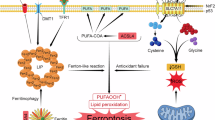
Abstract
The environmental pollutant Benzo[a]pyrene (B[a]P) and its ultimate metabolite B[a]P-7,8-diol-9,10-epoxide (BPDE) exhibit neurotoxic effects, yet the underlying molecular mechanisms remain enigmatic. Recently, ferroptosis has emerged as a potential player in B[a]P/BPDE-induced cellular damage. However, whether ferroptosis contributes to B[a]P/BPDE-induced neuron injury remains uncertain. We established a mouse model of learning and memory dysfunction through sub-chronic intragastric administration of B[a]P, with confirmation provided by behavioral alterations and pathological changes in the hippocampus. Furthermore, ferroptosis hallmarks, such as mitochondrial shrinkage, iron and glutamate metabolism disorders, and increased MDA level, were evident. Meanwhile, treatment with 1 mg/kg Ferrostatin-1 (Fer-1), a ferroptosis inhibitor, could effectively reduce hippocampal neuronal ferroptosis and improve learning and memory ability. HT22 cells were treated with 0, 0.5, and 0.75 µM BPDE, and ferroptosis was observed. Subsequent proteomic and transcriptomic analyses elucidated the underlying mechanisms of ferroptosis. Using integrated bi-omics and machine-learning algorithms, we delineated a network involving autophagy, ATP concentration, and molecule transport. Mechanistically, BPDE inhibited the transport of iron, amino acids and carbohydrates, decreased ATP content, increased ROS levels, and promoted autophagy, potentially leading to metabolic disorders and imbalance of oxidation and antioxidant systems, culminating in ferroptosis. These findings expand our understanding of B[a]P/BPDE neurotoxicity and pave the way for future investigations.
Similar content being viewed by others
Introduction
Benzo[a]pyrene (B[a]P), a representative of polycyclic aromatic hydrocarbons, is a common environmental toxicant produced by the incomplete combustion of organic matter and poses a significant health risk1,2. After a series of metabolic processes, it converts into B[a]P-7, 8-diol-9, 10-epoxide (BPDE), which affects various systems, such as the endocrine, immune, reproductive systems, and notably the nervous system, causing early damage upon exposure3,4. Epidemiological and experimental research have demonstrated the neurotoxic effects of B[a]P, including hippocampal damage, developmental disturbances within the central nervous system, developmental disorders of the central nervous system, alteration of neurotransmitter and synaptic plasticity, neurodegenerative disease-like pathological changes, and cognitive dysfunction5,6. The hippocampus, vital for learning and memory, is especially susceptible to B[a]P-induced neurotoxic effects7. Previous research has demonstrated that B[a]P-induced hippocampal injury is associated with changes in the expression of the N-methyl-D-aspartate receptor (NMDAR), the formation of DNA adducts, the generation of reactive oxygen species (ROS), and an increase in neuronal apoptosis4,8,9,10. However, the precise mechanism underlying the neurotoxic effects of B[a]P requires further exploration.
Ferroptosis is a non-apoptotic programmed cell death characterized by iron, amino acid and lipid metabolic disorders, and ROS accumulation11. The increased level of lipid peroxidation in cell membrane phospholipids makes cells more susceptible to ferroptosis12,13. The SLC7A11/GPX4 axis is a key endogenous mechanism to prevent lipid peroxidation. The glutamate-cystine anti-transport system (SLC7A11/SLC3A2) provides cysteine for glutathione (GSH) synthesis, and GPX4 utilizes GSH as a reducing substrate to convert lipid peroxides into lipoid alcohols14. Ferroptosis has been linked to multiple neurodegenerative conditions, encompassing Alzheimer’s disease and Parkinson’s disease15, and inhibition of ferroptosis can improve cognitive impairment16. B[a]P and BPDE are known to trigger ferroptosis in different cells by modulating the levels of GPX4, increasing iron levels17,18. However, whether B[a]P triggers neuronal ferroptosis remains uncertain.
In recent years, high-throughput omics has emerged as a powerful technology to study disease biomarkers, biological mechanisms, and molecular toxicity. Integrated multi-omics analysis offers a comprehensive and precise understanding of interconnected molecular networks in biological phenomena19. Furthermore, artificial intelligence methods represented by machine learning can generate unified networks incorporating transcriptomics, proteomics, and metabolomics by processing vast and diverse datasets, and have been applied to integrated analysis and prediction of omics data20. However, there is a paucity of omics studies concerning B[a]P neurotoxicity, which is mechanistically related to NMDAR perturbation, DNA methylation, alteration of tyrosine, and glycolytic metabolism changes9,21. Meanwhile, despite being an active metabolite of B[a]P, the neurotoxicity of BPDE remains underexplored, with no relevant omics study currently available. Given the complexity of hippocampal neurons, it is of great significance to use integrated multi-omics to reveal the potential molecular mechanism of BPDE-induced hippocampal neuron damage.
This study combined toxicological approaches with multi-omics methodologies to investigate the underlying mechanisms of B[a]P neurotoxicity at multiple levels. First, we used different doses of B[a]P and BPDE to prepare in vivo and in vitro neurotoxic effects models and found that B[a]P/BPDE induced hippocampal neuron ferroptosis. Subsequently, integrated bi-omics analysis, combined with machine learning, was used to reveal the underlying mechanism of BPDE-induced ferroptosis. Mechanistically, BPDE inhibits the transport of iron, amino acids and carbohydrates, as well as ATP content, while promoting autophagy. These disruptions could potentially instigate metabolic disorders and imbalance of oxidation and antioxidant systems, eventually leading to ferroptosis. Ours is the first study to reveal an indispensable role of ferroptosis in B[a]P-induced learning and memory dysfunction, providing fundamental insights for future investigations into mechanisms of B[a]P/BPDE neurotoxicity.
Results
B[a]P-induced learning and memory impairments in mice
The spatial learning and memory function of mice was evaluated by MWM test. We assessed motor deficits by measuring swimming speed and found no significant differences in moving speed among the groups, indicating no motor deficits in the mice. Thus, the observed impairments in spatial learning and memory can be attributed to B[a]P treatment rather than motor dysfunction (Fig. S1A). During the place navigation test (Fig. 1A), with the increase of training days, escape latency of mice in each group to find the hidden platform was gradually shortened (F = 59.338, P < 0.001), indicating that the mice acquire spatial cognition after repeated learning. A significant interaction between days and dose was observed, indicating varying improvements in the learning ability of mice across different time points (F = 1.728, P = 0.041). In contrast to the control group, the escape latency of B[a]P-exposed groups was prolonged with the dosage (F = 23.022, P < 0.001), signifying that the detrimental impact of B[a]P on learning capabilities was proportional to the dose administered. In the spatial exploration experiment, the high-dose group exhibited a marked decrease in the number of platform crossings compared to the control group (Fig. 1B, P < 0.01). Additionally, mice receiving medium and high doses of B[a]P required a greater duration to initially enter the platform (Fig. 1C, P < 0.05 and P < 0.01). The representative path maps were shown in Fig. 1D, the control group mice displayed purposeful swimming trajectories within the pool, whereas those exposed to B[a]P exhibited more convoluted and roundabout paths. However, ferroptosis inhibitor Fer-1 could significantly ameliorate these adverse effects induced by high-dose B[a]P exposure (P < 0.01), indicating its potential protective role.
