Abstract
Opisthorchiasis, a major foodborne parasitic zoonotic disease in Thailand and neighboring countries, is caused by the carcinogenic liver fluke Opisthorchis viverrini (OV). Accurate classification of OV infection is critical for timely intervention and public health management. In this study, we propose a reliable machine learning (ML) classification model based on peak current data from an electrochemical immunosensor and additional patient condition features, which can facilitate intuitive decision-making without the need for expert personnel. This is the first report to classify OV infection using a ML algorithm integrated with electrochemical biosensor data. A total of 531 urine samples from both OV-positive and OV-negative individuals in endemic areas were analyzed using the immunosensor. We evaluated the effectiveness of six different ML models through cross-validation. Among these models, the decision tree and AdaBoost classifiers demonstrated outstanding performance, each achieving the highest accuracy of 90.65% (95% CI 0.89–0.91). The decision tree model yielded an F1 score of 91%, sensitivity of 95%, and specificity of 83%, while the AdaBoost model achieved an F1 score of 90%, sensitivity of 94%, and a higher specificity of 86%. The neural network model also performed excellently, with an accuracy of 89.72% (95% CI 0.84–0.93), and an F1 score of 89%. The statistical comparison of the model’s performance highlighted the significant difference between the top-performing models and the rest. These results underscore the significance of incorporating sensor data and ML to accurately classify OV infections and enable early diagnosis and intervention. By using this ML model, the status of OV infection can be detected by interpreting sophisticated raw electrochemical data. This implies that patients or medical staff with no prior experience with electrochemical sensors can nevertheless comprehend the disease condition with confidence. The proposed ML models hold promise for enhancing disease surveillance and control strategies in endemic regions and could thus assist medical professionals in the decision-making process and in addressing the burden of opisthorchiasis infection.
Similar content being viewed by others
Introduction
Opisthorchis viverrini (OV) infection is a major foodborne parasitic disease and a neglected, clinically significant tropical disease1. Approximately 40 million people are infected with OV in the GMS region alone2,3, and more than 600 million people are at risk4. The life cycle of OV can take place in multiple hosts, including freshwater snails (Bythinia spp.), freshwater fish (Cyprinid spp.) and fish-consuming hosts such as humans, cats, or dogs. The primary mechanism of infection is the ingestion of raw or inadequately cooked fish cuisines in Thailand. Notably, opisthorchiasis is the most critical risk factor associated with cholangiocarcinoma (CCA) and is classified as a group 1 biological carcinogen5. Early detection, diagnosis and prevention of OV infection constitute an effective approach for addressing the high prevalence and control of the disease in endemic areas. Highly sensitive detection methods and precise classification of infections are urgently needed for timely intervention and surveillance of OV infections.
The gold standard for detecting OV infection is the formalin-ethyl acetate concentration technique (FECT), which is commonly used to quantify OV in fecal samples6. However, the FECT has limitations, including low sensitivity and specificity, extensive sampling requirements, and the need for a skilled microscopist. Additionally, the FECT may fail to detect mild infections, and biliary tract obstruction can impede OV egg flow into feces, leading to false-negative results7. Recent advancements in immunological and molecular diagnostic methods, such as enzyme-linked immunosorbent assay (ELISA), have been explored for the detection of somatic and excretory-secretory (ES) antigens8,9. Although these methods offer higher sensitivity and specificity over the FECT, their disadvantages include high testing costs, complex procedures, and the need for sophisticated instruments. Serological OV antibody testing is a highly accurate diagnostic technique but has drawbacks such as cross-reactivity and persistent antibodies post-treatment. Attempts to develop ELISA techniques for urine OV-ES antigen detection have shown promise but are hindered by detection limits and complex procedures, highlighting the need for a simple, cost-effective point-of-care biosensor for OV detection.
Since the successful implementation of the amperometric glucose sensor for diabetes monitoring, many electrochemical biosensors have been developed at the industrial level10. The development of point-of-care biosensors has revolutionized onsite detection and disease screening in the community. Electrochemical biosensors have strong advantages, including cost-effectiveness and a small size allowing portability for point-of-care testing. A typical bioelectrochemical sensor is an analytical device that integrates a biorecognition component termed a receptor with an electrochemical transducer11. Our group recently developed an electrochemical immunosensor for sensitive detection of OV antigens in urine samples12. The developed immunosensor exhibited the lowest limit of detection (LOD) of 1.5 ng mL− 1 for OV antigen detection in urine compared to 78 ng mL− 1 for detection by ELISA and 28.5 ng mL− 1 for detection by a rapid OV Ag detection test kit (OV-RDT). Compared with ELISA and OV-RDT, the developed electrochemical sensor demonstrated high diagnostic sensitivity and specificity (96% and 90%, respectively) for OV detection. However, the major challenges associated with electrochemical biosensors include low signal-to-noise (S/N) ratios, electrode fouling, chemical interference, complex signal processing, and matrix effects, which undermine the precision and accuracy of biosensors13.
The incorporation of AI into biosensors has emerged as a solution for overcoming their limitations. Machine learning (ML), a critical aspect of AI, has been employed as a powerful tool for data analysis and processing in materials science14. ML can effectively process big sensing data for complex matrices or samples and extract detectable analytical signals from noisy and low-resolution sensing data. Furthermore, proper implementation of ML methods can uncover correlations between sample parameters and sensing signals through data visualization and enable mining of interrelations between signals and bio events. ML-reinforced electrochemical biosensors can improve sensor performance by transforming unintelligible raw data into understandable and valuable information that can be applied in disease diagnosis, treatment evaluation, pathogen detection15, health monitoring16, and food safety17. The common primary machine learning algorithms include artificial neural networks (ANNs), convolutional neural networks (CNNs), decision trees (DTs), random forests (RFs), support vector machines (SVMs) and k-nearest neighbors (kNNs)13. Certain challenges associated with biosensors can be overcome by ML-based data analysis. Moreover, intelligent biosensors that can automatically predict concentrations of analytes or species can be optimized by incorporating ML.
