Abstract
Kawasaki disease (KD) is an acute pediatric systemic vasculitis of unknown origin, where emerging evidence suggests that immune dysregulation and alterations in peripheral immune cell populations play critical roles in its pathogenesis. In this study, we performed comprehensive bioinformatics analyses to identify KD-associated genes and develop an accurate diagnostic model for early detection. Two publicly available microarray datasets, GSE73461 (132 samples) and GSE68004 (126 samples), were obtained from the Gene Expression Omnibus database and analyzed. Immune-related differentially expressed genes (IMRDEGs) were identified by integrating differential expression profiling with Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway analysis. To select optimal biomarkers, we employed a multi-algorithm framework combining logistic regression, SVM-RFE, and LASSO regression. Among 12 IMRDEGs identified, five genes (ITGAM, CAMP, CD4, IL2RB, and IL1B) were identified as having potential diagnostic value, with ITGAM showing the strongest predictive value. Functional enrichment revealed these genes’ involvement in leukocyte activation and immune-related signaling pathways, including tuberculosis signaling. Comparative immune profiling highlighted significant differences in 14 immune cell populations between KD patients and healthy controls. Overall, our findings uncover critical immune-related genes linked to KD pathogenesis and propose a robust diagnostic model with promising clinical applicability.
Similar content being viewed by others

Introduction
KD is an acute, self-limited vasculitis of unknown etiology, first described by Tomisaku Kawasaki in 1967 as “mucocutaneous lymph node syndrome.” It primarily affects medium and small-sized arteries in children under five years of age, with a particularly high incidence in East Asian populations1,2. KD occurs approximately 1.5 times more often in boys than in girls of the same age group. This vasculitic disorder predominantly involves medium and small caliber arteries in pediatric populations under five years of age, with particularly high prevalence observed in East Asian regions1,2. Approximately 25% of affected individuals develop coronary artery lesions (CALs), which may lead to coronary artery aneurysms, stenosis, thrombosis, myocardial infarction, or even sudden death—making KD the most common cause of acquired heart disease in children from developed countries3,4.
The diagnosis of Kawasaki disease (KD) is predominantly established through clinical manifestations. Classical KD is identified when patients exhibit persistent fever for five or more days accompanied by four out of five cardinal clinical criteria: First, oral mucosal changes including lip erythema with fissuring, strawberry tongue, or pharyngeal erythema; Second, bilateral conjunctival congestion without purulent discharge; Third, polymorphic cutaneous eruptions (either maculopapular or diffuse erythematous patterns); Fourth, peripheral extremity changes characterized by erythema and swelling during the acute phase; and fifth cervical lymphadenopathy, typically unilateral, with lymph nodes exceeding 1.5 cm in diameter. Importantly, cases presenting with incomplete clinical features may still warrant a KD diagnosis when coronary artery anomalies are demonstrated through appropriate imaging modalities1,3.
The primary treatment for Kawasaki disease (KD) involves early administration of high-dose intravenous immunoglobulin (IVIG) combined with aspirin, ideally within the first 10 days of fever onset. However, approximately 10–20% of patients exhibit resistance to IVIG, increasing their risk of CALs5. Diagnosing KD can be challenging, as many of its symptoms overlap with those of other febrile illnesses in children. Missed, incorrect, or delayed diagnoses may lead to severe complications or even death. Emerging evidence indicates that immunological mechanisms are fundamentally involved in the development of KD, although specific molecular mechanisms remain unclear1,5. Therefore, investigating KD pathogenesis and identifying novel molecular biomarkers are critical for early and accurate diagnosis, treatment, and prevention of KD complications.
Recent advances in high-throughput microarray platforms, combined with computational biology approaches, have significantly enhanced our ability to identify genetic biomarkers associated with various pathological conditions. This synergistic methodology has revolutionized the systematic discovery of molecular signatures linked to disease pathogenesis. With the progression of computational technologies, numerous gene expression profiling studies have been conducted to elucidate the mechanisms underlying KD. However, due to high false-positive rates, single microarray analyses rarely yield reliable results6,7,8. Therefore, this study applied well-established bioinformatics techniques to investigate IMRDEGs and their roles in KD pathogenesis.
To identify DEGs between healthy controls and Kawasaki disease (KD) patients, we obtained two mRNA expression profiling datasets from the Gene Expression Omnibus (GEO) database. Following DEG identification, we conducted Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses to investigate the biological significance of these differentially expressed transcripts. The LASSO method, celebrated for its effectiveness in detecting variable associations and limiting overfitting, was selected to refine the detection of genes associated with distinct phenotypic traits, surpassing Cox or logistic regression models. By incorporating L1 regularization, the approach boosts variable selection, thereby enhancing its capacity to pinpoint biologically relevant genetic markers. This method aided in unearthing immunoregulatory pathways associated with the molecular markers. Precision and efficiency in variable selection define the technique’s strength, a trait highly beneficial in genetic research where the identification of significant associations is paramount. The application of this method in the current context has clarified the relationships between genetic elements and phenotypic traits, while also highlighting critical immune interactions associated with these molecular findings. The regularization feature of the method, which fortifies robustness, prevents model overcomplexity, and thus yields more reliable and interpretable results in genetic association studies. The regularization feature of the method, which fortifies robustness, prevents model overcomplexity, and thus yields more reliable and interpretable results in genetic association studies.
The study identified 12 immune-related differentially expressed genes (IMRDEGs) and evaluated their roles in immune signaling pathways during acute KD. We comprehensively profiled over 14 peripheral blood immune cell populations, analyzing their distribution patterns during both the acute infection phase and subsequent recovery. This in-depth analysis facilitated dynamic monitoring of immune cell composition and functionality across the disease course, offering critical insights into the immunological mechanisms underlying KD pathogenesis and resolution. Identified biomarkers for disease monitoring and therapeutic targets are presented, along with enhanced insights into the complex immune dysregulation of pediatric vasculitis. Furthermore, we explored the correlations between immune cell infiltration and hub genes, yielding novel insights into KD pathogenesis and laying the groundwork for the subsequent investigation of disease mechanisms and therapeutic targets.Furthermore, we explored the correlations between immune cell infiltration and hub genes, yielding novel insights into KD pathogenesis and laying the groundwork for the subsequent investigation of disease mechanisms and therapeutic targets.
