Abstract
Kidney renal cell carcinoma (RCC) is a highly invasive malignancy, where early-stage diagnosis can significantly reduce treatment complexity and mortality risk. This study aims to enhance diagnostic precision through a novel model, RCC-ProbNet, capable of identifying and classifying RCC across various stages. We introduce RCC-ProbNet, a hybrid deep learning model that incorporates an initial stage where features are extracted from medical imaging data, and a subsequent construction of a probabilistic feature model. This hybridized method could allow a more granular feature representation through incorporating a variety of features and enhance the model’s ability to differentiate different RCC stages. The model is integrated with a Logistic Regression (LR) classifier for the final stage prediction. We validate performance using k-fold cross-validation. Overall, we obtain a notable diagnostic accuracy of 99.93% by RCC-ProbNet + LR, surpassing the current state-of-the-art techniques. Next, the model shows strong stability in different cross-validation folds. The comparative results also demonstrate that our model outperforms traditional and deep learning methods for RCC classification tasks consistently. The outstanding performance and the invulnerability of RCC-ProbNet + LR make it a powerful classifier for early RCC screening and staging in clinical practice. Its high performance indicates it has the potential to assist in timely and personalised treatments. Its future iterations may broaden the use toward real-time clinical decision support systems and consequently optimise patients’ outcomes regarding renal oncology.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) accounts for approximately 90% of kidney cancers in adults1. It originates in the renal tubules of the kidney, which are responsible for filtering blood and producing urine. RCC begins when genetic mutations cause cells in the renal tubules to grow abnormally, forming masses or tumors that may expand locally and, if left untreated, metastasize to other parts of the body2. The most common subtype, clear cell RCC, appears as pale or clear cells under a microscope and is often more aggressive than other variants such as papillary RCC or chromophobe RCC3 . Risk factors for RCC include smoking, obesity, hypertension, and certain genetic conditions, such as von Hippel-Lindau disease. Symptoms may be subtle and can include hematuria (blood in the urine), unexplained back pain, fatigue or abdominal enlargement4. Diagnosis primarily involves imaging modalities like CT scan and MRI, while treatment choices are based on its stage and spread5. They could include surgery and targeted therapy, immunotherapy or – for advanced disease – systemic treatment designed to suppress metastasis. Early diagnosis has a significant positive impact on patient survival making routine screening all the more important, especially among people who are at a higher risk.
Renal cell carcinoma (RCC) is a deadly kidney cancer and it is a life threatening disease which has rapidly spreading consequences if not get diagnosed early6. The danger is that it can silently grow, usually without unusual symptoms7 until the damage is significant. By then it may have already spread to other parts of the body, including the lung, bone or liver, and hence, treatment becomes a lot more challenging and less effective8. Advanced RCC is characterized by severely reduced survival rates, with limited treatment options and disease control. Early detection, however, significantly increases the chances of a patient’s survival9. When diagnosed in the early stages, RCC is localised starting in the kidney and can generally be cured with a surgical resection of the tumour These surgeries can sometimes be curative10. Early detection results in an increase in life expectancy and a decrease in require for drastic treatment, improving the quality of life for these patients.
Current techniques, enhanced such as deep learning and machine learning, have7 the detection of RCC by increasing its efficiency, accuracy, and stage detectionrecognising. Some of the latest advances in the fields of deep learning and, more particularly, convolutional neural networks (CNNs)11 are capable of dissecting Medical images such as Computerized tomography (CT), functional magnetic resonance imaging (fMRI) scans where they might pick out the tumor characteristics that are rather unnoticed by those skilled in radiology. These models8 are capable of learning intricate patterns of disease states within imaging data so that they can distinguish between malignant and benign tissue, and even between the subtypes of RCC. It is supported by machine learning algorithms by processing patient data and recognizing the probabilities of deterioration of the disease. Unlike in traditional diagnoses that only use patient demographic data, the techniques use genetic markers, histological data, resulting in early diagnosis and hence treatment at a lower stage, resulting in better results. Furthermore, these methods can also be used to track cancer over time, and in doing so, the doctors are then able to determine treatment effectiveness and patient management. Consequently, machine learning and deep learning are emerging as a critical landscape across RCC diagnosis, improvement of staging assessments, and enabling improved management of RCC.
In this paper, we recommend an efficient workflow for feature extraction and probabilistic analysis in medical imaging using VGG-19 and a hybrid model of CNN. This motivation forms the basis for the entire study and should be made evident outright. VGG-19’s deep hierarchical architecture is good at capturing detailed spatial features (e.g., textures and shapes of tissues), which are important to differentiate between regions that are healthy versus potentially diseased. These spatial aspects are particularly useful in medical imaging, where slight differences can mean the presence of a disease or disease progression. We do not use spatial information only; we transform it to the probabilistic space by a hybrid method. Embracing both CNN and VGG-19, our model is capable of capturing high-level semantics, and meanwhile predicting probabilities for powerfully categorizing the stage/subtype of disease. This ability is particularly advantageous for RCC, which requires precise staging to effectively treat the patient. Our proposed mixed model not only has better detection performance than some popular models in outputting probabilities for each stage of RCC, but also can serve as a foundation for clinical decisions. Furthermore, this system is potentially scalable for use in other medical imaging applications and could help form the basis for AI-driven diagnostic tools in healthcare.
Key contributions
The key contributions of this manuscript are summarized as follows:
-
RCC-ProbNet We propose RCC-ProbNet, a novel deep learning framework for accurate diagnosis and stage classification of renal cell carcinoma (RCC). The model jointly leverages spatial representations from convolutional encoders and uncertainty-aware probabilistic descriptors to enhance stage discrimination.
-
Adaptive probabilistic fusion strategy Unlike naive concatenation, we introduce an uncertainty-driven fusion mechanism in which spatial embeddings are projected into a shared latent space and adaptively weighted by confidence measures derived from the probabilistic branch (e.g., entropy-calibrated softmax and Bayesian dropout). This approach enhances robustness against noisy and ambiguous samples.
-
Robust evaluation protocol The proposed model is rigorously evaluated using k-fold cross-validation and compared against state-of-the-art ML and DL baselines. Results demonstrate superior accuracy, stability, and generalization across tumor stages and heterogeneous data conditions.