In this study, we proposed a reliable ML classification model based on peak current data from an electrochemical immunosensor and additional patient condition features, which can facilitate intuitive decision-making without the need for expert personnel. This is the first report of classifying OV infection using a ML algorithm integrated with electrochemical biosensor data. Technically, mathematical calculations must be carried out by technicians on peak current changes obtained from sensors to translate these changes into concentrations at which doctors can interpret the status of OV infection. ML integration with biosensors can enable classification of the levels of OV infection based on raw sensor data, thereby obviating manual calculations. Moreover, intelligent OV classification can improve the accuracy of sensors and thus facilitate sensor measurements and data interpretation in a timely manner.
Materials and methods
Electrochemical setup
The electrochemical immunosensor was fabricated by immobilizing a graphene oxide-monoclonal antibody (GO-mAb) conjugate onto screen-printed carbon electrodes (SPCEs) for 90 min. Excess areas were blocked with 0.1% BSA solution for 30 min. Electrochemical analysis of OV antigens in urine samples was performed by introducing seven microliters of the samples on modified SPCEs, followed by incubation for 60 min at room temperature. The voltammetric signals of OV antigens were collected by square wave voltammetry (SWV) with an applied potential of -0.6 to 0.6 V, an amplitude of 2 mV, a step potential of 5 mV and a frequency of 8 Hz in the presence of 5 mM [Fe(CN)6]3−/4− as a redox indicator. The peak currents before (I0) and after (I1) OV antigen detection were recorded by SWV. Peak current changes (∆I = I0 – I1) were calculated to identify correlations with OV antigen levels in the samples.
Data labeling
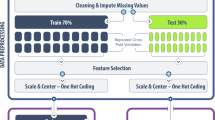
A total of 531 urine samples from both OV-positive and OV-negative individuals were collected from endemic areas in Northeastern Thailand and were analyzed using the immunosensor. Peak current changes (∆I) were extracted from square wave voltammograms. Demographic information such as age, sex, and urinalysis results (urine glucose, ketones, protein, pH, specific gravity and red blood cell levels) were collected as clinical parameters. The characteristics of the collected data are shown in Table 1. Missing values, redundant information, and incompatible data strongly impacted model performance. Therefore, the data were preprocessed by data cleaning, data integration, and feature engineering. The categorical variables, such as Sex, were transformed into a numerical format using label encoding. Each unique category was assigned an integer value, and the original variable was preserved alongside the encoded version for reference. To ensure data completeness and consistency, all rows containing missing data were removed prior to model training. Inputs (features) were selected based on their correlations with OV infection. For supervised classification, OV-positive and OV-negative data were labeled as confirmed by the conventional ELISA.
Exploratory data analysis
The dataset contained 531 rows of data. The labels that we used were OV-positive and OV-negative, representing the infection status of the patients. The number of patients with each label is depicted in Fig. 1a. A total of 204 patients were observed to have OV infection, and 327 individuals did not. The sex ratio is illustrated in Fig. 1b. A total of 135 in 204 OV-infected patients were female, accounting for 66% of the patients, and 69 male patients accounted for 34%. The OV-negative individuals included 95 males and 232 females.
The distributions of age, peak current changes, pH and specific gravity for each label class are depicted in a box plot (see Fig. 2). The mean age of the OV-infected patients (53.34 ± 14.03 years) was slightly greater than that of the noninfected individuals (45.19 ± 20.33 years) (Fig. 2a). Peak current changes were significantly different between the two groups at 2.21 ± 2.16 µA in the OV-positive patients and 1.10 ± 2.15 µA in the OV-negative patients (Fig. 2b). The urine pH and specific gravity data did not differ significantly between the classes (Fig. 2c,d). Urine glucose, ketone and protein levels were within the normal ranges for both groups, with some outliers that were not associated with the disease (data not shown).
Sample size determination
For classification models, the minimum sample size required was estimated based on the rule of thumb that at least 10 events per variable (EPV) are needed for developing a reliable model. Using 9 variables and binary classification in our full model, a minimum of 180 positive cases were required. Additionally, power analysis was performed assuming 80% power, a significance level of 0.05, and an expected AUC of 0.75, which indicated a minimum required sample size of 114 samples (57 per group). Our actual sample size of 531 (204 OV-positive and 327 OV-negative) was substantially larger than this requirement, providing adequate statistical power for our analyses.
Learning curve analysis
A learning curve approach was employed to estimate the adequate training sample size required to achieve a model performance target of AUC > 0.85. This approach allows us to evaluate how model performance scales with increasing training data and to determine the point at which performance metrics converge. Each model was trained and validated using 10-fold stratified cross-validation to ensure that both classes were proportionally represented in each fold. The minimum training sample size to achieve AUC > 0.85 was estimated by computing the model performance across ten training set sizes, incrementally increasing from 10% to 100% of the total dataset (531 samples) in equal steps. The learning curve for each model was plotted as mean AUC versus training set size, with a horizontal reference line at AUC = 0.85. The AUC increased with increasing sample size in all models, reaching a plateau at 40% of the sample size (~ 0.86 AUC). Therefore, a training set size of approximately 265–320 samples (i.e., ~ 40–50% of total data) was sufficient for decision tree, AdaBoost, and neural network models to achieve or exceed the target AUC of 0.85.
