Abstract
Early and accurate detection of sugarcane leaf diseases is critical for improving crop productivity and reducing economic losses in the agricultural sector. Timely interventions enable sustainable crop management and better resource use. In this study, we propose a deep learning-based approach for sugarcane leaf disease classification that leverages a novel architecture, the Multi-scale Attention-based Dense Residual Network (MADRN). The MADRN model integrates dense residual learning and multi-scale attention mechanisms to effectively capture fine-grained, disease-specific features and address challenges related to domain variability and complex data patterns. Two datasets are used to evaluate the model: a Kaggle dataset and a blended dataset created by combining Kaggle images with those from the Bangladesh Sugarcrop Research Institute (BSRI), simulating real-world conditions. All images undergo preprocessing steps, including resizing, normalization, and data augmentation, before training. Additionally, several baseline models (CNN, VGG16, MobileNetV2, and XceptionNet) are fine-tuned and compared with the MADRN model. Experimental results demonstrate that MADRN consistently outperforms baseline models in accuracy, precision, recall, and F1-score across both datasets, achieving up to 94.78% accuracy on the Kaggle dataset and 92.25% on the blended dataset. These findings highlight MADRN’s superior ability to learn discriminative features and generalize effectively across diverse data sources, making it a promising tool for precision agriculture and disease management. To facilitate practical implementation, a web-based application is developed, enabling real-time and user-friendly disease detection. This research lays a strong foundation for the development of accurate, scalable, and practical disease classification tools that can support sustainable agricultural practices.
Similar content being viewed by others
Introduction
Plant diseases have usually hampered plant growth and agricultural output in many regions of the globe, reducing human food supply. Among all commercially cultivated products, sugarcane holds significant economic importance in Bangladesh as one of the most profitable and widely cultivated crops across various regions. It contributes substantially to agricultural income and supports the production of key products, including sugar, bioethanol, and energy generation1. Globally, sugarcane cultivation spans over 100 countries, including major producers like India, Brazil, Australia, and China2. In Bangladesh, approximately 0.38 million acres of land are dedicated to sugarcane cultivation, yielding an annual production of nearly 5.5 million tons1. Despite its critical role in the agricultural economy, Bangladesh meets only about 5% of its total sugar demand through domestic production, with jaggery contributing 20% and the remaining 75% met through imports3. However, sugarcane farming in the country faces significant challenges, particularly due to the widespread occurrence of various leaf diseases. In recent decades, pest and disease infestations have adversely impacted sugarcane productivity, and rapid detection and remediation of afflicted sugarcane may greatly reduce economic damages4. The bulk of sugarcane disease manifestations are observed on the leaves, but due to their morphological similarities, identifying these diseases can be challenging even for expert farmers5. The common sugarcane leaf diseases are rust spots, yellow spots, ring spots, brown stripe, and downy mildew6. These diseases severely impact sugarcane yield and quality, with estimated losses of up to 40% of the crop annually. The morphological similarities between these diseases make them difficult to diagnose accurately, even for experienced farmers. Furthermore, traditional disease identification procedures, which frequently rely on visual inspection, are tedious, expensive, and vulnerable to human error, limiting their ability to provide timely and precise interventions.
With the advent of advanced technologies, artificial intelligence (AI) has developed as a reliable tool for addressing these challenges. The adoption of AI in farming practices could revolutionize disease management, enabling timely and precise interventions that could enhance crop health and productivity. By utilizing large datasets and complex algorithms, AI systems can analyze plant images with remarkable precision, enabling automatic disease identification and classification7. Among these technologies, Convolutional Neural Networks (CNNs) demonstrated tremendous potential in image-based applications, particularly in detecting and classifying diseases in plants. Deep learning applications in agricultural disease detection have the potential to not only increase the accuracy of disease diagnosis but also enhance the speed and scalability of disease monitoring, providing farmers with timely and actionable insights. Several studies have explored deep learning techniques for sugarcane disease detection. S. Srivastava et al.8 proposed a deep learning-based system for detecting sugarcane plant diseases using leaf, stem, and color features, employing VGG-16, Inception V3, and VGG-19 as feature extractors. They compared various models, including neural networks and hybrid AdaBoost, and used Orange software to evaluate performance using metrics like sensitivity, specificity, and AUC. S. D. Daphal et al.9 introduced an attention-based multilevel residual convolutional neural network (AMRCNN) for classifying sugarcane leaf diseases, combining spatial and channel attention mechanisms to enhance feature learning. They collected a dataset from diverse field and weather conditions, organized it into training, validation, and testing subsets, and used TensorFlow Lite tools for real-time deployment. Bangladesh is an agricultural country where sugarcane plays a vital role in both daily life and national economic stability. However, diseases in sugarcane present a substantial challenge, resulting in the devastation of harvests and considerable economic setbacks for smallholder farmers10. Traditional disease detection methods, primarily based on morphological analysis, are time-consuming, inefficient, and inadequate for large-scale monitoring. Institutions like the Bangladesh Sugarcane Research Institute (BSRI) still rely on traditional morphological analysis methods and lack comprehensive image datasets for research and innovation. These challenges motivated the development of an AI-driven deep learning solution aimed at improving sugarcane leaf disease detection.
Using Kaggle’s publicly available sugarcane leaf image dataset, combined with real-world samples from BSRI, this research seeks to build a model that facilitates timely and accurate diagnosis, enhances sugarcane health, and supports sustainable agricultural development. This study presents a comprehensive study aimed at improving sugarcane disease detection by incorporating deep learning techniques. In this study, we propose a Multi-Scale Attention-Based Dense Residual Network (MADRN) to boost accuracy and robustness by capturing both local and global features, enabling the model to handle intricate disease patterns and environmental variations effectively. Additionally, this research leverages a blended dataset that integrates local data from the BSRI with publicly available datasets.