Materials and methods
Data download
The gene expression datasets GSE734619,10 and GSE680046 were retrieved from the GEO database11 (https://www.ncbi.nlm.nih.gov/geo/) using the GEOquery8 package (v2.70.0) in R. Both datasets comprised peripheral blood samples collected from human subjects. The microarray platforms employed were GPL10558 for both GSE73461 and GSE68004. As detailed in Table 1, GSE73461 included 77 KD cases and 55 healthy controls, while GSE68004 contained 89 KD patients and 37 control subjects. All available samples from these datasets were incorporated in our analysis.
The immunomodulatory-related genes (IMRGs) were systematically retrieved from the GeneCards12 database (https://www.genecards.org/), a comprehensive repository of human genomic information. Using “Immunomodulatory” as the primary keyword and applying strict criteria specifically selecting protein-coding genes with relevance scores greater than 2, we initially identified 64 candidate IMRGs. To ensure comprehensive coverage, we conducted a parallel literature13,14,15,16 search in PubMed (https://pubmed.ncbi.nlm.nih.gov/) via the same keyword, which yielded an additional 88 immunoregulatory genes from published studies. Following the integration and deduplication of the two datasets, we finalized a curated set of 136 unique IMRGs. The full list of genes, along with detailed annotations, is presented in Supplementary Table S1. This dual strategy, combining database mining with literature curation, enhanced the robustness of our immunomodulatory gene selection while minimizing potential bias in gene inclusion.. The stringent relevance score threshold (score > 2) in GeneCards ensured the inclusion of only the most biologically significant candidates, whereas the PubMed search captured recently characterized genes that might not yet be fully annotated in genomic databases.
Batch effect correction for datasets GSE73461 and GSE68004 was performed via the sva17 package (v3.50.0) in R, resulting in a combined dataset comprising 166 KD samples and 92 healthy controls. The integrated GEO datasets were subsequently processed via the limma package18 (v3.58.1) for standardization and probe annotation normalization. To assess the effectiveness of batch effect removal, principal component analysis (PCA)19 was conducted on both pre- and post-processed expression matrices. Principal Component Analysis (PCA) extracts orthogonal feature vectors known as principal components from high-dimensional data. This enables the visualization of complex datasets in two or three-dimensional space while preserving as much of the original variance as possible.
Kawasaki disease-related immunomodulatory related differentially expressed genes
The samples from the combined datasets were categorized into two distinct groups: KD patients and healthy controls. Gene expression profiling was performed via the limma package (v3.58.1) in R to identify significant transcriptional changes between these groups. Stringent criteria were applied to define DEGs, requiring an absolute log2-fold change (|logFC|) greater than 1 and an adjusted p value (P.adj) below 0.05 following Benjamini‒Hochberg correction. Transcripts whose logFC > 1 with P.adj < 0.05 were classified as upregulated, whereas those whose logFC < -1 with P.adj < 0.05 were classified as downregulated. The differential expression patterns were visualized through volcano plots generated via ggplot2 (v3.4.4). To identify immunometabolism-related DEGs (IMRDEGs) potentially involved in KD pathogenesis, To identify immunometabolism-related DEGs (IMRDEGs) potentially involved in KD pathogenesis, we intersected the complete set of significant DEGs with known immunometabolism-related genes (IMRGs), with the overlap illustrated via Venn diagrams.. The expression profiles of these IMRDEGs were subsequently depicted in a heatmap constructed with the pheatmap package (v1.0.12), providing a comprehensive view of their regulatory patterns across sample groups.
Differential expression verification and correlation analysis of immunomodulation related differentially expressed genes
To investigate the differential expression patterns of IMRDEGs between KD patients and healthy controls across integrated GEO datasets, comparative expression profiles were generated by analyzing IMRDEG expression levels.
To further investigate potential relationships among IMRDEGs, we employed Spearman’s rank correlation method to assess pairwise associations between gene expression profiles within the combined datasets. The correlation patterns were visualized via ggplot2 (v3.4.4) to construct a comprehensive heatmap representation. On the basis of established statistical thresholds, we categorized correlation strengths as follows: negligible association (|r|< 0.3), modest correlation (0.3 ≤|r|< 0.5), substantial correlation (0.5 ≤|r|< 0.8), and highly significant correlation (|r|≥ 0.8). This systematic approach enables the quantitative evaluation of intergene relationships within the immune microenvironment.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis
Functional annotation analysis was conducted through Gene Ontology (GO) classification20, which systematically categorizes gene functions into three primary domains: Biological Processes (BP), Cellular Components (CC), and Molecular Functions (MF). For pathway analysis, we utilized the KEGG21 database, a comprehensive resource containing genomic, pathway, disease, and pharmacological data. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses of the identified IMRDEGs were performed using the cluster Profiler R package (v4.10.0)22. Statistical significance thresholds was determined using adjusted P-values (P.adj) and false discovery rate (FDR/q-values) thresholds of < 0.05, with multiple testing correction applied via the Benjamini–Hochberg (BH) procedure.
Establishment of a diagnostic model for KD
To develop diagnostic models for KD utilizing integrated datasets,a stepwise multi-algorithm framework integrating logistic regression, SVM-RFE, and LASSO regression was employed for progressive feature selection.In the SVM-RFE feature selection process, fivefold cross-validation was used (nfold = 5), and LASSO regression selected the optimal λ value through tenfold cross-validation (default parameters of cv.glmnet). SVM-RFE identified the most stable 8 gene features through multiple cross-validations, while LASSO regression automatically performed feature screening via regularization, ultimately retaining only 5 key genes, significantly reducing the risk of overfitting. All random processes were set with fixed seeds to ensure reproducibility. The calibration curve showed high consistency between predicted probabilities and actual probabilities, and decision curve analysis demonstrated that the model had significant clinical net benefit.We employed logistic regression analysis on immune-related differentially expressed genes (IMRDEGs). Given the binary nature of the outcome variable (KD cases versus healthy controls), this statistical approach was selected to evaluate potential associations between predictor variables and disease status. The study integrated genes showing statistically significant associations (P.adj < 0.05). These genes were incorporated into a logistic regression model, and their expression patterns were visualized using forest plots.Based on these findings, feature selection was further optimized using the upport vector machine (SVM) algorithm.This process evaluated the stability of different gene combinations through fivefold cross-validation, and the optimal gene subset was selected by balancing the goal of maximizing classification accuracy with minimizing error rates. The comprehensive methodology facilitated the meticulous and methodical identification of immune-related genes exhibiting differential expression profiles, potentially offering substantial diagnostic value for KD. The comprehensive methodology facilitated the meticulous and methodical identification of immune-related genes exhibiting differential expression profiles, potentially offering substantial diagnostic value for KD.