Training and validation of classification machine learning models
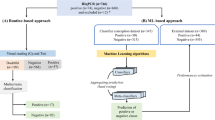
A variety of ML classifiers were applied in our study to ensure the robustness of our models in interpreting correlations between risk factors and infection outcomes. Six ML classifiers commonly used on datasets composed of heterogeneous features were trained, including a decision tree, k-nearest neighbors, an AdaBoost, a random forest, naïve Bayes, and an artificial neural network. The implementation of these six classification algorithms on the dataset was evaluated by several metrics, including accuracy, precision, recall, F1 score, and cross-validation score. Five-fold cross-validation was implemented to protect against overfitting and ensure the generalizability of the model performance. To account for the different number of samples in the OV-positive and OV-negative groups, the cross-validation was stratified. This ensures that the proportion of cases from each class was maintained in every fold, leading to a more reliable estimate of model performance. In the training phase, all models were optimized by fine-tuning their hyperparameters using a 5-fold cross-validation scheme and a grid search algorithm, with the weighted F1-score as the scoring metric. This approach was applied to all models except Naïve Bayes. For the decision tree, the maximum depth was varied from 1 to 5 to control model complexity. The k-Nearest Neighbors algorithm was evaluated using different numbers of neighbors: 3, 5, and 7, which influence the smoothness of the decision boundary. For AdaBoost, multiple parameters were tuned, including the number of estimators (50, 100, 200), learning rates (0.01, 0.1, 0.5, 1), and the maximum depth of the base decision tree estimators (1, 2, 3), to balance between bias and variance. Random Forest models were tuned by adjusting the number of trees (50, 100, 200) and their maximum depth (5, 10, 20). Lastly, for the Neural Network, different hidden layer architectures (50, 25), (100, 50), and (200, 100) were tested to determine the most effective network structure. These tuning strategies help ensure that each model is properly configured for optimal predictive accuracy. The code used to develop the models was written in Python and the open-source library scikit-learn. The overall ML framework is illustrated in Fig. 3.
Machine learning models
Decision tree
Decision tree consists of nodes that split the instance space into two or more subspaces according to a discrete function over the input attributes18. It works by recursively partitioning the dataset into subsets based on the values of input features, ultimately leading to a tree-like structure of decision nodes and leaf nodes. The decision tree model is prone to overfitting. Maximum depth governs the depth of the tree and can prevent overfitting. By using the grid search, a maximum depth of 1 was set to train the model.
k-nearest neighbors
K-nearest neighbors algorithm is a data-classification method of estimating the likelihood that a data point will become a member of one group based on what group the data point nearest to it belongs to. The optimal k-value of 7, determined through fine-tuning, was used to train the model.
AdaBoost
AdaBoost works by combining the predictions of weak learners, which are typically simple models that perform slightly better than random guessing. Commonly, decision trees with a single split (stumps) are used as weak learners in AdaBoost. The AdaBoost algorithm is an iterative procedure that attempts to approximate the Bayes classifier by combining multiple weak classifiers19. The AdaBoost classifier was implemented using the scikit-learn library to perform supervised classification. The base estimator was a decision tree classifier with a maximum depth of 3, allowing each weak learner to capture more complex patterns than a simple decision stump. The ensemble consisted of 200 weak learners, trained sequentially, with each learner focusing on the errors made by its predecessors. A learning rate of 0.01 was applied to reduce the contribution of each learner and mitigate the risk of overfitting, thereby promoting a more gradual and stable learning process.
Random forest
Random Forest is an ensemble classifier based on decision tree models. It developed many trees and bootstrap technique and applied it to every tree from a set of training data. Every tree is present as a forest, and then it votes individually for the class outcome. Random Forest will vote for the majority class of the outcome from every tree20. The optimized hyperparameters of 200 estimators and maximum depth 5 have been applied for training the model.
Naïve Bayes
Naïve Bayes is based on Bayes’ theorem and assumes independence between features. It calculates the probability of each class given the features and selects the class with the highest probability.
Artificial neural networks
ANNs are computational models inspired by the human brain. They consist of interconnected layers of nodes (neurons). Neurons receive inputs, apply weights, pass the result through an activation function, and send the output to the next layer. In this study, a feedforward multi-layer perceptron (MLP) architecture was implemented using Scikit-learn’s MLP Classifier. The proposed ANN model follows a multi-layer perceptron (MLP) architecture. This architecture involved an input layer with 3 neurons, two optimized hidden layers of 100, 50 neurons each, and an output layer. The hidden layers used the ReLU (Rectified Linear Unit) activation function, while the output layer employed a sigmoid activation function suitable for binary classification. The model was trained using the Adam optimizer, with a maximum of 500 iterations.
Model evaluation metrics
The performance of ML classifiers was evaluated using a range of metrics to gain a comprehensive understanding of their robustness and effectiveness. The common evaluation metrics for binary classification are accuracy, F1-score, sensitivity, and specificity, which express the percentage of correctly classified instances in the set of all the instances, the truly positive instances, the truly negative instances, or the instances classified as positive, respectively. Sensitivity is commonly referred to as recall21.
Recall (or sensitivity) measures the proportion of actual positives that were correctly identified by the model. Accuracy represents the overall percentage of correct predictions among all predictions made, giving an overall snapshot of model performance. F1-score is the harmonic mean of precision and recall, providing a balanced measure especially useful when the class distribution is uneven. Additionally, a Receiver Operating Characteristic (ROC) curve was generated, and the Area Under the Curve (AUC) was calculated. The ROC curve illustrates the trade-off between the true positive rate and the false positive rate across different classification thresholds. To further assess model robustness, cross-validation was employed, particularly useful when data is limited.
In practice, ML algorithms are evaluated using a confusion matrix that provides classes, such as the probability of true-negative (TN), true-positive (TP), false-negative (FN), and false-positive (FP). The confusion matrix was further utilized to evaluate the classification accuracy, precision, and recall. The significant performance indicators were obtained using the following Eqs. (1–4):
Confidence intervals for model performance metrics were estimated using the bootstrap percentile method with 1,000 resamples. The resulting 95% intervals, derived from the empirical distribution of the resampled statistics, were incorporated into the interpretation of model performance.