B[a]P exposed mouse displayed cognitive impairments. (A) The escape latency over the course of a 5-day training period. (B) Number of platform crossings, (C) time to first entry by each group of mice during the spatial probe trial. (D) Representative path maps for each group in MWM test. (E) Representative path maps in Y-maze test, the blue arm means the novel arm. (F) The number of times each group of mice entered the novel arm. (G) The proportion of time spent in the novel arm of the Y-maze by different groups during the test. Data are presented as mean ± SEM (n = 10), *P < 0.05, **P < 0.01 versus Control (Ctrl) group, #P < 0.05, ##P < 0.01 versus High-dose group.
The Y-maze uses mouse’s tendency to explore a novel arm to assess spatial memory. We assessed motor deficits by measuring the total distance traveled and found no significant differences in moving distances among the groups, indicating no motor deficits in the mice. Thus, the observed impairments in spatial learning and memory can be attributed to B[a]P treatment rather than motor dysfunction (Fig. S1B). In contrast to the control group, B[a]P-exposed mice showed reduced interest in exploring the novel arm (Fig. 1E), with a notable reduction in the number of entries into the novel arm observed in the high-dose group (Fig. 1F, P < 0.01). Moreover, mice exposed to medium and high doses of B[a]P had reduced duration in the novel arm compared to the control group (Fig. 1G, P < 0.05 and P < 0.01). Notably, Fer-1 treatment significantly counteracted these tendencies (P < 0.01). Overall, B[a]P exposure adversely affected learning and memory abilities in a dose-dependent manner, a phenomenon mitigated by ferroptosis inhibitor Fer-1.
B[a]P induces mouse hippocampal neuron injury by promoting ferroptosis
The hippocampus plays a significant role in managing emotions and memory functions, with particular importance to its CA1 and CA3 subregions22. HE staining was employed to examine lesions in the hippocampal microstructure. In the control group, neurons within the hippocampal CA1 and CA3 areas were neatly aligned and exhibited typical, regular morphology. In contrast, in the groups exposed to medium and high doses of B[a]P (Fig. 2A-C), we observed a decrease in the number of neurons and a disorganized arrangement. These changes reflect the overall impact of B[a]P exposure on tissue architecture and cell distribution. Nissl staining revealed a reduction in the number of Nissl bodies after B[a]P exposure (Fig. 2D-F). HE and Nissl staining indicated that exposure to medium and high doses of B[a]P led to a marked decrease in neuronal count in the hippocampal CA1 and CA3 regions when compared to the control group, with Fer-1 treatment showing ameliorative effects (P < 0.05). TEM findings indicated that, in comparison to the control group, the mitochondria in hippocampal neurons of the B[a]P-exposed group were atrophied, and cristae were broken or absent, with the high-dose group being particularly affected, while Fer-1 could improve the mitochondrial damage (Fig. 2G). Taken together, the above results indicate that B[a]P damages hippocampal structure and causes neuronal death in the hippocampus.
B[a]P triggered ferroptosis in the hippocampal neurons of mice. (A) Hematoxylin and eosin (HE) staining images of the hippocampal CA1 and CA3 areas from each group; scale bars = 50 μm. (B-C) The corresponding number of neurons in HE staining. (D) Nissl-stained images of the hippocampal CA1 and CA3 regions across various groups; scale bars = 50 μm. (E-F) The number of Nissl bodies in Nissl staining images. (G) Representative hippocampal ultrastructure image; the red box in the top image is enlarged in the bottom image. Scale bars = 2 μm, 1 μm. N, cell nucleus; Arrows, mitochondria. (H-K) Influences of B[a]P or Fer-1 on iron, GSH, GSH-Px, and MDA levels in the hippocampus tissue (n = 8). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01 versus Ctrl group, #P < 0.05, ##P < 0.01 versus High-dose group.
Figure 2H illustrated that B[a]P caused a pronounced, dose-dependent elevation in the iron content of the hippocampus relative to the control group, with the high-dose group being particularly affected (P < 0.01), while Fer-1 treatment effectively reversed the increase of iron level induced by B[a]P (P < 0.01). After B[a]P treatment, the GSH and GSH-Px enzyme activities of hippocampal neurons declined proportionally to the dosage administered, while Fer-1 treatment could improve this phenomenon (Fig. 2I and J). The lipid ROS biomarker MDA increased after B[a]P exposure, which was reversed by Fer-1 (Fig. 2K). Overall, these results indicated that B[a]P exposures induce hippocampal neuronal ferroptosis.
BPDE induced HT22 cell ferroptosis
B[a]P can cross the blood-brain barrier, and both it and its active metabolite BPDE are neurotoxic23. However, there is a lack of research on the neurotoxicity of BPDE. The HT22 cell line, derived from mouse hippocampal neurons and characterized by its immortalized nature, is extensively utilized as a beneficial model for research pertaining to memory functions24. Considering the complexity of neuronal components in the hippocampus, we established a BPDE-treated HT22 cell model to study the mechanism of neuronal death caused by B[a]P at the molecular level.
As shown in Fig. 3A, BPDE dose-dependently inhibited cell viability, with a minimum significant toxic dose of 0.5 µM and an LC50 close to 0.75 µM. The apoptosis inhibitor Z-VAD, necrosis inhibitor NEC-1, and ferroptosis inhibitor Fer-1 all reversed the decline of cell viability induced by 0.75 µM BPDE, and Fer-1 showed the strongest effect (P < 0.01). These results indicate that BPDE-induced cell death involves multiple forms of cell death mechanisms, and the primary mode of cell death in HT22 cells varies with different doses of BPDE treatment. We subsequently focus on exploring the mechanisms underlying ferroptosis induced by BPDE in HT22 cells. BODIPY 581/591 C11 and FerroOrange served as indicators for lipid ROS and intracellular iron (Fe2+) levels, respectively. Fluorescence quantification revealed that both intracellular ROS (Fig. 3B and C, P < 0.01) and Fe2+ (Fig. 3D and E, P < 0.01) rose significantly alongside escalating BPDE concentrations, which was reversed by Fer-1. As expected, we confirmed several additional biomarkers indicative of BPDE-induced ferroptosis, including the depletion of GSH, a decrease in GSH-Px enzyme activity, an accumulation of MDA, and an elevation in PTGS2 mRNA levels. Notably, Fer-1 reversed BPDE-induced changes in these indicators (Fig. 3F-I). In addition, TEM showed that BPDE-injured cells exhibited the characteristic morphological features of ferroptosis25, including mitochondrial shrinkage as well as reduced or absent mitochondrial crista, which was reversed by Fer-1 (Fig. 3J). The findings suggest that BPDE triggered ferroptosis in HT22 cells in a concentration-dependent fashion. To further explore its molecular mechanism, 0.5 µM and 0.75 µM BPDE were selected according to the above results for subsequent transcriptome and proteome studies.
BPDE induced HT22 cell ferroptosis. (A) Cell viability in groups exposed to various BPDE concentrations for 24 h following a 2-hour preincubation with 10 µM Fer-1, Z-VAD, or 20 µM NEC-1. (B) Typical fluorescent images of BODIPY 581/591 C11 and corresponding fluorescence intensity quantitative statistical plots (C). (D) Representative fluorescence images of FerroOrange and corresponding fluorescence intensity quantitative statistical plots (E). (F-H) Effects of BPDE and Fer-1 treatment on GSH (F), GSH-Px activity (G), and MDA levels (H) in HT22 cells. (I) mRNA expression of PTGS2. (J) Ultrastructure of cells was observed by a TEM; scale bars = 2 μm and 1 μm; N: cell nuclei; Arrows, mitochondria. Data are expressed as mean ± SEM, n = 3 independent repeats. **P < 0.01 versus control group, #P < 0.05, ##P < 0.01 versus 0.75 µM BPDE group.
