Abstract
Mitochondrial transplantation holds great promise as a therapeutic strategy; however, the mechanisms by which recipient cells interact with and internalize isolated mitochondria remain unclear. Therefore, in this study, we isolated functional mitochondria from mesenchymal stromal cells (MSCs) and characterized their biological activities and physicochemical properties. Additionally, effects of isolated mitochondria on MSC functions were evaluated. Treatment with isolated mitochondria promoted cell proliferation, improved cellular viability under stress conditions, and increased the oxygen consumption rate, indicating enhanced bioenergetic capacity. Uptake of isolated mitochondria by MSCs was visualized via fluorescence imaging and quantitatively assessed over time, showing progressive internalization within 24 h. To investigate the mechanism of mitochondrial uptake, endocytosis was chemically inhibited, which revealed that endocytic pathways contributed to the internalization of the isolated mitochondria. These findings suggest that MSCs incorporate isolated mitochondria via active uptake mechanisms and that the internalized mitochondria retain their functional activity. Collectively, our results provide critical evidence of mitochondrial internalization in MSCs and offer insights into the potential applications of mitochondrial therapy for various diseases.
Similar content being viewed by others
Introduction
Mitochondria are multifunctional organelles indispensable for cellular homeostasis maintenance1,2. In addition to their well-established role in ATP generation via oxidative phosphorylation, they participate in diverse processes, such as calcium buffering, reactive oxygen species regulation, lipid metabolism, and programmed cell death initiation3. Owing to their broad functional spectrum, mitochondrial integrity is fundamental for tissue health and survival. Disruption of mitochondrial function contributes to the pathogenesis of various human disorders4,5. Neurodegenerative diseases, such as Parkinson’s and Alzheimer’s diseases, are strongly linked to impaired mitochondrial dynamics and energy metabolism6,7. Similarly, mitochondrial dysfunction is involved in ischemia–reperfusion-related conditions, such as myocardial infarction and stroke8,9, metabolic diseases10,11, and inflammatory disorders12,13. Consequently, therapeutic strategies aimed at restoring or replacing defective mitochondria have attracted increasing interest. Among various proposed approaches, mitochondrial transplantation, which involves the administration of isolated mitochondria to compromised cells or tissues, has emerged as a promising strategy14,15. Unlike gene-based strategies and chemical agents indirectly influencing the mitochondrial activity, this approach directly supplements the recipient cells with functional mitochondria, aiding in the rescue of bioenergetic capacity and improvement of cellular viability across diverse pathological contexts16,17.
Although the beneficial effects of mitochondrial transplantation have been reported in various preclinical models18,19, cellular basis of these outcomes remains unclear. Experimental studies have shown that the administration of isolated mitochondria reduces myocardial damage following ischemia–reperfusion20,21,22, mitigates toxin-induced hepatic injury23,24,25, and improves neuronal performance in Alzheimer’s disease26,27,28 and traumatic brain injury29,30,31 models. These findings strongly suggest that isolated mitochondria support cellular recovery under pathological conditions. However, the specific interactions between isolated mitochondria and recipient cells remain ambiguous. Functional rescue possibly requires the internalization of mitochondria, enabling them to participate in recipient bioenergetic and signaling networks. However, experimental evidence supporting this uptake process is limited32, and the underlying mechanisms remain largely uncharacterized. Therefore, clarifying the mechanisms by which recipient cells incorporate isolated mitochondria is crucial for the reliable therapeutic application of mitochondrial transplantation.
Mesenchymal stromal cells (MSCs) are valuable models for mitochondrial transplantation assessment due to their unique biological properties and broad therapeutic relevance33,34. MSCs are multipotent cells capable of differentiating into various lineages that are widely used in regenerative medicine35,36. In addition to their differentiation capacity, these cells exert paracrine and immunomodulatory effects, contributing to tissue repair and protecting from inflammatory damage37. Importantly, MSCs are relatively easy to isolate and expand in culture, serving as practical experimental systems38. Mitochondrial dynamics are critical for MSC functions as their regenerative and immunomodulatory capacities are strongly influenced by mitochondrial activity39,40. Therefore, evaluation of the mechanisms by which MSCs interact with isolated mitochondria will provide insights into the cellular basis of mitochondrial transplantation and contribute to the development of MSC-based therapies. In this study, we evaluated the uptake of mitochondria isolated from MSCs. We further characterized their biological activity and physicochemical properties, evaluated their effects on MSC functions, and explored the underlying pathways using chemical inhibitors of endocytosis. Our findings provide new insights into the cellular uptake mechanisms of mitochondria, contributing to the development of improved mitochondrial transplantation strategies and novel MSC-based therapies.
Results
Characterization of the mitochondria isolated from MSCs
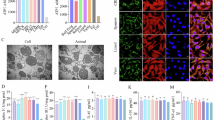
Mitochondria were isolated from the C3H10T1/2 mouse MSC line using streptolysin O (SLO), followed by differential centrifugation. Morphology was confirmed via fluorescence microscopy of carboxyfluorescein succinimidyl ester (CFSE)-labeled mitochondria (Fig. 1a). Dynamic light scattering analysis revealed a unimodal size distribution with a cumulant diameter of approximately 1000 nm and zeta potential of − 16 mV (Fig. 1b; Table 1), indicating relatively uniform negatively charged particles. A small population of large-size particles (105–106 nm) observed in the DLS profile likely represents aggregated mitochondria. The purity of the 0.6–7 × 103 × g fraction selected as the mitochondrial fraction was further validated via western blotting, which detected cytochrome c oxidase subunit IV as a mitochondrial marker. However, nuclear marker lamin B1 and cytoskeletal protein α-tubulin were almost undetectable (Fig. 1c). The isolated mitochondria retained their bioenergetic capacity, with ATP-generating capacity of approximately 4.6 µmol/107 cells and a protein yield of approximately 260 µg/107 cells (Table 1). These data confirmed that the isolated mitochondria were highly purified and metabolically active.
