Abstract
Tumor-associated macrophages (TAMs) with highly expressed secreted phosphoprotein 1 (SPP1) carry immunosuppressive property as a potential target for tumor metastasis. However, the mechanisms regulating SPP1 + TAMs in head and neck squamous cell carcinoma (HNSCC) remain poorly understood. This study employs a combination of single-cell and bulk RNA sequencing bioinformatics analysis to confirm the impact of TAMs with high levels of SPP1 on patient prognosis. Additionally, Key genes linked to SPP1 + macrophages were identified using weighted gene co-expression network analysis. A prognostic model was built using the Random Forest algorithm. Here we show, P4HA1 is strongly correlated with SPP1 + macrophages and holds significant value in predicting patient prognosis and diagnosis. In vitro experiments demonstrated that TAMs educated by HNSCC cells with knockdown P4HA1 expressed lower SPP1 level compared to the control group. Furthermore, Gene Set Variation Analysis and Gene Set Enrichment Analysis indicated that P4HA1 mediates the hypoxia pathway in HNSCC. In xenografts model, P4HA1 knockdown effectively suppressed tumor malignant progress, confirming that P4HA1 was positive correlation with SPP1 + TAMs and could mediate tumor hypoxia pathways. Overall, this study identified P4HA1 as a key gene involved in regulating SPP1 + TAMs through modulating hypoxia, providing a potential macrophage-centered therapeutic target in HNSCC.
Similar content being viewed by others
Introduction
Head and neck squamous cell carcinoma (HNSCC) accounts for approximately 90% of head and neck cancers (HNC) and is recognized as the sixth most common type of cancer worldwide1,2. Despite multidisciplinary treatment of locally advanced HNSCC by surgery combined with adjuvant chemoradiation or platinum-based concurrent chemoradiation, less than 50% of patients can be cured3. HNSCC is characterized by the heterogeneity of tumor tissues and the complexity of tumor microenvironment (TME)4. Tumor-associated macrophages (TAMs), the main components of infiltrating leukocytes in the TME, have a substantial promoting effect on malignant progression5,6. Further research on the molecular mechanism of TAMs in HNSCC holds great significance in terms of identifying therapeutic targets.
A large infiltration of macrophages in solid tumors not only contributes to treatment resistance but also has a negative impact on patient prognosis7. Generally speaking, monocytes are recruited to the ‘field effect in cancer’ by a variety of chemokines and cytokines released by tumor cells, and they are educated and polarized into TAMs, including the pro-inflammatory M1 and pro-tumorigenic M2 phenotypes8. However, with the advancement of single-cell sequencing (scRNA-seq) technologies, it has become increasingly clear that the traditional classification of TAMs into M1 (ITGAX, CD80) and M2 (CD163, MRC1) types is insufficient to account for the complexity9. In certain types of cancers, TAMs isolated from patients have been found to express both M1 and M2 markers, suggesting the presence of TAMs with high heterogeneity and dynamical phenotypes10. Therefore, the subtyping of TAMs based on scRNA-seq better reflects the complexity of the TME.
SPP1 (Secreted Phosphoprotein 1) is aberrantly expressed and exerts oncogenic roles in various cancers, being produced not only by tumor cells but also secreted by immune cells such as macrophages11,12. SPP1 + macrophages are closely associated with hypoxic microenvironments, with their high expression significantly correlated with poor patient prognosis, co-infiltration with FAP + fibroblasts, reduced lymphocyte infiltration, and accumulation of cancer stem cells13,14. Collagen prolyl 4-hydroxylase (P4H), an α-ketoglutarate (α-KG)-dependent dioxygenase, facilitates proline hydroxylation in collagen, thereby promoting collagen synthesis while releasing succinate as a byproduct15. P4H subunit alpha 1 (P4HA1) is a key component of the P4H family, which facilitates post-translational modifications crucial for protein folding and stability. As the main isoform of this enzyme, P4HA1 is present in multiple human tissues and is essential for promoting P4H activity16. Recent research on pancreatic ductal adenocarcinoma has identified P4HA1 as a vital regulator in glycolysis and various oncogenic processes through hypoxia-inducible factor-1α (HIF-1α), including cell proliferation, chemoresistance, and stem cell characteristics17. Regulating the levels of alpha-ketoglutarate (α-KG) and succinate can reduce prolyl hydroxylation on HIF-1α by modulating the expression of P4HA1 in breast cancer cells18. Despite these insights, the relationship between P4HA1 expression and SPP1 + macrophage infiltration in HNSCC remains unclear.
Here, we conducted a combined analysis of scRNA-seq and bulk RNA-seq (RNA-seq) data of HNSCC, revealing a subtype of macrophages with high expression SPP1 that has a detrimental impact on patient prognosis. Through weighted gene co-expression network analysis (WGCNA), genes associated with SPP1 + macrophages were identified. And a clinical prognostic model was established using various machine learning algorithms. P4HA1 was strongly associated with the abundance of SPP1 + macrophages and indicated poor prognosis for patients with HNSCC. More importantly, both in vitro and in vivo experiments revealed that tumor cell-derived P4HA1 orchestrates macrophage polarization toward the SPP1 + phenotype through hypoxia pathway. This study identified P4HA1 as a novel regulator of SPP1 + TAM subtype differentiation, which promotes the malignant progression of HNSCC through hypoxia pathway, thereby offering a promising immunotherapeutic target for HNSCC treatment.
Results
The impact of different macrophage subtypes on the prognosis of HNSCC patients
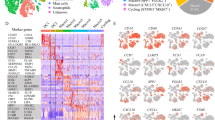
A dataset was acquired from the GEO database, which comprised scRNA-seq data from 52 patients with HNSCC, and was identified as GSE234933. Using the R package Seurat, a single-cell atlas for HNSCC cases was constructed by integrating data and applying stringent quality control measures, resulting in a refined dataset of 172,569 cells for further analysis. PCA was then employed to reduce the dimensionality of the dataset (Fig. S1). The cell types were visualized using UAMP, which displayed an extensive variety of cells in the dataset (Fig. 1A). Cell classification was finalized by annotating cells with marker genes (Fig. 1B). Next, the macrophages were further divided into 8 subtypes, and each subtype was named based on its specific marker genes (Fig. 1C and D).
The abundance of SPP1 + macrophages is negatively associated with patient prognosis in HNSCC. (A) UMAP visualization showed 172,569 cells from 52 HNSCC patients, categorized into 10 distinct cell types. (B) Heatmap exhibited the expression levels of marker genes for the 10 cell types. (C) UMAP plot showing 8 subtypes of macrophages. (D) The marker genes of macrophage subtypes are presented in a contour density plot. (E) The immune infiltration of the 8 macrophage subtypes in the TCGA-HNSCC cohort. (F) The box plot shows the infiltration ratios of eight macrophage subtypes in HNSCC and adjacent normal tissues. (G) Univariate Cox regression analysis of eight macrophage subtypes. (H) Kaplan-Meier survival analysis of high and low SPP1+ macrophage infiltration groups. *,p < 0.05,**, p < 0.01; ***.