The concluding analytical phase incorporated the glmnet package, version 4.1. The application of LASSO regression in R involved setting a random seed to 500 for reproducibility, and the binomial family distribution was used. The extension of linear regression with tenfold cross-validation is used to automatically select the optimal regularization parameter (lambda) is aimed at diminishing overfitting and enhancing the model’s predictive power on novel datasets. Selective application of LASSO regression was conducted on the IMRDEGs identified via our previous SVM analysis.Coefficient trajectory plots and diagnostic model visualizations were used to illustrate the results, ultimately facilitating the construction of a KD diagnostic model centered on the most informative IMRDEGs. For each sample, Risk Score was computed with regression coefficients [insert formula here], yielding an objective gauge of disease risk from the expression profiles of the genes. Risk Score was derived for each sample by regression coefficients [insert formula here], yielding an objective gauge of disease risk from the expression profiles of the genes.
Validation of the KD diagnostic model
Complex mathematical relationships between predictor variables are graphically illustrated by the nomogram, which utilizes multiple parallel scaled axes within a two-dimensional coordinate system. Employing the rms package, version 6.7. Utilizing R statistical software, we constructed a predictive nomogram grounded in logistic regression coefficients to delineate the interrelations among the identified model genes.. Additionally, we performed decision curve analysis (DCA) using the ggDCA package (version 1) to assess the clinical utility of our gene signature across the integrated GEO datasets. The integrated methodology ensures a dependable mechanism for determining the practical value of predictive models, diagnostic biomarkers, and molecular signatures in real-world clinical decision-making. The integrated methodology ensures a dependable mechanism for determining the practical value of predictive models, diagnostic biomarkers, and molecular signatures in real-world clinical decision-making.
The computational assessment of GO annotation similarities offers a robust quantitative approach for evaluating functional relationships between genes and genomes, serving as a fundamental component in numerous bioinformatics methodologies. In this study, we employed the GOSemSim23 package (v2.28.0) implemented in R to systematically quantify functional associations among Model Genes. These intergene relationships were subsequently characterized through functional similarity metrics (Friends analysis), providing insights into their biological relatedness at the molecular level.
Immune infiltration analysis of Kawasaki disease (CIBERSORT)
The CIBERSORT computational method24 utilizes linear support vector regression to deconvolute transcriptomic data, allowing for precise quantification of immune cell composition with heterogeneous tissue samples. By utilizing the LM22 signature gene matrix as a reference, we applied stringent quality control by excluding samples with nonpositive immune cell enrichment scores, thereby generating a robust immune infiltration matrix for the integrated datasets. These results were visualized through proportionally stacked bar plots to illustrate the relative abundance of distinct immune cell populations. Comparative analysis between the KD and control groups was conducted via the ggplot2 package (v3.4.4) to generate differential expression profiles of LM22-defined immune subsets across the combined GEO datasets. Statistically significant immune cell subtypes showing intergroup variation were identified for downstream investigation. Intercellular immune network relationships were evaluated through Spearman’s rank correlation analysis, with the results graphically represented as a heatmap via the pheatmap package (v1.0.12). Furthermore, we examined associations between model genes and immune cell fractions via Spearman’s method, retaining only those interactions demonstrating statistical significance (P.adj < 0.05). These gene‒immune cell correlations were subsequently visualized as bubble plots through the ggplot2 visualization toolkit (v3.4.4).
Statistical analysis
All statistical analyses in this study were performed using R software (version 4.3.3). For normally distributed continuous variables, group comparisons were conducted using independent Student’s t-tests. Non-normally distributed data were analyzed using the nonparametric Mann–Whitney U test (Wilcoxon Rank Sum Test). For comparisons involving three or more groups, the Kruskal–Wallis test was applied. Spearman’s rank correlation coefficient was used to quantify relationships between molecular variables. Unless otherwise specified, all statistical tests were two-tailed, with statistical significance defined asP.adj < 0.05.
Results
Technology roadmap
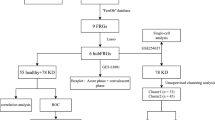
See Fig. 1.
Flow Chart for the Comprehensive Analysis of IMRDEGs. KD, Kawasaki Disease; DEGs, Differentially Expressed Genes; IMRGs, Immunomodulatory-Related Genes; IMRDEGs, Immunomodulatory-Related Differentially Expressed Genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; SVM, Support Vector Machine; LASSO, Least Absolute Shrinkage and Selection Operator.
Merging of Kawasaki disease datasets
Initially, batch effect correction was performed on datasets GSE73461 and GSE68004 using the sva package (v3.50.0) in R, resulting in an integrated combined dataset. To evaluate the effectiveness of this normalization procedure, we conducted comparative analyses through two complementary approaches. First, expression value distributions were visualized using boxplots (Fig. 2A,B), demonstrating substantial improvement in data consistency postcorrection. Second, PCA was employed to assess low-dimensional feature distributions (Fig. 2C,D), revealing significantly reduced batch-specific clustering patterns. These comprehensive quality control measures collectively indicated successful mitigation of technical batch effects in the processed dataset.
