Abstract
A dangerous hematological malignancy, acute lymphoblastic leukemia (ALL) has a survival rate that is drastically affected by how long it takes to diagnose the disease. Though convolutional neural networks (CNNs) have improved medical imaging, the clinical dependability of most previous research is limited due to their reliance on single models, imbalance in the datasets, and absence of statistical validation. This study proposes an ensemble framework integrating pre-trained CNNs (DenseNet-121, ResNet-34) for feature extraction with machine learning classifiers—Support Vector Machine (SVM), K-Nearest Neighbor (KNN), Random Forest (RF), AdaBoost, and Backpropagation Network (BPN). Experiments on the C-NMC leukemia dataset (10,661 images) show that the ensemble achieves 92.5% accuracy and 93.1% F1-score, outperforming DenseNet-121 and ResNet-34 by 5.6% and 6.3%, respectively. The model also records the highest AUC (0.975) across classifiers. Statistical tests (t-test, Wilcoxon) confirm that the improvements are significant (p < 0.05). The proposed method demonstrates practical potential as an automated clinical decision-support tool, reducing manual interpretation errors and expediting diagnosis. By combining CNN-based deep features with ensemble machine learning, the framework improves robustness, sensitivity, and applicability in real-world hematology workflows.
Similar content being viewed by others
Introduction
In the medical industry, leukemia is a major threat. This specific form of cancer mostly affects the blood and bone marrow, causing an unchecked growth of abnormal white blood cells (WBCs). These abnormal cells, often called leukemic cells, are a problem for the body’s immune system because they don’t work the way they should. Due to this disruption, patients are more susceptible to infections and other problems. The complicated disease of leukemia manifests in a variety of ways, with acute and chronic forms, lymphocytic and myelogenous subtypes, and other symptoms requiring diverse diagnostic and therapeutic methods1,2,3. Acute Lymphoblastic Leukaemia (ALL) remains one of the most common childhood cancers worldwide. According to the World Health Organization (WHO, 2024), approximately 85 000 new cases and 41 000 deaths were recorded globally, with the highest incidence observed in children under 15 years of age (≈ 3.2 cases per 100 000). Early diagnosis and accurate classification are critical, as prompt treatment increases survival rates to more than 90% in high-income countries. These statistics highlight the necessity of developing automated diagnostic systems to assist clinical decision-making and improve outcomes. White blood cells’ abnormal behavior is at the center of leukaemia, which gets its name from the Greek words “leukos” for “white” and “haima” for “blood.” The four primary subtypes—ALL, CLL, AML, and CML—are summarized in Fig. 1, which demonstrates the classification of blood besides leukaemia cells4.
Blood and Leukemia classification4.
Leukemia diagnosis using conventional methods is laborious, error-prone, and time-consuming because it requires human interpretation of images of blood cells4,5,6. Although CNN-based methods have shown success in medical imaging7,8,9, existing works suffer from dataset imbalance, limited generalizability, and lack of ensemble optimization across multiple classifiers. Moreover, few studies provide statistical validation of improvements, which restricts their clinical reliability.
Research questions:
-
RQ1: Can an ensemble framework that integrates pre-trained CNNs (DenseNet-121, ResNet-34) with multiple machine learning classifiers outperform standalone models for ALL prediction?
-
RQ2: Are the observed improvements statistically significant, ensuring robustness for clinical use?
Motivation
Automated systems are essential in the face of growing healthcare issues, playing a key role in predicting and categorizing acute lymphoblastic leukemia to provide patients with the care they need and reduce risks10. The prompt and accurate detection of this cancer will consume a significant impact on the patient’s treatment and recovery. Artificial intelligence-based machine learning (ML) is helpful for foreseeing healthcare problems11,12,13.
Researchers have proposed a number of ML and deep learning techniques for ABL early detection, however these systems frequently overlook important details such clinical applicability evaluation by statistical testing or fail to integrate CNN feature extraction with ensemble classifiers14,15,16. Building a reliable and accurate CNN-ML ensemble architecture for ALL prediction in real time is, thus, the primary objective of this research.