Transcriptomics features of the HT22 cell exposed to BPDE
A total of 36,269 genes were identified through bioinformatics analysis. Principal component analysis (PCA) of gene expression data revealed a clear separation between the three groups, indicating that our mass spectrometry method was able to distinguish the transcriptome profiles of BPDE-treated cells (Fig. S2A). The thresholds of differentially expressed genes (DEGs) were set as |log2 FC| > 0.58 and Padj < 0.05. 959 and 3132 DEGs were identified in the 0.5 µM and 0.75 µM BPDE groups compared to the control group, respectively, of which 818 and 2267 were up-regulated and 141 and 865 were down-regulated, and the overall distribution of DEGs was visualized by volcano plots (Fig. 4A and B). We found that BPDE could alter the cellular transcriptome profile and that the expression of most DEGs was upregulated with the increase of BPDE concentration; in particular, high doses of BPDE could significantly alter gene expression in HT22 cells. Our analysis identified key genes with substantial alterations in expression levels following BPDE exposure. The significantly upregulated genes included PM20D1 and ANKK1. PM20D1, involved in the PM20D1-NADA pathway, has been shown to protect against Parkinson’s disease26. ANKK1, a dopaminergic gene, has regulatory rare variants that may serve as potential risk factors for Parkinson’s disease27. The significantly downregulated genes included MsrA and RBMS3. MsrA, an antioxidant enzyme, may be a potential therapeutic target for organ and cell damage induced by ferroptosis28. RBMS3, whose expression is enhanced by metformin, has been shown to promote ferroptosis and suppress ovarian cancer progression29.
Transcriptomic analysis of HT22 cells treated with varying concentrations of BPDE. (A-B) The volcano plot of gene between the 0.5 µM BPDE and the control (CT) group, as well as between the 0.75 µM BPDE and the CT group. The gray dots: no significant changed genes; the blue dots: down-regulated DEGs; the red dots: up-regulated DEGs. (C) The KEGG enrichment analysis of significant pathways in 0.5 µM BPDE versus CT group and 0.75 µM BPDE versus CT group. (D) Heatmap of the identified ferroptosis-related DEGs. Purple to red colors represent the expression level of genes from low to high. (E) The trimmed transcriptomic network with prediction using IPA in 0.75 µM BPDE-treated HT22 cells.
We subsequently performed KEGG pathway enrichment analysis on the omics data30,31,32. The enriched pathways of DEGs mainly distinguished into six clusters, including material metabolism, cancer and immune disease, cell growth and death, endocrine system, nervous system signal transduction, and xenobiotics biodegradation and metabolism (Fig. 4C). Overall, 0.75 µM BPDE had a significantly stronger effect on cellular pathways than 0.5 µM BPDE, as the number of altered pathways increased in almost all six clusters. Specifically, the 0.75 µM concentration of BPDE significantly influenced signal transduction pathways, notably those associated with the HIF-1 signaling pathway, TNF signaling pathway, MAPK signaling pathway, PI3K-Akt signaling pathway, and ECM-receptor interaction. Additionally, other pathways included cancer, cell growth and death, nervous system, signal transduction, and signaling molecules and interaction, likely reflecting the stress directly caused by BPDE. Notably, in addition to causing the same changes in the p53 signaling pathway and apoptosis in the 0.5 µM group, 0.75 µM BPDE treatment triggered ferroptosis.
To gain further insights into the mechanisms underlying BPDE-induced ferroptosis, a comparison was made between DEGs and the ferroptosis-associated molecules listed in the FerrDb V2 database (http://www.zhounan.org/ferrdb/current/), a specialized repository for ferroptosis-related information33. The ferroptosis-related DEGs heat map showed that 0.5 µM and 0.75 µM BPDE induced 32 and 60 up-regulated and 12 and 39 down-regulated genes, respectively, and the higher the concentration of BPDE, the more the number of DEGs and the greater the change of expression level (Fig. 4D and Table S4). Subsequently, the transcriptomic profiles of ferroptosis-related DEGs in the 0.5 and 0.75 µM BPDE-exposed groups were examined through Ingenuity Pathway Analysis (IPA, http://www.ingenuity. com). The results of canonical pathways and biological functions associated with these genes are shown in Tables S5 and S6. Further examination revealed that these genes were associated with three key biological functions, encompassing autophagy, concentration of ATP, and transport of molecules, which were also used to predict biological functions. The transcriptome network was trimmed according to the degree of correlation between DEG and biological function, and the subsequent network predictions indicated the potential for autophagy activation and inhibition of ATP concentration and transport of molecules (Fig. 4E).
Proteomic features of the HT22 cell exposed to BPDE
There are 6127 proteins identified in our proteomics. The PCA results of the proteins show that BPDE had a significant effect on the proteome profile, and a clear separation between the different treatments was observed in the proteome (Fig. S2B). The criteria for identifying differentially expressed proteins (DEPs) were established as FC > 1.5 or FC < 0.667, and P < 0.05. The volcano plot showed that there were 615 and 588 DEPs (165 and 224 up-regulated and 450 and 364 down-regulated) in the 0.5 µM BPDE group and the 0.75 µM BPDE group, respectively, compared with the control group, suggesting that BPDE predominantly exerts an inhibitory effect on protein expression (Fig. 5A and B). The number and expression levels of DEPs and KEGG-enriched pathways were combined to create enrichment circles (Fig. 5C). Compared with the control group, 0.5 µM BPDE significantly changed environmental information processing and organismal systems, accompanied by downregulation of most proteins. However, 0.75 µM BPDE significantly altered the metabolic pathways and upregulated the expression of most proteins (Fig. 5D). Specifically, our analysis identified key proteins significantly altered by BPDE exposure. The most upregulated proteins included Krt76 and Lpin1. Krt76, with immunomodulatory roles and involvement in cellular stress responses34, showed a marked increase, likely due to BPDE-induced cellular damage. Lpin1, linked to hepatic ferroptosis and lipid metabolism, was also highly upregulated35. The most downregulated proteins included COL3A1 and COLEC12. COL3A1, potentially associated with M1 macrophages and ferroptosis in alcoholic hepatitis, exhibited a significant decrease36. COLEC12, involved in innate immune responses, also showed substantial downregulation, possibly contributing to the overall inflammatory state in the context of BPDE exposure37.
Proteomics analysis of HT22 cells exposed to different BPDE concentrations. (A-B) The volcano plot of proteins between 0.5 µM BPDE versus CT group, and 0.75 µM BPDE versus CT group. The gray dots: no significant changed proteins; the blue dots: down-regulated DEPs; the red dots: up-regulated DEPs. (C-D) The enriched circles of significant pathways in 0.5 µM BPDE versus CT group (C) and 0.75 µM BPDE versus CT group (D). The enrichment cycle diagram consists of four cycles. The first circle: different colors represent distinct enriched functional categories; Second circle: the quantity of proteins associated with each category and their respective P values; The third circle: a bar graph depicting the ratio of up-regulated (dark purple) to down-regulated (light purple) proteins; The fourth circle: Rich Factor values. (E) Heatmap of the identified ferroptosis-related DEPs. (F) The trimmed proteomic network with predictive analysis for three classes of biological functions.