To investigate the impact of subtype infiltration on patient survival outcomes in a large cohort study, we applied the scRNA-seq data to TCGA-HNSCC cohort using the Bisque RNA-seq deconvolution algorithm (Fig. 1E). The infiltration levels of Mac_SLC40A and Mac_SPP1 are significantly increased in head and neck cancer compared to normal tissue (Fig. 1F). Univariate Cox regression analyses indicated that SPP1 + macrophages were associated with an unfavorable prognosis in patients with hazard ratios of 4.978 (p < 0.001) (Fig. 1G). And survival curves confirmed that high infiltration of SPP1 + macrophage subtypes was significantly associated with poor prognosis in HNSCC patients (log-rank p < 0.001; Fig. 1H).
WGCNA identified SPP1+ macrophage related genes
We used the WGCNA algorithm to identify coding genes that may influence the abundance of SPP1 + macrophages in HNSCC. RNA-seq data of 502 HNSCC patients from TCGA was used for analysis. After removing the outliers, 471 high-quality samples remained (Fig. 2A). Then we chose the optimal soft-thresholding power (β) of 6. At the β value, an R2 > 0.85, specifically 0.921, indicated that the network’s topological structure conforms to scale-free characteristics (Fig. 2B). Following the gene network analysis, we categorized the genes within the network into 36 distinct modules, with each module containing at least 30 genes (Fig. 2C). Further evaluation using Pearson’s correlation analysis was performed to assess the relationship between each module and the proportion of SPP1 + macrophages (Fig. 2D). The results showed that two modules, named “midnightblue” and “brown,” exhibited significant correlation with coefficients exceeding 0.3. Therefore, genes from these two modules were selected for further investigation, with thresholds for module membership and gene significance deliberately established at 0.3 and 0.2, respectively, to minimize noise and ensure an adequate pool for selection (Fig. 2E). Next, we performed differential analysis using the TCGA-HNSCC cohort. This analysis identified 2872 differentially expressed genes between tumor and normal samples (Fig. 2F). Ultimately, 193 genes were identified at the intersection by comparing the genes from the modules with the differentially expressed genes (Fig. 2G). To establish a clinical prognostic model and investigate the most valuable genes for further research, we employed univariate Cox regression analysis, which identified 14 intersecting genes that were significantly associated with clinical prognosis (Fig. 2H).
WGCNA identifies module genes associated with SPP1 + macrophages. (A) Among 502 HNSCC-TCGA data instances, after excluding outliers (red line = 430), a total of 471 samples were analyzed. (B) Analysis of the scale-free fitting index (left) and average connectivity (right) for different soft-thresholding powers identified the optimal soft-thresholding power as 6 (R^2 = 0.921). (C) Dendrogram of 18,253 encoded genes based on dissimilarity measure (1-TOM) clustering, with each module containing at least 30 genes. (D) Heatmap depicting the correlation between module feature genes and SPP1 + macrophage abundance. (E) Scatter plot showing the association between midnight blue and brown module membership and gene expression correlation with Mac_SPP1 cell abundance. (F) Volcano plot of 2,872 differentially expressed genes between tumor and adjacent normal tissue. (G) Venn diagram presenting 193 overlapping genes between module genes and differentially expressed genes. (H) After applying Cox univariate regression analysis to the intersection genes. 14 genes were identified as significantly associated with patient survival prognosis (P < 0.05).
Prognosis model was established using the SPP1+ macrophages related genes
To validate the clinical prognostic value of the 14 key SPP1 + macrophage-related genes, we established a clinical prognosis model based on a training set from TCGA-HNSCC cohort (n = 502) and used GSE41613 (n = 97), GSE65858 (n = 270), and GSE117973 (n = 77) as testing sets. This model was developed by employing 12 classical machine learning methods integrated into 113 algorithmic combinations. PCA conducted before and after correction suggested that the effect of sequencing batch effects was adequately mitigated (Fig. S2). Among the 113 combined algorithms, the fusion of glmBoost and RF achieved the highest composite score 0.715 (Fig. 3A). Next, we employed glmBoost-RF for optimal feature selection, which identified the following nine genes as most predictive: P4HA1, SPOCK1, CCL26, STC1, TRIM9, PTPRN, TPM4, DGKG, and CLDN8 (Fig. 3B). Finally, a RF algorithm with ten-fold cross-validation was implemented. The error rate was minimized, and the model was stabilized when the number of trees was set to 1000 (Fig. 3C). Each feature was scored by calculating the mean decrease in accuracy and Gini impurities (Fig. 3D). To accurately assess the predictive and generalization capabilities of the final model, we utilized confusion matrices (CM) and Receiver Operating Characteristic Curve (ROC) to evaluate its performance on both training and validation datasets (Fig. 3E). The CM analysis revealed that the clinical prognostic model demonstrated a higher degree of predictive precision for living patients, whereas its predictive capacity for deceased individuals was less accurate. Evaluation of the AUC scores indicated that the model exhibited robust predictive accuracy on the TCGA training set and showed a reasonable degree of generalizability to the GSE41613 dataset. We then established a correlation between these 9 genes and the proportion of SPP1 + macrophages using Pearson’s correlation analysis (Fig. S3A). The correlation between expression of P4HA1 and infiltration of SPP1 + macrophage is 0.32 (p < 0.0001). We also evaluated the diagnostic potential of nine SPP1 + macrophage related genes using ROC curves in the TCGA-HNSCC and GSE6631 datasets (Fig. S3B). P4HA1 consistently exhibited the highest AUC scores of 0.89 and 0.91, highlighting its strong diagnostic capability.
Constructing a clinical prognosis model of SPP1 + macrophages related genes. (A) The heatmap shows the AUC scores of various machine learning ensemble prognostic models on the TCGA training set and different GEO datasets. (B) Importance scores of 15 feature genes evaluated using the glmBoost machine learning algorithm. (C) The line graph illustrates the error rates of three cross. (D) The scatter plot visually demonstrates the average decrease in accuracy and average decrease in Gini impurity of the 9 genes with glmBoost feature engineering scores greater than 0 in the random forest model. (E) The confusion matrix and ROC curve of RF model, including the TCGA dataset and the GSE41613, GSE65658, and GSE117973 datasets.