-
Generalizability and clinical applicability The proposed probabilistic fusion paradigm is architecture-agnostic and can be extended to other medical imaging tasks, making RCC-ProbNet a flexible and scalable framework for clinical decision support.
The structure of this study is outlined as follows: Section II reviews previous approaches, while Section III details the proposed methodology. Section IV analyses and discusses the experimental findings. Finally, Section V presents the conclusions drawn from the study.
Past research work
This review aims to provide a comprehensive overview of the current knowledge on the topic, focusing on identifying gaps, trends, and emerging themes that form the basis for the present study. Table 1 presents the research conducted in previous studies.
Hybrid model role in kidney cancer detection
Renal cell carcinoma (RCC) is a neoplasm originating in the renal cortex, the outer layer of the kidney, and includes several subtypes such as clear cell, papillary, and chromophobe renal cell carcinomas12. While detection is typically performed through imaging, differentiating between these subtypes remains a significant diagnostic challenge. To address these difficulties, Arthur has proposed a computational model called Kidney Ensemble-Net, designed to enable accurate classification of renal carcinoma subtypes. Kidney Ensemble-Net employs a Convolutional Neural Network (CNN) to extract two types of features–primarily spatial features from contrast-enhanced images–that capture fine-grained patterns associated with each carcinoma subtype. These spatial features are subsequently transformed into a probabilistic feature set, forming the foundation for an ensemble model that combines the strengths of Logistic Regression (LR), Random Forest (RF), and Gaussian Naïve Bayes (GNB) classifiers. The objectives of Kidney Ensemble-Net were achieved by integrating these models within a unified framework, where the Kidney Ensemble-Net + LR configuration achieved an impressive accuracy of 99.72%, demonstrating the superiority of the proposed methodology over existing approaches. Furthermore, the application of higher-order k-fold cross-validation reinforced the model’s robustness and generalizability across diverse datasets.
In the contemporary technological era, the diagnosis of illnesses plays a critical role in improving recovery rates among patients undergoing treatment13. Renal carcinoma is a common disease in the adult population, and early diagnosis significantly enhances treatment outcomes. However, traditional diagnostic methods, such as blood analysis and CT scan assessments performed by nephrologists, while accurate, tend to be time-consuming and labour-intensive. To address these limitations, researchers have increasingly focused on developing automated kidney tumour detection systems using machine learning (ML) and deep learning (DL) techniques. Models such as Convolutional Neural Networks (CNNs) and hybrid neural-fuzzy inference systems have been explored for both detection and segmentation of kidney tumours. Recent studies have also proposed combining CNNs with ML algorithms, yielding promising results in terms of imaging and tumour segmentation. One such approach demonstrated impressive performance, achieving 98.69% accuracy in tumour classification and a Dice coefficient of 0.8923 for segmentation, making it comparable to traditional diagnostic techniques, but significantly more efficient.
Role of deep learning in kidney cancer detection
Classical histology image-based analysis, considered the clinical gold standard for diagnosis, is nonetheless highly labour-intensive and manual8. As a result, there is growing interest in computer-aided diagnostic (CAD) applications that can assist pathologists in improving diagnostic efficiency and accuracy. In this context, deep learning (DL) has emerged as a promising approach. By leveraging the complementary strengths of these models, the ensemble approach enhances classification performance and enables more accurate and robust detection of kidney cancer. This work evaluates the proposed methods on both synthetic and publicly available benchmark datasets, including the Dartmouth Kidney Cancer Histology Dataset. The findings reveal that Vision Transformers (ViTs) achieved high detection accuracy–up to 99% average accuracy–even when applied to full-slide image patches. Specifically, transformer models such as CAiT, DeiT, ViT, and Swin outperformed traditional architectures like ResNet. Overall, the Vision Transformer models consistently achieved an average accuracy of 98.51% across five folds, demonstrating their strong generalization capabilities and effectiveness in extracting meaningful features from small histopathological image patches. These results indicate that the use of Vision Transformers in histopathological image analysis could significantly aid pathologists in diagnosing kidney cancer more accurately and earlier, thereby improving patient survival outcomes.
This study addresses the gap between conventional evaluation methods and deep learning approaches by demonstrating how the latter can significantly reduce diagnostic time, increase test accuracy, lower costs, and alleviate the workload on radiologists14. Clinicians sometimes struggle to distinguish between different types of kidney cancers in renal CT images, which can be critical during surgical planning, as the complexity and severity of the tumor influence treatment decisions. In this research, raw CT images are retrieved from conventional archives and processed using the developed 3D-Trans-Residual DenseUNet++ (3D-TR-DUnet++) model for kidney segmentation. This process assists clinicians in identifying normal versus pathological regions of the kidney. The segmented images are then passed to the classification stage, for which the study introduces a novel deep learning model called Adaptive and Attentive Residual DenseNet with Gated Recurrent Unit (AA-RD-GRU). Additionally, the ACC-MVNB configuration is optimised using the Modified Crayfish Optimisation Algorithm (MCOA) to enhance performance. The proposed strategy achieved a success rate of 97% on the first sample and 98% on the second, highlighting its effectiveness in improving diagnostic precision and decision-making in clinical kidney cancer analysis.
