This chart visualizes their relative performance across ten qualitative evaluation dimensions, normalized from 0 to 1 . The blue polygon (H&E) defines the upper diagnostic envelope. It serves as the gold-standard reference for morphological and color fidelity. The red polygon (CUTGAN + DCLGAN mean ) contracts sharply inward, especially along axes of mitotic fidelity, architecture, and nuclear atypia. This contraction illustrates perceptual and diagnostic deterioration mirrored by HSFI. Axes for artifacts (overstaining, hallucinations) invert the scale, where higher values indicate better performance. The red contraction toward the center highlights increased artifact frequency.
Mode collapse risk probability
Mode collapse is a common concern in GANs, occurring when the generator produces limited output variations that do not represent the full data distribution. This is particularly significant in virtual staining of histopathological images, where preserving fine textures and color variations is critical. We compared the probability of mode collapse risk based on epochs, risk level, and contributing factors. To measure mode collapse risk, we computed feature space entropy by extracting feature embeddings from real and generated H&E images using a pretrained Inception-v3 network. We then clustered these embeddings using k-means (k = 50, elbow-validated) and calculated the entropy of the cluster distributions55,56. Figure 12 highlights the relative risk probabilities of the considered GANs during training over 200 epochs. Hardware (NVIDIA A100 GPU, 80GB VRAM) and training setup play a major role in identifying the likelihood of mode collapse, as summarized in Table 9. We observed that CycleGAN exhibited the highest risk (~ 25%), CUTGAN had a moderate risk (~ 15%), and DCLGAN demonstrated the lowest mode collapse risk (~ 5–10%). Low feature space entropy (H) corresponds to reduced diversity and greater risk. CycleGAN had the lowest mean entropy (H = 2.1), indicating reduced diversity, while CUTGAN (H = 3.5) and DCLGAN (H = 3.8) showed moderate and mild risks, respectively. Table 10 presents the comparative entropy values for the GANs and H&E images, based on quantitative evaluations.
The image represents the evaluation of the mode collapse risk probability behavior of respective GANs across 200 epochs. CycleGAN demonstrates the highest risk initially (50 epochs) and follows a gentle downwards trend till convergence. CUTGAN initializes at 40% risk but gradually improves diversity before finishing at 15% risk. On the contrary, DCLGAN displays mostly stable behavior with the highest diversity and lowest mode collapse risk compared to other GANs.
Discussion
CycleGAN’s cyclic consistency loss enforces pixel-level reconstruction between original and synthetic images. This directly optimizes for structural preservation and aligns with SSIM/PSNR’s focus on pixel-wise accuracy, not semantic fidelity. Evidence57,58 shows cyclic consistency learning maximizes stain-specific intensity preservation in melanin-dense areas and helps disentangle features. On the other hand, contrastive methods (CUTGAN/DCLGAN) prioritize feature-level consistency over pixel alignment, sacrificing SSIM/PSNR for diversity. The lower mean SSIM and PSNR scores from CUTGAN and DCLGAN are associated with melanin-dense area artifacts, limiting the effectiveness of contrastive losses across complicated histologies. Specifically, melanin can create strong signals interfering with feature extraction, making it harder for contrastive models to capture underlying tissue structure59,60,61. Most importantly, CycleGAN’s mean SSIM (~ 0.93) and PSNR (~ 29 dB) are sufficient for clinical and structural realism, supporting melanoma grading62.
CycleGAN demonstrates superior distributional fidelity, highlighting its ability to replicate feature distributions. This capability is crucial for melanoma diagnosis. The adversarial loss of CycleGAN, combined with cyclic constraints, produces stains that closely match the average statistics of the target domain. This observation aligns with finding63 that cyclic consistency learning minimizes domain shifts in melanin-rich tissues36. Contrastive methods such as CUTGAN or DCLGAN offer greater diversity but sacrifice distributional tightness. CycleGAN’s FID (~ 28) and KID (~ 0.010) meet the requirements for clinical-grade synthetic histopathology64. Furthermore, the FID metric exhibits a strong inverse Pearson correlation (r = −0.98) across all GANs and H&E images. This ensures structural realism with diagnostic concordance, as reported in Table 4. CycleGAN stands out with exceptional perceptual and distributional fidelity. It effectively replicates critical textures and structures. This aligns with studies51,65 that cyclic consistency preserves nuclear granularity and melanin differentiation. CycleGAN’s cyclic consistency preserves mid-level features such as edges and textures that are critical for LPIPS. On the other hand, contrastive losses in CUTGAN and DCLGAN may alter feature distributions.