After overlaying DEPs with ferroptosis-related molecules from FerrDb V2, we found the total number of ferroptosis-related DEPs was similar in the two dose groups, with 8 and 18 upregulated and 21 and 12 downregulated proteins induced by 0.5 µM and 0.75 µM BPDE, respectively (Fig. 5E and Table S7). We subsequently analyzed canonical pathways and biological functions of ferroptosis-related proteins in HT22 cells exposed to 0.5 and 0.75 µM concentrations of BPDE using IPA software (Table S8 and S9). The proteomic network analysis indicated that the ferroptosis-related DEPs corresponded to the trio of biological functions previously identified within the transcriptomic network. We then trimmed the proteomic network based on the correlation of proteins with the biological functions, and the prediction of the network found activation of autophagy and inhibition of ATP concentration and transport molecules, which was consistent with the transcriptome prediction network (Fig. 5F).
Collectively, BPDE can cause significant changes in the cellular transcriptome and proteome profiles, and the molecular expression patterns may be different under different doses. Concretely, both concentrations of BPDE caused changes in the same pathways, including steroid biosynthesis, p53 signaling pathway, fluid shear stress and atherosclerosis, bile secretion, ECM-receptor interaction, and cytokine-cytokine receptor interaction. Importantly, when compared to 0.5 µM BPDE, the higher concentration of BPDE enriched ferroptosis.
Integrated bi-omics revealed the underlying mechanism of ferroptosis
To address the limitations of single omics approaches and find the core molecular mechanism of ferroptosis, we integrated the findings from transcriptomics and proteomics mentioned above to create an integrated bi-omics network (Fig. 6A)20. Subsequently, the genes and proteins from the integrated omics network, along with the three biological functions—autophagy, ATP concentration, and molecular transport—were analyzed using an unsupervised machine learning approach based on the k-means clustering algorithm (Table S10). Then, unsupervised analysis based on machine learning was performed on the integrated biological-related molecules and the three biological functions (autophagy, ATP concentration, and molecular transport) using the k-means algorithm. In Fig. 6B, individual data points correspond to distinct omics elements, each of which is implicated in at least one of the three biological functions under investigation. Based on the clustering results, we identified three clusters, and molecules in the same cluster indicated a stronger relationship despite their different biological functions. To further visualize the intersections between the two omics data and the interactions between the three biological functions, we plotted the molecular interaction networks of each of the three clusters using String (https://cn.string-db.org/). The molecules within the major cluster predominantly participate in molecular transport, including the transmembrane transport of amino acids mediated by the solute carrier (SLC) family, and the cellular iron uptake mediated by TFRC (Fig. 6C). The second cluster predominately related to autophagy (Fig. 6D), while those in the third cluster were mainly related to ATP concentration (Fig. 6E). This demonstrates the ability of machine learning approaches to resolute complex signatures within multi-omics and identify possible pathways affecting ferroptosis.
Cluster analysis of the integrated bi‑omics network. (A) The simple merged bi‑omics network of 0.75 µM BPDE‑treated HT22 cells. (B) PCA for bi‑omics and biological functions. Different colors represent different clusters, triangles represent proteins, and circles represent genes. Interactions of the molecules in cluster 1 (C), cluster 2 (D), and cluster 3 (E) are plotted with String. Proteins are in the first row; the rest are genes.
Specifically, genes involved in autophagy included ULIK1 and MAP1LC3A; SIRT3 and TXNIP are involved in ATP concentration, and SLC7A11 and SLC7A5 were involved in molecule transport. In addition, the proteins involved in three biological functions were SQSTM1 (p62) and SLC2A1, and the other proteins or genes were involved in at least one biological function. Each molecule can play a role in multiple pathways, and here, we clarified the roles of different molecules in molecular transport, autophagy, and ATP concentration-mediated BPDE-induced ferroptosis by machine learning. Of course, the complexity of cellular biological processes and the network of molecular interactions also suggested that these pathways were interconnected rather than independent (Fig. 7A). We further conducted experimental validation, as illustrated in Fig. 7B. with the increase of BPDE concentration, the protein expression levels of p62, SLC7A11, and GPX4 decreased, and Fer-1 significantly reversed the changes of the theses ferroptosis-related proteins (Fig. 7C-E, P < 0.05). Similar results were also observed in mice treated with B[a]P (Fig. 7F-I, P < 0.05).
The proposed mechanism of B[a]P/BPDE-induced ferroptosis. (A) The schematic representation of BPDE-induced ferroptosis in HT22 cells based on integrative omics. Red represents upregulation; green represents downregulation. According to the analysis results of integrated omics combined with machine learning algorithm, BPDE inhibited cellular molecular transport and energy metabolism pathways, and activated autophagy. Then, after the disorder of multiple metabolic processes, cellular iron overload results in a disturbed redox equilibrium, and the accumulation of lipid peroxides eventually leads to ferroptosis. (B) Western blot detection of p62, SLC7A11, and GPX4 within the HT22 cells (n = 3). Protein abundance was quantified in (C-E). (F) Western blot detection of p62, SLC7A11, and GPX4 within the hippocampus (n = 3). Protein abundance was quantified in (G-I). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01 versus Ctrl group, #P < 0.05, ##P < 0.01 versus High-dose group.
Collectively, these findings highlight that BPDE inhibits the transport of iron, amino acids and carbohydrates, as well as ATP concentration, while promoting autophagy, which may cause metabolic disorders and imbalance of oxidation and antioxidant systems, eventually leading to ferroptosis.
Discussion
B[a]P contamination represents a persistent environmental issue, with the potential to harm the nervous system due to excessive or prolonged exposure. Despite this, the specific mechanisms at play remain largely elusive. In this study, leveraging toxicological insights, an integrative approach was employed that merged multi-omics technologies with machine learning algorithms to investigate the molecular underpinnings of learning and memory impairment associated with B[a]P exposure from various perspectives.
In humans, over 99% of B[a]P exposure occurs via the gastrointestinal tract38. The lowest exposure doses of B[a]P range from 0.02 to 2 mg/kg, corresponding to levels experienced by smokers, individuals with occupational exposure, or those who frequently consume grilled or smoked foods. Doses between 20 and 200 mg/kg are typically considered toxic39,40. The average daily dietary intake of B[a]P in humans is approximately 55 ng/kg body weight per day41. The maximum allowable B[a]P concentration in China is 0.15 mg/m3, whereas in industrial settings, B[a]P concentrations can reach up to 100 µg/m342. The mean concentration of urinary 1-OHP among coke oven workers (3.61 ± 1.30 mmol/mol Cr) was significantly higher than that of controls (1.89 ± 0.84 mmol/mol Cr). Concentration levels of B[a]P and its hydroxylated metabolites have been reported at 100 ng/ml in serum and 100 pg/mg in brain tissue of B[a]P-exposed mice via oral administration at doses of 200 mg/kg for 11 consecutive days39. In this study, we established a subchronic exposure model in C57BL/6 mice using oral gavage with B[a]P at doses of 2.5, 5, and 10 mg/kg, which were selected based on the conversion of human exposure levels to equivalent murine treatment doses3,43.
Damage to the hippocampus and neuronal loss caused by B[a]P are typical features of impaired learning and memory ability and depression, and hippocampal neuron death is the key to B[a]P neurotoxicity. Apoptosis accounts for only a fraction of the neurotoxic effects attributed to B[a]P, suggesting that other mechanisms may also be at work in causing neuronal damage44. Considering the significant role that ferroptosis plays in the etiology and progression of neurological disorders, along with the found overlap with B[a]P’s toxic mechanisms, such as the accumulation of iron, we hypothesized that ferroptosis might be a contributing factor to the neurotoxicity induced by B[a]P. In this study, mice in the B[a]P neurotoxicity model showed decreased learning and memory function, including learning and memory impairment, especially at high-dose exposure, which is consistent with previous studies5,43. Fer-1, a synthetic antioxidant initially discovered through high-throughput screening, selectively inhibits ferroptosis45. Based on previous research, we selected a dose of 1 mg/kg Fer-1 for our animal model, which demonstrated significant neuroprotective effects by inhibiting ferroptosis without any observed off-target effects at the tested concentration46. Moreover, B[a]P-exposed mice showed typical ferroptosis features of hippocampal mitochondrial shrinkage, iron overload, oxidation and antioxidant imbalance, and lipid peroxide accumulation, which were inhibited by Fer-1, suggesting that B[a]P triggered ferroptosis in hippocampal neurons.