The expression of P4HA1 in HNSCC was positively correlated with SPP1+ macrophages
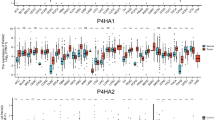
To explore the association between P4HA1 and SPP1 + macrophages, we conducted an analysis of P4HA1 mRNA expression in 502 HNSCC samples and 44 adjacent normal tissues in the TCGA-HNSCC. Our findings revealed a significantly elevated expression of P4HA1 in HNSCC tissues compared to the adjacent normal tissues (p < 0.0001, Fig. 4A). Next, we categorized HNSCC samples based on P4HA1 expression levels. The Wilcoxon rank-sum test demonstrated a significant correlation between high expression of P4HA1 (cutoff = 0.5) and an increased proportion of SPP1 + macrophages in patients (p < 0.0001; Fig. 4B). Kaplan-Meier analysis showed that patients with high P4HA1 expression had significantly lower 10-year overall survival compared to those with low P4HA1 expression (n = 260, p < 0.0001, log-rank test, Fig. 4C). Additionally, we induced M0 macrophages with conditioned media of CAL27 and SCC25 cells and then detected the expression of SPP1 in TAMs by IF, which was significantly higher in TAMs than that in M0 macrophages (Fig. 4D).
To further explore the effect of P4HA1 in SCC25, CAL27 and FaDu cells on the expression of SPP1 in TAMs, we used siRNA targeting the human P4HA1 sequence to knockdown P4HA1 in SCC25, CAL27 and FaDu cells, and the transfection efficiency was verified using RT-qPCR and western blotting (p < 0.05; Fig. S4A and B). Then, SCC25, CAL27 and FaDu cells were co-cultured with macrophages through a 0.4-µm pore Transwell chamber (Fig. S4C). Compared to the control group, 72 h after the co-culture of P4HA1-knockdown SCC25, CAL27 and FaDu cells with macrophages, the mRNA expression level of SPP1 in TAMs was significantly decreased (p < 0.05, Fig. S4D). And then the fluorescence intensity of SPP1 in TAMs induced by siP4HA1 tumor cells was detected by IF, which was significantly downregulated compared to the si-Control group (p < 0.05, Figs. 4E, S5A and S6A). TAMs can enhance the migration and invasion of tumor cells, while we found that this promoting effect was significantly reduced when macrophages were induced by SCC25, CAL27 and FaDu cells with siP4HA1 (p < 0.05, Figs. 4F and G, S5B and C, S6B and C). In summary, the expression of P4HA1 in tumor cells is positively correlated with the level of SPP1 in TAMs, which can mediate the migration and invasion of HNSCC cells by regulating SPP1 + TAMs.
The expression of SPP1 in HNSCC positively correlates with P4HA1. (A) The box plot illustrates the distribution of P4HA1 gene expression in normal tissues and HNSCC tissues from the TCGA-HNSCC cohort, and the differences were analyzed using the Wilcoxon rank-sum test (P < 0.001). (B) The box plot displays the abundance of SPP1 + macrophage infiltration in the high P4HA1 expression group and low P4HA1 expression group (cut-off = 0.5) among 502 TCGA-HNSCC patients, and the differences were analyzed using the Wilcoxon rank-sum test (P < 0.001). (C) Kaplan-Meier survival analysis was conducted on HNSCC samples (n = 499), and the log-rank test revealed a significant difference (P < 0.0001). (D) The macrophages were treated with SCC25 or CAL27 conditioned media for 48 h. And then the expression of SPP1 in the macrophages was detected using cell immunofluorescence (left panel), and the cell skeleton was labeled using F-actin (left panel). The proportion of macrophages with SPP1 expression was quantified relative to the total number of macrophages (right panel). (E) Immunofluorescence staining was performed to detect the levels of SPP1 in macrophages after 72 h of indirect co-culture with SCC25 (transfected with si-Ctrl or si-P4HA1), and the proportion of macrophages with SPP1 expression was quantified relative to the total number of macrophages. (F) Transwell migration and invasion assays were conducted to assess the number of tumor cells at the bottom of the chamber within 24 h after addition of conditioned medium of macrophages. (G) Wound healing assay was performed to detect the migration of tumor cells. Error bars, SEM, **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
P4HA1 mediates the SPP1+ TAM phenotype of macrophages by activating hypoxia pathway in HNSCC
To investigate the role of P4HA1 in the TME and elucidate how P4HA1 promotes the polarization of macrophages towards the SPP1 phenotype, gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA) were used to analyze TCGA-HNSCC cohorts. The results revealed that P4HA1 mediated cellular hypoxia within the TME (NES = 2.279, p.adjust < 0.001; Fig. 5A and B). According to the high and low P4HA1 expression groups, differential gene expression analysis conducted using the limma package identified 331 genes that varied significantly between the two groups (Fig. 5C). The 331 intersecting with 200 hypoxia-associated genes from the hallmark gene set yielded a common set of 26 intersecting hypoxia-related genes (Fig. 5D). The intersecting genes were subsequently imported into a protein-protein interaction (PPI) network, from which the proteins that interact with P4HA1 were chosen for further experimental validation (Fig. 5E and F). GAPDH, which served as a housekeeping gene, was excluded from the experiment. Likewise, P4HA2, owing to its functional similarity to P4HA1, was not subjected to validation assays. The other four proteins were detected at the mRNA level, and the results showed that knockdown of P4HA1 in SCC25 or CAL27 resulted in concomitant downregulation of ANGPTL4, COL5A1, SLC2A1, and NDRG1 transcripts (Figs. 5G, S7A). Next, reactive oxygen species (ROS) levels and HIF-1α protein expression were detected using western blotting and ROS assays, respectively. The results showed that a notable reduction in intracellular ROS accumulation and a decrease in HIF-1α protein levels were observed in the si-P4HA1 groups, substantiating the role of P4HA1 in promoting cellular hypoxia (Figs. 5H and I, S7B and C). Finally, macrophages treated with tumor cells and CoCl2 for 72 h showed a notable elevation in SPP1 expression relative to the control group (Figs. 5J, S7D). These results indicated that P4HA1 could activate the HIF-1α pathway in tumor cells to polarize SPP1 + macrophages in HNSCC.