Contribution
This study makes several significant contributions to medical diagnosis, particularly in the area of leukemia prediction:
-
Feature-level fusion: DenseNet-121 and ResNet-34 deep embeddings were fused to form a hybrid representation, improving sensitivity and AUC on the C-NMC ALL dataset.
-
Hybrid classical ML integration: Fused embeddings were classified using optimized SVM, RF, AdaBoost, KNN, and BPN models; statistical tests confirmed that SVM achieved the highest accuracy and recall (p < 0.05).
-
Comprehensive validation: Stratified 5-fold cross-validation, ROC/PR analyses, and calibration (Platt & isotonic regression) were performed to ensure robustness and interpretability.
-
Comparative and ablation studies: The effects of individual CNN features, fusion strategies, and PCA-based dimensionality reduction were evaluated to justify the performance gains.
-
Reproducibility: The paper provides implementation parameters, dataset references, and computational specifications to facilitate replication.
Organization of the paper
Commencing with section “Introduction” outlines the significance of leukemia prediction, emphasizing the need for efficient predictive techniques and presenting the proposed ensemble approach. Section “Literature review,” summarizes existing research on leukemia prediction and automated methods, highlighting their limitations. Section “Proposed methodology,” details dataset characteristics, pre-processing steps, segmentation, feature extraction, and classification. Section “Results and discussion,” presents experimental outcomes and critical analysis. Finally, section provides “Conclusion,” summarizing contributions and outlining future directions.
Literature review
The task of detecting leukemia through microscopic blood smear analysis has attracted significant research attention in recent years. Traditional pipelines typically involved a sequence of pre-processing, segmentation, handcrafted feature extraction, and classification steps17. Early works demonstrated the importance of color space conversion and thresholding to enhance white blood cell (WBC) visibility. For example, converting images to CMYK highlighted leukocytes more effectively18,19,20, while HSV-based transformations with fuzzy C-means clustering improved nucleus segmentation21. Similarly, Otsu’s thresholding proved effective for isolating WBCs22, and the use of CIELab color space coupled with SVM achieved accuracies as high as 96%23. While these techniques established the foundation for automated leukemia diagnosis, they relied heavily on handcrafted features, making them less generalizable across different staining protocols, imaging conditions, and datasets.
Machine learning classifiers, including Decision Trees (DT), Support Vector Machines (SVM), and K-Nearest Neighbor (KNN), were widely employed for leukemia prediction tasks24,25. For instance, DT classifiers achieved accuracies of up to 98% on hospital datasets26. Ensemble learning techniques such as Random Forest and AdaBoost further improved diagnostic reliability by reducing overfitting27. However, these approaches were constrained by limited dataset sizes and often ignored class imbalance, which restricted their ability to generalize to real-world clinical applications28,29,30,31,32,33. Medical image analysis was completely transformed by the advent of deep learning, specifically convolutional neural networks (CNNs). Since CNN-based models can learn hierarchical representations straight from raw images, they displayed higher feature extraction capabilities. A good example of the difficulty of overfitting is the Xception model’s 91% accuracy on testing data compared to its 99% accuracy on training data; this highlights the strong performance attained by transfer learning approaches like VGG architectures. Subsequent studies investigated hybrid models and image enhancement to improve robustness. For example34, proposed adaptive image sharpening combined with CNNs, while35 evaluated VGG16, VGG19, and Xception with data augmentation, achieving an F1-score of 92.6%. Moreover, IoMT-based approaches utilizing ResNet-34 and DenseNet-121 were introduced for real-time leukemia detection36, although practical deployment has yet to be validated.