This approach creates a more diverse and comprehensive training set, enhancing the models’ ability to generalize across various environmental conditions and disease manifestations. The integration of these datasets ensures improved adaptability and resilience of the models in practical applications. However, this study aims to improve the early detection and classification of sugarcane leaf diseases using advance deep learning model. objectives of this study are to flourish classification of sugarcane leaf diseases. For this reason, we proposed an attention-based model (MADRN) to enhance feature representation and disease classification performance. An integrated dataset from BSRI and Kaggle is used to improve the model’s robustness across diverse environmental conditions and disease variations. Additionally, a web-based application is developed for real-time, offering practical, user-friendly tools for farmers and researchers. By leveraging AI, this research supports timely disease identification and intervention, helping reduce crop losses, improve sugarcane health, and promote sustainable agricultural practices in Bangladesh and beyond.
The structure of this paper is as follows: "Introduction" provides an introduction to the study, "Related work" covers the literature review,"Materials and methodology" describes the dataset, materials, and proposed model, focusing on its architecture and classification approach, "Results and discussion" presents results and discussion, including the web-based implementation and a comparison with existing studies and "Conclusion and Future Work" offers the conclusion along with recommendations for future research.
Related work
Recent studies explore deep learning architectures for crop disease classification, with CNNs emerging as the preferred method for detecting and classifying plant diseases. S. Srivastava et al.8 proposed a deep learning system for detecting sugarcane plant diseases by examining leaves, stems, and color. This approach utilized three scenarios based on VGG-16, Inception V3, and VGG-19 feature extractors, comparing cutting-edge algorithms to deep learning models such as neural networks and hybrid AdaBoost. Orange software was used to calculate statistical measures like sensitivity, AUC, specificity, accuracy, and precision. Using VGG-16 as the feature extractor and SVM as the classifier, the scenario with the highest accuracy was chosen, and an AUC of 90.2% was attained. S. D. Daphal et al.9 proposed an attention-based multilevel residual convolutional neural network (AMRCNN) for accurately classifying sugarcane leaf diseases. The proposed architecture combines spatial and channel attention techniques to increase prominence detection, as well as traits from several layers to improve categorization. The researchers gathered a dataset of sugarcane leaf images from a variety of cultivated fields and weather conditions. There are five classes in the dataset: one healthy class and four disease classes (rust, mosaic, red rot, and yellow). Each class contains approximately 500 images, with a total of 2569 images and the dataset had been organized into training, validation, and testing subsets at a 70:15:15 ratios. Tested upon a self-created database, an AMRCNN achieved a classification accuracy of 86.53%, outperforming advanced models like XceptionNet, ResNet50, EfficientNet_B7, and VGG19. They used Tensor Flow Lite tools to improve the model for quantization in order to enable real-time application. C. Sun et al.11 proposed a SE-VIT hybrid network that combines ResNet-18 with squeeze-and-excitation (SE) attention, multi-head self-attention (MHSA), and 2D relative positional encoding for sugarcane leaf disease detection. To improve lesion segmentation, they compared thresholding, K-means, and SVM, with SVM providing the best results. Their model achieved 97.26% accuracy on the PlantVillage dataset and 89.57% accuracy with 90.19% precision on the private SLD dataset. N. Paramanandham et al.12 introduced LeafNet, a deep learning architecture for detecting six groundnut leaf classes Early Rust, Early Leaf Spot, Nutrition Deficiency, Rust, Late Leaf Spot, and Healthy Leaf using a dataset of 10,361 images. The model leverages residual networks and optimized weight initialization within an ensemble framework. To evaluate adaptability and generalizability, LeafNet was tested on multiple plant disease datasets, achieving 82.67% validation accuracy on the Sugarcane Leaf Disease dataset, along with 85.41% (Rice), 93.30% (Tomato), 89.35% (Maize), and 94.57% (Potato). Tamilvizhi, T. et al.13 proposed a Quantum-Behaved Particle Swarm Optimization-Based Deep Transfer Learning (QBPSO-DTL) model for accurate classification and detection of sugarcane leaf diseases, addressing the limitations of existing methods. The approach integrates pre-processing, optimal region growing segmentation for identifying affected regions, Squeeze Net for feature extraction, and a Deep Stacked Autoencoder (DSAE) for classification. Hyperparameter tuning was performed using the QBPSO algorithm to boost the model’s effectiveness.