P4HA1 polarizes macrophages into the SPP1 phenotype through mediating hypoxia pathway. (A) GSVA analysis revealed differential pathway activity between the low P4HA1 expression group and the high P4HA1 expression group in HNSCC. Higher t-values indicate higher pathway activity. (B) GSEA analysis revealed an association between P4HA1 and the hypoxia pathway (NES = 2.279, p.adjust < 0.001). (C) Volcano plot displayed the differentially expressed genes between the high and the low P4HA1 expression groups in HNSCC (abs (logFC) > 0.5, p.adjust < 0.05). (D) Venn diagram showed the intersection of 26 genes between the differentially expressed genes (left side) and the Harlmark hypoxia gene set (right side). (E) Protein-protein interaction network composed of the 26 intersection genes. (F) Protein-protein interaction network centered around P4HA1 and the intersection genes. (G) Rt-qPCR was used to measure the expression levels of ANGPTL4, COL5A1, NDRG1, and SLC2A1 genes in SCC25 cells. (H) Activity assay kit was used to measure the level of reactive oxygen species in SCC25 cells (transfected with si-Ctrl or si-P4HA1. (I) Western blot analysis was performed to detect the expression level of HIF-1α in SCC25 cells (transfected with si-Ctrl or si-P4HA1). (J) Western blot analysis was conducted to examine the expression of SPP1 in macrophages after 72 h of co-culture with SCC25 cells (with or without CoCl2). Error bars, SEM; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
P4HA1 knockdown decreases SPP1+ macrophages through regulating hypoxia pathway in vivo
Subcutaneously inoculating nude mice with SCC25 cells transfected with sh-P4HA1 and the negative control, and subsequently dissecting the tumors four weeks post-injection, revealed that P4HA1 knockdown significantly inhibited tumor growth in vivo (Fig. 6A and D). To investigate the impact of P4HA1 knockdown on SPP1 expression, we performed HE staining and IHC analysis across various experimental groups (Fig. 6E). Given that the expression levels of SPP1 were reduced in the sh-P4HA1 group compared to the NC group. By performing co-IF staining of SPP1 and CD68, we found a reduction in the number of SPP1-positive macrophages in the P4HA1 knockdown group (Fig. 6F). This indicates that the knockdown of P4HA1 inhibits tumor growth and reduces the level of SPP1 in vivo. Additionally, to validate that P4HA1 knockdown suppresses hypoxia in HNSCC tumors, we evaluated hypoxia markers HIF-1α using IHC and co-IF. The results indicated that in the P4HA1 knockdown group, HIF-1αexpressions were significantly reduced (Fig. 6E and G). And we analyzed the correlation of HIF-1α and SPP1 expression in different groups using Pearson’s test (Fig. 6G), demonstrating that tumor hypoxia induced by P4HA1 was positively correlated with SPP1 expression.
P4HA1 knockdown inhibited SPP1 + TAMs by regulating the hypoxia pathway in vivo. (A) Schematic diagram of the nude mouse tumor model using SCC25 cell line, grouped as NC-Ctrl, sh-P4HA1#1, and sh-P4HA1#2. (B) Tumor volume growth curves in nude mice for each group. (C) The image of xenografts in nude mice for each group. (D) Tumor weights in different groups. (E) HE and immunohistochemical staining for SPP1 and HIF-1α, along with statistical analysis of their mean optical density. (F) Fluorescent double staining of CD68 and SPP1 was performed, along with statistical analysis of the merging average optical density. (G) Following fluorescent double staining of HIF-1α and SPP1, the correlation was analyzed using Pearson’s test. Error bars, SEM; *, p < 0.05, **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
Discussion
TAMs play a pivotal role in the stroma of most cancers, responding to tumor stimuli with a unique set of factors and enzymes that influence tumor growth, angiogenesis, and possibly metastasis19,20. Within the tumor microenvironment (TME), the M1-like TAMs, which exert antitumor effects, and the M2-like, which fosters a pro-tumorigenic milieu, significantly shape the dynamics of cancer progression. The interplay between these M1/M2 subsets substantially affects the development of strategies aimed at enhancing antitumor immune responses10. However, with the advancement of scRNA-seq technologies, an increasing number of research indicates that traditional markers used to differentiate macrophage subsets are insufficient to effectively categorize macrophage populations effectively9,21. To gain a more nuanced understanding of macrophage diversity in the TME of HNSCC, we meticulously categorized macrophages from an scRNA-seq dataset of 52 HNSCC specimens into 12 distinct subtypes. Next, using deconvolution techniques, we ascertained the relative proportions of infiltration of the 12 macrophage subtypes across 502 cases in TCGA-HNSCC cohort. The top five subtypes with the highest cell infiltration rankings were selected based on the median values. Cox regression analysis, both in its univariate and multivariate forms, substantiated the profound influence of SPP1 + macrophages on patient prognosis.
Recent studies on TAMs have primarily utilized the infiltration abundance of M2 macrophages based on their signature genes as phenotypic data. WGCNA was employed and Pearson correlation analysis was conducted to identify genes associated with M2 macrophages22. In this study, we used trait data derived from highly prognostic and finely delineated macrophage subpopulations identified using scRNA-seq. WGCNA was then used to identify genes specifically associated with these distinct macrophage subsets. We further integrated more than ten classical machine learning algorithms with 113 random combinations to select the optimal model ensemble for predicting clinical outcomes based on genes associated with SPP1 + macrophages. The final model of the Random Forest algorithm was employed by leveraging a panel of nine SPP1 + macrophage-associated genes, particularly P4HA1. This gene signature provided enhanced prediction of patient prognosis across the TCGA test dataset and three additional GEO test datasets.
P4HA1 serves as a critical regulatory component of prolyl 4-hydroxylase (P4H), an enzyme imperative for the hydroxylation of procollagen and the subsequent synthesis and secretion of collagen23. In our bioinformatics analysis, there was a positive association between P4HA1 levels and the count of SPP1 + macrophages. Further analysis using immunohistochemical techniques revealed that within HNSCC tissues, the presence of P4HA1 was consistently linked to elevated SPP1 levels. The vitro and in vivo experiments indicated that P4HA1 knockdown in tumor cells effectively inhibited the polarization of SPP1 + macrophages, suggesting that P4HA1 mediates SPP1 expression in TAMs in HNSCC. Research reported that P4HA1 stimulated the HIF-1α signaling pathway by inhibiting the proteolysis of HIF-1α. This regulatory mechanism was facilitated by the metabolic byproducts of alpha-ketoglutarate(α-KG) and succinate18. The HIF-1α signaling cascade triggers a metabolic reprogramming and boosts the angiogenesis, both are pivotal in facilitating cancer progression24. The oxygen-dependent dioxygenases (PHDs) could hydroxylate HIF-1α, leading to HIF-1α ubiquitination and subsequent degradation25,26. However, P4HA1 might modulate the functionality of PHD through alterations in the levels of α-KG and succinate, thereby suppressing the hydroxylation of HIF-1α and enhancing HIF-1α stability.