Code availability
The custom code used to perform the analysis in this study is provided as part of the replication package. It is available at supplementary B.
Results
Electrochemical biosensing of urine samples
Field-collected urine samples with or without OV antigens were detected by the developed electrochemical immunosensor as detailed above. The resulting peak current with varying electrical potential was recorded. Examples of SWV curves for OV-positive and OV-negative individuals are shown in Fig. 4. In the OV-positive samples, as the OV antigen present in the urine bound to the immobilized antibody on the immunosensor surface, the relative peak current was significantly reduced (Fig. 4a). This phenomenon was caused by the antigen-antibody complex acting as a hindrance on the electrode surface, leading to less penetration of redox species and a subsequent reduction in the relative signal. Conversely, increasing peak current or no current changes were observed in the OV-negative samples due to the release of the immobilized antibody (Fig. 4b). The square wave voltammograms generated the raw data signal. The raw data graphs were automatically calculated to extract peak current changes (∆I) by Python programming. First, the desired peak current areas were defined by limiting the working voltage range from − 0.1 to 0.6 V. The index of peak points and the lowest points were located, and the baseline was drawn between the two lowest points. The distance between the peak point and the baseline is defined as the peak height of the sensor. The distance difference between peak 1(baseline) and peak 2 (Sample) indicated the peak current change (∆I/µA). The resulting ∆I is directly proportional to OV antigen concentration as described in our previous study12. A standard curve was constructed to translate the ∆I into an understandable OV antigen concentration. The new dataset can further be tested to extract the information about ∆I value and/or OV antigen concentration from a single raw-spectra. The requirement of complicated signal processing and post-processing data analysis were overcome by the automated calculation programming.
Feature selection
The associations between the collected input parameters and the labels were visualized by a correlation heatmap. The heatmap visualization can be seen in Fig. 5. The peak current changes obtained from the electrochemical sensor measurements were most strongly correlated with OV infection (60%), followed by patient age (21%) and urine pH (14%) (Fig. 5). The correlation between age and OV positivity was consistent with results from previous studies indicating that older age (> 60 years) is a risk factor for OV infection22,23.
Feature importance analysis
The permutation-based feature importance analysis quantifies how much model performance decreases when each feature’s values are randomly shuffled, disrupting the true relationship between the feature and the target variable24. The permutation feature importance analysis (Fig. 6) provides critical insights into the predictive power of each feature in our machine learning model for OV infection detection. The results demonstrate that peak current changes obtained from electrochemical sensor measurements were the most important predictor of OV infection, with a relative importance of approximately 0.68 (68%). This strongly supports our sensor-based approach, indicating that the electrochemical signals directly associated with OV infection biomarkers provide the most significant diagnostic information. This finding extends beyond simple correlation analysis by measuring the actual impact on model prediction accuracy when this feature is removed or altered.
Age emerged as the second most important feature with a relative importance of approximately 0.15 (15%), confirming our initial correlation analysis findings. Specific gravity ranked third in importance (approximately 6%), despite showing only a moderate correlation in the previous analysis. This elevated importance suggests that specific gravity may capture unique information about OV infection status that is not linearly correlated but becomes valuable when combined with other features in a complex model. Urine pH showed approximately 3% importance, which is substantially lower than its correlation coefficient (14%). This decrease in relative importance may indicate that while pH correlates with OV infection, much of its predictive power may be captured by other features, particularly peak current changes. The remaining features such as gender, protein, glucose, blood, and ketone, demonstrated minimal importance, validating our feature selection process that excluded these parameters from the final model. The permutation importance analysis strongly supports our three-feature model (peak current changes, age, and pH) while suggesting that specific gravity might also be considered in future model iterations. The clear dominance of peak current changes validates our electrochemical sensing approach as the primary diagnostic modality, with demographic and urinalysis parameters serving as valuable supplementary information.
In our ML model, the decision pathway can be explicated as a sequential process by which the algorithm evaluates key features to reach a prediction. When an input sample is processed, the model first heavily weighs the peak current changes, which represent electrochemical sensor responses to potential OV biomarkers in urine. Peak current changes above 0.11 µA strongly directed the model toward a positive OV infection classification, serving as the first and most critical decision point with the highest information gain. If the peak current changes are borderline, the model then considers age, with higher age (particularly > 60 years) contributing additional risk weight toward OV positivity. Urine pH and other urinalysis parameters (e.g., protein, glucose) are given less influence, and generally do not affect the classification outcome unless the primary features are inconclusive.
In this study, feature selection was based on a combination of correlation analysis, permutation-based feature importance analysis, and domain expertise. Features with a Pearson correlation coefficient above 0.10 with the target label were considered for model input. Peak current change, age, and urine pH emerged as the most relevant variables. Domain knowledge also informed selection: age is a recognized risk factor for OV infection, and urinalysis parameters can influence electrochemical sensor output. While mutual information was not directly applied, permutation-based importance was used to capture feature relevance, accounting for both linear and non-linear contributions to the model.
Classification performance and validation of ML models
The performance of ML classifiers using different input variables was described in Table 2. Most classifiers, such as decision trees, AdaBoost, naïve Bayes, and neural networks, achieved accuracy levels above 80% and excellent AUC values exceeding 0.84 when using only peak current changes as an input. Eliminating the peak current changes reduced accuracy by 31%, while removing the clinical feature decreased the accuracy by 2%, confirming the hierarchical importance of these features in the decision process. While the decision tree model can effectively distinguish between OV-positive and OV-negative samples using sensor features alone, its accuracy decreases when clinical features are employed without sensor data. There is no significant difference in performance between using 9 features and 3 features, but notably the 3 selected features provided the highest accuracy. The available clinical data are observed to be insufficient to enable the precise classification. As a result, the developed ML model demonstrated the significance of incorporating sensor data and machine learning to accurately classify OV infections and enable early diagnosis and intervention. This comprehensive analysis of model interpretability and decision pathways provides transparent insight into how our ML model utilizes the measured features to detect OV infection. The clear dominance of peak current changes in the decision process, supported by demographic and urinary parameters at specific decision thresholds, validates our sensor-based approach while providing clinicians with an understandable framework for interpreting model outputs in real-world diagnostic settings.