The HT22 cell line, derived from murine hippocampal neurons, is extensively employed in research on neurotoxins and neurodegenerative diseases24. In fact, we also confirmed BPDE-induced ferroptosis in primary hippocampal neurons for enhanced physiological relevance47. Given the balance between the convenience and reliability of HT22 cells and the need for physiological relevance, we chose HT22 cells for the comprehensive experiments in this study. There are four classes of markers for detecting ferroptotic cells: lipid peroxidation markers (e.g., MDA), gene expression changes (e.g., upregulation of PTGS2), TfR1 mobilization, and shrunken, dense mitochondria. It is recommended to use at least three markers or a suitable combination of two markers to distinguish ferroptosis from other stress conditions, as some markers may also be activated by other types of stress11. In this study, we used changes in mitochondrial ultrastructure, levels of lipid ROS, Fe2+, GSH-PX activity, and mRNA expression of PTGS2 and the protein expression of GPX4 and SLC7A11 as indicators of ferroptosis. Consistent with the in vivo results, ferroptosis was also observed in HT22 cells treated with BPDE. BPDE-induced ferroptosis shares similarities with classical ferroptosis inducers such as Erastin and RSL3, but also has distinct features. Erastin primarily depletes intracellular GSH to induce ferroptosis, while RSL3 directly inhibits GPX414. BPDE induces ferroptosis by reducing GPX4 levels and causing lipid ROS accumulation, similar to known ferroptosis inducers. Additionally, BPDE causes iron accumulation. Using integrated bi-omics and machine-learning algorithms, we delineated a network involving autophagy, ATP concentration, and molecule transport. The specific pathways and mechanisms through which BPDE achieves these effects highlighting the complexity of ferroptosis induction by various agents.
Integrative analysis of multi-omics has been applied in the field of toxicology to screen key molecules, mine meaningful metabolic pathways, and propose a coordinated molecular network at different biological layers19. There are only omics studies on B[a]P but not on BPDE. In this research, we designated HT22 cells exposed to 0.5 µM and 0.75 µM BPDE as the groups representing low and high dosages, respectively, for our transcriptomic and proteomic analyses, guided by the outcomes of cell viability assays. The omics data revealed that BPDE induced substantial alterations in the cells’ transcriptional and proteomic landscapes. The altered proinflammatory, tumor, cell growth and death, nervous system, signal transduction and signaling molecules, and interacting signaling pathways reflect the stress directly caused by BPDE. Shen et al. found that exposure to B[a]P led to notable metabolic disruptions, impacting cellular processes related to viability and migration48. The dysregulation of tyrosine metabolism and glycolysis can compromise the energy provision to the hippocampus and the production of neurotransmitters9, potentially contributing to the learning and memory deficits associated with B[a]P exposure.
Integrated transcriptomic and proteomic analysis based on machine learning showed that BPDE activates ferroptosis by inhibiting cellular molecule transport, ATP concentration, and promoting autophagy. To validate these bioinformatics results, we experimentally confirmed the protein expression changes of the autophagy-related marker p62 and the ferroptosis-related marker SLC7A11, providing evidence for the effects of B[a]P/BPDE. Our STRING analysis revealed three distinct molecular clusters related to ferroptosis. The major cluster involved molecular transport, with key molecules like SLC7A11 and TFRC contributing to cystine uptake and iron overload, respectively. The second cluster linked autophagy to ferroptosis through altered expression of p62 and MAP1LC3A, potentially increasing the labile iron pool. The third cluster highlighted ATP concentration, with BPDE reducing ATP levels and exacerbating energy stress. These findings underscore the multifaceted mechanisms underlying BPDE-induced ferroptosis and neurotoxicity. Future work should validate these pathways and explore their therapeutic potential.
Normal molecular transport is necessary to maintain biological processes such as material exchange and metabolism in cells49. Specifically, our findings indicated that BPDE modulated the expression levels of iron transporters such as TFRC, STEAP3, and FTH, reflecting intracellular iron overload. The SLC membrane transport family molecules are responsible for the transport of nutrients and ions between the cells and the outside50. Moreover, the System Xc − transporter complex, composed of SLC7A11 and SLC3A2 subunits, predominantly facilitates the uptake of cystine in the cell in exchange for glutamate that is already present inside the cell51. Our study found that BPDE treatment significantly reduced the expression of SLC7A11/SLC3A2, leading to decreased GSH synthesis and GPX4 levels (Fig. 6F). Mitochondria can regulate ferroptosis by regulating iron metabolism, energy metabolism, ROS production, and mitophagy, while some ferroptosis activators or inhibitors can also target mitochondria52. SIRT3 functions as a NAD (+)-dependent deacetylase of histones, and its dysregulation can disrupt various mitochondrial metabolic processes, potentially promoting ferroptosis53,54. TXNIP, by suppressing the function and expression of thioredoxin (Trx), can cause an imbalance in cellular redox status, increasing oxidative stress and thereby inducing ferroptosis in renal tubular epithelial cells55. Our omics results show that BPDE alters the levels of SIRT3 and TXNIP. Further investigation is needed to elucidate their role in BPDE-induced ferroptosis. Autophagy plays an important role in Ferooptosis56. It has been shown that FTH/NCOA4-mediated ferritinophagy and mitophagy can promote ferroptosis by increasing the labile iron pool (LIP)57. Our omics results demonstrate that BPDE significantly alters the levels of FTH and autophagy-related molecules such as p62 and MAP1LC3A. Future research can delve deeper into the underlying molecular mechanisms of ferroptosis.The imbalance between oxidative and antioxidant status in cells leads to ferroptosis. Previous studies have indicated that BPDE can trigger ferroptosis by promoting GPX4 ubiquitylation degradation, redox imbalance, and increasing iron levels18,58. Consistent with these findings, our omics results showed that. BPDE treatment leads to increased intracellular lipid peroxides and inhibited antioxidant systems, ultimately causing ferroptosis.
Our study demonstrated for the first time that B[a]P-induced hippocampal neuron ferroptosis leads to learning and memory impairment in mice. Additionally, we established a BPDE-exposed HT22 cell model and explored the potential molecular mechanisms of ferroptosis through transcriptomics and proteomics. Our integrated bi-omics analysis powered by machine learning showed that BPDE interfered with intracellular autophagy, energy metabolism, and molecular transport. These disturbances may precipitate metabolic dysregulation and oxidative-antioxidative imbalance, eventually resulting in ferroptosis. Our findings not only shed light on the mechanistic insights into B[a]P/BPDE-induced hippocampal neuron death, but also provide the experimental and theoretical support for the treatment of diseases related to ferroptosis.