Hypoxic environments in vitro lead to increased SPP1 expression in macrophages13. It is plausible that SPP1 + macrophage-associated genes have a potential regulatory relationship with hypoxia pathway. Cobalt chloride (CoCl2) can be used to mimic the stabilization of HIF-1α, thereby facilitating studies on cellular behavior under hypoxic conditions without the reducing oxygen concentration in the culture environment. In this study, after co-culturing HNSCC tumor cells with macrophages for 72 h, western blot analysis demonstrated an increase in SPP1 expression in macrophages from the CoCl2- treated group, in contrast to the levels observed in the control group. Xu et al. indicated that SPP1 mediates TAMs through the SPP1-CD44 ligand-receptor axis27, which promotes the secretion of more pro-tumorigenic and anti-inflammatory factors, such as TGF-β, IL-10, and VEGF. Therefore, we collected conditioned media from TAMs induced by tumor cells with high P4HA1 or low P4HA1 expression. Transwell and wound healing assays in HNSCC tumor cells demonstrated that TAMs induced by tumor cells with high P4HA1 significantly promoted the migration and invasion of tumor cells, indicating P4HA1 regulates SPP1 + macrophages through hypoxic pathways, thereby promoting tumor invasion and metastasis (Fig. 7). However, in the TME, tumor cells and macrophages interact to form a feedback loop, which promotes the malignant progression. So, whether inhibiting SPP1 in macrophages may affect P4HA1 expression in tumor cells, thereby affecting the hypoxic pathway in the TME, remains to be further explored.
Graphic abstract of P4HA1-mediated SPP1 + tumor-associated macrophages through activating the HIF-1α signaling pathway in HNSCC. The enhancement of P4HA1 affects the levels of α-KG and succinate, which in turn reduces PHD-induced hydroxylation of HIF-1α and leads to increasing its stabilization. This regulatory mechanism activates the hypoxic pathway, resulting in the upregulation of SPP1 expression in TAMs. The elevated secretion of SPP1 from TAMs binds to CD44 on tumor cells, facilitating their migration and invasion in HNSCC.
In this study, the integration of RNA-seq and scRNA-seq data revealed a positive correlation between the key factor P4HA1 and the infiltration of SPP1 + macrophages. While recent studies have increasingly utilized transcriptomic approaches to characterize the molecular features of SPP1 + macrophages, relying solely on transcriptomics remains insufficient to fully reflect the in vivo protein level and the protein activity situation within the complexity of the tumor microenvironment27,28,29,30. Therefore, multi-omics approaches, such as proteomics, metabolomics, epigenomics, and spatial transcriptomics, would be valuable to further validate the functional relationship between P4HA1 and SPP1 in HNSCC. Additionally, although we demonstrated that P4HA1 induces cellular hypoxia and HIF-1α stabilization in HNSCC tumor cells, which in turn increases the expression of SPP1 in TAMs at both the transcriptional and translational levels, the specific pathway by which P4HA1 regulates hypoxia and the exact mechanism of the secreted cytokines that result in the upregulation of SPP1 in TAMs require further investigation. Furthermore, while the in vivo experiments utilizing human cell lines in immunodeficient mice enabled us to evaluate cell-intrinsic mechanisms, they were unable to fully replicate the complexities of the human tumor immune microenvironment31, particularly limiting in-depth investigation of the interaction mechanisms between tumor cells and macrophages. Subsequent studies employing immunocompetent models or humanized patient-derived xenografts (PDXs)32 would help to confirm these findings in a more physiologically relevant context. Nonetheless, the current work provides a valuable foundation and clear direction for understanding the mechanisms of HNSCC progression.
In conclusion, we conducted a comprehensive bioinformatics analysis and in vitro and in vivo experiments to elucidate the role of the SPP1 + TAM subtype in the initiation and progression of HNSCC. Using WGCNA, we identified 14 genes that significantly correlated with SPP1 + macrophages and developed a clinical prognostic model using machine learning. Notably, P4HA1 played a critical role in inducing HIF-1α stabilization and tumor cell hypoxia, which in turn promoted the polarization of TAMs towards the SPP1 phenotype. These findings suggest that the biomarker P4HA1 is involved in targeting SPP1 + TAMs and is a promising therapeutic strategy for HNSCC.
Methods
Dataset source
RNA expression data and clinical annotation of The Cancer Genome Atlas (TCGA)-HNSCC were obtained from the UCSC Cancer Genome Atlas33. Additionally, RNA expression data of HNSCC (GSE4161334, GSE6585835, GSE11797336, and GSE663137) along with their corresponding clinical data were retrieved from the NCBI Gene Expression Omnibus (GEO) database. Raw scRNA-seq data for 52 patients of GSE23493313 were also acquired from NCBI GEO database. All datasets are summarized in Supplementary Table S1.
scRNA-seq data preparation and cell annotation
We processed the single-cell RNA sequencing data using Seurat (v4.3.0)38. Quality control filters were applied to remove low-quality cells (retaining those with 500-5,000 expressed genes and < 20% mitochondrial reads) and genes (requiring detection in ≥ 3 cells), resulting in 172,569 high-quality cells and 26,695 genes. After normalization, we identified the top 2,000 highly variable genes for downstream analysis. Dimensionality reduction was performed using Principal Component Analysis (PCA), followed by cell clustering with the FindNeighbors function, Uniform Manifold Approximation and Projection algorithm (UMAP)39 visualization. Cell types were annotated based on established marker genes from previous studies40.
Cellular deconvolution
The Bisque algorithm, implemented using the R package Bisque RNA (v1.0.5)41, was used to analyze scRNA-seq data from 8 macrophage subtypes and infer the infiltration proportion of macrophages TCGA-HNSCC cohort.
Survival analysis
Cox proportional hazards models (R survival v3.5-5) estimated the relative risk (RR) and 95% confidence intervals (CI) for cell infiltration and gene expression. Optimal stratification cutoffs were determined using the surv-cutpoint function (max-rank statistic), and Kaplan-Meier curves (survfit) compared Overall survival (OS) between groups, visualized with survminer (v0.4.9).
Construction of a WGCNA and selection of module genes
The initial phase of the analysis involved Spearman’s rank correlation to assess pairwise gene relationships using the WGCNA package (v1.69)42. We then determined the optimal soft-thresholding power to be 6, which was chosen to achieve a network topology that closely approximates a scale-free network, as evidenced by the index of scale-free fit and the average connectivity. The adjacency matrix was subsequently transformed into a topological overlap matrix43, which better reflected the intricate pattern of gene co-expression. We employed hierarchical clustering to identify modules of co-expressed genes, ensuring robustness and biological relevance by setting a minimum module size of 30.
Differential gene expression analysis
Differential analysis was performed using limma (v3.50.3)44 and visualized with ggVolcano (v2.8). Comparing 502 HNSCC and 44 normal samples (AveExpr > 10, |log2FC| >1, FDR < 0.05), we identified differentially expressed genes. The HNSCC cohort was then stratified by median P4HA1 expression, with SPP1 differences analyzed using relaxed thresholds (AveExpr > 5, |log2FC| >1, FDR < 0.05).