This paper presents a recent development15 of a method called CIDL-DSSKC (Convolutional Image Deep Learning with Dual Subspace Sparse Kernel Classifier) designed to aid the identification and detection of kidney cancer using renal CT images. The CIDL-DSSKC method involves several sub-procedures, including image pre-processing, feature extraction using the Xception model, parameter optimization with the Flower Pollination Algorithm (FPA), and pattern identification through a Variational Autoencoder. In this framework, FPA plays a crucial role in fine-tuning the parameters of the Xception model, thereby improving the overall system performance. The proposed CIDL-DSSKC approach was tested on a renal image dataset and demonstrated superior performance compared to existing state-of-the-art methods for kidney cancer detection. Comparative analysis further confirms the enhanced capability of the CIDL-DSSKC method. For future work, exploring more advanced ensemble learning techniques could provide additional improvements in classification accuracy, thereby increasing the effectiveness of the system for kidney cancer detection. The proposed approach achieved a performance score of 97.73
Role of machine learning in kidney cancer detection
The kidneys are vital organs involved in the filtration and homeostasis of body fluids in humans17. However, tumors originating in the kidney–caused by uncontrolled cell growth, can impair kidney function and, if not detected early, may prove fatal. Traditionally, diagnosis relies on manual inspection of medical images (MRI) by radiologists, which can lead to variable accuracy depending on the expertise of the detecting radiologist. To address these challenges, recent studies have explored the use of machine learning (ML) techniques to improve the accuracy and speed of renal tumor diagnosis. In one study, authors attempted to discriminate between normal and diseased kidney conditions using public datasets and tested classifiers such as SVM, AO, GB, and KNN. Feature extraction was performed using a dimensionality reduction approach to reduce model training time and enhance classifier performance. Among these, the KNN model achieved the highest diagnostic accuracy of 93.3% for the normal class (control kidney models), but its sensitivity for the abnormal (tumour) class was relatively low at 51.0%. Similarly, GB, ADA, and SVM models showed poor performance, approximately 55% accuracy, in tumour detection. These results highlight both the potential and the limitations of traditional ML methods for renal tumor classification and emphasize the need for more sophisticated approaches to provide reliable diagnostic predictions.
Persistent kidney cancer causes deterioration of the kidney’s circulatory system and has shown an increasing incidence in recent years16. Since chronic kidney cancer (CKC) often presents with non-specific symptoms, especially in its early stages, identifying efficient predictors for early detection is crucial. The goal of this research is to develop a machine learning-based approach for screening early-stage kidney cancer using several methods, including SVM, Decision Tree, Random Forest, KNN, Naïve Bayes, Logistic Regression, LGBM, CatBoost, and AdaBoost. Among these models, Random Forest (RF), AdaBoost (AB), and LGBM demonstrated strong performance, with an accuracy of up to 99% reported for numeric features. The study also employed data pre-processing and feature selection techniques to further enhance model accuracy. Consequently, the proposed method proved to be more efficient and accurate compared to eight other tested approaches. Additionally, predictions generated by the best-performing model have been integrated into a user-friendly web application, increasing the practical relevance and accessibility of this work in real-world medical settings.
This article presents the EFS-MLC model18 which has been used to select the best feature selection and classification algorithms to build an accurate COVID-19 prediction model. In this research work, six feature selection approaches–Chi-square, Recursive Feature Elimination (RFE), Genetic Algorithm (GA), Particle Swarm Optimization (PSO), and Random Forests–were used to extract the important features from COVID-19 datasets. The aforementioned features contribute to the performance of machine learning classifiers such as Decision Tree (DT), Naïve Bayes (NB), K-Nearest Neighbours (KNN), Multi-Layer Perceptron (MLP), and Support Vector Machine (SVM). Of these, the highest accuracy on the same dataset was obtained with the KNN model with the ensemble feature selection method. More specifically, the classifiers used in this system with the ensemble feature selection technique achieved an average accuracy of 88% based on using the symptom-based), and with all applications imposing the need to add forensic evidence to trace COVID-19 presence. The difficulties in solving these two databases were further addressed by empirical experiments, which show that the EFS-MLC system can discover the optimal features that affect the classifier’s behaviour. This demonstrates the utility of the ensemble feature selection method to enhance COVID-19 identification models.
Research gaps and limitations
After reviewing previous research, we discovered some gaps.
-
Although significant progress has been made in the detection of renal cell cancer (RCC), state-of-the-art models frequently suffer from the inability to capture spatial and probabilistic subtleties required for accurate classification of multistage cancer cases. All those methods, commonly based on binary classification or single feature extraction, also result in the weakness of the inclusiveness and the staging precision of those methods. To solve this, we introduce an RCC-ProbNet model that integrates both spatial and probabilistic information for enhanced RCC early and detailed staging diagnosis, and that provides an important basis for precise medical imaging diagnosis.
Proposed methodology
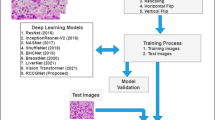
Our work begins with importing a dataset containing information on kidney renal cancer, which is fundamental for the subsequent analyses. In terms of image processing, enhancement methods are applied to adjust contrast, sharpness, and reduce noise, aiming to capture as many critical features of the images as possible. Figure 1 illustrates the methodology we adopted. This enhancement phase is designed to increase the utility of the images for the model, aiding in feature learning. Subsequently, we use a machine learning and deep learning-based model uniquely designed to extract newly generated probabilistic features from the data. This model integrates spatial feature information with probabilistic transformations, allowing for finer differentiation of the images–an important factor in distinguishing between cancer stages. The data was then split into training and testing sets, with 80% allocated for training and 20% for testing, to improve model performance evaluation and generate accurate performance estimates. We evaluated several candidate models to identify the one with the highest classification accuracy. Testing on the selected model enabled us to achieve highly accurate classification of the different stages of kidney cancer. This approach contributes not only to more precise staging but also to earlier detection, thereby facilitating more effective individualised treatment.
Kidney renal cell cancer image dataset
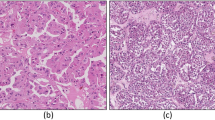
This study employs a publicly accessible benchmark dataset19 containing 277 images, specifically curated for the detection of kidney renal carcinoma. The dataset is organised into five classes, each representing a distinct stage of cancer progression: Stage 0, Stage 1, Stage 2, Stage 3, and Stage 4. Stage 0 corresponds to noncancerous cells, while Stages 1 through 4 represent increasing levels of cancer severity. A bar plot illustrating the class distribution is presented in Fig. 2, allowing for a comprehensive analysis of cancer progression across stages. This dataset structure effectively distinguishes between non-tumour and malignant stages, while providing valuable information about the gradual progression of the disease.
Image preprocessing
Our first approach uses simple image processing methods to accelerate the characterization of different stages of kidney renal cell cancer. The image analysis procedure is shown in Fig. 3 after pre-processing. This important first step preprocesses the input images for high quality and exposes all features required to identify cancer. The quality of the image is improved and standardized by a sequence of preprocessing steps, including image denoising, contrast stretching and normalization. These enhancements allow for better disease character progression to be detected and enhance the ability to distinguish between different cancer stages. This method provides an informative basis for further studies and model fitting leading to the accurate identification of kidney cancer.