The six ML classifiers, including decision tree, k-nearest neighbors, AdaBoost, naïve Bayes, random forest and neural network models, were compared. The evaluation metrics for assessing the performance of each ML model was described in Table 3. Almost all the models exhibited high accuracy for OV classification. The decision tree models exhibited excellent performance, with the highest accuracy of 90.65% (95% CI 0.89–0.91), an F1 score of 91% ( 95% CI 0.90–0.92), and a cross-validation score of 88.21% (95% CI 0.87–0.89) for predicting OV infection. The AdaBoost and neural network models also exhibited high accuracies of 90.65% (95% CI 0.89–0.91) and 89.72% (95% CI 0.84–0.93), respectively. The performance of all ML models and the confusion matrix of each ML model were illustrated (Fig. 7a–f). The AdaBoost model was observed to have a greater AUC (0.88) and specificity (0.86) than the decision tree model (0.86, 0.83) but had a lower F1 score and cross-validation score. We also created receiver operating characteristic (ROC) curves and calculated the area under the ROC curves (AUC) to assess the performance of the classification methods using the set of features and hyperparameters that produced the highest accuracy scores (see Fig. 7g).
Synthetic minority over-sampling technique
Synthetic Minority Over-sampling Technique (SMOTE) algorithm is an extended algorithm for imbalanced data proposed by Chawla25. SMOTE generates new synthetic instances by interpolating between a data point and one of its nearest neighbors from the same class, rather than duplicating existing samples. In our study, SMOTE was applied using the imblearn library to address class imbalance in the training data. This technique helps to expand the decision boundary of the minority classes in feature space. In our case, SMOTE adjusted the training set so that the number of positive and negative samples was equal. We verified the resampled dataset to ensure this balance was achieved. The oversampling process was configured with a random state of 42 to ensure reproducibility and an auto sampling strategy, which automatically increases the number of samples in each minority class to match the majority class (class 0). The k-neighbors parameter was set to 5, meaning that for each minority class sample, five nearest neighbors were considered during the generation of synthetic samples. We evaluated the performance of all machine learning models with and without SMOTE (see Table S1). The decision tree model, along with the other classifiers, showed no significant improvement in performance metrics following resampling. This indicates that the mild imbalance in our dataset did not adversely affect model performance.
Statistical test
For the statistical comparison of model performance, the Friedman non-parametric test was employed first to reject the null hypothesis, and then proceeded with a post-hoc analysis based on the Wilcoxon-Holm method as suggested by Ismail et al., 201926. The Friedman test utilizes a multi-model approach, simultaneously evaluating all models to provide a broader perspective on their relative performance. Across all metrics, a statistically significant difference (p < 0.001) was achieved. The Wilcoxon-Holm post hoc test with Critical Differences (CD) was conducted to have an overall statistical comparison, ensuring more robust results, addressing potential violations of normality assumptions, small sample sizes, and the influence of outliers. In the critical difference (CD) diagram for accuracy (Fig. 8), three distinct performance groups of ML models can be observed. The top-performing groups consist of AdaBoost, decision tree, and neural network (positioned on the right side of the scale), and they are not significantly different from each other. However, the bottom group includes random forest, k-nearest neighbors, and Naïve Bayes, indicating the worst performance. The overall performance of AdaBoost, decision tree, and artificial neural network are significantly better than random forest, k-nearest neighbors, and Naïve Bayes (p < 0.01).
Paired t-tests were further conducted to compare the best-performing classifiers. The results showed no statistically significant differences in accuracy: Decision Tree vs. AdaBoost (p = 0.888), Decision Tree vs. ANN (p = 0.081), and AdaBoost vs. ANN (p = 0.108). These findings support the conclusion that, despite achieving the highest performance metrics, the differences among these models are not statistically significant. This aligns with the CD diagram, which also identified all three as the best-performing models.
Discussion
Many studies have reported promising results in disease prediction using data-driven ML models. Intelligent electrochemical biosensors for pathogen detection, biomarker detection, and other medical applications have been constructed. Electrochemical biosensors can be classified into voltammetric, impedimetric and amperometric biosensors. Voltammetric sensors can detect biomolecules based on direct interaction with the electrical interface through which analyte information is obtained by varying a potential and then measuring the resulting current. Many forms of voltammetry exist based on potential variation, such as polarography (DC voltage)27, linear sweep, differential staircase, normal pulse, reverse pulse, and differential pulse, among others. Electrochemical impedance spectroscopy (EIS) measurements are more complicated and require circuit fitting for data processing. Recently, Wang et al. (2023) developed a machine learning-assisted electrochemical impedance sensor for the classification of three types of bacteria. They used six EIS parameters as inputs and trained the model with SVM, DT, random forest, naïve Bayes, and AdaBoost classifiers28. Ali et al. (2018) also applied an ANN, linear discrimination analysis (LDA) and the maximum likelihood method to classify Escherichia coli and Salmonella typhimurium strains using impedance signals15. In our study, we used SWV to measure OV antigen in clinical samples and extracted peak current changes as features for OV infection classification. The SWV peak current changes were found to be correlated with OV antigen concentrations and were ultrasensitive for OV infection detection in urine samples.