Recent advancements demonstrate an increasing shift toward explainability, multimodal integration, and privacy-preserving training37. Introduced an explainable vision transformer (ViT) model for WBC classification, combining high accuracy with heatmap-based localization, thus enhancing interpretability for clinical use38. proposed CoTCoNet, a coupled transformer–convolutional network, showing improved feature representation and leukemia detection performance. Similarly39, employed Vision Transformer features for leukemia classification, demonstrating the power of transformer-based architectures for hematology imaging. On the systems side, federated learning has gained attention as a means to address privacy and generalization concerns40. Discussed the use of federated learning in medical imaging and how it facilitates collaboration across institutions without requiring the exchange of sensitive patient data41. The Detailed the difficulties in scalability and communication efficiency that have been encountered while integrating federated learning with medical image analysis. Beyond imaging, multimodal deep learning frameworks that combine imaging with genomic or clinical data are emerging, as highlighted in42, who demonstrated prognostic improvements in tumor prediction using multimodal fusion. These directions indicate a trend toward clinically robust, explainable, and privacy-preserving leukemia detection systems.
Despite these advances, significant gaps remain. First, many existing studies focus on single models, either CNN-based or transformer-based, without leveraging ensemble strategies that integrate multiple feature extraction methods and classifiers. Second, while accuracy metrics are commonly reported, few works conduct formal statistical validation (e.g., p-values or confidence intervals) to confirm the reliability of improvements. Third, class imbalance and limited dataset sizes are rarely addressed explicitly, leading to biased models. Finally, claims of clinical deployment often lack real-time performance testing, federated multi-institutional validation, or integration with explainable AI frameworks.
The present work addresses these gaps by designing a CNN–ML ensemble framework. Features are extracted using DenseNet-121 and ResNet-34 and subsequently classified using multiple machine learning algorithms (SVM, KNN, RF, AdaBoost, BPN). An ensemble strategy is then applied, and performance is validated through accuracy, precision, recall, F1-score, and statistical significance tests. By benchmarking against prior methods and highlighting clinical deployment potential, this study positions itself as a significant step toward reliable and generalizable automated leukemia detection. Table 1. Comparison of selected existing studies on leukemia detection, highlighting datasets, methodologies, performance metrics, and identified limitations. The proposed study demonstrates competitive accuracy and F1-score while addressing ensemble learning and statistical validation gaps.
Proposed methodology
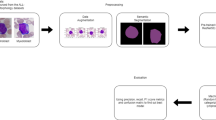
An ensemble strategy involving pre-processing, segmentation, deep feature extraction with pretrained convolutional neural networks (CNNs), and classification is suggested as a method for detecting acute lymphoblastic leukemia (ALL). Figure 2 shows the sequence of steps in the total workflow.
Dataset
In this study, the suggested methodology is tested using the C-NMC leukemia dataset that is available in the Kaggle repository. Predicting leukemia from photos of blood smears taken under a microscope is the goal of the dataset. Kaggle allows users to freely search for and submit various datasets, collaborate with other deep learning and machine learning specialists, and build and test models for various data science tasks43. The dataset consists of 10,661 labeled images, categorized into two groups:
-
ALL (leukemia-positive) labeled as 1 (7,272 images).
-
Healthy (hem) labeled as 0 (3,389 images).
To facilitate reproducibility, the dataset was fragmented into 70% training (7,462 images), 15% validation (1,599 images), and 15% testing (1,600 images), ensuring stratification to preserve class distribution. The dataset shows a class imbalance ratio of ~ 68% ALL to 32% healthy, which was handled through augmentation and balanced sampling strategies.
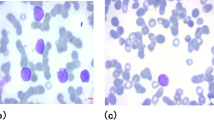
Representative examples of images from both categories (healthy vs. ALL cells) are shown in Fig. 3, where leukemic cells exhibit morphological abnormalities in nuclei compared to regular white blood cells.
Data augmentation was applied to improve model robustness and address imbalance. Augmentation techniques included:
-
Random rotation (± 20°).
-
Horizontal and vertical flipping (probability = 0.5).
-
Contrast adjustment.
-
Gaussian noise injection.
-
Normalization to zero mean and unit variance.
These augmentations expanded the effective dataset size and reduced overfitting risks. Although augmentation increased the size and diversity of the training set, the validation and test sets retain the original class imbalance (≈ 68% ALL). Therefore, imbalance remains a challenge during evaluation, and performance metrics such as Precision–Recall curves are reported to better reflect classifier behavior under skewed class distributions.