Proposed fusion mechanism and classifier design
In this paper, we propose RCC-ProbNet, which is formulated to specifically detect kidney renal cell cancer (RCC) by using deep learning and probabilistic features. RCC-ProbNet begins with spatial feature extraction using the VGG-19 architecture, as illustrated in Fig. 4. VGG-19, which consists of 19 convolutional layers, is particularly effective in generating high-level feature representations from imaging data. This capability is crucial given the detailed structures present in medical images of RCC. The algorithm 1 shows step by step feature engineering process. In the next step, RCC-ProbNet incorporates a probabilistic model centered around the CNN-based VGG-19. The convolutional neural networks employed in RCC-ProbNet leverage the spatial depth of VGG-19 for feature extraction. This hybrid approach not only enhances the network’s ability to encode spatial hierarchies but also transforms these spatial features into a probabilistic feature space. These modifications improve the model’s interpretability, reduce sensitivity to random fluctuations, and emphasize relationships between features that are most relevant for RCC detection. For better model selection, we apply a range of machine learning algorithms on the probabilistic feature set, performing a thorough feasibility assessment based on key performance indicators, including Accuracy, Precision, and Recall. This systematic evaluation allows us to benchmark models and select only the most suitable one for RCC detection. Beyond improving diagnostic accuracy, RCC-ProbNet leverages novel spatial and probabilistic feature extraction techniques combined with reliable model assessment, providing a stable and interpretable solution for fully automated RCC classification. This approach has the potential to enhance early diagnosis and intervention in clinical practice.
In RCC-ProbNet, we define probabilistic features as uncertainty-aware representations extracted via Monte Carlo (MC) Dropout applied to a VGG19-based feature extractor. Specifically, during inference, dropout layers are retained and multiple stochastic forward passes are performed on the same input image. Depth-wise dropout layers are kept in place as they would during inference, numerous stochastic forward passes performed over the identical input image. We also generate a distribution over activations from this process, and take the mean feature vector. This anticipated feature representation provides the model with a measure of its predictive uncertainty and is employed as the ultimate probabilistic feature vector. Unlike traditional probabilistic models like Gaussian Mixture Models (GMMs), this is principled in Bayesian Deep Learning, where the dropout serves as a variational approximation to deep Gaussian processes. Such probabilistic elements augment the spatial features learned from the CNN branch, making the model more reliable for representing various and ambiguous tissue patterns in RCC images.
Probabilistic feature modeling
In RCC-ProbNet, probabilistic features are extracted to capture the uncertainty and distributional characteristics of pixel intensities and spatial regions within tissue images. Specifically, we model the intensity values within a given image region as random variables and characterize their distribution using probability density functions (PDFs).
Let \(X = \{x_1, x_2, \dots , x_n\}\) represent the set of pixel intensities in a localized region of the tissue image. We assume that these intensities follow a certain probability distribution \(P(X \mid \theta )\), parameterized by \(\theta\) (e.g., mean and variance for a Gaussian distribution). For example, if we model \(X\) as Gaussian distributed:
where \(\mu\) and \(\sigma ^2\) denote the mean and variance of the pixel intensities, respectively.
From this distribution, probabilistic features such as the expected value \(E[X]\), variance \(\textrm{Var}[X]\), and higher-order moments can be extracted:
These probabilistic statistics capture essential texture and heterogeneity characteristics of the tissue, which are often indicative of malignancy stages. Furthermore, RCC-ProbNet combines these probabilistic features with spatial features derived from convolutional layers, which allows the model to benefit from both local spatial context and distributional uncertainty for more accurate classification. Introducing probability distributions instead of deterministic pixel values adds robustness against noise and variances typical to medical imaging data.
Applied learning schemes
In this study, a number of ML and DL approaches were explored to evaluate the efficiency of these models in diagnosing RCC. We applied a wide range of models, including conventional ML and DL methods, to identify the most optimal method for classifying RCC. To be specific, we used Convolutional Neural Networks (CNN), VGG-19, Stochastic Gradient Descent (SGD), Random Forest (RF), Logistic Regression (LR), Naive Bayes (NB), k-Nearest Neighbors Classifier (KNC), and Gaussian Naive Bayes (GNB). Through the benchmarking of different models with key indexes to evaluate the effects, we conducted a comprehensive comparison to select the suitable models capable enough to manage the various characteristics of RCC, so as to guarantee a high accuracy and effectiveness of the model for the detection task.
-
Convolutional Neural Network To detect and classify several different stages of kidney cancer, with the help of renal cell images captured from contrast-enhanced scans, we developed a CNN model20. The CNN is designed to learn specific visual cues for each stage of the evolution of the disease, which are essential patterns in tissue density, tumour boundaries, and vascular structures. With multiple convolution and pooling layers, the model learns effective stage-dependent features to differentiate the early, intermediate, and advanced stages of renal cell carcinoma. Using a cancer stage-labeled training dataset, the CNN can accurately classify it, correlated to diagnosis or early detection indications. This stage-directed focus helps the physician to tailor different treatment modalities according to the extent of cancer spread, resulting in more precise and efficient patient treatment.
-
VGG-19 For identifying and classifying21 varying levels of kidney cancer, a VGG-19 model is used with renal cell cancer images from contrast-enhanced scans. Renowned for its deep architecture, VGG-19 uses 19 layers to extract higher-level spatial features, which are fundamental for discriminating between the stages of cancer. The model achieves this by processing images of renal cells through a series of convolution and pooling layers to which extract density and tumor boundary at the level of early, intermediate, and advanced cancer cascade into features. Then, after training on an imageset with stage annotations, VGG-19 repeatedly encounters patterns that are cues to the stages and has satisfactory performance in stage classification of renal cell carcinomas. The feature selection and classification introduced in this paper help to stage the lung cancer and provide a wealth of information about cancer development to the doctor. Finally, early detection accuracy in kidney cancer patients is improved, and the patients are diagnosed, and then a reasonable treatment scheme is developed.