I. Kunduracıoğlu et al.14 conducted a study that explores the use of deep learning for the detection of sugarcane leaf diseases, circumventing the inefficiencies and inaccuracies of handcrafted diagnosis approaches. The researchers worked with the openly accessible Sugarcane Leaf Dataset, which consists of 6748 images and 11 illness classes, to train and assess models from the EfficientNetv1 and EfficientNetv2 architectures as well as other well-known convolutional neural networks (CNNs). For this dataset, there was no direct association between model depth, complexity, or accuracy. InceptionV4 and EfficientNet-b6 achieves the greatest accuracy rates of 93.10% and 93.39%, respectively, out of the 13 models that were tested. D. Bou et al.15 carried out a study integrating hyperspectral imaging (HSI) with deep learning techniques to facilitate the swift identification of sugarcane diseases, specifically sugarcane smut and sugarcane mosaic virus (ScMV). Hyperspectral imaging captured a broad spectral range, including near-infrared (NIR), which allowed the identification of disease-related features that were not visible to the human eye. The researchers created a high-resolution HSI dataset of healthy and diseased sugarcane plants, which they used to train and evaluate deep neural network models. Experimental results showed over 80% detection accuracy for sugarcane smut 8 weeks after inoculation and over 90% accuracy by 10 weeks, detecting symptoms a week earlier than visible signs. Within a week of inoculation, ScMV was identified with over 90% accuracy, which was far earlier than the onset of symptoms. Additionally, the study found that NIR and visible and near-infrared (VNIR) spectral ranges were superior to RGB images for disease detection. The models achieved over 90% accuracy across most datasets. N. Amarasingam et al.16 applied deep learning and UAV-based remote sensing for detecting White Leaf Disease (WLD) in the fields of sugarcane located at Sri Lanka’s Gal-Oya region. The standard WLD detection methodology includes acquiring RGB (red, green, blue) images using UAVs, dataset pre-processing, labeling, DL model tuning, and prediction. Four deep learning models— Faster R-CNN, YOLOv5, DETR, YOLOR and—were evaluated for their ability to detect WLD. Among them, YOLOv5 performed the best, with 95% precision, 92% recall, 93% mAP@0.50, and 79% mAP@0.95. DETR performed the worst, with respective metrics of 77%, 69%, 77%, and 41%. Militante and Gerardo17 explored the integration of various CNN frameworks within deep learning approaches to optimize the accuracy of sugarcane disease recognition and detection. Their model was trained using a database that included 14,725 photos of both diseased and healthy sugarcane leaf surfaces. The suggested approach achieved a peak accuracy of 95.40%. The CNN architectures employed for disease detection included LeNet, VGGNet, and StridedNet. D. Padilla et al.18 developed a portable device employing support vector machines (SVM) for recognizing yellow spot disease on sugarcane leaves. The study’s objective was to create an image processing apparatus that uses sugarcane leaf pictures to record and display them in a single integrated unit. The model was trained to characterize and classify the variations between diseased and healthy leaves by identifying yellow spots on the foliage. M. Ozguven et al.19 developed an enhanced Fast RCNN framework by modifying CNN parameters, presenting an automated system for recognizing leaf diseases in sugar beet. This optimized architecture improved the efficiency and accuracy of disease detection through advanced parameter adjustments. N. Hemalatha et al.20 designed a deep learning neural network (DL-NN) platform for forecasting various sugarcane diseases via trained a model using pictures of infected foliage. Diseases such as rust spots, red rot, yellow leaf disease, cercospora leaf spot, and helmanthospura leaf spot were all successfully detected by this approach. The approach employed a CNN architecture specifically trained for accurate image classification, enabling precise disease recognition. While previous studies demonstrate the effectiveness of CNNs, attention mechanisms, residual networks, and multispectral imaging in crop disease detection, they face limitations such as region-specific datasets, variations in image quality, and limited robustness across multiple disease classes. MADRN addresses these gaps by integrating multi-scale convolutional kernels, dense residual blocks, and both channel and spatial attention mechanisms into a unified framework. This comprehensive design enables more accurate and reliable sugarcane leaf disease classification, particularly under real-world conditions. Existing studies in Bangladesh remain limited, underscoring the need for models like MADRN for early diagnosis and treatment.
Materials and methodology
Proposed blended dataset description
In this research, a dataset sourced from Kaggle is utilized, containing 2521 images of sugarcane leaves21. The dataset is organized into five distinct categories, each representing a specific leaf health condition: ‘Red rot’, ‘Yellow’, ‘Rust’, ‘Mosaic’, ‘Healthy’. Approximately 500 images are available in each category, ensuring balanced representation across the different conditions. Figure 1 displays a sample from each class, Table 1 and exhibits the dataset’s statistical summary.
Figure 1a gives an example image from the healthy class, Fig. 1b illustrates the mosaic class, Fig. 1c represents the red rot class, Fig. 1d depicts an image from the rust class, and Fig. 1(e) depicts an image from the yellow class. In addition to the Kaggle dataset, a small dataset from the Bangladesh Sugarcane Research Institute (BSRI) has been used to test the model’s adaptability to real-world conditions. This dataset includes representative images of sugarcane leaves exhibiting different disease symptoms under varied environmental conditions. The BSRI dataset covers the following disease categories: ‘Mosaic’, ‘RedRot’, ‘White Leaf’, ‘Smut’, ‘Wilt’, ‘Healthy’. Figure 2 illustrates a sample image for every class from the BSRI dataset. The blended dataset is formed by combining images from both Kaggle and BSRI datasets, ensuring a diverse and representative sample for robust sugarcane leaf disease detection, as illustrated in Fig. 3. The Kaggle dataset contains 2521 high-resolution images representing five disease classes: mosaic, rust, red rot, yellow, and healthy with relatively balanced class distributions. The BSRI dataset provides images captured in real-world field conditions in Bangladesh, depicting additional disease classes such as white leaf, wilt, and smut. Challenges in the BSRI data include a limited number of images and environmental variations such as lighting conditions, plant growth stages, and humidity, all of which affect image quality and disease severity. To unify the datasets, common class labels are merged, while unique BSRI classes are set aside for future analysis. To address class imbalances, oversampling techniques such as synthetic image generation and image augmentation are employed, enhancing diversity and ensuring a balanced representation across classes. The datasets are combined in a way that maintains proportional representation, ensuring that the model remains robust and generalizes effectively across the diverse conditions present in both datasets. In order to maintain interoperability with deep learning networks, all images are scaled to 224 × 224 pixels.
To improve model convergence, pixel intensities are standardized by scaling those to the \(\:[0,\:1]\) range. To improve the diversity of the training samples and shrink overfitting, data augmentation techniques are used, such as random rotations (± 20°), zooming (± 20%), contrast modifications, and random flipping of the horizontal and vertical planes. Additionally, quality control measures are implemented by removing blurry, misclassified, or poor-quality images to improve the overall reliability of the dataset.
Methodology
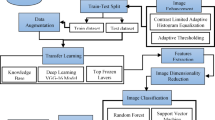
In this study, two datasets are employed to evaluate the performance of various deep learning models for sugarcane leaf disease classification. The first dataset is sourced from Kaggle, while the second referred to as the blended dataset is constructed by combining images from the Kaggle dataset and BSRI dataset to assess model generalization across diverse environments. Initially, images from both datasets are subjected to a series of preprocessing steps, including resizing, normalization, and data augmentation, to enhance quality and diversity. Following pre-processing, the datasets are split into training and testing subsets. Several deep learning models, including CNN, VGG16, MobileNetV2, and XceptionNet, are fine-tuned using transfer learning techniques to optimize performance for the specific classification task. To improve classification accuracy and robustness, a novel attention-based architecture named MADRN (Multi-scale Attention-based Dense Residual Network) is proposed. The MADRN model integrates multi-scale feature extraction, residual learning, and attention mechanisms to effectively capture discriminative patterns in leaf images. Extensive experiments are conducted using both datasets to evaluate and compare the performance of the proposed model against the fine-tuned conventional models. The evaluation metrics include accuracy, precision, recall, and F1-score. The overall methodology followed in this study is illustrated in Fig. 4, highlighting the systematic approach to developing an effective deep learning-based model for real-time sugarcane leaf disease classification.