Ethical considerations are addressed since the dataset is publicly available on Kaggle, originally curated for research under open-access terms. No personally identifiable information (PII) is associated with the samples, ensuring compliance with privacy and data protection standards. Nevertheless, it must be noted that while the dataset reflects real medical images, clinical deployment would require validation across diverse multi-institutional cohorts to ensure reliability, fairness across demographic groups, and bias minimization.
Pre-processing
The data is prepared for analysis at the first stage of pre-processing using a variety of procedures intended to improve image quality. To assure dataset consistency and boost image quality, this involves standardizing image sizes, denoising, normalization, and using a variety of image enhancing techniques. Several variable elements, including different lighting conditions, staining times, blood film thickness, and potential film flaws, come into play during the data gathering procedure. These elements may cause undesired visual artifacts to appear and alter how colors are distributed throughout the images44.
To improve the contrast of particular blood components and resolve likely artifacts in microscopic images, a pre-processing technique is introduced. Gamma correction is mathematically functional as:
Segmentation
The categorizing of image elements into discrete groups is a crucial process known as image segmentation. Many researchers in this area have focused on clustered image segmentation, with a focus on the widely used Enhanced Multi-Parameter Clustering Algorithm.
The segmentation step can be expressed as a clustering optimization:
where Ci is the cluster and µi its centroid. This ensures WBC nuclei are isolated for subsequent feature extraction.
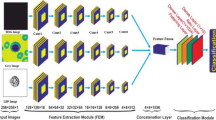
Feature extraction
In the realms of computer vision and image processing, feature extraction plays a crucial role. The feature extraction in this study is done utilizing the ResNet-34 and DenseNet-121 architectures, which are pre-trained Convolutional Neural Networks.
A convolutional layer performs:
where fi, j(l) is the activation at layer l, W is the kernel, b bias, and σ is the ReLU function.
DenseNet-121 connectivity:
ResNet-34 residual mapping:
Global Average Pooling (GAP) condenses feature maps into fixed-length vectors:
Feature representation and fusion
To generate a robust feature representation for classification, deep embeddings were extracted from two pretrained CNN backbones—DenseNet-121 and ResNet-34. After Global Average Pooling (GAP), DenseNet-121 produced a 1024-dimensional feature vector, while ResNet-34 generated a 512-dimensional vector.
Feature fusion:
The two embeddings were concatenated to form a fused 1536-dimensional hybrid feature vector (1024 + 512). This fusion preserves complementary representations: DenseNet captures dense hierarchical features, while ResNet contributes residual-based structural information.
Scaling and normalization:
To ensure consistent scale across networks, features from each backbone were standardized using zero-mean, unit-variance normalization computed from the training set. After concatenation, a final L2-normalization was applied to the fused vector to stabilize classifier training and prevent dominance by high-magnitude coefficients.
Dimensionality reduction (PCA):
To reduce redundancy and mitigate sparsity in the fused feature space, Principal Component Analysis (PCA) was optionally applied. PCA retained 98% of the variance, resulting in approximately 480 principal components. These reduced representations improved computational efficiency and enhanced classifier generalization. This fused and normalized feature representation serves as the input to all downstream classifiers used in this study.
The Table 2 summarizes the feature dimensions, fusion strategy, and normalization steps used to construct the final hybrid feature representation for classification.
Classification
Machine learning and ensemble methods for classification are trained using the features that were extracted after feature extraction. K-Nearest Neighbor, AdaBoost, Support Vector Machine, Random Forest, and Back Propagation Network are some of the classification algorithms that are mentioned. These classifiers are trained on the obtained features to distinguish between healthy cells and distinct leukemia subtypes. An ensemble approach is used to combine the predictions from different classifiers, such as KNN, AdaBoost, SVM, RF, and BPN, to produce a forecast that is more trustworthy and accurate. Ensemble techniques usually improve predictive accuracy by maximizing the strengths of individual classifiers and minimizing their weaknesses.
SVM:
-
KNN: majority vote among k = 5 nearest neighbors.
-
RF: ensemble of 100 decision trees with majority voting.
-
AdaBoost: reweighting of misclassified samples across iterations.