CycleGAN outperforms CUTGAN and DCLGAN in HSFI (~ 0.81 vs. ~ 0.59), excelling in structure and pixel fidelity. Its virtual stains deliver superior pixel accuracy while retaining perceptual and distributional coherence. This leads to robust diagnostic performance, accurately preserving nuclear morphology, tissue architecture, and stain consistency. With ~ 0.81 HSFI, CycleGAN demonstrates strong AI potential for melanoma grading66. The HSFI shows a very strong positive mean Pearson correlation (r = 0.97) with perceptual and reconstruction fidelity metrics. In contrast, the strong negative mean correlation (r = −0.94) with distributional realism indicates that higher HSFI values correspond to lower distributional realism. This high degree of accuracy and alignment with quantitative metrics shows that HSFI functions as a holistic composite fidelity index, aligning with theoretical expectations and practical diagnostic needs. Quantitative evaluations across virtual stains and H&E images empirically validate HSFI as a clinically interpretable proxy for both quantitative and perceptual image quality, as highlighted in Table 4.
CycleGAN demonstrates a superior visual Turing test success (~ 81%) and high diagnostic trustworthiness (~ 75%). This underscores its practical applicability in clinical workflows and shows clear outperformance of both CUTGAN and DCLGAN. The cyclic consistency constraints of CycleGAN suppress false artifact generation and enhance melanin differentiation fidelity. This finding validates prior work in melanocytic lesion evaluation67. In contrast, CUTGAN creates a substantial artifact burden (~ 25%) and only provides modest interrater agreement (κ ~ 0.58). These results indicate potential diagnostic hazards for sensitive histopathological tasks. DCLGAN moderately maintains robust structural fidelity due to its dual contrastive losses. However, this comes at the expense of accurate melanin representation, reducing its effectiveness in lesion characterization68. The HSFI metric shows consistently high and interpretable correlations (r ≥ 0.97) across qualitative measures and with tissue architecture (r = 0.88). This indicates strong construct validity as a clinical and diagnostic fidelity indicator. The strongest associations are seen with nuclear atypia (r = 0.995), Turing test (r = 0.992), and mitotic figure accuracy (r = 0.987). These results show that HSFI captures diagnostic-grade morphological fidelity and echoes the pathologist’s perceptual and interpretive accuracy. This is consistent with similar results where quantitative image features correlate with expert perceptual quality69. Strong negative correlations with overstaining and hallucinations show that HSFI penalizes generative artifacts that undermine interpretability70. The high correlation with inter-rater agreement (r = 0.973) shows HSFI consistency aligns with pathologists’ consensus, confirming reproducibility across raters71. The near-unity correlation between HSFI and Turing test success (r = 0.992), as highlighted in Table 6, validates HSFI as a perceptual and diagnostic proxy metric. It effectively predicts human diagnostic confidence in virtual stains3.
When restricted to H&E and CycleGAN, HSFI tracks declines in stain consistency, melanin differentiation, nuclear atypia, and tissue architecture, each a key determinant of histopathological interpretability72,73. Table 7 highlights inverse trends for overstaining and hallucinations, showing that HSFI quantitatively encodes perceptual distortions penalized by pathologists43,51. The parallel decline in inter-rater agreement and Turing test success confirms that HSFI reflects both expert agreement and perceptual realism, both core to diagnostic validity. In comparison with lower fidelity models (CUTGAN + DCLGAN), the strongest positive correlations emerge with H&E consistency, melanin differentiation, and nuclear atypia. This demonstrates that HSFI captures structural and cytological degradations affecting diagnostic adequacy69. Consistent declines in overstaining and hallucinations further show that HSFI penalizes perceptual and morphological artifacts, clarifying its link to perceptual and diagnostic realism. Collectively, these findings indicate that HSFI acts as a unified surrogate fidelity index, integrating perceptual realism, morphological accuracy, and diagnostic interpretability for synthetic histology. CycleGAN, with minimal artifacts, reliable pigment discrimination, positive Turing test rates, and strong HSFI alignment across qualitative metrics, emerges as a top candidate for AI-driven diagnostic pipelines in melanoma and dermatopathology74.
Medical stain translation prioritizes structural accuracy over diversity. This makes CycleGAN’s cyclic mappings preferable. Artifact hallucination is a critical concern in histopathology, and CycleGAN’s cyclic constraints help address this issue75. CycleGAN usually requires longer training (~ 48.33 h) than CUTGAN (~ 36.67 h) and DCLGAN (43.33 h). This is mainly due to three factors: two full generators and two discriminators, enforced cyclic consistency at every step, and doubled per-iteration translation and memory needs compared to one-sided methods like CUTGAN. In contrast, DCLGAN uses dual encoders and extra patch losses for more stable, high-quality outputs. While this increases computational demands over CUTGAN, DCLGAN still avoids full cycle reconstruction, so its training time sits between CUTGAN and CycleGAN. Since training is performed offline, this trade-off is acceptable for CycleGAN’s improved structure modeling and reduced artifacts. Efficient inference is essential to integrating patch-based tissue modeling into mainstream digital pathology. Because WSIs often reach gigapixel dimensions, they are partitioned into computationally manageable patches/tiles for AI analysis76. Once models are trained and frozen, they no longer perform gradient computation, loss backpropagation, or weight updates. Their value relies exclusively on optimized forward-pass speed and minimal-latency outputs. These characteristics are decisive for digital pathology. Pathologists require real-time visualization to flexibly investigate multiple diagnostic possibilities. Even millisecond-scale lags can disrupt workflow and diagnostic accuracy. Our evaluated GANs process images in approximately 1.72–1.96 min. This duration may fall short of the rapid feedback essential for histopathological workflows and may render high-throughput screening impractical. Reducing inference time from minutes to seconds per slide is imperative for clinical viability. Accelerating inference upholds interactive diagnosis and delivers AI insights at the point of care77.