-
Gaussian Naive Bayes In22, a GNB model is used to differentiate stages of kidney cancer by renal cell kidney images in contrast-enhanced scans. This probabilistic model uses the statistical distribution of pixel intensities and spatial structures that exist in the state-labelled renal cell images. GNB assumes Gaussian distributed features for each stage of cancer; therefore, it can readily differentiate early, intermediate, and advanced stages based on changes in tissue density & tumour morphology. The probability for an image being in one of the stage classes is estimated, then attributed to the most likely stage. Considering the efficiency of computational cost and the capability of high-dimensional data processing, GNB may present a quick yet effective approach to individual cancer stage identification. This methodology is useful for early detection and stage-specific analysis, and it has the potential to provide information for medical diagnosis for treating kidney cancer patients.
-
K-Nearest Neighbours Classifier The KNC model is applied to differentiate various23 phases of kidney cancer using renal cell images from contrast-enhanced scans. This model categorises images based on similarity and determines the characteristics of the stage to which an image belongs, given a set of labelled renal cell images and cancer stages. By comparing the “k” most similar images to the input image, the KNC model can distinguish early, intermediate, and advanced states of tissue structure, density, or tumor margin characteristic of kidney cancer. This similarity-based approach enables the model to capture progressive changes in features that differentiate stages while analyzing new cases, thus improving staging classification. It proves to be an easy yet effective method to help identify cancer progression and assist clinicians in understanding each patient’s cancer stage and the optimal treatment options.
-
Stochastic Gradient Descent This linear classifier24 trained using gradient descent is suitable for high-dimensional features and effectively extracts markers that define different stages of kidney cancer. Due to parameter updates based on small batches during training, the SGD classifier learns features about tissue density, tumor size, and boundary definition to distinguish between early, intermediate, and advanced cancer stages. The model rapidly converges to an optimum solution, making it uniquely suited for high-dimensional problems as staging predictions can be computed efficiently. This fast and effective method improves diagnostic accuracy and provides significant, interpretable stage designations that may enable appropriate treatment strategies for kidney cancer patients.
-
Naive Bayes Authors in25 use a Naiye bayes model to predict and categorise various levels of kidney cancer originating from renal cell images that are taken from contrast-enhanced computed tomography (CT) scans. This probabilistic model is very useful in the case of multivariate data where the outcome is based on individual feature values. For the stage of kidney cancer, Naive Bayes has been used to estimate pixel intensities and texture feature class-conditional probability distributions at three stages of the tumour development: early, intermediate, and late. It helps the model to classify images precisely and promptly by considering the contradictions in terms of tumour density, size, and shape among the stages. Due to its simple implementability and fast computation, Naive Bayes is more desirable in low-resource settings, assisting clinicians to understand the progression of cancer and contributing to personalised treatment planning accordingly.
-
Logistic Regression The LR model is used for detecting different stages of kidney cancer based on contrast-enhanced renal cell scan imaging26. By modelling the probability density function of an image’s likelihood of belonging to a certain cancer stage, Logistic Regression differentiates stages based on features such as tissue density, tumour size, and boundary sharpness. Its ability to compute weights for each image feature allows the model to predict the probability of each stage using a learned linear boundary. Although a basic method, Logistic Regression is easily interpretable, efficient, and well-suited for data with numerous features. This probabilistic output places clinicians in a good position to understand stage progression and make accurate, timely stage-specific diagnoses. The model aids quick and precise cancer stage identification and is helpful for providing short-term forecasts that support treatment planning and monitoring cancer progression.
-
Random Forest The Random Forest model is used27 to identify various stages of kidney cancer using renal cell images derived from contrast-enhanced CT scans. Random Forest is an ensemble learning technique that builds numerous decision trees during training and makes predictions based on the majority vote during testing, offering robust and reliable results. Each tree is grown from a random sample of features and instances, enabling the model to capture complex dependencies associated with early, intermediate, and advanced stages of kidney cancer. By utilizing diversified trees, Random Forest minimizes overfitting and improves predictive accuracy, making it well-suited for inherently noisy medical imaging data. This model allows evaluation of relationships between tumor morphology, tissue density, and vascular structures, making cancer staging more nuanced. Ultimately, Random Forest assists clinicians in early kidney cancer diagnosis and supports treatment decisions tailored to distinct cancer stages.
Hyperparameter tunning
In the applied models, we performed hyperparameter tuning to enhance their performance. Table 2 summarizes the hyperparameters used. Optuna was employed to optimize key hyperparameters for each model, including–but not limited to–learning rate, batch size, number of estimators, and regularization values. By thoroughly exploring various configurations, Optuna enabled us to select the best values, resulting in improved performance, greater stability, and faster training. This approach helped each model perform better and increased its ability to generalize to other datasets. The chosen hyperparameters represent a balance between model performance, training efficiency, and resource constraints. We set the number of epochs to 10 to avoid overfitting while allowing sufficient learning, as initial tests showed convergence within this range. A batch size of 32 was selected as a typical tradeoff between fast training and stable gradient updates. The Adam optimizer was used due to its adaptive learning rate and fast convergence, which are particularly beneficial for deep learning models like VGG-19. The loss function was categorical cross-entropy, suitable for the multiclass RCC stage classification. Finally, the input shape of (224, 224, 3) matches the model’s pre-trained configuration, ensuring compatibility with ImageNet weights and optimal feature extraction performance.
Results and discussions
In this section, we report the evaluation results of our proposed model and compare it with other existing models applied to the same datasets, to provide complete measures of their effects. We also evaluate the comparison among models on the criteria of accuracy, precision, recall, and F1 score to discuss their relative advantage further for the task. Such a comparative study not only validates the efficacy of our model but also shows the diverse ways to address the challenges of this dataset so that we can refine and improve in future work.
Experimental setup
For this experiment, we used Google Colab as a cloud environment to meet the computational demands of our tasks and provide a convenient interface for our machine learning operations. Leveraging Colab’s capabilities, we were able to execute our models efficiently, making extensive use of GPU resources for training and evaluation. Table 3 presents the setup information. Additionally, Colab’s collaboration features facilitated sharing and exchanging files and content within the system, enabling our team to communicate easily and work together on model fine-tuning, machine learning, and data analysis. Overall, using Google Colab was highly beneficial for running our experiments and producing accurate results.
CNN model results
we used a simple Convolutional Neural Network (CNN) in our experiments. However, the model failed to produce promising accuracy, achieving a rating of only 32%. This poor performance raises questions about the complexity of the architecture, which may not be sufficient to capture the underlying patterns in the data. It also highlights the need to explore more elaborate architectures or methods that could significantly enhance the model’s learning capacity and classification performance, given the training data.