Batch Effects Removal of GSE73461 and GSE68004. (A) Boxplots of GEO Combined Datasets distribution before batch removal. (B) Post-batch integrated GEO Datasets (Combined Datasets) distribution boxplots. (C) PCA plot of the datasets before debatching. (D) Go to the PCA map of the Combined GEO Datasets after batch processing. PCA, Principal Component Analysis; KD, Kawasaki Disease. The KD dataset GSE73461 is in orange and GSE68004 is in green.
KD-related immunomodulatory genes
The integrated GEO datasets were systematically categorized into KD specimens and healthy control specimens. To explore transcriptional differences between these two groups, we conducted differential gene expression analysis using the limma package in R. This computational approach identified 2,136 significantly altered genes (|logFC |> 1with P.adj < 0.05) in the combined datasets. Among these differentially expressed genes (DEGs), 1,414 exhibited significant upregulation (logFC > 1, BH-adjusted p < 0.05), while 722 genes showed marked downregulation (logFC < -1, BH-adjusted p < 0.05). The Benjamini‒Hochberg procedure was employed for multiple testing correction. The differential expression patterns were subsequently visualized through a volcano plot (Fig. 3A), which effectively illustrates the magnitude and statistical significance of gene expression changes between the KD and control groups.
Differential Gene Expression Analysis. (A) Volcano plot of differentially expressed genes analysis of Kawasaki disease (KD) samples and Control samples in the Combined GEO Datasets. (B) Differentially expressed genes (DEGs) and immunomodulatory related genes (IMRGs) Venn diagram in the integrated GEO Datasets (Combined Datasets). (C) Heat map of immunomodulatory related differentially expressed genes (IMRDEGs) in the integrated GEO Datasets (Combined Datasets). KD, Kawasaki Disease; DEGs, Differentially Expressed Genes; IMRGs, Immunomodulatory-Related Genes; IMRDEGs, Immunomodulatory-Related Differentially Expressed Genes. Green is the Control sample, and orange is the Kawasaki disease (KD) sample. In the heat map, red represents high expression and blue represents low expression.
To identify IMRDEGs, we intersected all differentially expressed genes (DEGs) that met the criteria of |log₂FC|> 1 and adjusted p-value < 0.05 with known immune-related genes (IMRGs). The overlap was visualized using a Venn diagram (Fig. 3B). This analysis yielded 12 immune regulatory genes: ITGAM, IL1RN, IL1B, TLR4, TLR2, STAT3, CAMP, CD274, ITGB2, IL2RB, CD4, and CD40LG. Subsequently, we examined the expression patterns of these IMRDEGs across different sample groups in the combined datasets. The differential expression profiles were graphically represented using heatmaps generated with the pheatmap R package (Fig. 3C).
Differential expression verification and correlation analysis of differentially expressed genes related to immune regulation
To investigate the distinct expression patterns of immunomodulatory-related differentially expressed genes (IMRDEGs) in the merged GEO datasets, we performed a comprehensive comparative analysis between Kawasaki disease (KD) patients and healthy controls. The comparative analysis (Fig. 4A) revealed significant differential expression of twelve key IMRDEGs, including ITGAM, IL1RN, IL1B, TLR4, TLR2, STAT3, CAMP, CD274, ITGB2, and IL2RB, in the integrated dataset. Notably, CD4 exhibited particularly striking differences in expression between the KD and control groups, with statistical significance reaching p < 0.001. Additionally, we performed correlation analyses among the 12 identified IMRDEGs using the combined GEO datasets and visualized their interrelationships through a heatmap (Fig. 4B). The analysis demonstrated predominantly positive intergenic correlations, suggesting coordinated regulation of these immunomodulatory factors in the context of KD. This pattern of coexpression may indicate potential functional interactions or shared regulatory mechanisms among these genes during disease pathogenesis.
Correlation Analysis of IMRDEGs. (A) group comparison diagram of the expression differences of immunomodulatory related differentially expressed genes (IMRDEGs) in the Combined GEO Datasets. (B) Correlation heat map of 11 immunomodulatory related differentially expressed genes (IMRDEGs) in the integrated GEO Datasets (Combined Datasets). KD; Kawasaki Disease; IMRDEGs, Immunomodulatory-Related Differentially Expressed Genes. * represents p value < 0.05, statistically significant; *** represents p value < 0.001, highly statistically significant. The absolute value of correlation coefficient (r value) below 0.3 was weak or no correlation, between 0.3 and 0.5 was weak correlation, between 0.5 and 0.8 was moderate correlation, and above 0.8 was strong correlation. Control group (green), Kawasaki disease group (orange). Red represents a positive correlation and blue represents a negative correlation.
Gene Ontology (GO) and pathway (KEGG) enrichment analysis
Elucidation of the functional attributes of the 12 IMRDEGs in KD was achieved via a comprehensive GO and KEGG pathway analysis. Systematic investigation of the biological processes, cellular components, molecular functions, and signaling pathways linked to the identified IMRDEGs was performed via bioinformatics methodologies. The enrichment analysis demonstrated notable correlations with pivotal immunological processes, specifically concerning the enhancement of leukocyte activation and interleukin-10 biosynthesis. The enrichment analysis yielded significant correlations with critical immunological processes, notably by modulating leukocyte activation and interleukin-10 biosynthesis positively. The IMRDEGs demonstrated prominent involvement in critical biological functions including the regulation of leukocyte adhesion processes and immune response activation. At the cellular level, these genes were predominantly localized to specialized membrane structures such as plasma membrane signaling complexes, lipid rafts, and specific granules. Molecular function analysis identified their participation in protein–protein interactions, notably in amyloid-beta binding, integrin recognition, and interleukin-1 receptor engagement. Pathway analysis uncovered substantial enrichment in several infectious disease pathways (tuberculosis, malaria, legionellosis, leishmaniasis) and immune checkpoint regulation, particularly the PD-L1/PD-1 signaling axis in oncological contexts. The analytical outcomes were graphically represented using comparative bar charts (Fig. 5A) and multidimensional bubble plots (Fig. 5B), with detailed quantitative results presented in Table 2. These findings collectively suggest that the 12 IMRDEGs play pivotal roles in modulating immune responses through diverse molecular mechanisms in KD pathogenesis.