While CycleGAN requires more computational power and slower inference speeds, its clinical value must be assessed beyond training time. Deployment feasibility is also crucial; as high computing needs may limit its use in low-resource settings. To address this, we evaluated several optimization models: pruning78,79,80, knowledge distillation81,82,83, quantization84, coarser patches/tiles 28,49, and runtime engine conversion85 against baseline CycleGAN for realizing GAN-based virtual staining systems to integrate seamlessly into time-sensitive clinical environments86, and identifying suitable avenues to reduce computational demands. Ablation experiments showed that structured pruning (removing filters or channels) yields about 2× faster runtime and 30% lower memory and FLOPs with simpler deployment. By integrating fine-tuning, it cuts latency by about 28%, with minimal loss in structure preservation (SSIM/HSFI). The knowledge distillation reduces training parameters and FLOPs by 60% and halves latency and memory. However, it incurs minor loss in SSIM/HSFI, slightly more than pruning because smaller capacity limits fine texture recovery. Quantization with calibration offers about 2.5× faster runtime, negligible fidelity loss (SSIM/HSFI), and halves activation memory, but does not change training FLOPs or training parameters owing to symmetric quantization. Coarser tiles give the model more spatial context per inference. We find that larger context cuts tile-boundary artifacts, lowers per-image latency by 25%, and boosts fidelity. Lastly, TensorRT reduces latency fourfold, cuts memory by about 60%, and results in minimal fidelity loss. Overall, these optimization models decrease CycleGAN’s computational demands and latency, clarifying trade-offs in fidelity and deployment efficiency.
CUTGAN relies on one generator with contrastive learning, making it lightweight and efficient in memory usage. For fast results, choose CUTGAN. On the other hand, DCLGAN incorporates dual contrastive learning. This adds modest computational overhead compared to CUTGAN, but enhances feature diversity and model stability. For applications demanding staining diversity with robust convergence, DCLGAN balances variation and reliability. In comparison, CycleGAN employs two generator-discriminator pairs with cyclic consistency and adversarial losses. This doubles the inference workload for each iteration, making it the most resource-intensive option. When resources permit, CycleGAN delivers the richest, most detailed virtual stains. Ultimately, while CUTGAN is fastest, DCLGAN presents a balanced compromise among stability, diversity, and efficiency.
CycleGAN faces the highest mode collapse risk (~ 25–30%), due to stringent cycle consistency that compels exact reconstruction and inadvertently limits generative diversity. Although this approach favors dominant tissue fidelity but limits to homogenize subtle pathology diversity, and increases potential false negatives in borderline samples. In comparison, CUTGAN’s one-sided mapping shows a moderate collapse risk (~ 15–20%): though it converges quickly and is computationally efficient, it misses fine morphological details, reducing feature diversity. Meanwhile, DCLGAN has a mild collapse risk (~ 5–10%) due to its dual contrastive learning and strong regularization, which encourages the generator to explore varied stains and maintain fidelity. By using context and tissue information, DCLGAN avoids repetitive outputs and preserves key histological features. Several strategies could help reduce mode collapse in CycleGAN while maintaining diagnostic fidelity. First, patch-wise contrastive learning enforces local correspondence between source and output patches and may discourage degenerate mappings7. Second, using contrastive terms in both translation directions with two generator–discriminator pairs reduces hallucinations and improves staining fidelity. Thus, contrastive hybrids may be a strong line of defense against CycleGAN’s mode collapse8. Third, using conditional latent sampling in generators enables structured stochastic mapping and varied outputs. This increases sample diversity and allows controlled generation of stain variants87,88.
CycleGAN’s performance across VISGAB demonstrates readiness for clinical virtual H&E staining, as supported by perceptual, distributional, HSFI, Turing test, and inter-rater agreement results. Higher HSFI scores reflect superior image and diagnostic fidelity, with modest inference time presenting a practical trade-off between fidelity and deployment efficiency. Despite CycleGAN’s low artifact rate, subtle distortions in melanin-rich regions and inflammatory cases may compromise diagnostic accuracy in borderline scenarios. To address this challenge, explainability tools can meaningfully increase clinician trust in virtual-staining systems. However, such explanations must not only pass algorithmic sanity and faithfulness checks, but also earn rigorous validation through active clinician collaboration, ensuring both reliability and acceptance in real-world diagnostic practice. Efficiency may be increased with methods like pruning and knowledge distillation, each affecting performance differently. CycleGAN has the highest mode collapse risk among the utilized GANs, but it prioritizes preserving critical tissue features over generative diversity, a trade-off89 justified by the low interclass variance in our histopathological data. In contrast, CUTGAN offers reasonable structural fidelity and fastest training/inference, but its moderate mode collapse risk, suboptimal melanin differentiation, and higher artifact rate may limit diagnostic safety in melanocytic lesions. DCLGAN occupies the middle ground, with satisfactory diagnostic sufficiency, good feature retention, the lowest mode collapse risk, and moderate computation time. Although CycleGAN leads in clinical sufficiency and minimal artifacts, DCLGAN balances stability, diversity, and efficiency. CycleGAN’s compliance with clinical thresholds90 and minimal artifact will make it well-suited for regulatory approval in melanoma diagnostics91, effectively aligning advanced generative modeling with clinical demands.