Visual geometry group-19
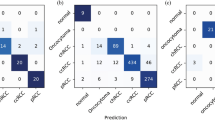
After using the VGG-19 model, it was observed that it performed significantly better than our simple CNN model. The accuracy score achieved by the VGG-19 model was 78%, clearly demonstrating its ability to capture the complex features and patterns within the dataset. Figure 5 illustrates the model’s evaluation results, while Fig. 6 presents the confusion matrix heatmap. This improved performance further supports the idea that deep and complex architectures are more effective for handling challenging tasks. The findings reveal that incorporating the VGG-19 model led to enhanced classification accuracy and provided a more robust platform for the experiments.
Performance analysis using spatial features
As part of our systematic analysis, we applied various machine learning models to the spatial features extracted from the dataset, resulting in a range of outcomes. Table 4 presents the complete classification report for all models tested. The K-Nearest Neighbors (KNC) model achieved an accuracy of 63%, while the Stochastic Gradient Descent (SGD) model performed better with an accuracy of 71%. The Random Forest (RF) model proved to be the most effective, delivering the highest accuracy of 91%. Additionally, the Naive Bayes (NB) model reached an accuracy of 83%. However, the Gaussian Naive Bayes (GNB) model performed poorly, with accuracy dropping to only 20%, indicating near model failure. These results highlight the varied performance of different algorithms on this task, demonstrating a mixed but informative performance landscape.
Performance analysis of proposed RCC-ProbNet model
We recreated the probabilistic features using the same feature engineering techniques and retrained the machine learning models on this newly derived feature set. The accuracy scores observed showed significant improvements over the earlier setup, providing strong evidence that refined features enhance model performance. The highest accuracy achieved by LR at 99.93%, highlighting its strong capacity to classify optimally using the selected features. KNC closely followed with 99.62% accuracy, demonstrating its effectiveness in handling complex data patterns. The complete evaluation report is provided in Table 5. The Random Forest (RF) model also delivered excellent results, achieving 99.42% accuracy, which reflects its ability to learn complex data relationships through ensemble learning. Naive Bayes (NB) achieved an accuracy of 98%, slightly lower than the other models, but still demonstrating how well the newly derived features align with probabilistic modelling. The GNB model, which was previously among the weaker performers, showed a substantial improvement to 99% accuracy, indicating that the new features strengthened models relying on Gaussian assumptions. Similarly, the Stochastic Gradient Descent (SGD) model reached 99% accuracy, confirming its suitability for the refined feature set through its fast and efficient convergence.
Figure 7 presents a bar chart comparing the performance using spatial features and the newly proposed features. In conclusion, these findings demonstrate that the newly obtained features significantly enhanced the classification performance of each model. The results shown in the last two sub-figures indicate that high accuracy was consistently achieved regardless of the chosen algorithm, suggesting that the feature engineering process successfully extracted important characteristics of the data and made them more generalizable and effective across different machine learning techniques. This consistency in performance provides strong evidence that the refined features can perform equally well in other applications, thereby improving the stability and flexibility of the models.
Kfold validations analysis
We selected 10-fold cross-validation to achieve a balanced trade-off between bias and variance in performance estimation, while ensuring that each subset of the data was sufficiently large for stable evaluation given the dataset size. For ROC analysis, we determined the classification threshold using Youden’s Index, which identifies the point that maximizes the difference between the true positive rate and the false positive rate. This approach was chosen to optimize both sensitivity and specificity, which is especially important in medical diagnostics where the cost of false negatives can be high. These clarifications have been added to ensure transparency and to strengthen the reproducibility and interpretability of our evaluation strategy.
To analyse the performance and stability of different classes of machine learning models, we used 10-fold cross-validation. Table 6 shows a 10-fold validation report. The outcomes pointed to the fact that the RF model offered a considerable amount of mean accuracy of 0.9963 with a standard deviation of ± 0.0111, which gave a top accuracy and reliability. In the same regard, the LR model has another evaluation measurement, including a mean accuracy of 0.9932 with no observed variability of ± 0.0000. The Naive Bayes (NB) has a mean accuracy of 0.973, but with a moderately higher Variability (±0.0227). The SGD model reached the mean accuracy of 99.09% while the variability of the samples was ±1.82% an The KNC had the highest accuracy of 0.9964, and this was accompanied by a very low variability, with a standard deviation of ±0.00107, which shows the stability of the models. The RF model achieved a mean accuracy of 0.9963 with a standard deviation of ±0.0111, demonstrating high performance and consistency. Similarly, the LR model achieved a mean accuracy of 0.9932 with no observed variability (±0.0000). The Naive Bayes (NB) model yielded a mean accuracy of 0.973 with a slightly higher variability (±0.0227), indicating moderate performance. The SGD model showed a mean accuracy of 0.9909 with a variability of ±0.0182. Figure 8 shows the confusion matrix heatmap graph. The KNC achieved the highest accuracy of 0.9964 with a very low variability of ±0.00107, highlighting its robustness. Finally, in the case of the GNB model, an average accuracy of 0.9857 with a standard deviation of ±0.0237 was achieved, which was at par with other models used in this study. These results help to confirm the usefulness of the tested models and show that KNC and RF are the most accurate and stable.
Computational complexity analysis
A computational cost comparison was conducted to assess the amount of time taken by different machine learning algorithms to complete. The NB model demonstrated the lowest computational cost, completing in just 0.00403 seconds, followed closely by the GNB model at 0.00630 seconds. Table 7 shows the time complexity analysis. The KNC took slightly longer at 0.00813 seconds, and the SGD model averaged 0.01123 seconds. Logistic Regression (LR) exhibited a moderate computational cost of 0.01295 seconds. In previous evaluations, the RF model showed high accuracy but required the longest computational time, taking 0.44943 seconds. These results highlight that simpler models, such as NB and GNB, require the least time, whereas more complex models like RF demand significantly more computational resources.