GO and KEGG Enrichment Analysis for IMRDEGs. (A) bar chart of gene ontology (GO) and pathway (KEGG) enrichment analysis results of immunomodulatory differentially expressed genes (IMRDEGs) (B) Bubble plot of gene ontology (GO) and pathway (KEGG) enrichment analysis results of immunomodulation-related differentially expressed genes (IMRDEGs). The ordinate is the GO terms and KEGG terms. (C–F) Gene ontology (GO) and pathway (KEGG) enrichment analysis results of immunomodulatory differentially expressed genes (IMRDEGs): BP (C), CC (D), MF (E) and KEGG (F). The orange nodes represent items, the green nodes represent molecules, and the lines represent the relationship between items and molecules. IMRDEGs, Immunomodulatory-Related Differentially Expressed Genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, Biological Process; CC, Cellular Component; MF, Molecular Function. The bubble size in the bubble plot represents the number of genes, and the color of the bubble represents the size of the adj. P-value, the reder the color, the smaller the adj. P-value, and the bluer the color, the larger the adj. P-value. The screening criteria for gene ontology (GO) and pathway (KEGG) enrichment analysis were P. adj < 0.05 and FDR value (q value) < 0.05, and the p value correction method was Benjamini-Hochberg (BH).
Furthermore, the functional enrichment analysis results were visualized through network diagrams depicting biological processes (BP), cellular components (CC), molecular functions (MF), and KEGG pathways (Fig. 5C–F). These graphical representations illustrate the relationships between identified molecules and their functional annotations, with node size proportional to the number of associated molecules within each category.
Establishment of a diagnostic model for Kawasaki disease
To evaluate the diagnostic potential of 12 IMRDEGs in KD, we initially developed a logistic regression model incorporating these molecular markers. The forest plot analysis (Fig. 6A) demonstrated statistically significant associations (P < 0.05) for all 12 genes: ITGAM, IL1RN, IL1B, TLR4, TLR2, STAT3, CAMP, CD274, ITGB2, IL2RB, CD4, and CD40LG.Subsequently, we implemented a support vector machine (SVM) algorithm to optimize gene selection, identifying an optimal subset of 8 IMRDEGs (ITGAM, CAMP, CD4, IL2RB, STAT3, TLR2, TLR4, and IL1B) that maximized predictive accuracy (Fig. 6B,C). This refined gene panel achieved showed good diagnostic performance when incorporating these eight markers. Further refinement through LASSO regression analysis yielded a parsimonious 5-gene signature (ITGAM, CAMP, CD4, IL2RB, and IL1B) for KD diagnosis, as visualized in the coefficient profiles (Fig. 6D) and variable trajectory plots (Fig. 6E). This sequential modeling approach systematically identified the most robust immunomodulatory biomarkers for KD detection.
Diagnostic Model of KD. Genes were progressively filtered by logistic regression, refined by SVM-RFE, and finalized through LASSO regression to obtain the five diagnostic genes. (A) Forest Plot of 12 immunomodulatory related differentially expressed genes (IMRDEGs) included in the Logistic regression model in the KD diagnostic model. (B–C) The number of genes with the lowest error rate (B) and the number of genes with the highest accuracy (C) obtained by the SVM algorithm are visualized. D-E. Diagnostic model plot (D) and variable trajectory plot (E) of LASSO regression model. KD, Kawasaki Disease; IMRDEGs, Immunomodulatory-Related Differentially Expressed Genes; SVM, Support Vector Machine; LASSO, Least Absolute Shrinkage and Selection Operator.
Validation of the diagnostic model for KD
To validate the clinical applicability of the KD diagnostic models, we constructed nomograms incorporating Model Genes via integrated GEO datasets (Fig. 7A). Our analysis revealed that ITGAM exhibited superior discriminatory power compared with other variables in the diagnostic model, demonstrating the highest predictive value. In contrast, IL2RB showed significantly lower diagnostic utility than the other model components did. Nomogram visualization effectively demonstrated the relative contributions of each Model Gene, with ITGAM emerging as the most influential biomarker for KD identification. These findings suggest that ITGAM expression levels may serve as a robust diagnostic indicator, whereas IL2RB appears to have limited discriminative capacity in this model framework.
Diagnostic and validation analysis of KD. (A) Nomograms of model genes in combined GEO datasets in Kawasaki disease (KD) diagnostic models. (B,C) Calibration curve plot (B) and decision curve analysis (DCA) plot (C) of Model genes in combined datasets for Kawasaki disease (KD) diagnostic model. (D) Box plot of functional similarity (Friends) analysis results of model genes. The ordinate of decision curve analysis (DCA) is the net benefit, and the abscissa is the probability threshold or threshold probability. KD, Kawasaki disease; DCA, Decision curve analysis.
To evaluate the predictive accuracy and discriminative capacity of the KD diagnostic model, we conducted a calibration analysis by generating a calibration curve (Fig. 7B). This analysis assessed the concordance between the model’s predicted probabilities and the observed event rates across different risk strata. The calibration plot demonstrated that while the predicted probabilities (represented by the dashed line) showed minor deviations from the ideal diagonal line, they maintained reasonable concordance with actual outcomes. Furthermore, we conducted decision curve analysis (DCA) to evaluate the clinical applicability of the KD diagnostic model derived from Model Genes in the integrated GEO datasets (Fig. 7C). The DCA results revealed that across a clinically relevant threshold probability range, the model’s net benefit curve consistently surpassed both the “treat-all” and “treat-none” strategies. This indicates superior clinical utility of our model compared to these extreme approaches, with the model providing greater net benefit over a wide range of decision thresholds.
Functional similarity analysis (Friends analysis) was conducted to identify key genes involved in the pathogenesis of KD, as illustrated in Fig. 7D. The results indicated that ITGAM emerged as a pivotal gene n KD pathogenesis, exhibiting the highest functional similarity score, which closely approached the predefined threshold (cut-off = 0.75). This finding suggests ITGAM’s potential significance in the disease’s biological mechanisms.