Despite the development of promising ML models integrated with electrochemical biosensors, their practical use on clinical samples remains challenging. Most studies have been conducted in spiked samples rather than in real clinical settings. Moreover, the data used to train models are often retrospectively collected and preselected. The scarcity of prospective clinical trials and validation on available datasets rather than on actual patients in the clinical setting has led to limited confidence in the validity of AI for real-world applications31. We tested urine samples collected from endemic areas of opisthorchiasis, ensuring an actual clinical setting for the development of the ML model.
Various machine learning approaches have been applied to identify OV infection using clinical features and sensor data. Logistic regression (LR) was reported to outperform other ML classifiers in terms of predictive tasks in several previous studies32,33,34. Herein, we demonstrated that the decision tree, AdaBoost and neural network methods achieved greater accuracy than the LR method. Moreover, the ML approach for OV detection enhanced the accuracy of the electrochemical immunosensor from 84% to 91%, signifying the potential utility of the ML model.
The permutation-based importance analysis supports the biological and clinical relevance of the selected features. The dominant influence of peak current changes confirms the biosensor’s efficacy in detecting OV-related biomarkers in urine. The decision-making process in the model follows a hierarchical weighting strategy, where dominant features drive primary decisions, while less important features refine or validate the classification. This layered decision pathway mirrors clinical reasoning, where strong diagnostic signals (e.g., a highly elevated biomarker) are supported by demographic context (e.g., patient age) and auxiliary findings. These findings have important implications for the real-world implementation of our diagnostic method in resource-limited settings. The overwhelming importance of the sensor-based measurement suggests that even in cases where complete patient data might be unavailable, the electrochemical readings alone could provide substantial diagnostic value. However, the meaningful contribution of age and, to a lesser extent, specific gravity and pH, indicates that incorporating these readily available parameters can enhance diagnostic accuracy. Overall, the feature importance results provide transparency into the model’s decision-making process and reinforce confidence in its applicability for field-deployable OV infection screening. Future work will include broader clinical features to further enhance model robustness.
In northeastern Thailand, where the samples were collected, raw fish consumption is common, resulting in the highest prevalence rates of OV infection and OV infection-associated CCA worldwide35. Hence, a critical solution to address the burden of opisthorchiasis in Thailand requires not only efficient diagnosis and treatment methods but also multisectoral and multidisciplinary involvement36. Early diagnosis of OV infection plays a major role in first-line defense strategies. However, due to limited numbers of health care workers and technicians, mass screening in endemic areas may be challenging, potentially resulting in delayed diagnoses. Therefore, the proposed ML model is intended to assist health care practitioners (HCPs) in decision-making processes, especially in uncertain situations. In our study, samples were collected from diverse demographics, including both urban and rural areas within endemic regions, to enhance model generalizability. Training with data from multiple endemic areas improved the model’s applicability beyond localized settings. However, the lack of data from non-endemic areas and other populations is limited.
The formalin-ethyl acetate concentration technique (FECT) is considered the gold standard for detecting Opisthorchis viverrini infection, particularly in cases of moderate to heavy infection. However, FECT has notable limitations such as a lack of sensitivity in detecting light infections, inability to detect in case of bile-duct obstruction, low specificity in areas where morphologically similar flukes (MIFs) are endemic (such as the Mekong Basin), and the requirement of extensive sample collection37. Additionally, FECT detects infection only at the egg stage, which typically appears four weeks post-infection, thus limiting its utility for early detection38. Molecular techniques provide high specificity for OV detection, but the sensitivity can be variable depending on the number of eggs in the faeces. Moreover, this method is costly, requires a specialized technician, and projects the risk of false negatives due to PCR inhibitors in feces39. In contrast, antigen detection methods, such as ELISA, enable earlier diagnosis and intervention, which is essential for reducing the risk of severe complications, including cholangiocarcinoma. A comparison with PCR or FECT would provide additional context for predictive capabilities. However, these tests were not concurrently performed in this study due to practical constraints, including cost and sample handling limitations. Using ELISA as a reference method for OV classification aligned the most with our study’s objective of enabling timely detection and intervention. However, the use of ELISA as the gold standard for labeling may introduce sampling and label bias due to its limited sensitivity and specificity. Misclassifications, particularly near detection thresholds or weak antigen expression, can lead to systematic labeling errors that affect model training and evaluation. If the training set is biased toward samples that ELISA classifies more easily, this may reduce generalizability. Therefore, the reliability of ELISA-based labels should be critically assessed, and complementary validation methods considered where feasible.
Real-world deployment strategy
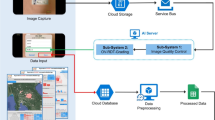
During recent years, research on the application of machine learning models in the healthcare sector has grown tremendously. It aims to develop and improve disease prediction, diagnosis, prognosis, treatment, and personalized medicine40. Establishing a time-saving and accurate diagnostic method is crucial in the surveillance, prevention, and control of parasitic diseases. Several implementation strategies are proposed to facilitate the real-world deployment of the proposed ML model for OV infection classification. A modular machine learning (ML) deployment architecture for disease classification tasks will be implemented to facilitate real-time clinical decision support. The system architecture, illustrated in Fig. 9, consists of five primary components: data acquisition, user interface, API integration, ML inference server, and model training pipeline. The user can input the clinical and sensor data manually or data will be collected directly from point-of-care devices into the application. This interface ensures usability and accessibility in clinical environments. ML server will apply ML algorithm locally or through cloud computing and return disease classification outputs via a secure API. The web application will provide the infection status to the user in a laboratory setting, assisting in decision-making. The public health professionals and non-expert users in rural areas will be empowered by the accessible mobile application. However, several challenges may be interfaced, including the requirement for standardization and improving the generalizability of ML models across different populations. Before widespread implementation, concerns around data protection, internet connectivity, and compliance with regulatory standards must be addressed. Regardless of these drawbacks, the proposed system holds significant potential tools to assist healthcare providers in decision making, particularly in resource-limited or high-prevalence settings.