Evaluation on KiTS21 dataset
We additionally confirmed the generalizability and robustness of RCC-ProbNet using the open-source KiTS21 dataset(Kidney Tumour Segmentation Challenge 2021). It is a dataset constituted of a large number of CT scans with annotations on kidney tumours, making it an extremely tough and varied reference for renal cancer classification and segmentation missions. Finally, to test the capability of our RCC-ProbNet model, we performed experiments by fine-tuning it on the KiTS21 dataset and testing it with different evaluation metrics, including Accuracy, Precision, Recall, F1-Score, Specificity, AUC and Cohen’s Kappa Score. We selected these metrics to measure classification quality in terms of trade-offs between true positives, false positives, and overall agreement with ground truth labels. As shown in Table 8, RCC-ProbNet achieves outstanding performance across all metrics on the KiTS21 dataset, achieving a near-perfect accuracy of 99.88%. This strong performance further confirms the model’s ability to generalize effectively beyond small-scale datasets, thus mitigating the risk of overfitting.
State of the art studies comparisons
We compared the proposed RCC-ProbNet approach with previously used techniques in this context and achieved higher accuracy. Table 8 presents the state-of-the-art comparisons. For reference, previous methods reached a performance level of 99.72%, demonstrating their effectiveness. However, our proposed approach, RCC-ProbNet, achieved the highest accuracy score of 99.93%, surpassing these methods. Thus, the RCC-ProbNet classifier outperforms all evaluated classifiers in terms of classification accuracy, setting a new benchmark for future work and highlighting the great potential of our approach as a highly accurate solution in this field (Table 9).
Feature space analysis
To examine the correlation between features extracted from the RCC-ProbNet model, we used a 3D feature space analysis28. This analysis required employing modern visualization methods. By projecting the data into a three-dimensional space, we aimed to enhance the understanding of how features correlate and segregate according to the different stages of renal cell carcinoma (RCC). Fine-tuning the utilization of the 3D map helped identify distinct patterns by revealing how features within various cancer stages exhibit differential distribution. This facilitates the identification of important features that significantly improve the model’s performance, providing deeper insights into the biology of RCC. Furthermore, this approach assists in the preliminary detection of potential overlaps or ambiguities between stages, which is crucial for fine-tuning the model and enhancing classification performance. Figure 9 shows the feature space analysis. The 3D feature space analysis serves as an effective diagnostic tool for evaluating the efficiency of the selected feature extraction methods. Thus, we confirm the effectiveness of the proposed approach and demonstrate that the chosen features are sufficiently discriminative to visually differentiate between the identified cancer stages. The insights derived from this analysis contribute not only to understanding the data structure in this study but also improve the interpretation of the model itself. These findings can support potential improvements and optimization of RCC detection techniques in the future. Finally, the integration of 3D feature space analysis enriches the overall theoretical framework of RCC-ProbNet diagnosis, thereby enhancing the model’s diagnostic capability.
Clinical implications
As a promising tool for improving the diagnosis process of RCC, RCC-ProbNet makes stage-specified classification much more reliable in medical images. Early and accurate diagnosis of RCC is very important for early intervention and to minimise the negative quality of life consequences to the patient. Through the combination of deep spatial feature extraction (using VGG-19) and probabilistic reasoning (exploited in a hybrid CNN), RCC-ProbNet provides a framework that can potentially help radiologists and oncologists in recognising features that may not be visually detectable by manual inspection. The model’s capacity to produce probabilistic outputs is also valuable for decision-making in the clinic and could lead to less challenging diagnoses. After further validation, RCC-ProbNet may be incorporated into computer-aided diagnosis systems to facilitate routine, efficient, and affordable screening and surveillance for RCC across varying clinical settings.
Conclusion
In this work, we proposed RCC-ProbNet, an original model for the detection of kidney renal cell carcinoma (RCC), a deadly cancer for which early diagnosis plays an important role in increasing survival rates. Through the combination of spatial feature extraction and a hybrid probabilistic feature modelling technique, RCC-ProbNet using Logistic Regression (LR) achieved a high accuracy of 99.3%, which outperforms previous methods and sets a new state-of-the-art performance. The performance of the model was highly evaluated by using the k-fold cross-validation, indicating that the model was of high reliability and stability for RCC detection. A comparative study also demonstrated that RCC-ProbNet + LR has better performance in comparison to the previous method, indicating great progress in RCC detection. The application of the model has apparent clinical benefits in the diagnosis, greatly improved, and could early intervention, and finally to improve the survival rate of the RCC patients. This work provides a solid underpinning for future development and application in RCC detection and therapy.
Future work
Future work in this field will also concern the extension of RCC-ProbNet towards other functions and its application in the context of clinical practice. One such area of improvement includes adapting the model to make it more suitable for larger and more diverse datasets (eg, multi-institutional imaging data) to improve transferability across different patient populations. In addition, this work will also further investigate utilizing RCC-ProbNet with more complicated imaging modalities and histopathology, which may bring the model more diagnostic cues and increase the performance. Another avenue for research development pertains to the extension of the types of kidney cancer that the model can distinguish and the application of the model within the field of oncology. Finally, attempts will be continued to assist in the clinical application of RCC-ProbNet by implementing a user-friendly GUI as well as investigating potential computational speed enhancements. These innovations will facilitate the integration of RCC-ProbNet into clinical practices so healthcare providers can make diagnoses more timely and accurately, improving patient care.
Discussions and limitations
The high accuracy of our approach in the cancer stage classification demonstrates that RCC-ProbNet indeed has potential power in terms of cancer diagnosis. However, we are adding more sources of clinical information, such as patient history or laboratory tests. Finally but importantly, the clinical application of the model can bring about questions with respect to the computational requirement of high-performing models, as the latter often demand strong computational resources and cannot be conducted in community-based medical systems that are not supported by such hardware. Overcoming this constraint in subsequent studies and uses will be key to the broadening of applications of RCC-ProbNet for the diagnosis, prognosis, and treatment of kidney renal cell carcinoma.
Data Availability
All data and code supporting the findings of this study are available from the corresponding authors upon request.
References
Kadhim, D. A. & Mohammed, M. A. Advanced machine learning models for accurate kidney cancer classification using CT images. Mesopotamian J Big Data 2025, 1–25 (2025).
Doe, J. & Smith, J. Fine-tuned deep learning models for early detection and classification of kidney tumors. Sci. Rep. 12, 1234–1245 (2025).
Lee, A. & Kumar, R. Digital pathology and ensemble deep learning for kidney cancer detection. Appl Biosci 4, 8 (2025).