Characterization of mitochondria isolated from mesenchymal stromal cells. (a) Fluorescence microscopy of carboxyfluorescein succinimidyl ester (CFSE)-labeled mitochondria isolated from C3H10T1/2 cells. Scale bar, 20 µm. (b) Scattered intensity distribution measured via dynamic light scattering (DLS). Results of three independent measurements are shown. (c) Western blotting of differential centrifugation fractions obtained after streptolysin O (SLO) treatment and pipetting (< 0.6, 0.6–7, and 7–20 × 103 × g) and probing for lamin B1, α-tubulin, and cytochrome c oxidase subunit IV (COXIV). Full-length, uncropped blots are provided in the Supplementary Information (Supplementary Fig. S4).
Effects of the isolated mitochondria on cellular functions
To determine whether isolated mitochondria modulate cellular functions, we examined cell proliferation, stress resistance, and bioenergetic capacity following mitochondrial treatment. Addition of isolated mitochondria significantly enhanced the proliferation of C3H10T1/2 cells in a concentration-dependent manner, with the most potent effects observed at a mitochondrial protein concentration of 50 mg/L (Fig. 2a). Hydrogen peroxide (H2O2) scavenging assays further revealed that the isolated mitochondria effectively decreased the residual H2O2 levels in a concentration-dependent manner (Fig. 2b). To assess the protective effects under stress conditions, we treated carbon tetrachloride (CCl4)-exposed Hepa 1–6 mouse hepatocellular carcinoma cells as hepatocyte injury models with isolated mitochondria. Notably, release of hepatic injury biomarker alanine aminotransferase was markedly reduced, indicating the attenuation of hepatocellular damage (Fig. 2c). To further determine the impact on cellular bioenergetics, oxygen consumption rate (OCR) was measured. Mitochondrial treatment increased the basal respiration rate, ATP production rate, and maximal respiratory capacity of MSCs in a concentration-dependent manner (Fig. 2d and e). These findings suggest that the isolated mitochondria not only preserve their intrinsic bioenergetic activity but also exert proliferative and cytoprotective effects on MSCs and hepatocytes.
Effects of the isolated mitochondria on mesenchymal stromal cell functions. (a) Proliferation of C3H10T1/2 cells treated with the isolated mitochondria was evaluated using the cell counting kit-8 assay. *p < 0.05 (Dunnett’s test vs. 0 mg/L). (b) Hydrogen peroxide (H2O2) scavenging activity of the isolated mitochondria was quantified based on fluorescence intensity. *p < 0.05 (Dunnett’s test vs. H2O2 only group). (c) Alanine aminotransferase (ALT) activity was measured to evaluate the cytoprotective effects of the isolated mitochondria on carbon tetrachloride (CCl4)-treated Hepa 1-6 cells. *p < 0.05 (Dunnett’s test vs. CCl4 only group). (d) Oxygen consumption rate (OCR) of C3H10T1/2 cells was measured after incubation with the isolated mitochondria. Dotted vertical lines indicate the sequential addition of oligomycin, FCCP, and rotenone/antimycin A. (e) Non-mitochondrial respiration rate, basal respiration rate, maximal respiration rate, ATP production rate, and spare capacity were measured using OCR data. **p < 0.01, and ***p < 0.001 (Dunnett’s test vs. 0 mg/L). ns, not significant. All data are represented as the mean ± standard deviation (SD); n = 3 for (a–c) and n = 6 for (d, e).
Time-dependent dynamics of mitochondrial uptake by MSCs
Internalization of the isolated mitochondria into MSCs was assessed via fluorescence microscopy and flow cytometry. Confocal imaging of CFSE-labeled mitochondria revealed progressive intracellular accumulation in MSCs, beginning as early as 0.5 h post-treatment and becoming more prominent at later time points, with robust intracellular localization observed at 6 h (Fig. 3a and b). Quantitative analysis via flow cytometry confirmed these findings. Moreover, geometric mean fluorescence intensity of CFSE-labeled mitochondria steadily increased over time, peaked at 6 h, and subsequently declined by 24 h (Fig. 3c). Time-lapse imaging using a digital microscope revealed the dynamic uptake of CFSE-labeled mitochondria by MSCs, visualizing their progressive incorporation and redistribution under live-cell conditions (Supplementary Fig. S1). To further visualize the uptake of unlabeled mitochondria by MSCs, time-lapse imaging was performed using a three-dimensional optical wave field microscope (3D-OWFM)41 (Fig. 4; Supplementary Fig. S2). In the absence of cells, isolated mitochondria gradually adhered to the bottom of the culture plate (Fig. 4, upper panels). When added to MSCs, isolated mitochondria were gradually incorporated and redistributed intracellularly during 12 h of continuous monitoring (Fig. 4, lower panels). Taken together, these results provide critical evidence that MSCs actively internalize the isolated mitochondria in a time-dependent manner, with maximal uptake occurring within 24 h. To further confirm the intracellular localization of isolated mitochondria, transmission electron microscopy (TEM) was performed after mitochondrial treatment. As shown in Supplementary Fig. S3, electron-dense vesicular structures similar in size to isolated mitochondria were observed within the cytoplasm of C3H10T1/2 cells, occasionally enclosed by vesicular membranes, suggesting endocytic internalization.
Uptake of the isolated mitochondria by mesenchymal stromal cells. (a) Confocal fluorescence images of C3H10T1/2 cells treated with CFSE-labeled mitochondria for 0.5, 1, 3, 6, and 24 h. Nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; blue). Mitochondria internalized or adsorbed onto the cell surface are shown in green. Scale bar, 20 µm. (b) CFSE fluorescence intensity in C3H10T1/2 cells treated with labeled mitochondria was analyzed via flow cytometry. NT, non-treated control. (c) Mitochondrial uptake was quantified using the geometric mean fluorescence intensity (MFI). Data are represented as the mean ± SD (n = 3). *p < 0.05, **p < 0.01, and ***p < 0.001 (Dunnett’s test vs. NT).