Immune infiltration analysis of KD cells (CIBERSORT)
The immune cell infiltration profiles across 22 immune cell subtypes in the integrated GEO datasets were quantitatively assessed via the CIBERSORT algorithm. A comprehensive visualization of immune cell distribution patterns within the combined datasets is presented as a proportional bar chart (Fig. 8A). Subsequent comparative analysis revealed statistically significant differences (P < 0.05) in the infiltration levels of 14 distinct immune cell populations, including activated B cells, both resting and activated dendritic cell subsets, M0 and M2 macrophage phenotypes, resting mast cells, monocytes, neutrophils, resting and activated natural killer cells, central memory CD8 + T cells, resting and activated central memory CD4 + T cells, and gamma-delta T cells, between KD patients and healthy controls (Fig. 8B). Intercellular correlation analysis, depicted in a heatmap (Fig. 8C), demonstrated significant associations among these immune cell populations. Notably, neutrophils presented the strongest positive correlation with M0 macrophages (r = 0.54) but the most pronounced negative correlation with central memory CD8 + T cells (r = -0.75). Furthermore, bubble plot analysis (Fig. 8D) revealed significant relationships between model genes and immune cell infiltration patterns. Specifically, CD4 expression was positively correlated with the number of resting NK cells (r = 0.46, P < 0.05) but negatively associated with monocyte infiltration (r = − 0.47, P < 0.05). These findings collectively underscore the intricate immune landscape and its molecular correlates in the pathogenesis of KD.
Combined datasets immune infiltration analysis by CIBERSORT algorithm. (A,B) The proportion of immune cells in the integrated GEO datasets (Combined Datasets) bar graph (A) and group comparison graph (B). (C) Correlation heatmap of immune cell infiltration abundance in the integrated GEO Datasets (Combined Datasets). (D) Bubble plot of Model Genes’ correlation with immune cell infiltration abundance in the combined GEO Datasets. KD, Kawasaki Disease. * represents P-value < 0.05, indicating statistical significance; ** represents P-value < 0.01, highly statistically significant; *** represents P-value < 0.001 and highly statistically significant. The absolute value of correlation coefficient (r value) below 0.3 was weak or no correlation, between 0.3 and 0.5 was weak correlation, and between 0.5 and 0.8 was moderate correlation. Control samples are green, and KD samples are orange. Blue is negative correlation, red is positive correlation, and the depth of color represents the strength of correlation.
Discussion
A major challenge in KD management is the lack of reliable early diagnostic tools, which often results in delayed diagnosis and suboptimal clinical outcomes. The principal cause of pediatric acquired heart disease, KD, exerts a substantial public health burden, yet diagnostic and therapeutic strategies are constrained by the challenges of early detection and the scarcity of specific biomarkers. This study uses bioinformatics to uncover novel molecular markers and their pathological correlates, with the goal of improving diagnostic capabilities and therapeutic strategies25,26,27. This study employs bioinformatics approaches to identify novel molecular markers and elucidate their pathological mechanisms, aiming to improve diagnostic precision and inform therapeutic strategies25,26,27.
The health impact of KD is considerable, particularly in the pediatric demographic, and it is a predominant cause of acquired heart disease. To advance diagnostic and therapeutic strategies, this study employed a multifaceted bioinformatics toolkit, featuring batch effect correction, differential expression analysis, functional enrichment assessment, and machine learning-based predictive modeling. The study focused on identifying IMRDEGs to clarify their association with the pathophysiology of Kawasaki Disease. Through systematic analysis and integration of multiple datasets, we identified 12 significant IMRDEGs and developed a robust diagnostic model that demonstrated strong validation performance. The study’s results expand our grasp of KD’s foundational mechanisms and unveil a hopeful avenue for early clinical diagnosis. The study’s results expand our grasp of KD’s foundational mechanisms and unveil a hopeful avenue for early clinical diagnosis.
Significant biological relevance is exhibited by the 12 IMRDEGs, which encompass ITGAM, IL1RN, IL1B, TLR4, TLR2, STAT3, CAMP, CD274, ITGB2, IL2RB, CD4, and CD40LG, TLR4 and TLR2, functioning as pattern recognition receptors, are associated with the mediation of inflammatory responses stemming from bacterial or viral infections, which may be linked to KD development28. Furthermore, the regulation of cytokines by IL1B and IL1RN is essential, impacting immune cell activation and function29,30 The genes identified not only reflect the pathophysiological changes associated with KD but also highlight promising therapeutic targets. Investigating their involvement across a spectrum of autoimmune disorders may help elucidate the complex mechanisms underlying KD. Studying their role across various autoimmune disorders may reveal the intricate mechanisms at play in KD.
Enrichment analysis of function demonstrated the participation of 12 IMRDEGs in critical biological processes, such as leukocyte activation regulation, which underscores their role in the progression of Kawasaki disease via key signaling pathways. The NF-κB and JAK/STAT pathways interaction is highlighted as crucial for inflammatory response mediation, as indicated by available research.The TLR2/TLR4–NF-κB–IL1B inflammatory signaling pathway and JAK/STAT immune regulation are crucial, closely related to the functions of the ITGAM, IL1B, CD4, IL2RB, and CAMP genes. TLR2 and TLR4 recognize relevant molecules to activate NF-κB, promoting IL1B expression and exacerbating inflammation; upregulation of ITGAM enhances leukocyte adhesion, aggravating inflammation. In JAK/STAT regulation, cytokines activate JAK kinases, and STAT enters the nucleus to regulate gene expression, with IL2RB participating in signal transduction and affecting CD4 + T cell function. CAMP is influenced by both mechanisms, exhibiting antibacterial and immunomodulatory effects31,32. The evidence suggests that further investigation into related pathways may uncover promising therapeutic targets, paving the way for more precise treatments that improve clinical outcomes. The delineation of these molecular mechanisms forms the groundwork for the development of enhanced intervention strategies in the management of Kawasaki disease33,34.