Limitations and future work
The promising performance of CycleGAN across VISGAB is tempered by several important caveats. First, testing on a single-institution cohort of 87 WSIs restricts generalizability, especially for rare skin conditions and institutions using different scanners or staining protocols; scanner heterogeneity can dramatically affect performance92. Second, the combined inference latency burden and mode collapse risks reveal a trade‐off, potentially limiting deployment in resource-constrained environments. Third, while general artifact rates are low, subtle distortions in melanin-rich regions and inflammatory cases may compromise diagnostic accuracy in borderline scenarios93. To meet these challenges, future work will prioritize multicenter dermatopathology networks to address diverse scanner and stain conditions94 utilize few-shot learning to improve robustness for rare lesion types95. Examine neural architecture search (NAS) and next-generation optimization models to fine-tune CycleGAN for edge-device compatibility on mobile GPUs96 and adopt discriminator and training regularizers97,98 that directly address mode collapse. In addition, integrating explainability tools such as heat maps will support clinician decision-making by providing insights about a model’s predictions and confidence99. Finally, prospective, real-time evaluation in clinical workflows will assess both clinical utility and compliance with regulatory standards, including the TRIPOD-AI checklist.
Conclusion
This paper institutes VISGAB, a comprehensive benchmarking framework for assessing GAN-based virtual staining of skin tissue histology. VISGAB combines rigorous quantitative evaluations (e.g., SSIM, PSNR, FID, KID, LPIPS, and HSFI) with qualitative evaluations such as pathologist-driven visual Turing tests and inter-rater agreement, collectively addressing perceptual quality, distributional fidelity, contrast accuracy, and diagnostic trustworthiness. By providing a unified platform for image-to-image translation assessment in digital pathology, VISGAB identifies CycleGAN as the leading candidate for clinical integration among utilized GANs for virtual H&E staining. The cyclic consistency constraints of CycleGAN ensure high structural and perceptual realism, as reflected in higher HSFI scores and strongly positive visual Turing test, resulting in diagnostic-grade virtual H&E stains. Nonetheless, CycleGAN’s heavier inference footprint, risk of mode collapse, and occasional subtle distortions in melanin-rich and inflammatory cases underscore the need for clinician vigilance in challenging scenarios. By addressing critical challenges in melanin differentiation and artifact reduction, VISGAB paves the way for future enhancements that reconcile computational rigor and output diversity with clinical efficiency. In line with FDA guidelines on AI as a medical device, future work will prioritize validation across multiple institutions, optimize architectures via edge-friendly NAS, integrate explainable heat maps, and implement targeted strategies to mitigate mode collapse. These efforts collectively aim to advance scalable, interpretable, and high-fidelity AI tools for precision histopathology.
Data availability
The E-Staining DermaRepo (Skin Histology) dataset was collected under Advarra IRB Protocol No. Pro00035376 and includes skin histopathology data. All procedures involving human participants, followed the Declaration of Helsinki according to institutional and national guidelines. Written informed consent was obtained from all participants. Patient privacy and data handling complied with HIPAA standards. The dataset is publicly available at https://biomisa.org/index.php/downloads/. Currently, the research relies on data collected from a single IRB protocol. To the best of our knowledge, there is no publicly available dataset for skin histopathology that contains data from multicenters. We plan to initiate collaborations with additional institutions and integrate new data sources to establish a multicenter dataset in the future. For data assistance, contact Muhammad Usman Akram (usmakram@gmail.com).
References
Bancroft, J. D. & Gamble, M. Theory and Practice of Histological Techniques, (6th ed). (Elsevier Health Sciences, 2008). https://ci.nii.ac.jp/ncid/BA10321241
Dabbs, D. J. (ed) Diagnostic Immunohistochemistry 5th edn (Elsevier, 2019).
Goodfellow, I. J. et al. Generative adversarial networks. Advances Neural Inform. Process. Systems, 27, 2672–2680, (2014).
Isola, P., Zhu, J. Y., Zhou, T. & Efros, A. A. Image-to-image translation with conditional adversarial networks. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, pp. 5967–5976., IEEE, pp. 5967–5976. (2017). https://doi.org/10.1109/CVPR.2017.632 (2017).
Zhu, J. Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks, In. IEEE Conference on Computer Vision (ICCV), IEEE, pp. 2223–2232. (2017). https://doi.org/10.1109/ICCV.2017.244 (2017).
Rivenson, Y. et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomedical Eng. 3 (6), 466–477. https://doi.org/10.1038/s41551-019-0362-y (2019).