Time-lapse observation of isolated mitochondrial uptake by mesenchymal stromal cells. Label-free mitochondria were added to C3H10T1/2 cells and observed over 12 h using a three-dimensional optical wave field microscope (3D-OWFM) via quantitative phase imaging. Upper panels show the label-free mitochondria alone, and lower panels show C3H10T1/2 cells incubated with label-free mitochondria. Pseudo-color indicates the optical phase delay. Scale bar, 200 µm.
Pathways of mitochondrial uptake by MSCs
To elucidate the mechanisms underlying mitochondrial internalization, MSCs were cultured at low temperatures and treated with endocytosis inhibitors. Flow cytometric quantification revealed that incubation at 4 °C, which typically inhibits energy-dependent endocytic processes, considerably suppressed mitochondrial uptake compared to that in the cells incubated at 37 °C (Fig. 5a). Pretreatment with dynasore, a dynamin-dependent endocytosis inhibitor, significantly reduced mitochondrial uptake compared to that in the non-treated group (Fig. 5b). As dynasore inhibits both clathrin-mediated endocytosis (CME) and fast endophilin-mediated endocytosis (FEME)42,43, these results suggest the involvement of dynamin-dependent pathways in mitochondrial uptake. Disruption of lipid rafts by methyl-β-cyclodextrin (MβCD) also markedly decreased mitochondrial internalization (Fig. 5c). MβCD impairs caveolae-mediated endocytosis (CavME) and the clathrin-independent carriers/GPI-enriched endocytic compartments (CLIC/GEEC) pathway, both of which depend on intact lipid rafts42,44. Inhibition of macropinocytosis with 5-(N-ethyl-N-isopropyl)-amiloride (EIPA) also attenuated mitochondrial uptake, albeit to a lesser extent (Fig. 5d). Together, these results suggest that MSCs internalize the isolated mitochondria via active energy-dependent endocytic pathways involving multiple mechanisms, including dynamin-mediated (CME and/or FEME), lipid raft-dependent (CavME and/or CLIC/GEEC), and micropinocytosis processes.
Pathway of isolated mitochondrial uptake by mesenchymal stromal cells. (a) C3H10T1/2 cells were pre-incubated at 4 °C for 30 min and treated with CFSE-labeled mitochondria at 4 °C for 1 h. *p < 0.05 (Welch’s t-test vs. 37 °C). (b–d) Effects of dynasore (DYN; 50 µM), methyl-β-cyclodextrin (MβCD; 20 mM), and 5-(N-ethyl-N-isopropyl)-amiloride (EIPA; 25 µM) on mitochondrial uptake were evaluated in C3H10T1/2 cells pretreated with each inhibitor and incubated with CFSE-labeled mitochondria for 6 h. Internalization was quantified via flow cytometry. Control indicates the cells treated with vehicle without any inhibitors. *p < 0.05 (Welch’s t-test vs. control). ns, not significant. All data are represented as the mean ± SD (n = 3).
Discussion
Mitochondrial transplantation is a promising therapeutic approach for diseases characterized by mitochondrial dysfunction. Although the beneficial effects of isolated mitochondria have been demonstrated in ischemia, neurodegeneration, and metabolic injury models, whether these outcomes require direct internalization remains controversial18,19,20,21,22,26,27,28,45. Recent studies on the endocytic uptake of isolated mitochondria derived from non-MSC sources have suggested macropinocytosis as a major pathway, with additional contributions from dynamin-dependent endocytosis and lipid raft-associated mechanisms17,46,47,48. However, some studies have challenged the involvement of macropinocytosis, resulting in no clear consensus on the dominant uptake pathway49,50. Previous studies have mainly focused on intercellular transfer via tunneling nanotubes and extracellular vesicles45,51,52. To our knowledge, no study has characterized the specific endocytic uptake mechanisms of isolated mitochondria to date. Therefore, in this study, we directly visualized and quantified mitochondrial internalization by MSCs and found that mitochondrial uptake was an active energy-dependent process mediated by multiple endocytic pathways.
Mitochondria isolated using SLO retained both their morphological integrity and bioenergetic activity (Fig. 1; Table 1), as the SLO-based isolation method described by Shibata et al. enabled the preparation of mitochondria with more structural and functional completeness53 compared to conventional isolation techniques. The size distribution data obtained by DLS occasionally included large-size peaks (105–106 nm), which were attributable to aggregated mitochondria rather than contaminants. As is well known in DLS measurements, large aggregates can cause excessive light scattering and result in apparent overestimation of the particle-size values. No additional purification steps were performed to remove such aggregates, as they were considered to represent the physiological state of isolated mitochondria.
Structural preservation of mitochondria is critical for their functional efficacy. Mitochondria disrupted by sonication or freeze–thaw cycles show markedly reduced bioenergetic capacity and proliferative effects compared to intact mitochondria17,20. In this study, treatment with isolated intact mitochondria enhanced proliferation, increased the OCR of C3H10T1/2 cells, and improved Hepa 1–6 cell resistance to chemical stress (Fig. 2a–e). OCR analysis revealed that mitochondrial treatment increased the basal respiration rate, ATP-linked respiration rate, and maximal respiratory capacity in a concentration-dependent manner (Fig. 2d and e), confirming that the isolated mitochondria preserved intrinsic metabolic activity and contributed to recipient cell bioenergetics, consistent with previously reports17,54. Although intact mitochondria are sufficient for enhancing cellular functions, their internalization mechanisms remain unclear. Our study found that cells internalized intact mitochondria via multiple pathways, suggesting that such uptake plays an important role in mediating their functional effects.