Materials and methods
Animal model
Four-week-old male C57BL/6J mice, sourced from the Animal Experiment Center of Shanxi Medical University in China, were randomly assigned into six groups (n = 12): the control group (0 mg/kg), the low-dose group (2.5 mg/kg), the medium-dose group (5 mg/kg), the high-dose group (10 mg/kg), the high-dose + Ferrostatin-1 (Fer-1) group, and the Fer-1 group (1 mg/kg). B[a]P (B1760, Sigma, USA) was prepared in corn oil, while Fer-1 (S7243, Selleck, USA) was dissolved in saline. The control group received an equal volume of vehicle treatment. B[a]P was administered via intragastric gavage every other day, and Fer-1 was injected intraperitoneally on alternate days for a total of 90 days, with dose selection predicated on the conversion of human exposure levels to equivalent murine treatment doses3,43,46. Behavioral tests were conducted to detect the learning and memory capabilities of mice, with 10 mice used per group for treatment. Throughout the experiment, utmost care was exercised to ensure that the mice suffered as little as possible. The animal experimental procedures were performed in accordance with the relevant guidelines and regulations and approved by the Institutional Animal Care and Use Committee of Shanxi Medical University. All methods are reported in accordance with ARRIVE guidelines.
Morris water maze (MWM)
The circular pool of MWM (RWD Life Science Co., LTD., China) was 120 cm in diameter and 35 cm in height and included four quadrants, each with a different geometric reference object on the pool wall. A platform with a diameter of 5 cm was set in the center of one quadrant, and an amount of water (changed daily) was injected into the circular pool with a water level 1 cm above the platform. The experiment includes two parts: a navigation (1–5 days) and a space exploration experiment (day 6), and the whole process was performed under dim light. The mice were given free swimming the day before the test as fitness training. During the experiment, the mice were promptly dried to avoid stress after each time out of the water. In the navigation experiment, the mice were free swimming from a predetermined location and were given 60 s to search for a hidden platform. The trajectory and time of finding the platform (escape latency) were captured by a video tracking system. Each mouse underwent 4 trials per day for a period of 5 days. On the 6th day, the hidden platform was removed from the space exploration experiment. Mice were granted 60 s to navigate and find the platform, with the frequency of crossing over the platform was recorded to detect their spatial memory ability.
Y-maze test
The Y-maze device, provided by the Institute of Materia Medica Chinese Academy of Medical Science was composed of three identical arms with a length of 30 cm, a width of 8 cm, and a height of 15 cm. There was a camera positioned above the maze to automatically record and analyze the movements of the mice. Three arms were randomly divided into starting arms, familiar arms, and novel arms, and a different colored 2 cm × 2 cm sticker was affixed to the end of each arm 1 inch from the ground. During the training experiment, the passage to the novel arm was blocked with a movable opaque door, and each mouse was introduced at the end of the fixed starting arm towards a sticker, and permitted to explore the starting and familiar arm for 10 min. Following a 30-minute interval post-training, the memory test experiment was carried out, the opaque door was lifted, and the mouse was returned to the initial starting point, now with the freedom to explore all three arms of the maze for a period of 5 min. The number of entries each group of mice made into the novel arm, as well as the proportion of time they spent in the novel arm, were recorded to assess their spatial memory abilities.
Sample collection
After the behavioral tests, the mice were anesthetized with isoflurane and then decapitated. Brains (n = 3) were preserved in 4% polyformaldehyde for subsequent hematoxylin-eosin (HE) and Nissl staining procedures. The hippocampal regions were carefully extracted, with a portion (n = 3) being fixed in 2.5% glutaraldehyde solution for examination under transmission electron microscopy (TEM), while the remainder was preserved at −80 °C for future experimental use.
Hematoxylin and Eosin staining
H&E staining was employed to examine lesions in the hippocampal microstructure, providing a broad assessment of tissue architecture and overall cellular morphology. Paraffin sections were dewaxed, sequentially stained with hematoxylin and eosin, then cleared with xylene and mounted with neutral gum before being observed and imaged under a microscope.
Nissl staining
Nissl staining revealed a reduction in the number of Nissl bodies after B[a]P exposure, specifically highlighting changes in neuronal density. The paraffin sections were dewaxed, washed with distilled water, then immersed in preheated 1% toluidine blue staining solution at 65 °C for 45 min, followed by another wash with distilled water. The sections were sequentially placed in 70%, 80%, and 95% ethanol for differentiation, dehydrated, cleared with xylene, and mounted with neutral gum before being observed and imaged under a microscope.
Cell culture and cytotoxicity assays
HT22 mouse hippocampal neurons, obtained from the National Infrastructure of Cell-line Resources in Beijing, China, were grown in Dulbecco’s Modified Eagle Medium (DMEM; Corning, USA) with 4.5 g/L glucose, supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum from TransGen, China. The cells were maintained at 37 ℃ in an atmosphere of 5% CO2. After cells were cultured to 60% confluence, they were treated with complete medium containing different doses of BPDE (B287550, Toronto Research Chemicals, Canada) or Fer-1.
HT22 (8 × 103 per well) were plated onto 96-well plates and allowed to incubate for 24 h. Subsequently, they were exposed to various reagents according to the experimental content, with six replicates for each treatment condition. The study assessed cytotoxicity using a CCK-8 assay kit from MedChemExpress (HY-K0301).
Lipid peroxide determination
Malondialdehyde (MDA) levels were measured by adhering to the guidelines provided with the MDA assay kit (S0131, Beyotime™, Nanjing, China). After sample protein extraction, MDA levels were measured at an absorbance of 532 nm. To observe lipid peroxidation of cells, HT22 cells (1 × 105/mL) were plated in confocal dishes, washed three times with PBS at the end of the treatment, followed by incubation with 2 µM BODIPY 581/591 C11 (Invitrogen, USA) for a duration of 30 min. Finally, fluorescence images were taken by confocal microscope (Leica TCS SP8, Germany) after three washes with PBS. The green and red fluorescence were obtained by dual-wavelength excitation (excitation light 488 and 568 nm, emission light 565–581 nm and 585–591 nm). Mean fluorescence intensity values were measured by ImageJ software.
GSH, GSH-Px, and iron level determination
The content of GSH and the enzymatic activity of GPX were detected by dedicated commercial kits provided by the Nanjing Jiancheng Bioengineering Institute in China. The iron content in mice was quantified with a tissue iron determination assay from the same institute. For assessing intracellular Fe2+ levels, FerroOrange reagent from Dojindo™ in Kumamoto, Japan, was utilized. HT22 cells were cultured in confocal dishes, and upon completion of the treatment, they were treated with a 1 µM working solution of FerroOrange for 30 min, followed by three PBS washes to remove excess reagent. Fluorescence imaging was conducted through confocal microscopy. Single wavelength excitation (excitation light 561 nm, emission light 570–620 nm) was used to obtain the red fluorescence of the FerroOrange probe. Mean fluorescence intensity values were measured using ImageJ software.
Transcriptomics and proteomics analyses
HT22 cells were subjected to various concentrations of BPDE (0, 0.5, and 0.75 µM) for a period of 24 h. The total RNA was isolated using RNAiso Plus from Takara, Japan, for subsequent transcriptome analysis. To collect proteomic samples, HT22 cells underwent a similar treatment at the same concentrations for 24 h and were lysed with RIPA containing 1% PMSF. Both RNA and protein samples contained three biological replicates and transcriptomic and proteomic analyses were conducted by Shanghai Applied Protein Technology Co., LTD. A detailed description is provided in the Supplementary methods and Tables S1, S2, and S3.
Unsupervised principal component analysis (PCA)
To access the relevance and to trim the bi-omics network constructed based on transcriptomics and proteomics, PCA was conducted. The data from each omic was standardized to a Z-score to ensure the data were at a comparable level of change for cluster analysis. The multi-dimensional Z-scores of differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) were projected into a two-dimensional (2D) plane. In this plane, both the first and second dimensions represent weighted linear combinations of the original values. The unsupervised K-means clustering algorithm can divide the 2D data obtained from PCA into ‘K’ cluster groups, where K is set to 359. In this study, the machine learning tool sklearn was used to run the K-means algorithm in Python to determine the largest cluster in the PCA figure20.