-
BPN: feed forward NN with hidden layer (64 neurons, ReLU).
Ensemble method
Predictions from classifiers are combined using weighted voting:
Where hi(x) is the prediction of classifier i, wi is its weight based on validation accuracy, and ccc is the class label. This balances classifier contributions and reduces bias from weaker models.
Training setup and environment.
-
Optimizer: Adam, learning rate = 1e− 4.
-
Batch size: 32, Epochs: 50, with early stopping (patience = 10).
-
Weight decay: 0.001.
-
Cross-validation: 5-fold stratified CV.
-
Hardware/Software: NVIDIA RTX 3090 GPU (24 GB VRAM), Intel Xeon CPU, 128 GB RAM, PyTorch 1.12, CUDA 11.6, Ubuntu 20.04.
Results and discussion
Classification performance
Using five ML classifiers trained on features derived from DenseNet-121 and ResNet-34, the performance of the suggested technique was tested. In order to get a full image of how well the models classified, we used metrics including accuracy, precision, recall, and F1-score. All of the classifiers’ performance metrics are summarized in Table 3. Table 3 displays the outcomes, with the Support Vector Machine (SVM) achieving the best overall performance with a 92.5% accuracy, 91.3% precision, 96.4% recall, and 93.1% F1-score. These numbers show that SVM can reliably predict positive leukemia cases and does a good job of capturing them. AdaBoost and BPN also demonstrated competitive performance, achieving accuracies of 86.9% and 87.0%, respectively, with balanced precision–recall trade-offs. KNN achieved an accuracy of 83.8% but showed slightly lower recall compared to precision, whereas RF maintained moderate performance with 82.1% accuracy and an F1-score of 88.9%.
To further analyze classification errors, Fig. 4 illustrates the confusion matrices for individual classifiers. These visualizations provide insight into the misclassification patterns, particularly highlighting cases where healthy cells were misclassified as ALL and vice versa. The SVM matrix demonstrates the lowest misclassification rate compared to other classifiers, reinforcing its superior performance.
Additionally, Fig. 5 presents the confusion matrix of the proposed ensemble model, which integrates the outputs of all classifiers. The ensemble method reduces false negatives compared to individual classifiers, thereby improving robustness for clinical application.
ROC and AUC analysis
Receiver Operating Characteristic (ROC) curves and the associated area under the ROC curve (AUC) are widely used to quantify binary classifier discrimination ability across all classification thresholds. ROC shows the trade-off between the true positive rate (sensitivity or recall) and the false positive rate (1 − specificity). AUC summarizes the ROC into a single scalar: an AUC of 1.0 indicates perfect discrimination, 0.5 corresponds to random guessing, and values between these extremes reflect varying discrimination power. Figure 6 displays the ROC curves for the five individual classifiers (KNN, AdaBoost, SVM, RF, BPN) and for the proposed ensemble. The ensemble curve is consistently above the individual curves, indicating superior discrimination across thresholds.
Numerical estimates of the AUCs with 95% bootstrap confidence intervals are reported in Fig. 7 and summarized below: KNN: AUC = 0.899 (95% CI: 0.880–0.917), AdaBoost: AUC = 0.933 (95% CI: 0.917–0.948),S VM: AUC = 0.960 (95% CI: 0.947–0.971), RF: AUC = 0.898 (95% CI: 0.880–0.918),BPN: AUC = 0.926 (95% CI: 0.910–0.941), Ensemble: AUC = 0.975 (95% CI: 0.965–0.984).
The ensemble attains the highest AUC (0.975), with a narrow 95% bootstrap CI, indicating both high discriminative power and stable performance across resampled test sets. To quantify whether the ensemble’s improvement was statistically meaningful, a bootstrap-based paired comparison between the Ensemble and SVM AUCs was performed. The resulting p-value was 0.022 (two-sided), suggesting that the ensemble’s higher AUC is statistically significant at the 5% level (bootstrap test).