Park, T., Efros, A. A., Zhang, R. & Zhu, J. Y. Contrastive learning for unpaired image-to-image translation. Proc. Eur. Conf. Comput. Vis. (ECCV). 776–794. https://doi.org/10.1007/978-3-030-58545-7_19 (2020).
Asaf, M. Z. et al. Dual contrastive learning based image-to-image translation of unstained skin tissue into virtually stained H&E images. Sci. Rep. 14, 2335. https://doi.org/10.1038/s41598-024-52833-7 (2024).
Patrawala, S. et al. Discordance of histopathologic parameters in cutaneous melanoma: clinical implications. J. Am. Acad. Dermatol. 74 (1), 75–80. https://doi.org/10.1016/j.jaad.2015.09.008 (2016).
Katz, I. et al. Variability in the histopathological diagnosis of nonmelanocytic lesions excised to exclude melanoma. Dermatology Practical Concept. 11 (4), e2021094. https://doi.org/10.5826/dpc.1104a94 (2021).
Asaf, Z. et al. E-Staining DermaRepo. Mendeley Data, v1, (2024). https://doi.org/10.117632/gxgg933ny3.1
Lillie, R. D. & Conn, H. J. & biological stain commission. HJ Conn’s biological stains: A handbook on the nature and uses of the dyes employed in the biological laboratory, (9th ed). Baltimore: Williams & Wilkins, (1977).
Kiernan, J. Histological and Histochemical Methods 5th edn (Scion Publishing Ltd, 2015).
Greenwald, S. E., Brown, A. G., Roberts, A. & Club, F. J. Jr Histology and staining. In J. E. Moore Jr. & G. W. Palmer, Biomedical Technology and Devices Handbook, (Boca Raton, FL: CRC Press, (2004).
Horobin, R. W. & Bancroft, J. D. Troubleshooting Histology Stains (Springer, 1998).
Titford, M. The long history of hematoxylin. Biotech. Histochem. 80 (2), 73–78 (2005).
Reinhard, E., Adhikhmin, M., Gooch, B. & Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 21 (5), 34–41. https://doi.org/10.1109/38.946629 (2002).
Shorten, C. & Khoshgoftaar, T. M. A survey on image data augmentation for deep learning. J. Big Data. 60. https://doi.org/10.1186/s40537-019-0197-0 (2019). 6.
Macenko, M. et al. A method for normalizing histology slides for quantitative analysis. In 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, IEEE, pp. 1107–1110. (2009). https://doi.org/10.1109/ISBI.2009.5193250
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image. Anal. 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005 (2017).
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology-new tools for diagnosis and precision oncology. Nat. Reviews Clin. Oncol. 16 (11), 703–715. https://doi.org/10.1038/s41571-019-0252-y (2019).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In 18th International Conference on Medical Image Computing and Computer-assisted Intervention (MICCAI), Springer, 9351, 234–241. (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Shaban, M. T., Baur, C. & Navab, N. & Albarqouni. Staingan: Stain style transfer for digital histological images. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI), IEEE, pp. 953–956. (2019). https://doi.org/10.1109/ISBI.2019.8759515
Deng, S. et al. Deep learning in digital pathology image analysis: a survey. Front. Med. 14, 470–487. https://doi.org/10.1007/s11684-020-0757-3 (2020).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. man. Cybernetics. 9 (1), 62–66. https://doi.org/10.1109/TSMC.1979.4310076 (1979).
Nyúl, L. G., Udupa, J. K. & Zhang, X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging. 19 (2), 143–150. https://doi.org/10.1109/42.836373 (2000).
Ye, H., Su, K. & Liu, Y. Image enhancement method based on bicubic interpolation and singular value decomposition. In 2021 IEEE 4th Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), IEEE, 4, pp. 1972–1976. (2021). https://doi.org/10.1109/IMCEC51613.2021.9482303
Afshari, M. et al. Single patch superresolution of histopathology whole slide images: A comparative study. J. Med. Imaging. 10 (1). https://doi.org/10.1117/1.JMI.10.1.017501 (2023). 017501-1-10.
Wang, J. & Perez, L. The effectiveness of data augmentation in image classification using deep learning. Convolutional Neural Networks Visualization Recognit. 11, 1–8. https://doi.org/10.48550/arXiv.1712.04621 (2017).
Micikevicius, P. et al. Mixed precision training. ArXiv Preprint arXiv:1710 03740. https://doi.org/10.48550/arXiv.1710.03740 (2017).
Mao, X. et al. Least squares generative adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), IEEE, pp. 2794–2802. (2017). https://doi.org/10.1109/ICCV.2017.304
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. In International Conference on Machine Learning, PmLR, pp. 1597–1607. (2020). https://doi.org/10.5555/3524938.3525087
Baudisch, P. & Gutwin, C. Multiblending: displaying overlapping windows simultaneously without the drawbacks of alpha blending. In Proceedings of the SIGCHI Conference on human factors in computing systems, SIGCHI, pp. 367–374. (2024). https://doi.org/10.1145/985692.985739
Wang, Z., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13 (4), 600–612. https://doi.org/10.1109/TIP.2003.819861 (2004).