Attention mechanism
A crucial aspect of the human visual system (HVS) is its capacity to focus attention. Instead of processing the whole scene at once, the HVS selectively concentrates on important regions to gather vital information and make informed judgments22. Inspired by this selective attention capability, this study incorporates attention mechanisms to enhance the performance of the proposed sugarcane disease detection model by focusing on disease-relevant image features. Specifically, Channel Attention (CA) and Spatial Attention (SA) are employed for effective saliency detection and feature enhancement.
Channel attention (CA)
Channel attention captures relationships between feature map channels to determine their importance for classification23. It determines “what” is important in the scene, and each channel serves as a feature detector. Given an input feature map \(\:F\in\:{R}^{C\times\:H\times\:W}\).To efficiently compute channel attention, spatial information is pooled using both average pooling and maximum pooling, which capture different statistical features. These pooled features are then passed through a shared multi-layer perceptron (MLP) with hidden activations of size \(\:{R}^{C/r}\times\:1\times\:1\) to generate the channel attention map\(\:\:{M}_{c}\in\:{\mathbb{R}}^{C}\times\:1\times\:1\)24. Figure 5 illustrates the detailed process of channel attention. The mathematical computation of the channel attention map is presented in Eq. (1),
Where, \(\:F\) represents the input feature map, \(\:\sigma\:\) represents the sigmoid activation function, \(\:{F}_{avg}\) and \(\:{F}_{max}\) present features obtained through average pooling and max pooling, respectively, and \(\:{W}_{0}\) and \(\:{W}_{1}\) are learnable weights of the MLP layers and the ReLU activation function is followed by \(\:{W}_{0}\). By utilizing these techniques, CA expands the network’s ability to prioritize relevant features, hence boosting overall representational efficiency.
Spatial attention
Spatial attention focuses on identifying “where” disease-related information is located in an image by highlighting significant regions and suppressing irrelevant areas, complementing channel attention’s focus on feature channels25. The spatial attention map \(\:{M}_{s}\left(F\right)\in\:{\mathbb{R}}^{H\times\:W}\) is generated by performing average-pooling and max-pooling along the channel axis to create two 2D maps, as illustrated in Fig. 6. These pooled maps are concatenated and passed through a 7 × 7 convolution layer, followed by a sigmoid activation to produce the spatial attention map. The formula for computing spatial attention is expressed mathematically in Eq. (2):
Here, \(\:{f}^{7\times\:7}\) represents a convolutional layer with a filter size of\(\:\:7\times\:7\), \(\:AvgPool\left(F\right)\in\:{\mathbb{R}}^{1\times\:H\times\:W}\) and \(\:MaxPool\left(F\right)\in\:{\mathbb{R}}^{1\times\:H\times\:W}\) are the average-pooled and max-pooled features, respectively. This approach enables the model to dynamically focus on disease-specific regions by emphasizing areas critical for classification and ignoring less informative regions.
Proposed multi-scale attention-based dense residual network (MADRN)
In this study, we propose a novel Multi-Scale Attention-Based Dense Residual Network (MADRN) for sugarcane disease classification, as illustrated in Fig. 7. The architecture integrates dense residual blocks with multi-scale attention mechanisms to improve feature extraction and enhance disease classification performance while addressing the limitations of conventional CNNs. Traditional CNNs, when stacking multiple convolutional layers, often face challenges such as increased parameters, computational complexity, and gradient vanishing issues as the network depth increases. To overcome these challenges, the proposed architecture introduces a feature extraction module employing dense residual blocks and multi-scale convolutional kernels (3 × 3, 5 × 5, and 7 × 7) to capture features at varying resolutions. This enables the network to extract both local and global disease-specific features. These extracted features are processed through concatenation, followed by Dense Residual Blocks that progressively increase filter sizes (#128, #256, and #512), ensuring comprehensive learning of low-level, mid-level, and high-level features. The initial layers of the network capture low-level features, such as simple textures, edges, and color patches, which serve as the foundation for extracting more complex disease-specific patterns. Mid-level features, learned in the second Dense Residual Block, represent textures and object shapes relevant to identifying sugarcane diseases, while high-level features captured in the third Dense Residual Block provide global representations essential for distinguishing between disease classes. Unlike conventional CNNs that primarily emphasize basic features, the proposed architecture carefully combines local and global features from each block to create a more informative feature representation. To enhance feature learning, Dense Residual Blocks utilize dense connections in which every layer is associated with every additional layer in the block, ensuring feature reuse and efficient gradient flow. These connections are mathematically expressed as:
Where, \(\:{x}_{i}\) represents the outputs from previous layers, and \(\:F\left(x\right)\) is the transformation function applied to the input\(\:\:x\)
This helps avoid the degradation problem in deep networks by learning residual functions rather than direct mappings, facilitating better convergence and model optimization. Attention mechanisms are embedded after each Dense Residual Block to further refine feature learning. A channel attention (CA) module emphasizes essential feature channels, while a spatial attention (SA) module focuses on disease-relevant regions. These modules play a vital role in accurately localizing disease spots in input images by enhancing significant features and suppressing redundant or noisy information, thereby improving recognition accuracy and robustness to variations in image quality. The attentive features extracted from all three Dense Residual Blocks are combined using element-wise addition operations and downsampled using convolution and max-pooling to maintain consistent feature map sizes across the network. This down sampling ensures the extraction of more intuitive features relevant to sugarcane disease detection while maintaining spatial consistency for effective feature fusion. The fused features are passed through two fully connected layers with 512 and 256 neurons, followed by a softmax activation function for five-class classification (healthy, mosaic, red rot, rust, yellow).