Pimpalkar, A. et al. Fine-tuned deep learning models for early detection and classification of kidney conditions in CT imaging. Sci. Rep. 15, 10741 (2025).
Gogoi, P. & Valan, J. A. Machine learning approaches for predicting and diagnosing chronic kidney disease: Current trends, challenges, solutions, and future directions. Int. Urol. Nephrol. 57, 1245–1268 (2025).
Ghosh, A. & Chaki, J. Fuzzy enhanced kidney tumor detection: integrating machine learning operations for a fusion of twin transferable network and weighted ensemble machine learning classifier. IEEE Access (2025).
Sharma, G. et al. Transfer learning empowered multi-class classification of kidney diseases: A deep learning approach. In: 2024 2nd International Conference on Advancement in Computation & Computer Technologies (InCACCT), 240–245 (IEEE, 2024).
Jain, M. N., Al-Heejawi, S. M. A., Azzi, J. R. & Amal, S. Digital pathology and ensemble deep learning for kidney cancer diagnosis: Dartmouth kidney cancer histology dataset. Appl Biosci 4, 8 (2025).
Jaddi, N. S. et al. Optimizing heart-kidney interaction for cancer detection through physiological process simulation as a decision support system. IEEE Access (2025).
Ślusarczyk, A. et al. Prediction of glomerular filtration rate following partial nephrectomy for localized renal cell carcinoma with different machine learning techniques. Cancers 17, 1647 (2025).
Habchi, Y. et al. Deep transfer learning for kidney cancer diagnosis. arXiv preprint arXiv:2408.04318 (2024).
Akram, Z., Munir, K., Tanveer, M. U., Rehman, A. U. & Bermak, A. Kidney ensemble-net: Enhancing renal carcinoma detection through probabilistic feature selection and ensemble learning. IEEE Access (2024).
Rao, P. S., Bheemavarapu, P. K., Swapna, D., Polamuri, S. R. & Subramanyam, M. M. Hybrid neural network-based fuzzy inference system combined with machine learning to detect and segment kidney tumor. In Computational Intelligence in Medical Decision Making and Diagnosis, 101–111 (CRC Press, 2023).
Patel, V. V., Yadav, A. R., Jain, P. & Cenkeramaddi, L. R. A systematic kidney tumor segmentation and classification framework using adaptive and attentive-based deep learning networks with improved crayfish optimization algorithm. IEEE Access 12, 85635–85660 (2024).
Hasan, M. A. Deep learning model based decision support system for kidney cancer on renal images. Central Asian J Math Theory Comput Sci 5, 82–97 (2024).
Uddin, K. M. M. et al. Machine learning-based chronic kidney cancer prediction application: A predictive analytics approach. Biomed Mater Dev 2, 1028–1048 (2024).
Al-taie, R. H. & Hussein, N. J. Renal tumor classification and detection based on artificial intelligence. In Third International Conference on Communications, Information System, and Data Science (CISDS 2024), vol. 13519, 107–114 (SPIE, 2025).
Hossen, M. J., Ramanathan, T. T. & Al Mamun, A. An ensemble feature selection approach-based machine learning classifiers for prediction of covid-19 disease. Int. J. Telemed. Appl. 2024, 8188904 (2024).
Majumdar, A. kidney cancer (2024). https://www.kaggle.com/datasets/atreyamajumdar/kidney-cancer/data.
Naseer, A. et al. A novel transfer learning approach for detection of pomegranates growth stages. IEEE Access 12, 27073–27087 (2024).
Raza, A., Munir, K., Almutairi, M. S. & Sehar, R. Novel transfer learning based deep features for diagnosis of down syndrome in children using facial images. IEEE Access 12, 16386–16396 (2024).
Yamini, B. et al. Machine learning algorithms for predicting of chronic kidney disease and its significance in healthcare. Int J Adv Technol Eng Explor 11, 388 (2024).
Islam, M. S. & Rony, M. A. T. CDK: A novel high-performance transfer feature technique for early detection of osteoarthritis. J Pathol Inform 15, 100382 (2024).
Xu, C. et al. Bass: A blockchain-based asynchronous signsgd architecture for efficient and secure federated learning. IEEE Trans Depend Secure Comput 21(6), 5388–5402 (2024).
Tanveer, M. U., Munir, K., Raza, A. & Almutairi, M. S. Novel artificial intelligence assisted landsat-8 imagery analysis for mango orchard detection and area mapping. PLoS ONE 19, e0304450 (2024).
Khan, H. et al. Heart disease prediction using novel ensemble and blending based cardiovascular disease detection networks: Enscvdd-net and blcvdd-net. IEEE Access. 12, 109230–109254 (2024).
Munir, K., Tanveer, M. U., Alyamani, H. J., Bermak, A. & Rehman, A. U. Pneux-net: An enhanced feature extraction and transformation approach for pneumonia detection in x-ray images. IEEE Access (2025).
Liu, K., Xiao, X., You, J. & Zhou, Y. Robust discriminative t-linear subspace learning for image feature extraction. IEEE Trans Circuits Syst Video Technol. 34(8), 7315–7327 (2024).
Funding
This work was supported in part by Multimedia University under the Research Fellow Grant MMUI/250008, and in part by Telekom Research and Development Sdn Bhd under Grant RDTC/241149 and RDTC/231095.
Author information
Authors and Affiliations
Contributions
Zaib Akram (Z.A.): Conceptualization, formal analysis, investigation, writing–original draft. Kashif Munir (K.M.): Supervision, project administration, resources, writing–review & editing. Muhammad Usama Tanveer (M.U.T.): Formal analysis, validation, methodology, visualization, writing–original draft. Yawar Abbas Abid (Y.A.A.): Software development, data curation. Syed Rizwan Hassan (S.R.H.): Supervision, writing–review & editing. Muhammad Sheraz (M.S.): Conceptualization, writing–review & editing. Teong Chee Chuah (T.C.C.): writing–review & funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Akram, Z., Munir, K., Tanveer, M.U. et al. A novel hybrid approach for multi stage kidney cancer diagnosis using RCC ProbNet. Sci Rep 15, 45281 (2025). https://doi.org/10.1038/s41598-025-29742-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-29742-4