Taha, A. & Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging. 15 (1), 1–28. https://doi.org/10.1186/s12880-015-0068-x (2015).
Liu, K. et al. VSGD-Net: Virtual staining guided melanocyte detection on histopathological images. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), IEEE, pp. 1918–1927. (2023). https://doi.org/10.1109/WACV56688.2023.00196
Hore, A. & Ziou, D. Image quality metrics: PSNR vs. SSIM. In IEEE 20th international Conference on Pattern Recognition (ICPR), IEEE, pp. 2366–2369., IEEE, pp. 2366–2369. (2010). https://doi.org/10.1109/ICPR.2010.579 (2010).
Setiadi, D. R. I. M. PSNR vs SSIM: imperceptibility quality assessment for image steganography. Multimedia Tools Appl. 80 (6), 8423–8444. https://doi.org/10.1007/s11042-020-10175-3 (2021).
Xie, Z., Du, S., Huang, D., Ding, Y. & Ma, L. A unified fidelity optimization model for global color transfer. In Image and Graphics: 8th International Conference, ICIG Proceedings, Part I 8, Springer, pp. 504–515. (2015). https://doi.org/10.1007/978-3-319-21969-6_4
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, pp. 2818–2826. (2016). https://doi.org/10.1109/CVPR.2016.308
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in Gastrointestinal cancer. Nat. Med. 25 (7), 1054–1056. https://doi.org/10.1038/s41591-019-0462-y (2019).
Jayasumana, S. et al. Towards a better evaluation metric for image generation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition(CVPR), IEEE, pp. 9307–9315. (2024). https://doi.org/10.1109/CVPR52733.2024.00889
Bińkowski, M., Sutherland, D. J., Arbel, M. & Gretton, A. Demystifying mmd gans. arXiv preprint arXiv:1801.01401. (2018). https://doi.org/10.48550/arXiv.1801.01401
Yu, W. et al. Visualizing and comparing AlexNet and VGG using deconvolutional layers. In Proceedings of the 33rd International Conference on Machine Learning, 3, pp. 43–76, (2016).
Wen, S. et al. A methodology for texture feature-based quality assessment in nucleus segmentation of histopathology image. J. Pathol. Inf. 8 (1), 38. https://doi.org/10.4103/jpi.jpi_51_17 (2017).
Thanh-Tung, H. & Tran, T. Catastrophic forgetting and mode collapse in GANs. In International Joint Conference on Neural Networks (IJCNN), IEEE, pp. 1–10., IEEE, pp. 1–10. (2020). https://doi.org/10.1109/IJCNN48605.2020.9207367 (2020).
Nečasová, T., Burgos, N. & Svoboda, D. Validation and evaluation metrics for medical and biomedical image synthesis. In Biomedical Image Synthesis and Simulation, Academic, 573–600. https://doi.org/10.1016/B978-0-12-824482-1.00022-2 (2022).
Sornapudi, S. et al. Deep learning nuclei detection in digitized histology images by superpixels. J. Pathol. Inf. 9 (1), 5. https://doi.org/10.1016/jpi.jpi_74_17 (2018).
Vahadane, A. et al. Structure-preserved color normalization for histological images. In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), IEEE, pp. 1012–1015. (2015). https://doi.org/10.1109/ISBI.2015.7164042
Pantanowitz, L. et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the college of American pathologists pathology and laboratory quality center. Arch. Pathol. Lab. Med. 137 (12), 1710–1722. https://doi.org/10.5858/arpa.2013-0093-cpt (2013).
Zhang, R., Isola, P., Efros, A. A., Shechtman, E. & Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, pp. 586–595. (2018). https://doi.org/10.1109/CVPR.2018.00068
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B. & Hochreiter, S. GANs trained by a two time-scale update rule converge to a local Nash equilibrium. Advances Neural Inform. Process. Systems, 30, 6626–6637, (2017).
Sun, S., Goldgof, G., Butte, A. & Alaa, A. M. Aligning synthetic medical images with clinical knowledge using human feedback. Adv. Neural. Inf. Process. Syst. 36, 13408–13428. https://doi.org/10.5555/3666122.3666711 (2023).
Evans, A. J. et al. Validating whole slide imaging systems for diagnostic purposes in pathology. Arch. Pathol. Lab. Med. 146 (4), 440–450. https://doi.org/10.5858/arpa.2020-0723-CPT (2022).
Kossale, Y., Airaj, M. & Darouichi, A. Mode collapse in generative adversarial networks: An overview. In 8th International Conference on Optimization and Applications (ICOA), IEEE, pp. 1–6., IEEE, pp. 1–6. (2022). https://doi.org/10.1109/ICOA55659.2022.9934291 (2022).
Chi, Z. & Yan, H. Feature evaluation and selection based on an entropy measure with data clustering. Opt. Eng. 34 (12), 3514–3519. https://doi.org/10.1117/12.212977 (1995).