The novelty of MADRN lies in its integration of multi-scale feature extraction, dense residual learning, and attention mechanisms into a single framework. While multi-scale kernels capture both fine-grained local features and broader global patterns, dense residual connections enable efficient feature reuse and prevent gradient vanishing in deep networks. Attention modules further refine these features by emphasizing the most disease-relevant channels and spatial regions, suppressing irrelevant information. This combination allows MADRN to learn richer, more discriminative features than conventional CNNs or models using these strategies in isolation, resulting in improved accuracy and robustness for sugarcane disease classification. By focusing on both local and global features, the system can more accurately detect and classify sugarcane diseases while reducing the loss of critical feature information. This comprehensive approach ensures resilience to real-world conditions, improving disease classification rates and enabling timely, accurate diagnoses. The attention-based dense residual structure plays a pivotal role in mitigating gradient vanishing issues and optimizing network learning, enabling robust performance and improved recognition rates for sugarcane disease detection .
Fine tuned parameters of MADRN
All critical network configuration settings are carefully selected to highlight the flexibility and generalization capability of the proposed architecture in Table 2, regardless of the specific characteristics of the input data or application domain. The Adam optimizer, with default initialization settings, is used to efficiently update the network weights. Categorical cross-entropy serves as the loss function to address the multi-class classification task.
The learning rate is fixed at 0.0001 to ensure stable and consistent training. Additionally, LeakyReLU activation with an alpha value of 0.02 is applied in all convolutional and fully connected layers to maintain gradient flow even in inactive neurons, supporting effective learning across the network.
Evaluation metrics
For model development and evaluation, the dataset is split into 70% for training, 15% for validation, and 15% for testing. This allocation allows for robust model training, hyperparameter tuning, and final performance assessment. To evaluate the classification performance of the model, standard metrics derived from the confusion matrix are used, including Accuracy, Precision, Recall, and F1-Score. These metrics provide a comprehensive assessment of the model’s predictive performance, particularly in the presence of class imbalance.
Results and discussion
Comparative analysis of model performance
Table 3 presents a comprehensive performance comparison of four deep learning models-CNN, VGG16, MobileNetV2, XceptionNet, and the proposed MADRN (Multi-scale Attention-based Dense Residual Network) evaluated on both the Kaggle dataset and a blended dataset comprising images from the Kaggle and BSRI datasets.
The results indicate that the proposed MADRN model consistently outperformed the baseline models across all evaluation metrics, including accuracy, precision, recall, and F1-score. Specifically, on the Kaggle dataset, MADRN achieves the highest accuracy of 94.78%, with a precision of 91.00%, recall of 93.00%, and F1-score of 92.00%. XceptionNet emerges as the closest competitor, achieving 92.97% accuracy, but with slightly lower precision (90.25%), recall (89.56%), and F1-score (91.36%). Traditional CNN and lightweight architectures such as MobileNetV2 exhibit lower performance, indicating a limited ability to extract complex features required for accurate classification. When evaluated on the blended dataset, which merges the variability and complexity of both the Kaggle and BSRI sources to test the models’ generalization capabilities, MADRN continues to deliver robust results with 92.25% accuracy, 93.00% precision, 92.00% recall, and 92.00% F1-score. In contrast, XceptionNet’s performance drops significantly to 79.67% accuracy, revealing a potential sensitivity to domain variation. VGG16 and MobileNetV2 maintain relatively stable yet inferior performance across datasets. The superior performance of MADRN is attributed to its architectural design, which seamlessly integrates dense residual connections and multi-scale attention mechanisms. This combination enables the model to capture subtle, disease-specific features and complex data patterns, resulting in enhanced feature extraction and improved generalization. Figure 8 shows the confusion matrix of the proposed MADRN model on both the Kaggle and blended datasets, demonstrating its accurate classification across multiple disease categories. Figure 9 illustrates the ROC curve for the proposed MADRN model on the Kaggle dataset, further validating the model’s robust discriminative capability and balanced sensitivity-specificity trade-off. This comprehensive evaluation highlights MADRN’s effectiveness as a robust and scalable solution for real-world agricultural disease detection challenges. Figure 10 illustrates the accuracy and loss curves for the proposed MADRN model on the blended sugarcane leaf disease dataset and Fig. 11.
depicts the confusion matrices for various baseline models on the same dataset for comparative analysis. These findings demonstrate that MADRN not only excels at learning discriminative features from a single-source dataset but also generalizes effectively to diverse, real-world data—making it a highly promising model for accurate and scalable sugarcane leaf disease classification in practical agricultural applications.
Statistical significance and robustness of the proposed MADRN model
To further validate the superiority of the proposed MADRN model, we have performed paired t-tests on the results of 5-fold cross-validation using the Kaggle dataset. The goal is to verify whether the performance improvements of MADRN over baseline models are consistent and not due to random variation.