Through complementary modalities, including confocal fluorescence microscopy, flow cytometry, and label-free quantitative phase imaging using 3D-OWFM, we directly captured the time-resolved internalization of isolated mitochondria by C3H10T1/2 cells. The signal became detectable within 0.5 h, intensified by 6 h, and approached a plateau by 24 h (Fig. 3a–c; Supplementary Fig. S1), and 3D-OWFM revealed gradual intracellular accumulation over 12 h (Fig. 4; Supplementary Fig. S2). These observations were further supported by TEM analysis, which revealed mitochondria-like electron-dense structures enclosed by vesicular membranes within the cytoplasm (Supplementary Fig. S3). These ultrastructural findings provide direct evidence that isolated mitochondria were internalized through endocytic processes rather than surface adherence. These kinetics are consistent with an energy-dependent endocytic cascade rather than instantaneous membrane fusion. Moreover, mitochondrial internalization was markedly suppressed upon incubation at 4 °C, consistent with a previous report that isolated mitochondrial uptake is inhibited under low-temperature conditions55. Inhibitor experiments further confirmed the active nature of this process; dynasore (dynamin inhibitor affecting both CME and FEME) and MβCD (disrupts lipid rafts and impairs CavME and the CLIC/GEEC pathway) significantly reduced mitochondrial uptake, whereas EIPA (macropinocytosis inhibitor) only caused non-significant inhibition (Fig. 5). Generally, uptake of particles larger than 200 nm is attributed to macropinocytosis, which is the predominant mechanism for the internalization of such large structures in non-phagocytic cells42. Although CME, CavME, CLIC/GEEC, and FEME are generally reported to internalize particles smaller than 200 nm, Rejman et al. demonstrated the caveolae-mediated internalization of particles up to 500 nm in size56. This finding remains an exception to the general paradigm; however, such size flexibility is particularly relevant to mitochondrial uptake, which can vary depending on the recipient cell type and physiological state. Collectively, these results suggest that dynamin-dependent endocytosis and lipid raft-associated mechanisms are the primary contributors to mitochondrial internalization, whereas macropinocytosis plays a supportive role in MSCs in specific contexts. Simultaneous engagement of multiple distinct pathways suggests that mitochondrial uptake relies on a flexible and redundant network, ensuring the efficient incorporation of exogenous mitochondria under diverse cellular conditions. This redundancy possibly explains the consistent therapeutic benefits reported in mitochondrial transplantation studies.
Our findings have important implications for future studies. Functional integration of isolated mitochondria into MSCs supports the rationale for mitochondrial transplantation as a means of restoring cellular bioenergetics in pathological settings. Furthermore, identification of endocytic pathways revealed potential targets to modulate mitochondrial uptake and enhance the efficiency of mitochondrial transplantation. However, some limitations should be addressed. First, our experiments were conducted primarily in vitro using an MSC murine cell line; therefore, whether similar mechanisms operate across diverse cell types and in vivo environments warrants further investigation. As the membrane composition and metabolic demands vary among cardiomyocytes, neurons, hepatocytes, and immune cells57,58, systematic comparisons are essential to determine whether the mechanisms identified in this study are universally applicable. Addressing these issues is essential for translating mitochondrial transplantation into robust clinical practice. Further comparative analyses of different cell populations and in vivo disease models and long-term tracking of transplanted organelles will aid in clarifying the generalizable principles of mitochondrial uptake.
Conclusions
In conclusion, this study provides crucial evidence of isolated mitochondria internalization by MSCs via multiple endocytic pathways, leading to enhanced cellular bioenergetics and stress resistance. Our findings clarify a key aspect of mitochondrial transplantation, contributing to the development of novel MSC-based biomedicines and disease treatment strategies.
Methods
Materials
CH3COOK, MgCl2, dithiothreitol (DTT), HEPES, sucrose, Tris-HCl, SLO recombinant solution, H2O2, CCl4, and CultureSure dimethyl sulfoxide (DMSO) were obtained from FUJIFILM Wako Pure Chemical Corp. (Osaka, Japan). CFSE was obtained from Cayman Chemical Co. Inc. (Ann Arbor, MI, USA). Dynasore was obtained from Selleck Chemicals LLC (Houston, TX, USA). EIPA was obtained from R&D Systems, Inc. (Minneapolis, MN, USA). Ethylenediaminetetraacetic acid (EDTA), MβCD was obtained from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). All other chemicals used were of the highest grade available commercially.
Cell culture
C3H10T1/2 murine MSC line was obtained from Dr. Hiroki Kagawa (Department of Cell Biology, Kyoto Pharmaceutical University, Kyoto, Japan) and cultured in the Dulbecco’s modified Eagle’s medium (DMEM; Shimadzu Corp., Kyoto, Japan) supplemented with 15% heat-inactivated fetal bovine serum (Gibco, Inc., Waltham, MA, USA), 0.15% sodium bicarbonate, and 1% penicillin–streptomycin–L-glutamine solution (FUJIFILM Wako Pure Chemical Corp.). The Hepa 1-6 murine hepatoma cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA; Cat. No. CRL-1830) and cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 0.15% sodium bicarbonate, and 1% penicillin–streptomycin–L-glutamine solution. These cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.