Limitations
The major limitations of the study are the small number of features associated with the disease and the lack of external validation. More features associated with OV infection, such as laboratory results, epidemiological data and socioeconomic status, will be further applied to train the models. Furthermore, the size of our dataset limited the model’s predictive capabilities, which can be mitigated by increasing the number of patients in each class for further validation procedures. At present, external validation is not feasible due to the limited availability of independent datasets and the fact that electrochemical biosensing for Opisthorchis viverrini infection is currently confined to our testing center. As such, the peak current data were limited to other populations. While external validation on an independent dataset would further strengthen the results, it was not performed in this study as it is a single-center study. Instead, five-fold cross-validation was applied to provide a more reliable and unbiased estimate of model performance than a single hold-out split, with classifier parameters carefully optimized for each model. Furthermore, our study was based on field testing rather than retrospective clinical data, which made it challenging to include external samples from outside the endemic areas. Future multicenter collaborations will be conducted to enable broader data collection and enhance the generalizability and reliability of our model. The sensor’s variability and instability over time can potentially affect model performance. To ensure consistent electrochemical signals, regular calibration and the use of internal standards are essential. Although our current dataset did not exhibit significant evidence of performance drift, addressing this issue is critical for long-term or real-world applications. Future work may include the implementation of drift detection and correction algorithms, as well as periodic recalibration protocols, to sustain the reliability and robustness of the model over time.
Conclusion
In conclusion, this is the first study to classify OV infection using an ML approach based on electrochemical sensor data and patient information. The interpretability of model is demonstrated by the clear dominance of peak current changes in the decision process, supported by demographic and urinary parameters at specific thresholds. The application of this ML model allows for the interpretation of complex raw electrochemical signals to determine OV infection status. This means that medical personnel or patients without prior knowledge of electrochemical sensors can easily understand the disease condition. The decision tree classifier outperformed all other models, achieving an accuracy of 90.65%, a F1 score of 91%, recall of 96%, specificity of 83%, and ROC-AUC of 0.86. Similarly, the AdaBoost model demonstrated excellent performance, with a higher specificity of 86% and a superior ROC-AUC of 0.88 compared to the decision tree. The statistical analyses evaluated the significant difference in the model’s performance. The ML model can be trained to continuously learn and adapt to new data, improving its predictive capabilities over time. Moreover, the proposed ML model is not limited to OV infection and can be applied for the classification of other parasitic infections. A mobile or web application for real-world deployment was proposed in this study. In the future, this study can be used as a prototype to develop a health care strategy for OV-infected patients. The proposed ML model holds promise for enhancing disease surveillance and control strategies and assisting HCPs in decision-making processes in endemic regions.
Data availability
The datasets used and/or analysed during the current study belong to Cholangiocarcinoma Research Institute, Khon Kaen University, Thailand. The datasets used to test the model are available in the supplementary.
References
Ahmad, A. et al. Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete,Rhodococcusspecies. Nanotechnology 14, 824–828 (2003).
Parkin, D. M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 118, 3030–3044 (2006).
Lun, Z. R. et al. Clonorchiasis: a key foodborne zoonosis in China. Lancet Infect. Dis. 5, 31–41 (2005).
Sripa, B. et al. Opisthorchiasis and Opisthorchis-associated cholangiocarcinoma in Thailand and Laos. Acta Trop. 120 (Suppl 1), S158–168 (2011).
Sithithaworn, P., Yongvanit, P., Duenngai, K., Kiatsopit, N. & Pairojkul, C. Roles of liver fluke infection as risk factor for cholangiocarcinoma. J. Hepato-Biliary-Pancreat Sci. 21, 301–308 (2014).
Qian, M. B. et al. Accuracy of the Kato-Katz method and formalin-ether concentration technique for the diagnosis of clonorchis sinensis, and implication for assessing drug efficacy. Parasit. Vectors. 6, 314 (2013).
Johansen, M. V., Sithithaworn, P., Bergquist, R. & Utzinger, J. Towards improved diagnosis of zoonotic trematode infections in Southeast Asia. Adv. Parasitol. 73, 171–195 (2010).
Wongratanacheewin, S., Sermswan, R. W. & Sirisinha, S. Immunology and molecular biology of opisthorchis Viverrini infection. Acta Trop. 88, 195–207 (2003).
Haswell-Elkins, M. R. et al. Immune responsiveness and parasite-specific antibody levels in human hepatobiliary disease associated with opisthorchis Viverrini infection. Clin. Exp. Immunol. 84, 213–218 (1991).
Bettazzi, F., Marrazza, G., Minunni, M., Palchetti, I. & Scarano, S. Chapter one - biosensors and related bioanalytical tools. In Comprehensive Analytical Chemistry (eds Palchetti, I., Hansen, P.-D. & Barceló, D.) 77 1–33 (Elsevier, 2017).
Menon, S., Mathew, M. R., Sam, S., Keerthi, K. & Kumar, K. G. Recent advances and challenges in electrochemical biosensors for emerging and re-emerging infectious diseases. J. Electroanal. Chem. 878, 114596 (2020).
Aye, N. N. S. et al. Functionalized graphene oxide–antibody conjugate-based electrochemical immunosensors to detect opisthorchis Viverrini antigen in urine. Mater. Adv. https://doi.org/10.1039/D3MA01075A (2024).
Cui, F., Yue, Y., Zhang, Y., Zhang, Z. & Zhou, H. S. Advancing biosensors with machine learning. ACS Sens. https://doi.org/10.1021/acssensors.0c01424 (2020).
Ramprasad, R., Batra, R., Pilania, G., Mannodi-Kanakkithodi, A. & Kim, C. Machine learning in materials informatics: recent applications and prospects. Npj Comput. Mater. 3, 1–13 (2017).
Ali, S. et al. Disposable all-printed electronic biosensor for instantaneous detection and classification of pathogens. Sci. Rep. 8, 5920 (2018).