Table 4 summarizes the paired t-test results comparing the accuracy of MADRN with four baseline deep learning models (CNN, MobileNetV2, VGG16, and XceptionNet). The reported metrics include mean accuracies with standard deviations (SD), 95% confidence intervals (CI), t-statistics, and p-values. The paired samples t-test results provide compelling evidence that the proposed MADRN model consistently outperforms all baseline deep learning models across five folds, with improvements that are statistically significant in every comparison. Specifically, CNN achieves a mean accuracy of 76.64 ± 2.66% (95% CI: [73.34, 79.94]), while MADRN reaches 90.64 ± 2.31%. The paired t-test yields t = 7.9941 and p = 0.0007, strongly rejecting the null hypothesis and confirming that the nearly 14% improvement is highly significant and not due to chance. Against MobileNetV2, which achieves 86.32 ± 1.14% (95% CI: [84.90, 87.74]), MADRN shows a mean improvement of 4.32%, with t = 6.3745 and p = 0.0016, indicating strong statistical evidence of superiority. Similarly, for VGG16 with 84.92 ± 0.14% (95% CI: [84.75, 85.09]), MADRN achieves an improvement of 5.72%, and the paired t-test result (t = 5.2561, p = 0.0031) confirms that the improvement is statistically significant. Finally, even against XceptionNet, which records 88.80 ± 2.94% (95% CI: [85.15, 92.45]), MADRN secures an additional 1.83% improvement, with t = 4.6559 and p = 0.0048, demonstrating that the performance gain is still significant despite being relatively smaller. Importantly, in all cases, the p-values are less than 0.01, which according to conventional thresholds indicates strong statistical significance. Moreover, the 95% confidence intervals for the mean differences do not cross zero, further confirming that the observed improvements are genuine and not due to random variation. The consistently small SD values across folds also suggest that MADRN’s performance is stable and reliable. Similar statistical validation on the Blended dataset is planned for future work, which will further confirm the generalizability and robustness of the proposed model. Overall, the large t-statistics, very small p-values (p < 0.01), and positive confidence intervals collectively demonstrate that MADRN delivers consistent, reliable, and statistically significant improvements over all baseline deep learning models. This establishes MADRN as a robust and superior architecture for the given datasets.
Leaf disease prediction using web based system
To enhance the accessibility and practical usability of the proposed technology, a web-based application is developed using the Streamlit framework, providing a user-friendly platform for real-time sugarcane disease detection, shown in Fig. 12. This application allows users, such as farmers and agricultural professionals, to upload images of sugarcane leaves and receive instant predictions on disease classification. It leverages proposed MADRN models on a combined dataset from Kaggle and BSRI. Key features of the application include an intuitive interface for image uploading, real-time prediction capabilities, and seamless integration of high-performing models to ensure accuracy and reliability. Rigorous testing with the BSRI dataset confirms the application’s robustness and adaptability, achieving exceptional accuracy across diverse datasets. In terms of computational performance, the model processes a 224 × 224 input image in approximately 3.0–5.0 s on a standard GPU enabled server. While running the full MADRN model directly on mobile devices is challenging due to memory and processing constraints, the current architecture supports server based inference, allowing farmers to upload images via mobile devices or web browsers and receive rapid, high-accuracy predictions. This setup is scalable, supports multiple concurrent users, and provides a robust solution suitable for deployment in agricultural settings. In the future, lightweight or optimized versions of MADRN could enable fully offline, real-time mobile deployment, further extending its usability in field conditions. By enabling timely interventions and optimizing resource allocation, this scalable solution reduces dependency on traditional manual inspection methods, which are often time-consuming and error-prone. With its potential for large-scale deployment in agricultural settings, the application offers an efficient and impactful tool for improving sugarcane disease management and advancing precision agriculture.
Benchmarking with similar existing studies
Benchmarking is an essential method of comparing and assessing the performance of the proposed models in this research against similar studies in the field. For this study, the focus is on evaluating the performance of the sugarcane leaf disease detection models against various recent studies that address plant disease classification using deep learning techniques Srivastava et al.26 proposed a deep learning system for sugarcane disease detection using features from leaves, stems, and color. They evaluated VGG-16, Inception V3, and VGG-19 as feature extractors, comparing them with algorithms like neural networks and hybrid AdaBoost. Using VGG-16 with SVM, the best accuracy and an AUC of 90.2% were achieved, with statistical metrics calculated via Orange software. Daphal et al.9 proposed an attention-based multilevel residual CNN (AMRCNN) for classifying sugarcane leaf diseases. The architecture integrates spatial and channel attention to enhance feature detection and classification. Using a dataset of 2569 images across five classes (healthy, rust, mosaic, red rot, and yellow), divided into 70:15:15 for training, validation, and testing, AMRCNN achieved 86.53% accuracy, outperforming XceptionNet, ResNet50, EfficientNet_B7, and VGG19. TensorFlow Lite tools were used for model quantization to enable real-time applications. C. Sun et al.11 proposed a SE-VIT hybrid network combining ResNet-18 with SE attention, multi-head self-attention, and 2D positional encoding for sugarcane leaf disease detection. Using SVM for lesion segmentation yielded the best results, achieving 97.26% accuracy on PlantVillage and 89.57% accuracy with 90.19% precision on a private SLD dataset. N. Paramanandham et al.12 introduced LeafNet, a deep learning model for six groundnut leaf classes, leveraging residual networks within an ensemble. Tested across multiple datasets, LeafNet achieved 82.67% validation accuracy on the Sugarcane Leaf Disease dataset and high performance on Rice (85.41%), Tomato (93.30%), Maize (89.35%), and Potato (94.57%). Kunduracıoğlu et al.14 investigated deep learning for sugarcane leaf disease detection, addressing inefficiencies in traditional diagnostic methods. Using the Sugarcane Leaf Dataset with 6748 images across 11 classes, they evaluated EfficientNetv1, EfficientNetv2, and other CNNs. Among 13 tested models, InceptionV4 and EfficientNet-B6 achieved the highest accuracies of 93.10% and 93.39%, respectively .
Bou et al.15 used hyperspectral imaging (HSI) and deep learning to detect sugarcane diseases like smut and ScMV. HSI, leveraging NIR and VNIR spectral ranges, enabled early detection, achieving over 90% accuracy for ScMV within a week and smut a week before visible symptoms. HSI outperformed RGB imaging for disease detection. Padilla et al.18 developed a portable device using SVM to detect yellow spot disease on sugarcane leaves. The integrated unit processed images to classify healthy and diseased leaves based on yellow spot features, achieving 86% accuracy. Our proposed intensive statistical exploration for sugarcane leaf disease detection, utilizing advanced deep learning models XceptionNet, ResNet101 for feature extraction and fine-tuning, achieved the highest accuracy score of 98.22% and an AUC score of 95.00%. Furthermore, our proposed MADRN achieved a notable accuracy of 94% and an AUC score of 99%, further validating the effectiveness of our approach. These results surpass those from previous studies in the field of plant disease detection, underscoring the robustness and precision of our methodology. However, our research emphasizes the importance of thoroughly analyzing statistical data and image features before applying machine learning algorithms. Deep learning-based approaches consistently outperformed conventional machine learning techniques in the context of sugarcane leaf disease classification, showcasing their significant potential for agricultural research and early disease detection. Table 5 provides a detailed comparison of benchmarking studies alongside our proposed model’s results.