Isolation of mitochondria
Mitochondria were isolated from C3H10T1/2 cells as previous described53, with some modifications. Briefly, the cells were cultured under standard conditions, detached with 2.5 g/L trypsin/1 mmol/L EDTA solution (Nacalai Tesque, Inc., Kyoto, Japan), and neutralized with complete DMEM. After harvesting via centrifugation at 300 × g for 3 min, the cells were washed once with phosphate-buffered saline (PBS; Shimadzu Corp.) and centrifuged again under the same conditions. The cell pellet was resuspended in the HEPES-CH3COOK buffer (115 mM CH3COOK, 2.5 mM MgCl2, 1 mM DTT, and 25 mM HEPES [pH 7.4]), and SLO (final concentration: 1 mg/L) was added. The suspension was incubated on ice for 10 min to allow SLO binding to the plasma membrane, followed by centrifugation at 600 × g for 3 min at 4 °C, and the resulting pellet was washed twice with Tris-sucrose buffer (250 mM sucrose, 0.5 mM EDTA, and 10 mM Tris-HCl [pH 7.4]) to eliminate the residual SLO. To form SLO-induced small pores in the plasma membrane, the suspension was incubated at 37 °C for 10 min. To enlarge these pores and achieve selective disruption of the plasma membrane while minimizing mechanical damage to mitochondria, the suspension was gently pipetted 200 times per mL using a micropipette (Nichiryo Co., Ltd., Tokyo, Japan). The sample was centrifuged at 600 × g for 10 min at 4 °C to remove the unbroken cells, nuclei, and large cell debris. This centrifugation step was repeated until no visible pellet remained. The combined supernatants were further centrifuged at 7000 × g for 10 min at 4 °C to pellet the mitochondria. The resulting mitochondrial pellet was resuspended in an appropriate buffer, depending on the downstream application. Finally, protein concentration was measured using the BCA protein assay kit (Nacalai Tesque, Inc.), and mitochondrial preparations were used for subsequent analyses.
Characterization of the isolated mitochondria
Mitochondrial ATP production was evaluated using the CellTiter-Glo 3D Cell Viability Assay (Promega Corp., Madison, WI, USA), according to the manufacturer’s instructions. The isolated mitochondria were suspended in DMEM at a concentration of 60 mg/L, and luminescence was measured using the GloMax Discover System (Promega Corp.). Then, size distribution, cumulant diameter, and zeta potential of the mitochondria (15 mg/L in PBS) were measured via dynamic light scattering (DLS) using an electrophoretic light scattering analyzer (ELSZ-2000ZS; Otsuka Electronics Co., Ltd., Osaka, Japan). For fluorescence imaging, CFSE-labeled mitochondria (300 mg/L in PBS) were placed on glass slides and imaged using a fluorescence microscope (BZ-X800; Keyence Corp., Osaka, Japan). H2O2 scavenging activity was assessed by incubating the isolated mitochondria (final concentration: 2, 10, and 50 mg/mL, suspended in PBS) with H2O2 (final concentration: 800 μM) at ambient temperature (approximately 22–25 °C) for 30 min. Residual H2O2 concentration was quantified using the Red Hydrogen Peroxide Assay Kit (Enzo Life Sciences, Inc., Farmingdale, NY, USA) and expressed as a percentage of the initial H2O2 concentration.
Western blotting analysis
Mitochondrial pellets were lysed in the radioimmunoprecipitation assay buffer (Nacalai Tesque, Inc.) supplemented with a Protease Inhibitor Cocktail Set III, EDTA-free, DMSO solution (× 100; FUJIFILM Wako Pure Chemical Corp.) on ice. The lysates were centrifuged at 16,000 × g for 15 min at 4 °C, and protein concentration in the supernatant was determined using a BCA protein assay kit. Briefly, 30 μg of protein was mixed with 4 × Laemmli sample buffer (250 mM Tris-HCl [pH 6.8], 8% SDS, 40% glycerol, and 0.04% bromophenol blue), and 100 mM DTT was added immediately before use. The samples were boiled at 95 °C for 3 min and loaded onto gels. Electrophoresis was performed using 12% polyacrylamide gels prepared with the TGX FastCast Acrylamide Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 150 V. Subsequently, the proteins were transferred onto Immobilon-P polyvinylidene difluoride membranes (Merck Millipore, Burlington, MA, USA) at 100 V. The membranes were blocked with 5% skim milk in PBS containing 0.1% Tween-20 and incubated with primary antibodies against lamin B1 (12987-1-AP), α-tubulin (11224-1-AP), and cytochrome c oxidase subunit IV (11242-1-AP; all from Proteintech Group, Inc., Rosemont, IL, USA) for 1 h at room temperature (RT). After washing, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (#7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 1 h at RT Protein bands were visualized using the Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) and detected using the iBright Imaging System (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Assessment of cell proliferative effects
C3H10T1/2 cells were seeded at a density of 1 × 103 cells/well in a 96-well plate (Costar, Corning, NY, USA) and cultured overnight. The medium was replaced with DMEM containing isolated mitochondria at final protein concentrations of 2, 10, and 50 mg/L. After 48 h, cell proliferation was assessed using the Cell Counting Kit-8 (Dojindo Laboratories, Inc., Kumamoto, Japan), with background correction performed using mitochondria-containing blank wells.
Treatment of hepatic injury model cells
Hepa 1–6 cells were seeded at a density of 3 × 104 cells/well in a 96-well plate and cultured overnight. To induce hepatocellular damage, the culture medium was replaced with DMEM containing 30 mM CCl4 in DMSO. After 3-h incubation, the cells were washed with PBS, and the medium was replaced with DMEM containing mitochondria (10 and 50 mg/L). After 24-h incubation, the culture supernatant was collected and centrifuged at 300 × g for 3 min to remove any debris. Alanine aminotransferase activity in the supernatant was measured via Transaminase CII Test (FUJIFILM Wako Pure Chemical Corp.).