Zeng, Z. et al. Nonintrusive monitoring of mental fatigue status using epidermal electronic systems and Machine-Learning algorithms. ACS Sens. 5, 1305–1313 (2020).
Uwaya, G. E., Sagrado, S. & Bisetty, K. Smart electrochemical sensing of xylitol using a combined machine learning and simulation approach. Talanta Open. 6, 100144 (2022).
Baghdadi, N. A. et al. Advanced machine learning techniques for cardiovascular disease early detection and diagnosis. J. Big Data. 10, 144 (2023).
Hastie, T., Rosset, S., Zhu, J. & Zou, H. Multi-class adaboost. Stat. Interface. 2, 349–360 (2009).
Kumari, S., Kumar, D. & Mittal, M. An ensemble approach for classification and prediction of diabetes mellitus using soft voting classifier. Int. J. Cogn. Comput. Eng. 2, 40–46 (2021).
Rainio, O., Teuho, J. & Klén, R. Evaluation metrics and statistical tests for machine learning. Sci. Rep. 14, 6086 (2024).
Srithai, C., Chuangchaiya, S., Jaichuang, S. & Idris, Z. M. Prevalence of opisthorchis Viverrini and its associated risk factors in the phon Sawan district of Nakhon phanom Province, Thailand. Iran. J. Parasitol. 16, 474–482 (2021).
Thaewnongiew, K. et al. Prevalence and risk factors for opisthorchis Viverrini infections in upper Northeast Thailand. Asian Pac. J. Cancer Prev. APJCP. 15, 6609–6612 (2014).
Nirmalraj, S. et al. Permutation feature importance-based fusion techniques for diabetes prediction. Soft Comput. https://doi.org/10.1007/s00500-023-08041-y (2023).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority Over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002).
Ismail Fawaz, H., Forestier, G., Weber, J., Idoumghar, L. & Muller, P. A. Deep learning for time series classification: a review. Data Min. Knowl. Discov. 33, 917–963 (2019).
Heyrovský, J. The development of Polarographic analysis. Analyst 81, 189–192 (1956).
Wang, C. et al. Machine learning-assisted cell-imprinted electrochemical impedance sensor for qualitative and quantitative analysis of three bacteria. Sens. Actuators B Chem. 384, 133672 (2023).
Worasith, C. et al. Comparing the performance of urine and copro-antigen detection in evaluating opisthorchis Viverrini infection in communities with different transmission levels in Northeast Thailand. PLoS Negl. Trop. Dis. 13, e0007186 (2019).
Worasith, C. et al. Effects of day-to-day variation of opisthorchis Viverrini antigen in urine on the accuracy of diagnosing opisthorchiasis in Northeast Thailand. PLOS ONE. 17, e0271553 (2022).
Thomas, L. B., Mastorides, S. M., Viswanadhan, N. A., Jakey, C. E. & Borkowski, A. A. Artificial intelligence: review of current and future applications in medicine. Fed. Pract. Health Care Prof. VA. DoD PHS. 38, 527–538 (2021).
Christodoulou, E. et al. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 110, 12–22 (2019).
van der Ploeg, T., Austin, P. C. & Steyerberg, E. W. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med. Res. Methodol. 14, 137 (2014).
Jiang, Y. et al. Cardiovascular disease prediction by machine learning algorithms based on cytokines in Kazakhs of China. Clin. Epidemiol. 13, 417–428 (2021). <\/p>
Alsaleh, M. et al. Characterisation of the urinary metabolic profile of liver Fluke-Associated cholangiocarcinoma. J. Clin. Exp. Hepatol. 9, 657–675 (2019).
Sripa, B. et al. Toward integrated opisthorchiasis control in Northeast thailand: the Lawa project. Acta Trop. 141, 361–367 (2015).
Worasith, C. et al. Advances in the diagnosis of human opisthorchiasis: development of opisthorchis Viverrini antigen detection in urine. PLoS Negl. Trop. Dis. 9, e0004157 (2015).
Maitongngam, K., Tipayamongkholgul, M. & Kosaisavee, V. Diagnostic accuracy of opisthorchis Viverrini antigen methods for human opisthorchiasis: systematic review and Meta-analysis. Thai J. Public. Health. 53, 465–481 (2023).
Khuntikeo, N. et al. Current perspectives on opisthorchiasis control and cholangiocarcinoma detection in Southeast Asia. Front. Med. 5, (2018).
Panch, T., Szolovits, P. & Atun, R. Artificial intelligence, machine learning and health systems. J. Glob Health. 8, 020303 (2018).
Acknowledgements
The author would like to express gratitude for a Khon Kaen University ASEAN-GMS scholarship and sample resources from Cholangiocarcinoma Research Institute.
Funding
This research was supported by NSRF under the Basic Research Fund of Khon Kaen University through the Cholangiocarcinoma Research Institute: CARI-BRF64-47 and the Fundamental Fund of Khon Kaen University and the National Sience, Research, and Innovation Fund (FF2566).
Author information
Authors and Affiliations
Contributions
Conceptualization, N.N.S.A, R.P, and J.D; Formal analysis, N.N.S.A; Investigation, N.N.S.A; Methodology, N.N.S.A, R.P ; Supervision, P.M, R.P, and J.D; Writing—original draft, N.N.S.A ; Writing—review and editing, N.N.S.A, S.D, P.T, A.T, P.M, P.S, R.P, and J.D. Resources—P.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Institutional review board
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Centre for Ethics in Human Research, Khon Kaen University (HE664025).
Informed consent
Written informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aye, N.N.S., Daduang, S., Tippayawat, P. et al. Machine learning approach using electrochemical immunosensor data for precise classification of Opisthorchis viverrini infection. Sci Rep 16, 234 (2026). https://doi.org/10.1038/s41598-025-29347-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-29347-x