Development of a prognostic mode
We systematically evaluated twelve classical machine learning algorithms (Table S2) and an ensemble of 113 prediction models45 to construct clinical prognostic models for 14 SPP1 + macrophage-related genes. Using a rigorous 10-fold cross-validation framework, we calculated AUC values for all models across validation datasets, enabling comprehensive performance comparison. An ensemble consisting of glmboost and RF was identified as the optimal combination with a superior average performance score of 0.715.
GSVA and GSEA
Both Gene Set Variation Analysis (GSVA)46 and Gene Set Enrichment Analysis (GSEA) used the hallmark gene sets downloaded from the Broad Institute’s Molecular Signatures Database (MSigDB, GSEA, gsea-msigdb.org)47. Then the limma package was utilized to calculate the statistical significance between the two groups and the R package clusterProfile48 was employed to analysis GSEA.
PPI network
To assess the hypoxia-related genes mediated by P4HA1, the PPI network was constructed using STRING database (https://string-db.org/)49. Subsequently, the Cytoscape plugin CytoHubba was used to identify genes closely associated with P4HA1 within the PPI network50.
Cell transfection
Cells were added to six-well plates (1.5 × 105 cells per well) until they reached 70–80% confluence. Next, si-P4HA1 (A10001, GenePharma, Shanghai, China) was transfected into CAL27, SCC25 and FaDu cells using Lipofectamine 3000 Reagent (L3000015, Thermo Fisher Scientific, USA). The transfection process was carried out for a duration of 8 h at a temperature of 37 °C. Further experimental procedures were initiated 24 h post-transfection. The specific sequences for the siRNA are detailed in Table S3.
Stable cell line construction
We first generated lentivirus by transfecting 293T cells with P4HA1-targeting shRNA vectors using Lipofectamine 3000. After 48 h, we collected and filtered the viral supernatant, then used it to infect SCC25 cells in the presence of polybrene (6–8 µg/mL). Following another 48-hour incubation, we selected transduced cells with puromycin for 1–2 weeks to establish stable lines. The specific shRNA target sequences are provided in Table S4.
Cell cultures
The human tongue squamous carcinoma cell lines SCC25 (CVCL_1682) and CAL27 (CVCL_1107) and FaDu (CVCL_1218) were obtained from ATCC and cultured in DMEM, while THP-1 monocytes (CVCL_0006) from Cellcook were maintained in RPMI-1640. All cells were grown at 37 °C with 5% CO2 in medium supplemented with 10% FBS and 1% penicillin-streptomycin. For macrophage differentiation, THP-1 cells were treated with 100 nM PMA (Sigma 524400) for 24 h. Mycoplasma-free status was confirmed for all cell lines.
Migration and invasion assays
Cell migration and invasion assays were performed using 24-well transwell plates (8 μm pore size; Corning, NY, USA), with invasion assays using Matrigel-coated inserts (BD, NJ, USA). Cells (5 × 10³/well) in 250 µL serum-free medium were seeded in upper chambers, with 700 µL 20% FBS medium in lower chambers as chemoattractant. After 24 h incubation, non-migrated cells were removed. Migrated/invaded cells were fixed with 4% paraformaldehyde (Solarbio) for 20 min, stained with 1% crystal violet (Coolibo Technology, China) for 15 min, and counted in five random fields per membrane using an inverted microscope.
Wound healing assay
SCC25, CAL27 and FaDu cell lines were seeded in 6-well plates at a density of 5 × 105 cells per well. When the cell density reached 75–80%, a 200 µl pipette tip was used to create scratches in the cell monolayer. After washing with PBS, different groups (si-NC and si-P4HA1) of macrophage-conditioned media were added. Images were obtained using a light microscope (Olympus Corporation) at 0 and 24 h to record the wound area. Finally, the area of cell migration was quantified using Image-Pro Plus software.
RT-qPCR
Total RNA was extracted using RNAkey reagent (SM139-02, Seven, Beijing, China), followed by reverse transcription with SevenFast Two-Step RT&qPCR Kit (SM143-01, Seven). qPCR was performed using ChamQ Universal SYBR Green Master Mix (SM143-01, Seven) on a Dice Real-Time System (TP800, Takara, Japan), with GAPDH as reference. Reactions were run in triplicate, and relative expression was calculated using the 2^(-ΔΔCt) method. Primer sequences are listed in Table S5.
Western blotting
Cells were lysed on ice using lysis buffer (RIPAR0010; Solarbio), and protein concentration was determined with a dicysteine assay kit (BCAP0009; Beyotime, Shanghai, China). After denaturation (95 °C, 5 min), proteins were separated by SDS-PAGE and transferred to PVDF membranes (Millipore, Merck KGaA, Darmstadt, Germany). Membranes were blocked with 5% skim milk (1 h, RT), then incubated with primary antibodies (4 °C overnight) and HRP-conjugated secondary antibodies (1 h, RT). Protein bands were visualized using ECL (PE0010, Solarbio) and imaged with a FluorChem system (Bio-Rad Laboratories, Hercules, CA, USA). Antibody details are provided in Table S6.
ROS detection
Using a previously established transfection protocol, either si-Ctrl or si-PH4A1 was introduced into SCC25 or CAL27 cells and allowed to express for 48 h. After the original culture medium was removed, a medium containing a green fluorescent redox probe (CA1410,10 µmol/L, Solarbio) was added. After incubation at 37 °C for 30 min, the cells were rinsed with DMEM on three separate occasions. The Reactive Oxygen Species (ROS) level of tumor cells was detected and photographed under a fluorescence microscope at 10x magnification.
Hypoxia induction by CoCl2
SCC25 and CAL27 were seeded at a density of 5 × 105 cells/well in a 6-well plate. On the subsequent day, the culture medium was refreshed, followed by the addition of CoCl2 to reach a final concentration of 800µM for 48 h, thus creating a chemically induced hypoxic environment.
Animal experiment
To establish the tumor model, 15 female BALB/c nude mice (4 weeks, 18–20 g) from Dalian Medical University were randomized into sh-control, sh-P4HA1#1, and sh-P4HA1#2 groups (n = 4). SCC25 cells (5 × 106) transfected with respective shRNAs were suspended in 100 µl Matrigel and injected subcutaneously. Mouse weights were recorded biweekly (Table S7), and tumor volume was calculated (length×width²/2). Xenografts were harvested at day 28 post-injection for analysis. At the experimental endpoint, mice were intraperitoneally injected with an appropriate dose of tribromoethanol (250 mg/kg; TargetMol, Shanghai, China) based on their body weights. Following the confirmation of deep anesthesia, as determined by the absence of foot pinch and corneal reflexes, euthanasia was performed by cervical dislocation. The study is reported in accordance with ARRIVE guidelines. And all the animal experiments were approved by the Institutional Animal Care and Use Committee of Dalian Medical University (No. AEE23070).