OCR measurement
OCR of C3H10T1/2 cells was measured using the Seahorse XF96 Analyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The cells (3 × 104 cells/well) were seeded in a 24-well plate (Costar) and incubated overnight at 37 °C in a humidified incubator with 5% CO2. The isolated mitochondria were added to the culture medium at concentrations of 0, 10, and 50 mg/L and incubated for 24 h. To eliminate the non-internalized mitochondria, the cells were washed with PBS, treated with trypsin–EDTA, and collected via centrifugation. The cells were further seeded in XF96 Cell Culture Microplates (Agilent Technologies, Inc.) at a density of 1.5 × 104 cells/well and incubated overnight. On the day of the assay, the culture medium was replaced with the Seahorse XF assay medium (supplemented with 10 mM glucose, 1 mM pyruvate, and 2 mM glutamine [pH 7.4]), and the cells were incubated at 37 °C in a non-CO2 incubator for 30 min. OCR was measured via sequential injections of 0.5 µM oligomycin, 2 µM FCCP, and 0.5 µM rotenone/antimycin A. Basal respiration rate, ATP-linked respiration rate, maximal respiration rate, and spare respiratory capacity were calculated using the Wave software (Agilent Technologies, Inc.).
Observation of mitochondrial uptake
C3H10T1/2 cells were seeded at a density of 5 × 104 cells in a 35-mm glass-bottom dish (27-mm glass diameter; Matsunami Glass Ind., Ltd., Osaka, Japan) and incubated overnight. The culture medium was replaced with DMEM containing 10 mg/L CFSE-labeled mitochondria. At the designated time points (0.5, 1, 3, 6, and 24 h), the cells were washed with PBS and fixed with 4% paraformaldehyde (PFA). The cells were mounted with the VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratories, Inc., Newark, CA, USA). Fluorescence images were acquired using the TCS SP8 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) equipped with a 63 × oil-immersion objective lens.
Time-lapse imaging of mitochondrial uptake
C3H10T1/2 cells were seeded at a density of 1 × 105 cells in a 35-mm glass-bottom dish. After overnight incubation, CFSE-labeled mitochondria were added to the culture medium at a final concentration of 10 mg/L. Time-lapse fluorescence imaging was performed using the APEXVIEW APX100 Benchtop Fluorescence Microscope (EVIDENT Co., Ltd., Tokyo, Japan) equipped with an internal incubation system maintaining the cells at 37 °C and 5% CO2. Images were continuously captured every 30 s for a total of 12 h.
Flow cytometry
C3H10T1/2 cells were seeded at a density of 1 × 105 cells/well in a 24-well plate and incubated overnight. The medium was replaced with DMEM containing CFSE-labeled mitochondria (20–50 mg/L), and the cells were incubated for 0.5, 1, 3, 6, and 24 h. After washing with PBS, the cells were treated with trypsin–EDTA to remove any surface-bound mitochondria and distinguish them from the internalized mitochondria. The cells were fixed with 4% PFA, and geometric mean of CFSE fluorescence intensity was measured using the BD FACS Lyric flow cytometer (Becton, Dickinson and Co., Franklin Lakes, NJ, USA) to quantify mitochondrial uptake. To assess the inhibitory effect on mitochondrial uptake, the cells were pre-incubated at 4 °C for 30 min prior to the addition of CFSE-labeled mitochondria and further incubated for 1 h at 4 or 37 °C for comparison. Alternatively, the cells were pretreated with specific inhibitors (50 μM dynasore, 20 mM MβCD, or 25 μM EIPA), followed by incubation with mitochondria for 6 h. Vehicle-treated cells (DMSO or PBS) were used as controls. Geometric mean fluorescence intensity was expressed as a percentage relative to that in the control group.
Non-labeled observation of mitochondrial uptake
C3H10T1/2 cells were seeded at a density of 5 × 105 cells in a 35-mm culture dish without a glass bottom (Costar) and cultured overnight. The cells were treated with mitochondria (250 mg/L in DMEM), and mitochondrial dynamics were monitored using a 3D-OWFM (MINUK; Otsuka Electronics Co., Ltd.). Time-lapse images were acquired every 30 s for 12 h using an external incubation system (SV-70A; BLAST Co., Inc., Kawasaki, Japan) to maintain the cells at 37 °C and 5% CO2. Data were acquired and analyzed using the MINUK software (version 2.7.0.0) and MINUK Viewer (version 2.13.2.0), respectively.
TEM imaging of mitochondrial uptake
C3H10T1/2 cells were seeded at a density of 1 × 105 cells in a 35-mm glass-bottom dish. After overnight incubation, isolated mitochondria were added to the culture medium to a final concentration of 10 mg/L. After 6 h, cells were washed with PBS and fixed in 4% PFA. Post-fixation, processing, and imaging were performed by Tokai-EMA Inc. (Nagoya, Japan) using standard protocols as follows. Unless otherwise specified, all steps were performed at 4 °C. Prefixation in 2% PFA and 2% glutaraldehyde in 0.1 M phosphate buffer (PB) pre-warmed to culture temperature, followed by cooling; additional fixation in 2% glutaraldehyde in 0.1 M PB overnight; rinsing with 0.1 M PB; postfixation in 2% osmium tetroxide in 0.1 M PB for 1 h; graded ethanol dehydration (50% and 70% for 10 min each; 90% for 10 min at RT; absolute ethanol for 10 min, three changes, at RT); resin infiltration in a 1:1 mixture of ethanol and Quetol-812 (Nisshin EM Co., Tokyo, Japan) for 1 h at RT; embedding in Quetol-812 at 60 °C for 48 h. Ultrathin sections (70 nm) were prepared on an Ultracut UCT ultramicrotome (Leica Microsystems GmbH) with a diamond knife, collected on copper grids, stained with 2% uranyl acetate for 15 min at RT and lead stain solution (Sigma-Aldrich Co. LLC., St. Louis, MO, USA) for 3 min at RT, and examined on a JEM-1400Plus TEM (JEOL Ltd., Tokyo, Japan) operated at 100 kV with a charge-coupled device camera (EM-14830RUBY2).