A LASSO regression model has been constructed with the inclusion of five immune-related genes (ITGAM, CAMP, CD4, IL2RB, and IL1B) for KD diagnosis, yielding positive results. The model’s performance was robust, with statistically significant results (P.adj < 0.05), suggesting its potential as a screening instrument for the identification of high-risk patients in clinical scenarios35.Further empirical study is needed to ascertain the model’s efficacy across diverse ethnic and age groups. Incorporating additional clinical variables could enhance the model’s precision and utility36,37. The viability and economic implications of integrating this model into real-world healthcare systems should be a priority in future research38. Subsequent studies must evaluate the practicality and economic efficiency of adopting this model within real-world healthcare settings38.
In this investigation, we reported a significant upregulation of CD4⁺, which may have important biological significance with respect to immune regulation in Kawasaki disease (KD). CD4⁺ is a glycoprotein expressed primarily on CD4⁺ helper T cells, which are required for coordinating an immune response. CD4⁺ helper T cells have the ability to recognize antigens presented by antigen presenting cells, secrete cytokines to activate B cells and cytotoxic T cells, and modulate immune balance within the overall immune system. As such, the notable upregulation of CD4⁺ in this cohort could represent an immune response to counter-regulate aberrant immune activation in KD, potentially by increasing the number or function of CD4⁺ helper T cells to restore immune homeostasis. We also noted a positive correlation between CD4 and resting NK cells, as well as a negative correlation with monocytes. The positive correlation with resting NK cells may suggest that CD4⁺ T cells help maintain NK-cell quiescence. CD4⁺ cells could secrete cytokines that maintain quiescence in NK cells during a less severe immune challenge, and then activate the NK cells during acute immune responses to minimize tissue damage associated with overactivation in KD. The negative correlation with monocytes may indicate that CD4⁺ T cells inhibit monocyte recruitment or function. Monocytes can differentiate into macrophages and dendritic cells in response to antigen stimulation, which are potent antigen presenting cells. By down-modulating monocytes, CD4⁺ could fine-tune antigen presentation, inhibit hyperactivation, and restore immune homeostasis in KD. These relationships together elucidate a complicated network in which CD4 is a key immune regulator of the immune response in KD.
This study provides valuable insights into KD and identifies potential biomarkers for future investigation; however, several limitations warrant recognition. First, the findings are based solely on data from public databases lacking laboratory verification, the data sources mainly rely on public databases, with a lack of standardized collection and processing, high heterogeneity, deviations in accuracy and consistency, and no rigorous validation,which may limit the reliability of the results. Second, the relatively small sample size unable to cover all patient characteristics,resulting in insufficient representativeness and generalizability of the findings.Third, lack of experimental validation prevents determination of the biomarker’s actual function and application feasibility.The absence of long-term follow-up data restricts a thorough evaluation of the identified markers’ impact on patient outcomes.Currently, the diagnostic model has not been compared and validated with other gene combinations or independent datasets. “Subsequent validation of diagnostic efficacy will be further conducted through cross-validation and performance comparisons with different models.Future research will conduct multicenter sample studies to increase sample size and reduce heterogeneity; perform rigorous experimental studies to validate the functional mechanisms of biomarkers; strengthen long-term follow-up efforts to assess their prognostic value, in order to verify the effectiveness and clinical applicability of the diagnostic model.Despite these limitations, the study represents a meaningful advancement in KD research, establishing a strong foundation for the development of more targeted and personalized therapeutic strategies in future work25,39,40,41,42.
Our results also suggest possible functions for ITGAM and IL1B in immune-related inflammation and vasculitis. ITGAM may help leukocytes adhere to and migrate within the tissue during inflammatory scenarios, promoting the recruitment of immune cells, and the start progression of immune-mediated vascular injury. IL1B is an important proinflammatory cytokine that can initiate an inflammatory cascade, activating the endothelial cells and compromising vascular integrity, and potentially leading to its contribution to vasculitis. These observations provide new insights into the specific molecular mechanisms involved with this immune-related vasculitis.
Moving forward we plan to perform protein–protein interaction (PPI) analyses using databases such as STRING, to confirm the predicted associations/modifications for molecular interaction with the targets that we identified. Once we define the relevant interaction networks we will have a more robust understanding of the regulatory framework that is involved with immune inflammation and vasculitis, and this understanding will assist in developing specific intervention strategies.
Conclusion
The study identified a set of immune-modulatory genes with differential expression in KD, culminating in the development of a robust diagnostic tool. This advancement provides novel insights into KD pathogenesis and lays the foundation for more targeted therapeutic strategies. However, several limitations must be acknowledged. Although multiple datasets were integrated and various machine learning algorithms (Logistic regression, SVM-RFE, LASSO) were employed to enhance model robustness, this study did not validate the diagnostic model using an independent external validation set. Therefore, the model’s generalizability still requires further evaluation.These include the exclusive reliance on publicly available datasets without experimental (wet-lab) validation, a relatively limited sample size that may affect the generalizability of the results, and the absence of long-term follow-up data to assess the clinical relevance of the identified biomarkers. In future phases, the study will be expanded to include a larger cohort, incorporate clinical validation trials, and investigate additional molecular pathways. These steps aim to address the current limitations and deepen the understanding of KD pathophysiology.
Data availability
The datasets generated and/or analyzed during the current study are available in the GEO repository, GSE73461, GSE68004.
References
Marek-Iannucci, S. et al. Targeting IRE1 endoribonuclease activity alleviates cardiovascular lesions in a murine model of Kawasaki disease vasculitis. JCI insight 7(6), e157203 (2022).
Sakurai, Y. Autoimmune aspects of Kawasaki disease. J. Investig. Allergol. Clin. Immunol. 29(4), 251–261 (2019).
Sundel, R. P. Kawasaki disease. Rheum. Dis. Clin. North Am. 41(1), 63–73 (2015).
McCrindle, B. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A scientific statement for health professionals from the American heart association. Circulation 135(17), e927–e999 (2017).
Kobayashi, T. et al. Revision of diagnostic guidelines for Kawasaki disease (6th revised edition). Pediatr. Int. Off. J. Jpn. Pediatr. Soc. 62(10), 1135–1138 (2020).
Jaggi, P. et al. Whole blood transcriptional profiles as a prognostic tool in complete and incomplete Kawasaki Disease. PLoS ONE 13(5), e0197858 (2018).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets--update. Nucl. Acids Res. 41, D991–D995 (2013).