Nerrienet, N. et al. Standardized cyclegan training for unsupervised stain adaptation in invasive carcinoma classification for breast histopathology. J. Med. Imaging. 10 (6), 067502–067502. https://doi.org/10.1117/1.JMI.10.6.067502 (2023).
Liu, S. et al. Generating Seamless Virtual immunohistochemical whole slide images with content and color consistency. In 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI), IEEE, pp. 1–5. (2025). https://doi.org/10.1109/ISBI160581.2025.10981306
Andonian, A. et al. Contrastive feature loss for image prediction. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), IEEE, pp. 1934–1943. (2021). https://doi.org/10.1109/ICCV48922.2021.00491
Lee, H. et al. Contrastive learning for unsupervised image-to-image translation. Appl. Soft Comput. 151, 111170. https://doi.org/10.1016/j.asoc.2023.111170 (2024).
Zhuang, X. The influences of color and shape features in visual contrastive learning. ArXiv Preprint arXiv:2301 12459. https://doi.org/10.48550/arXiv.2301.12459 (2024).
Lee, J. C., Park, H. W. & Kang, Y. N. Feasibility study of structural similarity index for patient-specific quality assurance. J. Appl. Clin. Med. Phys. 26 (3), e14591. https://doi.org/10.1002/acm2.14591 (2025).
Kynkäänniemi, T. et al. The role of imagenet classes in fréchet inception distance. ArXiv Preprint arXiv:2203 06026. https://doi.org/10.48550/arXiv.2203.06026 (2025).
Jeevan, P., Nixon, N., Patil, A. & Sethi, A. Evaluation metric for quality control and generative models in histopathology images. In 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI), IEEE, pp. 1–4. (2025)., April https://doi.org/10.1109/ISBI160581.2025.10981064 (2025).
Olugbara, O. O., Taiwo, T. B. & Heukelman, D. Segmentation of melanoma skin lesion using perceptual color difference saliency with morphological analysis. Math. Probl. Eng. 2018 (1), 1524286. https://doi.org/10.1155/2018/1524286 (2018).
Cazzato, G. & Rongioletti, F. Artificial intelligence in dermatopathology: Updates, strengths, and challenges. Clin. Dermatol. 42 (5), 437–442. https://doi.org/10.1016/j.clindermatol.2024.06.010 (2024).
de Coelho, P. A. Guidelines for diagnosis and pathological report of melanocytic skin lesions-recommendations from the Brazilian society of pathology. Surg. Experimental Pathol. 8 (1), 3. https://doi.org/10.1186/s42047-025-00178-4 (2025).
Ke, J. et al. Artifact detection and restoration in histology images with stain-style and structural preservation. IEEE Trans. Med. Imaging. 42 (12), 3487–3500. https://doi.org/10.1109/TMI.2023.3288940 (2023).
Wang, Q., Tweel, J. E., Reza, P. H. & Layton, A. Pathology-guided virtual staining metric for evaluation and training. arXiv preprint arXiv:2507.12624. (2025). https://doi.org/10.48550/arXiv.2507.12624
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25 (1), 24–29. https://doi.org/10.1038/s41591-018-0316-z (2019).
Fleiss, J. L. Measuring nominal scale agreement among many raters. Psychological bulletin, 76(5), 378–382. (1971). https://doi.org/10.1037/h0031619 (1971).
Komura, D. & Ishikawa, S. Machine learning methods for histopathological image analysis. Computational and structural biotechnology journal, 16, 34–42. (2018). https://doi.org/10.1016/j.csbj.2018.01.001
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25 (8), 1301–1309. https://doi.org/10.1038/s41591-019-0508-1 (2019).
Brown, N. A., Carey, C. H. & Gerry, E. I. FDA releases action plan for artificial intelligence/machine learning-enabled software as a medical device (SaMD). Retrieved from (2021). https://www.fda.gov/media/145022/download
Guan, H. & Liu, M. Domain adaptation for medical image analysis: a survey. IEEE Trans. Biomed. Eng. 69 (3), 1173–1185. https://doi.org/10.1109/TBME.2021.3117407 (2021).
Jenkinson, E. & Arandjelović, O. Whole slide image understanding in pathology: What is the salient scale of analysis? BioMedInformatics, 4(1), 489–518. (2024). https://doi.org/10.3390/biomedinformatics4010028
Chen, P. H. C. et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med. 25, 1453–1457. https://doi.org/10.1038/s41591-019-0539-7 (2019).
Han, S., Pool, J., Tran, J. & Dally, W. Learning both weights and connections for efficient neural network. Adv. Neural. Inf. Process. Syst. 28, 1135–1143 (2015).
Li, H., Kadav, A., Durdanovic, I., Samet, H. & Graf, H. P. Pruning filters for efficient Convnets. ArXiv Preprint arXiv:1608 08710. https://doi.org/10.48550/arXiv.1608.08710 (2017).
Cheng, Y., Wang, D., Zhou, P. & Zhang, T. A survey of model compression and acceleration for deep neural networks. ArXiv Preprint arXiv:1710 09282. https://doi.org/10.48550/arXiv.1710.09282 (2017).