Statistical analyses
Statistical analyses were conducted using the R software (version 4.3.2; R Foundation for Statistical Computing, Vienna, Austria). The official URL for R software is https://www.R-project.org/. Welch’s t-test was used to compare two unpaired groups, whereas Dunnett’s test was used for multigroup comparison against a single control. Analysis of variance and F-tests were not performed to avoid issues related to multiple comparisons. Statistical significance was set at p < 0.05 (*p < 0.05, **p < 0.01, and ***p < 0.001).
Use of large language models (LLMs)
We used ChatGPT (GPT-5, OpenAI, accessed August–September 2025) for language editing and refinement of the manuscript. The LLM was applied solely to improve grammar and readability and to draft figure legends, without generating scientific content or analyzing data. The final manuscript content was reviewed, validated, and approved by all authors.
Data availability
All datasets used/analyzed in this study are available upon request from the corresponding author.
References
Suomalainen, A. & Nunnari, J. Mitochondria at the crossroads of health and disease. Cell 187, 2601–2627 (2024).
Palikaras, K., Lionaki, E. & Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022 (2018).
Monzel, A. S., Enríquez, J. A. & Picard, M. Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nat. Metab. 5, 546–562 (2023).
Nunnari, J. & Suomalainen, A. Mitochondria: In sickness and in health. Cell 148, 1145–1159 (2012).
Schapira, A. H. Mitochondrial disease. Lancet 368, 70–82 (2006).
Johri, A. & Beal, M. F. Mitochondrial dysfunction in neurodegenerative diseases. J. Pharmacol. Exp. Ther. 342, 619–630 (2012).
Pagano Zottola, A. C. et al. Potentiation of mitochondrial function by mitoDREADD-G. Nat Neurosci (2025).
Hayashida, K. et al. Mitochondrial transplantation therapy for ischemia reperfusion injury: A systematic review of animal and human studies. J. Transl. Med. 19, 214 (2021).
Chouchani, E. T. et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431–435 (2014).
Patti, M. E. & Corvera, S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr. Rev. 31, 364–395 (2010).
Goncalves, R. L. S. et al. CoQ imbalance drives reverse electron transport to disrupt liver metabolism. Nature 643, 1057–1065 (2025).
Marchi, S., Guilbaud, E., Tait, S. W. G., Yamazaki, T. & Galluzzi, L. Mitochondrial control of inflammation. Nat. Rev. Immunol. 23, 159–173 (2023).
Hong, Y. The role of mitochondria-related proteins in inflammation and autoimmune diseases: A causal analysis using Mendelian randomization and colocalization. Health Inf. Sci. Syst. 13, 42 (2025).
Zong, Y. et al. Mitochondrial dysfunction: mechanisms and advances in therapy. Signal Transduct Target Ther 9, 124 (2024).
Brestoff, J. R. et al. Recommendations for mitochondria transfer and transplantation nomenclature and characterization. Nat. Metab. 7, 53–67 (2025).
King, M. P. & Attardi, G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 246, 500–503 (1989).
Bhattacharya, D., Slavin, M. B. & Hood, D. A. Muscle mitochondrial transplantation can rescue and maintain cellular homeostasis. Am. J. Physiol. Cell Physiol. 325, C862–C884 (2023).
Miao, X., Jiang, P., Wang, Z., Kong, W. & Feng, L. Mitochondrial Transplantation: A Novel Therapeutic Approach for Treating Diseases. MedComm 6, e70253 (2025).
Perrier, Q. et al. Therapeutic transplantation of mitochondria and extracellular vesicles: Mechanistic insights into mitochondria bioenergetics, redox signaling, and organelle dynamics in preclinical models. Free Radic. Biol. Med. 238, 473–495 (2025).
McCully, J. D. et al. Injection of isolated mitochondria during early reperfusion for cardioprotection. Am. J. Physiol. Heart Circ. Physiol. 296, H94–H105 (2009).
Cowan, D. B. et al. Intracoronary delivery of mitochondria to the ischemic heart for cardioprotection. PLoS ONE 11, e0160889 (2016).
Doulamis, I. P. et al. Mitochondrial transplantation normalizes transcriptomic and proteomic shift associated with ischemia reperfusion injury in neonatal hearts donated after circulatory death. Sci. Rep. 14, 31236 (2024).
Zhao, Z., Hou, Y., Zhou, W., Keerthiga, R. & Fu, A. Mitochondrial transplantation therapy inhibit carbon tetrachloride-induced liver injury through scavenging free radicals and protecting hepatocytes. Bioeng. Transl. Med. 6, e10209 (2021).
Shi, X. et al. Treatment of acetaminophen-induced liver injury with exogenous mitochondria in mice. Transl. Res. 196, 31–41 (2018).
Ulger, O. et al. The effects of mitochondrial transplantation in acetaminophen-induced liver toxicity in rats. Life Sci. 279, 119669 (2021).
Nitzan, K. et al. Mitochondrial Transfer ameliorates cognitive deficits, neuronal loss, and gliosis in Alzheimer’s disease mice. J. Alzheimers Dis. 72, 587–604 (2019).
Sweetat, S. et al. The Beneficial Effect of Mitochondrial Transfer Therapy in 5XFAD Mice via Liver-Serum-Brain Response. Cells 12, 1006 (2023).
Gupta, T. et al. Mitotherapy Restores mitochondrial function and improves cognitive deficits in Alzheimer’s disease. Mitochondrion 85, 102077 (2025).
Zhang, B. et al. Effects of Brain-derived mitochondria on the function of neuron and vascular endothelial cell after traumatic brain injury. World Neurosurg. 138, e1–e9 (2020).
Zhao, J. et al. Mitochondria transplantation protects traumatic brain injury via promoting neuronal survival and astrocytic BDNF. Transl. Res. 235, 102–114 (2021).
Bamshad, C. et al. Human umbilical cord-derived mesenchymal stem cells-harvested mitochondrial transplantation improved motor function in TBI models through rescuing neuronal cells from apoptosis and alleviating astrogliosis and microglia activation. Int. Immunopharmacol. 118, 110106 (2023).