Histological staining
We processed tumor tissue of mice by fixing in 10% formalin for 24 h followed by standard paraffin embedding. Six-micrometer sections were dewaxed and underwent antigen retrieval in citrate buffer. After blocking endogenous peroxidase activity with 3% H2O2 (10 min), we incubated sections with primary antibodies overnight at 4 °C and corresponding secondary antibodies for 1 h at room temperature. Detection was performed using DAB with hematoxylin counterstaining.
For immunofluorescence staining, antibody incubations were conducted at 37 °C (2 h for primaries, 1 h for secondaries) followed by DAPI nuclear staining. All images were acquired and analyzed using Image-Pro Plus software. Detailed antibody information is provided in Table S6.
Statistical analysis
All statistical analyses were performed using R (v4.1.3) and GraphPad Prism 7.0. Group comparisons used Wilcoxon test (two groups) or t-test, with p < 0.05 considered significant. Survival analysis employed log-rank test and Cox proportional hazards (Wald test). Correlations were assessed by Pearson’s test. Data are presented as mean ± SD.
Data availability
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Abbreviations
- AUC:
-
Area under the curve
- Cocl2:
-
Cobalt chloride 2
- GEO:
-
Gene expression omnibus
- GSEA:
-
Gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- HNC:
-
Head and neck cancers
- HIF-1α:
-
Hypoxia-inducible factor 1 alpha
- P4HA1:
-
Prolyl 4-hydroxylase subunit alpha 1
- PLS-GLM:
-
Partial least squares generalized linear models
- PCA:
-
Principal component analysis
- PDXs:
-
Patient-derived xenografts
- ROS:
-
Reactive oxygen species
- SVM:
-
Support vector machine
- SPP1:
-
Secreted phosphoprotein 1
- SGLM:
-
Stepwise generalized linear model
- TAMs:
-
Tumor-associated macrophages
- TME:
-
Tumor microenvironment
- TCGA:
-
The cancer genome atlas
- UMAP:
-
Uniform manifold approximation and projection algorithm
- WGCNA:
-
Weighted gene co-expression network analysis
- PHDs:
-
Oxygen-dependent dioxygenases
- α-KG:
-
Alpha-ketoglutarate
- CA9:
-
Carbonic anhydrase 9
- CM:
-
Confusion matrices
- ROC:
-
Receiver operating characteristic curve
References
Chow, L. Q. M. Head and neck cancer. N Engl. J. Med. 382 (1), 60–72. https://doi.org/10.1056/NEJMra1715715 (2020).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. https://doi.org/10.3322/caac.21660 (2021).
Ferlay, J. et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 144 (8), 1941–1953. https://doi.org/10.1002/ijc.31937 (2019).
Fitzmaurice, C. et al. Global, Regional, and National cancer Incidence, Mortality, years of life Lost, years lived with Disability, and Disability-Adjusted life-years for 32 cancer Groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 3 (4), 524–548. https://doi.org/10.1001/jamaoncol.2016.5688 (2017).
Baharom, F., Hermans, D., Delamarre, L. & Seder, R. A. Vax-Innate: improving therapeutic cancer vaccines by modulating T cells and the tumour microenvironment. Nat. Rev. Immunol. 25 (3), 195–211. https://doi.org/10.1038/s41577-024-01091-9 (2025).
Ghebremedhin, A. et al. Tumor-Associated macrophages as major immunosuppressive cells in the tumor microenvironment. Cancers (Basel). 16 (19). https://doi.org/10.3390/cancers16193410 (2024).
Zhu, Y. et al. Tissue-Resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 47 (2), 323–338. https://doi.org/10.1016/j.immuni.2017.07.014 (2017). .e326.
Wu, K. et al. Redefining tumor-Associated macrophage subpopulations and functions in the tumor microenvironment. Front. Immunol. 11, 1731. https://doi.org/10.3389/fimmu.2020.01731 (2020).
Nalio Ramos, R. et al. Tissue-resident FOLR2(+) macrophages associate with CD8(+) T cell infiltration in human breast cancer. Cell 185 (7), 1189–1207e1125. https://doi.org/10.1016/j.cell.2022.02.021 (2022).
Shi, Y. et al. Decoding the multicellular ecosystem of Vena caval tumor thrombus in clear cell renal cell carcinoma by single-cell RNA sequencing. Genome Biol. 23 (1), 87. https://doi.org/10.1186/s13059-022-02651-9 (2022).
Pang, X. et al. SPP1 promotes enzalutamide resistance and Epithelial-Mesenchymal-Transition activation in Castration-Resistant prostate cancer via PI3K/AKT and ERK1/2 pathways. Oxid. Med. Cell. Longev. 2021, 5806602. https://doi.org/10.1155/2021/5806602 (2021).
Zhang, Y., Du, W., Chen, Z. & Xiang, C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp. Cell. Res. 359 (2), 449–457. https://doi.org/10.1016/j.yexcr.2017.08.028 (2017).
Bill, R. et al. CXCL9:SPP1 macrophage Polarity identifies a network of cellular programs that control human cancers. Science 381 (6657), 515–524. https://doi.org/10.1126/science.ade2292 (2023).
Fan, G. et al. Single-cell and Spatial analyses revealed the co-location of cancer stem cells and SPP1 + macrophage in hypoxic region that determines the poor prognosis in hepatocellular carcinoma. NPJ Precis Oncol. 8 (1), 75. https://doi.org/10.1038/s41698-024-00564-3 (2024).
Xu, Y., Xia, D., Huang, K. & Liang, M. Hypoxia-induced P4HA1 overexpression promotes post-ischemic angiogenesis by enhancing endothelial Glycolysis through downregulating FBP1. J. Transl Med. 22 (1), 74. https://doi.org/10.1186/s12967-024-04872-x (2024).
Zhou, H. et al. Comprehensive analysis revealed P4Hs as new biomarkers for prognosis and immunotherapy in head and neck cancer. Sci. Rep. 14 (1), 12234. https://doi.org/10.1038/s41598-024-62678-9 (2024).
Cao, X. P., Cao, Y., Li, W. J., Zhang, H. H. & Zhu, Z. M. P4HA1/HIF1α feedback loop drives the glycolytic and malignant phenotypes of pancreatic cancer. Biochem. Biophys. Res. Commun. 516 (3), 606–612. https://doi.org/10.1016/j.bbrc.2019.06.096 (2019).
Xiong, G. et al. Collagen Prolyl 4-hydroxylase 1 is essential for HIF-1α stabilization and TNBC chemoresistance. Nat. Commun. 9 (1), 4456. https://doi.org/10.1038/s41467-018-06893-9 (2018).
Lin, Y., Xu, J. & Lan, H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J. Hematol. Oncol. 12 (1), 76. https://doi.org/10.1186/s13045-019-0760-3 (2019).