Hinton, G., Vinyals, O. & Dean, J. Distilling the knowledge in a neural network. ArXiv Preprint arXiv:1503 02531. https://doi.org/10.48550/arXiv.1503.02531 (2015).
Sanh, V., Debut, L., Chaumond, J. & Wolf, T. DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter. arXiv preprint arXiv:.01108. (1910). https://doi.org/10.48550/arXiv.1910.01108 (2019).
Zagoruyko, S. (ed Komodakis, N.) Paying more attention to attention: improving the performance of convolutional neural networks via attention transfer. ArXiv Preprint arXiv:1612 03928 https://doi.org/10.48550/arXiv.1612.03928 (2016).
Jacob, B. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, pp. 2704–2713. (2018). https://doi.org/10.1109/CVPR.2018.00286 (2018).
Hamid, N. A. W. A. & Singh, B. High-performance computing based operating systems, software dependencies and IoT integration. In High performance computing in biomimetics: Modeling, architecture and applications. Springer Nat. Singap. 175–204. https://doi.org/10.1007/978-981-97-1017-1_8 (2024).
Plekhanov, A. et al. Histological validation of in vivo assessment of cancer tissue inhomogeneity and automated morphological segmentation enabled by optical coherence elastography. Sci. Rep. 10, 11781. https://doi.org/10.1038/s41598-020-68631-w (2020).
Huang, X., Liu, M. Y., Belongie, S. & Kautz, J. Multimodal unsupervised image-to-image translation. In Proceedings of the European conference on computer vision (ECCV), pp. 172–189. (2018). https://doi.org/10.1007/978-3-030-01219-9_11
Lee, H. Y., Tseng, H. Y., Huang, J. B., Singh, M. & Yang, M. H. Diverse image-to-image translation via disentangled representations. In Proceedings of the European conference on computer vision (ECCV), pp. 35–51. (2018). https://doi.org/10.1007/978-3-030-01246-5_3
Dar, S. U. et al. Image synthesis in multi-contrast mri with conditional generative adversarial networks. IEEE Transactions Med. Imaging. 38 (10), 2375–2388. https://doi.org/10.1109/TMI.2019.2901750 (2019).
Latonen, L., Koivukoski, S., Khan, U. & Ruusuvuori, P. Virtual staining for histology by deep learning. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2024.02.029 (2024).
Jackson, B. R. et al. The ethics of artificial intelligence in pathology and laboratory medicine: principles and practice. Acad. Pathol. 8, 2374289521990784. https://doi.org/10.1177/2374289521990784 (2021).
Torbati, M. E. et al. Multiscanner harmonization of paired neuroimaging data via structure preserving embedding learning. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), IEEE, pp. 3284–3293. (2021). https://doi.org/10.1109/ICCVW54120.2021.00367
Petrie, T. C. et al. Quantifying acceptable artifact ranges for dermatologic classification algorithms. Skin. Health Disease. 1 (2). https://doi.org/10.1002/ski2.19 (2021). ski2-19.
Shafi, S. & Parwani, A. V. Artificial intelligence in diagnostic pathology. Diagn. Pathol. 18 (1), 109. https://doi.org/10.1186/s13000-023-01375-z (2023).
Su, R. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, pp. 363–368. (2024). https://doi.org/10.1109/BIBM62325.2024.10821940 (2024).
Zhang, F. et al. Privacy-preserving federated neural architecture search with enhanced robustness for edge computing. IEEE Trans. Mob. Comput. 24 (3), 2234–2252. https://doi.org/10.1109/TMC.2024.3490835 (2024).
Salimans, T. et al. Improved techniques for training Gans. Advances Neural Inform. Process. Systems, 29, 2234–2242, (2016).
Lin, Z., Khetan, A., Fanti, G., Oh, S. & Pacgan The power of two samples in generative adversarial networks. Advances Neural Inform. Process. Systems, 31, 1498–1507, (2018).
Ma, Y., Jamdade, S., Konduri, L. & Sailem, H. AI in histopathology explorer for comprehensive analysis of the evolving AI landscape in histopathology. Npj Digit. Med. 8 (1), 156. https://doi.org/10.1038/s41746-025-01524-2 (2025).
Acknowledgements
This research was assisted by domain experts from the Biomedical Image and Signal Analysis (BIOMISA) Research Group (https://biomisa.org). We also thank Dr. Babar Yasin (bmes360058@gmail.com) for feedback on skin histology-generated virtual stains.
Author information
Authors and Affiliations
Contributions
M.A.W. conceived the main concept. M.A.H. and M.U.A. implemented and refined GANs. M.A.H., M.U.A., and M.Z.A. collected and analyzed data. F.S. and A.J. gave expert qualitative feedback. M.A.H., M.A.W., and M.U.A. prepared the manuscript; M.J.K., S.O.G., and F.H. helped articulate the main findings. All authors reviewed and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hussain, M.A., Waris, M.A., Akram, M.U. et al. VISGAB: Virtual staining-driven GAN benchmarking for optimizing skin tissue histology. Sci Rep 15, 42430 (2025). https://doi.org/10.1038/s41598-025-26493-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26493-0