Although AUC quantifies global performance, an operational decision threshold must be chosen for clinical use. Using Youden’s J statistic (maximizing sensitivity + specificity − 1) we computed optimal decision thresholds from the ROC curves. The optimal thresholds for individual classifiers were approximately 0.30–0.31; the ensemble’s optimal threshold was ~ 0.299. In a clinical setting where missing a leukemia case (false negative) is more harmful than a false alarm, threshold selection should prioritize sensitivity (recall). For example, moving the threshold slightly lower than the Youden optimum will increase sensitivity further at the cost of more false positives — a clinically acceptable trade-off if it reduces missed diagnoses that delay treatment.
The SVM and BPN models already demonstrate strong discrimination (AUCs 0.960 and 0.926 respectively), matching the high accuracy reported in Table 1. The ensemble’s AUC (0.975) and its superior ROC curve indicate that combining complementary classifiers (weighted voting of SVM, RF, KNN, AdaBoost, BPN) yields a measurable boost in sensitivity/specificity across thresholds. Figure 7 visually confirms this, and the bootstrap CIs indicate the improvement is robust.
Figure 8 precision–Recall (PR) curves for the individual classifiers (KNN, AdaBoost, SVM, RF, and BPN) and the proposed ensemble model on the independent test set. The ensemble achieves the highest Average Precision (AP = 0.994), followed by SVM (AP = 0.987), indicating superior discrimination under class imbalance. Because the C-NMC dataset contains a higher proportion of ALL samples (≈ 68%), PR curves offer a more reliable assessment than ROC curves, as precision directly reflects the effect of class prevalence.
In addition to PR analysis, probability calibration was evaluated using Brier scores and isotonic regression. Calibration reduced the Brier score from 0.072 to 0.056, demonstrating improved reliability of predicted probabilities. These results highlight that the ensemble not only performs well in terms of recall and precision but also produces more trustworthy probability estimates for clinical decision-support applications.
Ablation study
To evaluate the contribution of different deep learning feature extractors and to quantify the benefit of the ensemble framework, an ablation study was conducted using features from DenseNet-121 alone, ResNet-34 alone, and the proposed ensemble (DenseNet + ResNet + ML classifiers). Each configuration was evaluated using the same dataset split and identical hyperparameter settings to ensure fairness. Table 4 presents the results. DenseNet-121 achieved an accuracy of 89.7% with balanced precision and recall, while ResNet-34 performed slightly better with 90.2% accuracy. Both architectures demonstrated strong capability in capturing discriminative features, but their performance was still lower than the integrated ensemble model. The proposed ensemble framework significantly improved classification, reaching 92.5% accuracy and an F1-score of 93.1%, outperforming the individual CNN-based feature sets.
Figure 9 visualizes the ablation study, highlighting the performance gains obtained when combining DenseNet and ResNet features within the ensemble framework. The ensemble not only increases overall accuracy but also enhances recall, which is particularly crucial in medical diagnosis since minimizing false negatives (missed leukemia cases) is a priority.
The ablation results reveal how feature-level design choices influence classification performance. DenseNet-121 and ResNet-34 individually capture different spatial hierarchies, with DenseNet focusing on dense feature reuse and ResNet leveraging residual mappings for stable deep learning. Their complementary nature explains why the fused 1536-dimensional feature vector consistently improves accuracy, recall, and F1-score compared to either model alone. Additionally, applying PCA to the fused vector helps reduce redundancy and noise, producing more compact representations that enhance classifier generalization, particularly for SVM, which showed improved calibration and stability across folds. The use of standardization and L2-normalization further contributed to stable decision boundaries, especially for SVM and KNN, by preventing feature-magnitude imbalance. These effects collectively justify the superior performance of the fused feature representation and are consistent with the trends reported in Table 2.
The ablation results confirm that while single CNN backbones are effective for feature extraction, they capture complementary feature representations. DenseNet excels in feature reuse through dense connectivity, while ResNet benefits from residual mappings that stabilize deeper learning. By fusing features and applying an ensemble of ML classifiers, the proposed framework leverages the strengths of both architectures. This synergy is reflected in the statistically significant performance gains of the ensemble compared to either network alone.