Dataset limitations
Although this study utilizes two datasets (Kaggle and BSRI) to provide diverse and representative samples for sugarcane leaf disease detection, certain limitations remain. The Kaggle dataset, while relatively balanced across five classes, still shows minor discrepancies in class counts, and the BSRI dataset is relatively small with additional disease categories, which may introduce variability in training and evaluation. Environmental factors in the BSRI images, such as lighting conditions, plant growth stages, and humidity, can introduce noise and affect image quality. Furthermore, some disease classes are underrepresented, potentially impacting the generalizability of the model to rare conditions. While oversampling and data augmentation help mitigate these imbalances, future studies could benefit from larger, more diverse datasets with standardized imaging conditions to further enhance model robustness and reliability. Additionally, our model relies solely on an attention mechanism to focus on diseased regions, without using explicit segmentation. While effective in most cases, attention may occasionally fail to capture all diseased regions when they are small, patchy, or sparsely distributed, potentially leading to false positives or false negatives. Incorporating explicit segmentation techniques in future work could help mitigate these limitations and further improve model performance.
Impact of domain shift and model generalization
One of the key challenges in plant disease detection is domain shift, arising from differences in image characteristics across datasets. The Kaggle dataset contains high resolution images captured under controlled conditions, where disease symptoms are clearly visible. In contrast, the BSRI dataset reflects real-world field environments in Bangladesh, where irregular lighting, shadows, overlapping leaves, varying growth stages, and humidity effects introduce significant complexity and variability. Baseline models such as CNN, VGG16, and MobileNetV2 showed reduced performance on the blended dataset, particularly for mosaic when leaf textures overlapped. In comparison, the proposed MADRN demonstrated greater adaptability by combining multi-scale feature extraction with attention mechanisms, enabling it to capture both fine-grained and contextual disease patterns while suppressing background noise and irrelevant variations. While traditional CNN and transfer learning models showed accuracy drops of 4–7% when moving from Kaggle to the blended dataset, MADRN maintained higher robustness, achieving 92.25% accuracy on the blended dataset with consistent precision and recall across classes. This demonstrates MADRN’s robustness and ability to generalize under real-world variability, making it suitable for practical precision agriculture applications.
Conclusion and future work
This research successfully presents a comprehensive deep learning-based solution for sugarcane leaf disease detection, leveraging the innovative MADRN architecture. Through extensive experimentation and evaluation on both the Kaggle dataset and a blended dataset incorporating images from the BSRI, MADRN consistently demonstrates superior performance in terms of accuracy, precision, recall, and F1-score compared to baseline models. The key strength of MADRN lies in its ability to effectively extract discriminative features by combining dense residual learning and multi-scale attention mechanisms, which not only enhances its performance on a single-source dataset but also enables robust generalization across diverse real-world conditions. This outcome reinforces the importance of using advanced architectural techniques for addressing the challenges inherent in agricultural disease detection. However, the study is limited by the dataset size and environmental variations that may affect field level generalization. In future work, we plan to enhance model explain ability through saliency and Grad-CAM visualizations, and to integrate the trained MADRN model into IoT-based monitoring systems for real time disease classification. Expanding the approach to other crops and diseases will further improve its applicability in precision agriculture.
Data availability
The data supporting the findings of this study are publicly available and can be accessed through the following sources. Kaggle Sugarcane Leaf Disease Dataset [https://www.kaggle.com/datasets/nirmalsankalana/sugarcane-leaf-disease-dataset](https:/www.kaggle.com/datasets/nirmalsankalana/sugarcane-leaf-disease-dataset) (accessed Jan. 15, 2025). Bangladesh Sugarcrop Research Institute (BSRI): Data available upon request. For BSRI data inquiries, please contact the corresponding author, Dr. Md. Shamim Reza.
References
Annual-Reports - Bangladesh Sugarcrop Research Institute-Government of the People’s Republic of Bangladesh. https://bsri-gov-bd.translate.goog/site/view/annual_reports/Annual-Reports?_x_tr_sl=bn (2025).
Sugar Producing Countries. 2023 - Wisevoter. https://wisevoter.com/country-rankings/sugar-producing-countries/ (2025).
Rahman, M. S., Khatun, S. & Rahman, M. K. Sugarcane and sugar industry in bangladesh: an overview. Sugar Tech. 18 (6), 627–635. https://doi.org/10.1007/S12355-016-0489-Z (2016).
Gonçalves, M. C., Pinto, L. R., Pimenta, R. J. G. & da Silva, M. F. Sugarcane. Viral Dis. F Hortic. Crop. 193–205. https://doi.org/10.1016/B978-0-323-90899-3.00056-2 (2024).
Li, X. et al. SLViT: Shuffle-convolution-based lightweight vision transformer for effective diagnosis of sugarcane leaf diseases. J. King Saud Univ. - Comput. Inf. Sci. 35 (6), 101401. https://doi.org/10.1016/J.JKSUCI.2022.09.013 (2023).
Huang, Y. K., Li, W. F., Zhang, R. Y. & Wang, X. Y. Diagnosis and Control of Sugarcane Important Diseases, Color Illus. Diagnosis Control Mod. Sugarcane Dis. Pests, Weeds, pp. 1–103, https://doi.org/10.1007/978-981-13-1319-6_1 (2018).
Rani, R., Sahoo, J., Bellamkonda, S., Kumar, S. & Pippal, S. K. Role of artificial intelligence in agriculture: an analysis and advancements with focus on plant diseases. IEEE Access. 11, 137999–138019. https://doi.org/10.1109/ACCESS.2023.3339375 (2023).