Kürten, C. H. L. et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 12 (1), 7338. https://doi.org/10.1038/s41467-021-27619-4 (2021).
Gao, J., Liang, Y. & Wang, L. Shaping polarization of Tumor-Associated macrophages in cancer immunotherapy. Front. Immunol. 13, 888713. https://doi.org/10.3389/fimmu.2022.888713 (2022).
Xu, Y. et al. TRAF2 promotes M2-polarized tumor-associated macrophage infiltration, angiogenesis and cancer progression by inhibiting autophagy in clear cell renal cell carcinoma. J. Exp. Clin. Cancer Res. 42 (1), 159. https://doi.org/10.1186/s13046-023-02742-w (2023).
Chen, L. et al. Human prolyl-4-hydroxylase alpha(I) transcription is mediated by upstream stimulatory factors. J. Biol. Chem. 281 (16), 10849–10855. https://doi.org/10.1074/jbc.M511237200 (2006).
Paredes, F. & Williams, H. C. San Martin, A. Metabolic adaptation in hypoxia and cancer. Cancer Lett. 502, 133–142. https://doi.org/10.1016/j.canlet.2020.12.020 (2021).
Jaśkiewicz, M. et al. The transition from HIF-1 to HIF-2 during prolonged hypoxia results from reactivation of phds and HIF1A mRNA instability. Cell. Mol. Biol. Lett. 27 (1), 109. https://doi.org/10.1186/s11658-022-00408-7 (2022).
Yan, Q. et al. Tumor-associated macrophage-derived Exosomal miR21-5p promotes tumor angiogenesis by regulating YAP1/HIF-1α axis in head and neck squamous cell carcinoma. Cell. Mol. Life Sci. 81 (1), 179. https://doi.org/10.1007/s00018-024-05210-6 (2024).
Xu, L. et al. Tumor-associated macrophage subtypes on cancer immunity along with prognostic analysis and SPP1-mediated interactions between tumor cells and macrophages. PLoS Genet. 20 (4), e1011235. https://doi.org/10.1371/journal.pgen.1011235 (2024).
Cui, X., Liu, S., Song, H., Xu, J. & Sun, Y. Single-cell and Spatial transcriptomic analyses revealing tumor microenvironment remodeling after neoadjuvant chemoimmunotherapy in non-small cell lung cancer. Mol. Cancer. 24 (1), 111. https://doi.org/10.1186/s12943-025-02287-w (2025).
Matsubara, E. et al. SPP1 derived from macrophages is associated with a worse clinical course and chemo-resistance in lung adenocarcinoma. Cancers (Basel). 14 (18). https://doi.org/10.3390/cancers14184374 (2022).
Matsubara, E. et al. The significance of SPP1 in lung cancers and its impact as a marker for protumor tumor-associated macrophages. Cancers (Basel). 15 (8). https://doi.org/10.3390/cancers15082250 (2023).
Ingersoll, M. A. et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115 (3), e10-19. https://doi.org/10.1182/blood-2009-07-235028 (2010).
Invrea, F. et al. Patient-derived xenografts (PDXs) as model systems for human cancer. Curr. Opin. Biotechnol. 63, 151–156. https://doi.org/10.1016/j.copbio.2020.01.003 (2020).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38 (6), 675–678. https://doi.org/10.1038/s41587-020-0546-8 (2020).
Lohavanichbutr, P. et al. A 13-gene signature prognostic of HPV-negative OSCC: discovery and external validation. Clin. Cancer Res. 19 (5), 1197–1203. https://doi.org/10.1158/1078-0432.Ccr-12-2647 (2013).
Wichmann, G. et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int. J. Cancer. 137 (12), 2846–2857. https://doi.org/10.1002/ijc.29649 (2015).
Mock, A. et al. EGFR and PI3K pathway activities might guide drug repurposing in HPV-Negative head and neck cancers. Front. Oncol. 11, 678966. https://doi.org/10.3389/fonc.2021.678966 (2021).
Keck, M. K. et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin. Cancer Res. 21 (4), 870–881. https://doi.org/10.1158/1078-0432.Ccr-14-2481 (2015).
Stuart, T. et al. Comprehensive integration of Single-Cell data. Cell 177 (7), 1888–1902e1821. https://doi.org/10.1016/j.cell.2019.05.031 (2019).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. https://doi.org/10.1038/nbt.4314 (2018).
Choi, J. H. et al. Single-cell transcriptome profiling of the Stepwise progression of head and neck cancer. Nat. Commun. 14 (1), 1055. https://doi.org/10.1038/s41467-023-36691-x (2023).
Jew, B. et al. Accurate Estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 11 (1), 1971. https://doi.org/10.1038/s41467-020-15816-6 (2020).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559. https://doi.org/10.1186/1471-2105-9-559 (2008).
Botía, J. A. et al. An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks. BMC Syst. Biol. 11 (1), 47. https://doi.org/10.1186/s12918-017-0420-6 (2017).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. https://doi.org/10.1093/nar/gkv007 (2015).
He, H. et al. Combined analysis of single-cell and bulk RNA sequencing reveals the expression patterns of circadian rhythm disruption in the immune microenvironment of alzheimer’s disease. Front. Immunol. 14, 1182307. https://doi.org/10.3389/fimmu.2023.1182307 (2023).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14 (7). https://doi.org/10.1186/1471-2105-14-7 (2013).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell. Syst. 1 (6), 417–425. https://doi.org/10.1016/j.cels.2015.12.004 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 102 (43), 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607-d613. https://doi.org/10.1093/nar/gky1131 (2019).
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27 (3), 431–432. https://doi.org/10.1093/bioinformatics/btq675 (2011).
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81802706), the Basic Scientific Research Project of Educational Department of Liaoning Province (Grant No. JYTMS20230569) and the Natural Science Foundation of Liaoning Province (Grant No. 2023-MSLH-024).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Yiding Liu, Bing Yan and Hui Dong conceived the study, ordered reagents, designed and conducted experiments, analyzed data, and wrote the manuscript. Yi Li and Yuying Zhang performed experiments and analyzed data. Yi Wang performed a portion of the cell experiments. Lu Gao and Ni Kou are the guarantors of this work and responsible for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
All the procedures were conducted following the ARRIVE guidelines. And this study was approved by the Ethics Committee of Dalian Medical University (Approval No. 2023004).
Animal ethics statement
All the animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Dalian Medical University (No. AEE23070).
Consent for publication
Consent to publish has been obtained from all authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Yan, B., Dong, H. et al. P4HA1 facilitates SPP1+ tumor-associated macrophages by activating the hypoxia pathway in head and neck squamous cell carcinoma. Sci Rep 16, 356 (2026). https://doi.org/10.1038/s41598-025-29651-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-29651-6