Statistical analysis
GraphPad Prism8 and SPSS 21.0 software were used for data visualization and statistical analysis, respectively. Data were presented as the mean with the standard error of the mean (SEM) indicated. The normality of the data was tested using the Shapiro-Wilk method. For data structured with repeated measures, the analysis was carried out using Repeated Measures ANOVA. When comparing multiple groups, a standard one-way ANOVA followed by the Bonferroni correction was used to assess the differences among groups. P < 0.05 was set as the threshold for statistical significance.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Kazerouni, N., Sinha, R., Hsu, C. H., Greenberg, A. & Rothman, N. Analysis of 200 food items for benzo[a]pyrene and Estimation of its intake in an epidemiologic study. Food Chem. Toxicol. 39, 423–436. https://doi.org/10.1016/s0278-6915(00)00158-7 (2001).
Boström, C. E. et al. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Perspect. 110 Suppl. (3), 451–488. https://doi.org/10.1289/ehp.110-1241197 (2002).
Chepelev, N. L., Moffat, I. D., Bowers, W. J. & Yauk, C. L. Neurotoxicity May be an overlooked consequence of benzo[a]pyrene exposure that is relevant to human health risk assessment. Mutat. Res. Rev. Mutat. Res. 764, 64–89. https://doi.org/10.1016/j.mrrev.2015.03.001 (2015).
Bukowska, B., Mokra, K. & Michalowicz, J. Benzo[a]pyrene-Environmental Occurrence, human Exposure, and mechanisms of toxicity. Int. J. Mol. Sci. 23, 6348. https://doi.org/10.3390/ijms23116348 (2022).
Liu, D. et al. Benzo(a)pyrene exposure induced neuronal loss, plaque deposition, and cognitive decline in APP/PS1 mice. J. Neuroinflammation. 17, 258. https://doi.org/10.1186/s12974-020-01925-y (2020).
Chongying Qiu, B. P., Shuqun Cheng, Y. X. & Tu, B. The Effect of occupational exposure to Benzo[a]pyrene on neurobehavioral function in coke oven workers. Am. J. Ind. Med. 56, 347–355. https://doi.org/10.1002/ajim.22119 (2013).
Yang, C. et al. Lactational exposure to environmentally relevant benzo(a)pyrene causes astrocytic activation and anxiety-like behavior in male mice. Chemosphere 221, 67–74. https://doi.org/10.1016/j.chemosphere.2019.01.006 (2019).
Stowers, S. J. & Anderson, M. W. Formation and persistence of benzo(a)pyrene metabolite-DNA adducts. Environ. Health Perspect. 62, 31–39. https://doi.org/10.1289/ehp.856231 (1985).
Wang, J. et al. Integrated Epigenetics, Transcriptomics, and metabolomics to analyze the mechanisms of Benzo[a]pyrene neurotoxicity in the hippocampus. Toxicol. Sci. 166, 65–81. https://doi.org/10.1093/toxsci/kfy192 (2018).
Nie, J. S., Zhang, H. M., Zhao, J., Liu, H. J. & Niu, Q. Involvement of mitochondrial pathway in benzo[a]pyrene-induced neuron apoptosis. Hum. Exp. Toxicol. 33, 240–250. https://doi.org/10.1177/0960327113493301 (2014).
Stockwell, B. R. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell 185, 2401–2421. https://doi.org/10.1016/j.cell.2022.06.003 (2022).
Doll, S. et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98. https://doi.org/10.1038/nchembio.2239 (2017).
Yang, W. S. et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U S A. 113, E4966–4975. https://doi.org/10.1073/pnas.1603244113 (2016).
Yang, W. S. et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331. https://doi.org/10.1016/j.cell.2013.12.010 (2014).
Song, X. & Long, D. Nrf2 and ferroptosis: A new research direction for neurodegenerative diseases. Front. Neurosci. 14, 267. https://doi.org/10.3389/fnins.2020.00267 (2020).
Yang, S. et al. Salidroside alleviates cognitive impairment by inhibiting ferroptosis via activation of the Nrf2/GPX4 axis in SAMP8 mice. Phytomedicine 114, 154762. https://doi.org/10.1016/j.phymed.2023.154762 (2023).
Wang, Z. et al. Comparison of quantitation methods in proteomics to define relevant toxicological information on AhR activation of HepG2 cells by bap. Toxicology 448, 152652. https://doi.org/10.1016/j.tox.2020.152652 (2021).
Tian, P. et al. BPDE induces human trophoblast cell ferroptosis by up-regulating iron metabolism and promoting GPX4 proteasomal degradation. Ecotoxicol. Environ. Saf. 228. https://doi.org/10.1016/j.ecoenv.2021.113028 (2021).
Canzler, S. et al. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 94, 371–388. https://doi.org/10.1007/s00204-020-02656-y (2020).
Shin, T. H. et al. Silica-coated magnetic-nanoparticle-induced cytotoxicity is reduced in microglia by glutathione and citrate identified using integrated omics. Part. Fibre Toxicol. 18, 42. https://doi.org/10.1186/s12989-021-00433-y (2021).
Chepelev, N. L. et al. Transcriptional profiling of the mouse hippocampus supports an NMDAR-mediated neurotoxic mode of action for benzo[a]pyrene. Environ. Mol. Mutagen. 57, 350–363. https://doi.org/10.1002/em.22020 (2016).
Zhang, H., Jiao, W., Cui, H., Sun, Q. & Fan, H. Combined exposure of alumina nanoparticles and chronic stress exacerbates hippocampal neuronal ferroptosis via activating IFN-γ/ASK1/JNK signaling pathway in rats. J. Hazard. Mater. 411 https://doi.org/10.1016/j.jhazmat.2021.125179 (2021).
Das, M., Seth, P. K. & Mukhtar, H. Distribution of benzo(a)pyrene in discrete regions of rat brain. Bull. Environ. Contam. Toxicol. 35, 500–504. https://doi.org/10.1007/bf01636545 (1985).
Liu, J., Li, L. & Suo, W. Z. HT22 hippocampal neuronal cell line possesses functional cholinergic properties. Life Sci. 84, 267–271. https://doi.org/10.1016/j.lfs.2008.12.008 (2009).
Wang, H., Liu, C., Zhao, Y. & Gao, G. Mitochondria regulation in ferroptosis. Eur. J. Cell. Biol. 99, 151058. https://doi.org/10.1016/j.ejcb.2019.151058 (2020).
Yang, Y. et al. The PM20D1-NADA pathway protects against parkinson’s disease. Cell. Death Differ. 31, 1545–1560. https://doi.org/10.1038/s41418-024-01356-9 (2024).
Pérez-Santamarina, E. et al. Regulatory rare variants of the dopaminergic gene ANKK1 as potential risk factors for parkinson’s disease. Sci. Rep. 11, 9879. https://doi.org/10.1038/s41598-021-89300-6 (2021).
Yang, L. et al. Methionine sulfoxide reductase A deficiency aggravated ferroptosis in LPS-induced acute kidney injury by inhibiting the AMPK/NRF2 axis and activating the CaMKII/HIF-1α pathway. Free Radic Biol. Med. 234, 248–263. https://doi.org/10.1016/j.freeradbiomed.2025.04.040 (2025).