Cowan, D. B. et al. Transit and integration of extracellular mitochondria in human heart cells. Sci. Rep. 7, 17450 (2017).
Liu, Z., Sun, Y., Qi, Z., Cao, L. & Ding, S. Mitochondrial transfer/transplantation: An emerging therapeutic approach for multiple diseases. Cell Biosci. 12, 66 (2022).
Malekpour, K., Hazrati, A., Soudi, S. & Hashemi, S. M. Mechanisms behind therapeutic potentials of mesenchymal stem cell mitochondria transfer/delivery. J. Control Release 354, 755–769 (2023).
Murphy, M. B., Moncivais, K. & Caplan, A. I. Mesenchymal stem cells: Environmentally responsive therapeutics for regenerative medicine. Exp. Mol. Med. 45, e54 (2013).
Han, X. et al. Mesenchymal stem cells in treating human diseases: molecular mechanisms and clinical studies. Signal Transduct. Target Ther. 10, 262 (2025).
Spees, J. L., Lee, R. H. & Gregory, C. A. Mechanisms of mesenchymal stem/stromal cell function. Stem Cell Res. Ther. 7, 125 (2016).
Keating, A. Mesenchymal stromal cells. Curr. Opin. Hematol. 13, 419–425 (2006).
Yang, F. et al. Leptin increases mitochondrial OPA1 via GSK3-mediated OMA1 ubiquitination to enhance therapeutic effects of mesenchymal stem cell transplantation. Cell Death Dis. 9, 556 (2018).
Piekarska, K. et al. Mesenchymal stem cells transfer mitochondria to allogeneic Tregs in an HLA-dependent manner improving their immunosuppressive activity. Nat. Commun. 13, 856 (2022).
Matsumoto, S. et al. Cell observation and analysis with a three-dimensional optical wave field microscope. Biosensors 15, 515 (2025).
Rennick, J. J., Johnston, A. P. R. & Parton, R. G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 16, 266–276 (2021).
Macia, E. et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10, 839–850 (2006).
Kilsdonk, E. P. et al. Cellular cholesterol efflux mediated by cyclodextrins. J. Biol. Chem. 270, 17250–17256 (1995).
Lin, R. Z. et al. Mitochondrial transfer mediates endothelial cell engraftment through mitophagy. Nature 629, 660–668 (2024).
Kitani, T., Kami, D., Matoba, S. & Gojo, S. Internalization of isolated functional mitochondria: Involvement of macropinocytosis. J. Cell Mol. Med. 18, 1694–1703 (2014).
Kesner, E. E., Saada-Reich, A. & Lorberboum-Galski, H. Characteristics of mitochondrial transformation into human cells. Sci. Rep. 6, 26057 (2016).
Patel, D. et al. Macropinocytic entry of isolated mitochondria in epidermal growth factor-activated human osteosarcoma cells. Sci. Rep. 7, 12886 (2017).
Pacak, C. A. et al. Actin-dependent mitochondrial internalization in cardiomyocytes: Evidence for rescue of mitochondrial function. Biol. Open 4, 622–626 (2015).
Levoux, J. et al. Platelets facilitate the wound-healing capability of mesenchymal stem cells by mitochondrial transfer and metabolic reprogramming. Cell Metab. 33, 283-299.e289 (2021).
Islam, M. N. et al. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 18, 759–765 (2012).
Phinney, D. G. et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat. Commun. 6, 8472 (2015).
Shibata, T., Yamashita, S., Hirusaki, K., Katoh, K. & Ohta, Y. Isolation of mitochondria by gentle cell membrane disruption, and their subsequent characterization. Biochem. Biophys. Res. Commun. 463, 563–568 (2015).
Ali Pour, P., Kenney, M. C. & Kheradvar, A. Bioenergetics consequences of mitochondrial transplantation in cardiomyocytes. J. Am. Heart Assoc. 9, e014501 (2020).
Ber, R., Stauver, M. G. & Shay, J. W. Use of isolated mitochondria to transfer chloramphenicol resistance in hamster cells. Isr. J. Med. Sci. 20, 244–248 (1984).
Rejman, J., Oberle, V., Zuhorn, I. S. & Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 377, 159–169 (2004).
Symons, J. L. et al. Lipidomic atlas of mammalian cell membranes reveals hierarchical variation induced by culture conditions, subcellular membranes, and cell lineages. Soft Matter 17, 288–297 (2021).
Thorp, E. B. & Karlstaedt, A. Intersection of immunology and metabolism in myocardial disease. Circ. Res. 134, 1824–1840 (2024).
Acknowledgements
The authors are grateful to Shimon Matsumoto, Sota Okamoto, Shuhei Goto, Kensaku Shimoda, and Sota Shioya (Otsuka Electronics Co., Ltd.) for helpful advice on the three-dimensional optical wave field microscope (3D-OWFM). The authors would also like to thank Editage for English language editing.
Funding
This work was supported by Grants-in-Aid for Scientific Research (B; grant numbers: 23H03749 and 23K28437) and Challenging Research (exploratory; grant number: 23K18595) from the Japan Society for the Promotion of Science (JSPS), Canon Foundation, Leading Pioneers Science Foundation, and Greater Tokyo Innovation Ecosystem (GTIE) supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and Japan Science and Technology Agency (JST).
Author information
Authors and Affiliations
Contributions
M.K., M.G., and K.K. conceived, designed, and performed the experiments, analyzed the data, and wrote and edited the manuscript. S.I. and M.N. analyzed the data and edited the manuscript. All authors have reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary Material 2
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kanai, M., Goto, M., Itakura, S. et al. Uptake mechanisms and functions of isolated mitochondria in mesenchymal stromal cells. Sci Rep 15, 44799 (2025). https://doi.org/10.1038/s41598-025-28494-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-28494-5