Davis, S. & Meltzer, P. S. GEOquery: A bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics 23(14), 1846–1847 (2007).
Kuiper, R. et al. Bridging a diagnostic Kawasaki disease classifier from a microarray platform to a qRT-PCR assay. Pediatr. Res. 93(3), 559–569 (2023).
Wright, V. J. et al. Diagnosis of Kawasaki disease using a minimal whole-blood gene expression signature. JAMA Pediatr. 172(10), e182293 (2018).
Barrett, T. et al. NCBI GEO: Archive for functional genomics data sets--update. Nucl. Acids Res. 41, D991–5 (2013).
Stelzer, G. et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protocols Bioinf. 54(1), 1–30 (2016).
Doifode, T. et al. Biomarkers unveiling the interplay of mind, nervous system, and immunity. Methods Mol. Biol. 2868, 73–90 (2025).
Lotfi, N. et al. Schizophrenia etiological factors and their correlation with the imbalance of the immune system: An update. Galen Med. J. 12, e3109 (2023).
Szabo, A. et al. Elevated levels of peripheral and central nervous system immune markers reflect innate immune dysregulation in autism spectrum disorder. Psychiatry Res. 342, 116245 (2024).
Wang, P. et al. The immune regulatory mechanism of ketamine-induced psychiatric disorders: A new perspective on drug-induced psychiatric symptoms. Prog. Neuropsychopharmacol. Biol. Psychiatry 136, 111194 (2025).
Leek, J. T. et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6), 882–883 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 43(7), e47 (2015).
Ben Salem, K. & Ben, A. A. Principal component analysis (PCA). La Tunisie Med. 99(4), 383–389 (2021).
Mi, H. et al. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucl. Acids Res. 47(D1), D419–D426 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28(1), 27–30 (2000).
Yu, G. et al. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16(5), 284–287 (2012).
Yu, G. et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26(7), 976–978 (2010).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5), 453–457 (2015).
Almeida, B., Gleeson, P., Chawla, K. & Brogan, P. Kawasaki disease: a case report and overview of symptoms, signs, investigations and treatment. Br. J. Hosp. Med. 73(2), C26–C28 (2012).
Kawasaki, T. Kawasaki disease. Mucocutaneous lymph node syndrome or MCLS. Acta Pathol. Jpn 32, 63–72 (1982).
Bayers, S., Shulman, S. T. & Paller, A. S. Kawasaki disease: part I Diagnosis, clinical features, and pathogenesis. J. Am. Acad. Dermatol. 69(4), 501-e1 (2013).
Rodríguez-Suárez, C. et al. Increase in transcript accumulation of Psy1 and e-Lcy genes in grain development is associated with differences in seed carotenoid content between durum wheat and tritordeum. Plant Mol. Biol. 84(6), 659–673 (2014).
Krejčí, K. et al. Protocol biopsy of a transplanted kidney as a tool for monitoring adequacy of immunosuppressive therapy: 10 years of experience from a single transplant center. Transpl. Proc. 43(5), 1576–1582 (2011).
Lin, H. H. et al. A modelling framework to support the selection and implementation of new tuberculosis diagnostic tools. Int. J. Tuberc. Lung Dis. 15(8), 996–1004 (2011).
Saint-Dizier, M. et al. Composing the early embryonic microenvironment: physiology and regulation of oviductal secretions. Int. J. Mol. Sci. 21(1), 223 (2019).
Xu, X. N., Wang, M. L. & Zhang, D. Effects of Different Inflammatory Factors on Hepatocyte Kinase Receptors and Ligands in Human Periodontal Ligament Fibroblasts. Zhongguo yi xue ke xue Yuan xue bao. Acta Academiae Medicinae Sinicae 41(3), 300–306 (2019).
Westphal, M. & Brannath, W. Evaluation of multiple prediction models: A novel view on model selection and performance assessment. Stat. Methods Med. Res. 29(6), 1728–1745 (2020).
Baranov, A. et al. The Russian child health care system. J. Pediatr. 177S, S148–S155 (2016).
Nishikimi, T. et al. A case of robot-assisted adrenalectomy performed for an adrenal tumor (anastomosing hemangioma) exceeding 7 cm. IJU Case Rep. 5(6), 469–473 (2022).
Nieboer, D., van der Ploeg, T. & Steyerberg, E. W. Assessing discriminative performance at external validation of clinical prediction models. PLoS ONE 11(2), e0148820 (2016).
Weitzman, M. et al. Housing and child health. Curr. Probl. Pediatr. Adolesc. Health Care 43(8), 187–224 (2013).
Sousa, S. et al. Machine learning models’ assessment: trust and performance. Med. Biol. Eng. Compu. 62(11), 3397–3410 (2024).
Yanagitani, Y. & Fujita, M. Avascular necrosis of the femoral head associated with mucocutaneous lymph node syndrome. J. Pediatr. Orthop. 6(1), 107–109 (1986).
Bitsadze, V. O. et al. Novel coronavirus infection and Kawasaki disease. J. Matern. Fetal. Neonatal. Med. 35(16), 3044–3048 (2022).
Berry, S. D., Kiel, D. P. & Colón-Emeric, C. Hip fractures in older adults in 2019. JAMA 321(22), 2231–2232 (2019).
Vega-Moreno, D. et al. Distribution and transport of microplastics in the upper 1150 m of the water column at the Eastern North Atlantic Subtropical Gyre, Canary Islands, Spain. Sci. Total Environ. 788, 147802 (2021).
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Mengjia Zhao,and Ruihua Yang were involved in the study concept and design. Mengjia Zhao contributed to data acquisition and analysis. Mengjia Zhao and Ruihua Yang were major contributor in writing of the manuscript. Mengjia Zhao mainly revised in the manuscript. Ruihua Yang had the final responsibility for the decision to submit for publication. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, M., Yang, R. Bioinformatics analysis of immune-related differentially expressed genes in Kawasaki disease. Sci Rep 16, 1052 (2026). https://doi.org/10.1038/s41598-025-30624-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30624-y