Zhao, Y., Wang, Y., Zhang, X., Han, S. & Yang, B. Metformin-induced RBMS3 expression enhances ferroptosis and suppresses ovarian cancer progression. Reprod. Biol. 25, 100968. https://doi.org/10.1016/j.repbio.2024.100968 (2025).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–d677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Zhou, N. et al. FerrDb V2: update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 51, D571–D582. https://doi.org/10.1093/nar/gkac935 (2023).
Sequeira, I. et al. Immunomodulatory role of keratin 76 in oral and gastric cancer. Nat. Commun. 9, 3437. https://doi.org/10.1038/s41467-018-05872-4 (2018).
Zhou, Z. et al. Adipose-Specific Lipin-1 overexpression renders hepatic ferroptosis and exacerbates alcoholic steatohepatitis in mice. Hepatol. Commun. 3, 656–669. https://doi.org/10.1002/hep4.1333 (2019).
Hou, S., Wang, D., Yuan, X., Yuan, X. & Yuan, Q. Identification of biomarkers co-associated with M1 macrophages, ferroptosis and Cuproptosis in alcoholic hepatitis by bioinformatics and experimental verification. Front. Immunol. 14, 1146693. https://doi.org/10.3389/fimmu.2023.1146693 (2023).
Chang, L. L. et al. Stromal C-type lectin receptor COLEC12 integrates H. pylori, PGE2-EP2/4 axis and innate immunity in gastric diseases. Sci. Rep. 8, 3821. https://doi.org/10.1038/s41598-018-20957-2 (2018).
Gorchev, H. G. & Ozolins, G. WHO guidelines for drinking-water quality. WHO Chron. 38, 104–108 (1984).
Cherif, L. S. et al. Assessment of 9-OH- and 7,8-diol-benzo[a]pyrene in Blood as Potent Markers of Cognitive Impairment Related To benzo[a]pyrene Exposure Toxics, 9 (An Animal Model Study, 2021). https://doi.org/10.3390/toxics9030050
Chen, Y. H. et al. Evaluation of benzo[a]pyrene in food from China by high-performance liquid chromatography-fluorescence detection. Int. J. Environ. Res. Public. Health. 9, 4159–4169. https://doi.org/10.3390/ijerph9114159 (2012).
Benford, D. et al. Application of the margin of exposure (MOE) approach to substances in food that are genotoxic and carcinogenic. Food Chem. Toxicol. 48, S2–S24. https://doi.org/10.1016/j.fct.2009.11.003 (2010).
Qiu, C., Peng, B., Cheng, S., Xia, Y. & Tu, B. The effect of occupational exposure to benzo[a]pyrene on neurobehavioral function in coke oven workers. Am. J. Ind. Med. 56, 347–355. https://doi.org/10.1002/ajim.22119 (2013).
Zhao, T. et al. Metformin alleviates the cognitive impairment induced by benzo[a]pyrene via glucolipid metabolism regulated by FTO/FoxO6 pathway in mice. Environ. Sci. Pollut Res. Int. 30, 69192–69204. https://doi.org/10.1007/s11356-023-27303-8 (2023).
Lin, S. et al. Oxidative stress and apoptosis in Benzo[a]pyrene-Induced neural tube defects. Free Radic Biol. Med. 116, 149–158. https://doi.org/10.1016/j.freeradbiomed.2018.01.004 (2018).
Scott, J. et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death, Cell, 149 1060–1072. (2012). https://doi.org/10.1016/j.cell.2012.03.042
Yu, Y. et al. Hepatic transferrin plays a role in systemic iron homeostasis and liver ferroptosis. Blood 136, 726–739. https://doi.org/10.1182/blood.2019002907 (2020).
Ji, T. et al. BPDE induces ferroptosis in hippocampal neurons through ACSL3 suppression. Neurotoxicology 107, 11–21. https://doi.org/10.1016/j.neuro.2025.01.002 (2025).
Shen, Y. T. et al. Metabolomics and proteomics study reveals the effects of benzo a pyrene on the viability and migration of KYSE-150 esophageal cells. Sci. Total Environ. 824. https://doi.org/10.1016/j.scitotenv.2022.153761 (2022).
Cho, I., Jackson, M. R. & Swift, J. Roles of Cross-Membrane transport and signaling in the maintenance of cellular homeostasis. Cell. Mol. Bioeng. 9, 234–246. https://doi.org/10.1007/s12195-016-0439-6 (2016).
César-Razquin, A. et al. A call for systematic research on solute carriers. Cell 162, 478–487. https://doi.org/10.1016/j.cell.2015.07.022 (2015).
Sato, H., Tamba, M., Ishii, T. & Bannai, S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 274, 11455–11458. https://doi.org/10.1074/jbc.274.17.11455 (1999).
Liu, Y. et al. The diversified role of mitochondria in ferroptosis in cancer. Cell. Death Dis. 14, 519. https://doi.org/10.1038/s41419-023-06045-y (2023).
Liu, L. et al. SIRT3 inhibits gallbladder cancer by induction of AKT-dependent ferroptosis and Blockade of epithelial-mesenchymal transition. Cancer Lett. 510, 93–104. https://doi.org/10.1016/j.canlet.2021.04.007 (2021).
Han, D. et al. SIRT3 deficiency is resistant to autophagy-dependent ferroptosis by inhibiting the AMPK/mTOR pathway and promoting GPX4 levels. J. Cell. Physiol. 235, 8839–8851. https://doi.org/10.1002/jcp.29727 (2020).
Guo, H. et al. ChREBP-β/TXNIP aggravates frucose-induced renal injury through triggering ferroptosis of renal tubular epithelial cells. Free Radic. Biol. Med. 199, 154–165. https://doi.org/10.1016/j.freeradbiomed.2023.02.013 (2023).
Lee, S. et al. Autophagy mediates an amplification loop during ferroptosis. Cell Death Dis. 14, 464. https://doi.org/10.1038/s41419-023-05978-8 (2023).
Yu, F. et al. Dynamic O-GlcNAcylation coordinates ferritinophagy and mitophagy to activate ferroptosis. Cell. Discov. 8, 40. https://doi.org/10.1038/s41421-022-00390-6 (2022).
Lyu, Y., Ren, X. K., Guo, C. C., Li, Z. F. & Zheng, J. P. Benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide induces ferroptosis in neuroblastoma cells through redox imbalance. J. Toxicol. Sci. 47, 519–529. https://doi.org/10.2131/jts.47.519 (2022).
Pedregosa, V. G. et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830. https://doi.org/10.5555/1953048.2078195 (2011).
Funding
This work was supported by the National Natural Science Foundation of China (30872137), Shanxi 1331 Project (2021-5-2-2-B1), Natural Science Foundation of Shanxi Province, China (202203021211106, 202203021211246, 20210302124301), Changzhi Medical College Innovation Team (CX202001) and “1+1” Research Collaboration Project (HZZD202404), and Key Laboratory of Shanxi Province for Aging Mechanism Research and Transformation.
Author information
Authors and Affiliations
Contributions
Conceptualization: Jinping Zheng. Methodology: Zhirui Ma, Xingdi Guo, Tingyu Ji. Formal analysis and investigation: Hui He, Shihan Ding, Chaoli Zhou. Writing - original draft preparation: Hui He. Funding acquisition: Jinping Zheng, Yi Lyu. Resources: Xiaohui Li. Supervision: Jinping Zheng, Yi Lyu, Fengjie Tian. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, H., Ding, S., Zhou, C. et al. Integrated transcriptomics and proteomics reveal ferroptosis induced by B[a]P and BPDE in mouse hippocampal neurons. Sci Rep 15, 41739 (2025). https://doi.org/10.1038/s41598-025-26928-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26928-8