Srivastava, S., Kumar, P., Mohd, N., Singh, A. & Gill, F. S. A novel deep learning framework approach for sugarcane disease detection. SN Comput. Sci. 1 (2). https://doi.org/10.1007/S42979-020-0094-9 (2020).
Daphal, S. D. & Koli, S. M. Enhanced deep learning technique for sugarcane leaf disease classification and mobile application integration. Heliyon, 10, 8, p. e29438, https://doi.org/10.1016/j.heliyon.2024.e29438
Militante, S. V., Gerardo, B. D. & Medina, R. P. Sugarcane Disease Recognition using Deep Learning, 2019 IEEE Eurasia Conf. IOT, Commun. Eng. ECICE 2019, pp. 575–578, https://doi.org/10.1109/ECICE47484.2019.8942690 (2019).
Sun, C., Zhou, X., Zhang, M. & Qin, A. SE-VisionTransformer: hybrid network for diagnosing sugarcane leaf diseases based on attention mechanism. Sens. (Basel). 23 (20). https://doi.org/10.3390/s23208529 (2023).
Paramanandham, N., Sundhar, S. & Priya, P. Enhancing Disease Detection with Weight Initialization and Residual Connections Using LeafNet for Groundnut Leaf Diseases, IEEE Access, vol. 12, no. July, pp. 91511–91526, https://doi.org/10.1109/ACCESS.2024.3422311 (2024).
Tamilvizhi, T., Surendran, R., Anbazhagan, K. & Rajkumar, K. Quantum Behaved Particle Swarm Optimization-Based Deep Transfer Learning Model for Sugarcane Leaf Disease Detection and Classification, Math. Probl. Eng., vol. 2022, https://doi.org/10.1155/2022/3452413 (2022).
Kunduracıoğlu, İ. & Paçal, İ. Deep Learning-Based Disease Detection in Sugarcane Leaves: Evaluating EfficientNet Models, J. Oper. Intell., vol. 2, no. 1, pp. 321–235, https://doi.org/10.31181/JOPI21202423 (2024).
Bao, D. et al. Early detection of sugarcane Smut and mosaic diseases via hyperspectral imaging and spectral-spatial attention deep neural networks. J. Agric. Food Res. 18, 101369. https://doi.org/10.1016/J.JAFR.2024.101369 (2024).
Amarasingam, N., Gonzalez, F., Salgadoe, A. S. A., Sandino, J. & Powell, K. Detection of white leaf disease in sugarcane crops using UAV-Derived RGB imagery with existing deep learning models. Remote Sens. 14 (23). https://doi.org/10.3390/rs14236137 (2022).
Militante, S. V. & Gerardo, B. D. Detecting Sugarcane Diseases through Adaptive Deep Learning Models of Convolutional Neural Network, ICETAS –2019 6th IEEE Int. Conf. Eng. Technol. Appl. Sci., Dec. 2019, Dec. 2019, (2019). https://doi.org/10.1109/ICETAS48360.2019.9117332
Padilla, D. A. et al. Portable Yellow Spot Disease Identifier on Sugarcane Leaf via Image Processing Using Support Vector Machine, 5th Int. Conf. Control. Autom. Robot. ICCAR 2019, pp. 901–905, Apr. 2019, pp. 901–905, https://doi.org/10.1109/ICCAR.2019.8813495 (2019).
Ozguven, M. M. & Adem, K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys. Stat. Mech. Its Appl. 535, 122537. https://doi.org/10.1016/J.PHYSA.2019.122537 (2019).
Hemalatha, N. K. et al. Sugarcane leaf disease detection through deep learning. Deep Learn. Sustain. Agric. 297–323. https://doi.org/10.1016/B978-0-323-85214-2.00003-3 (2022).
Sugarcane Leaf Disease Dataset. https://www.kaggle.com/datasets/nirmalsankalana/sugarcane-leaf-disease-dataset (Accessed Jan 15. (2025).
Filipe, S. & Alexandre, L. A. RETRACTED ARTICLE: from the human visual system to the computational models of visual attention: a survey. Artif. Intell. Rev. 43 (4), 601. https://doi.org/10.1007/S10462-012-9385-4 (2015).
Hu, J., Shen, L. & Sun, G. Squeeze-and-Excitation Networks, Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., pp. 7132–7141, https://doi.org/10.1109/CVPR.2018.00745. (2018).
Woo, S., Park, J., Lee, J. & Kweon, I. S. Convolutional_Block_Attention, Eccv, p. 17, (2018).
Wang, Q. et al. ECA-Net: Efficient channel attention for deep convolutional neural networks, Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., pp. 11531–11539, https://doi.org/10.1109/CVPR42600.2020.01155 (2020).
Srivastava, S., Kumar, P., Mohd, N., Singh, A. & Gill, F. S. A Novel Deep Learning Framework Approach for Sugarcane Disease Detection, SN Comput. Sci., vol. 1, no. 2, pp. 1–7, https://doi.org/10.1007/S42979-020-0094-9/METRICS (2020).
Acknowledgements
We sincerely acknowledge the Deep Statistical Learning and Research Lab, Department of Statistics, Pabna University of Science and Technology (PUST), Pabna, Bangladesh, for their valuable technical support and for facilitating this research.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Jannatul Mauya: Conceptualization, methodology, conducted the research, data analysis, writing the original draft, and correspondence. Ruhul Amin: Investigation, and review of the manuscript. Dr. Md. Shamim Reza & Dr. Sabba Ruhi: Supervision, guidance on modelling, and critical review of the final manuscript. Dr. Md. Imam Hossain: Dataset labelling and preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mauya, J., Amin, R., Hossain, M. et al. Improved multiscale attention based deep learning approach for automated sugarcane leaf disease detection using BSRI data. Sci Rep 15, 45474 (2025). https://doi.org/10.1038/s41598-025-28947-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-28947-x