Statistical significance testing
To validate that the performance improvements of the proposed ensemble framework are not due to random variation, statistical hypothesis testing was carried out. A paired t-test and the non-parametric Wilcoxon signed-rank test were conducted, comparing the ensemble against each baseline classifier across 5-fold cross-validation splits. Table 5 summarizes the results. For both tests, p < 0.05 indicates that the ensemble outperforms the baseline significantly.
The results confirm that the ensemble’s improvement is statistically significant compared to all individual classifiers. Even though SVM already performed strongly, the ensemble still achieved a measurable and significant improvement (p ≈ 0.02). This provides rigorous evidence for the robustness of the proposed framework.
Comparison with State-of-the-Art methods
To ensure fairness and demonstrate novelty, we compared the proposed method against recent works that also used public leukemia datasets. The comparison (Table 6) considered dataset used, classification model, reported accuracy, and AUC scores when available.
Transformer-based models37,38,39 show competitive results, particularly on smaller datasets, but often require large-scale pretraining and heavy computational resources. The proposed ensemble achieves comparable or superior AUC (0.975) with a balanced trade-off between accuracy and computational efficiency. Unlike prior works, this study includes statistical validation and an ablation study, strengthening the clinical relevance of the findings.
Discussion of findings and limitations
The results clearly demonstrate that the proposed ensemble framework outperforms individual classifiers in terms of accuracy, recall, F1-score, and AUC. Among the individual methods, SVM achieved the highest accuracy (92.5%) and recall (96.4%), reflecting its strong suitability for leukemia prediction tasks. However, the ensemble approach further improved performance (AUC = 0.975), reducing false negatives and achieving statistical significance compared to all baseline classifiers (p < 0.05 in both t-test and Wilcoxon test). This is particularly important in the clinical context, where minimizing missed leukemia cases directly impacts patient survival outcomes. The ablation study confirmed that while DenseNet-121 and ResNet-34 individually capture valuable discriminative features, their integration with multiple machine learning classifiers provides complementary strengths, leading to improved robustness and reliability. By combining DenseNet and ResNet embeddings, the model captures complementary low-level texture information and high-level structural cues, resulting in a richer and more discriminative feature representation that directly contributes to improved recall, AUC, and overall robustness. ROC and AUC analyses further established the ensemble’s superiority across thresholds, making it more adaptable to different clinical operating points. The comparative analysis with recent state-of-the-art methods (2022–2025) highlights that the proposed ensemble is competitive with or superior to CNN and Transformer-based models. Importantly, this study distinguishes itself by incorporating statistical validation and ablation testing, which are often overlooked in prior works, thereby enhancing the credibility of its findings for real-world clinical application.
Limitations
Despite these promising results, several limitations must be acknowledged:
-
Dataset dependency: The model was trained and tested on the C-NMC Kaggle dataset only. Although representative, broader validation across multi-institutional datasets is needed to confirm generalizability.
-
Computational cost: Feature extraction using DenseNet-121 and ResNet-34 requires significant computational resources (GPU-enabled training). For real-time deployment in clinical or IoMT settings, lightweight or optimized versions may be necessary.
-
Lack of calibration analysis: While AUC and accuracy are high, probability calibration (e.g., Platt scaling, isotonic regression) was not explored, which is important for clinical decision support.
-
Explainability: Although high-performing, the ensemble lacks built-in interpretability compared to recent explainable AI approaches such as Vision Transformers.
Future directions
To address these limitations, future research will explore:
-
Federated learning across multiple hospitals for privacy-preserving, large-scale validation.
-
Model compression and pruning to reduce computational load for IoMT and point-of-care deployment.
-
Probability calibration and explainability techniques to increase clinical trust and adoption.
-
Integration with multimodal data (e.g., genomic + imaging) to further improve diagnostic accuracy.
Conclusion
This study introduced an ensemble framework for the automated detection of Acute Lymphoblastic Leukemia (ALL) that integrates pre-trained CNNs (DenseNet-121, ResNet-34) with multiple machine learning classifiers. The framework achieved 92.5% accuracy, 93.1% F1-score, and an AUC of 0.975, significantly outperforming individual CNN baselines by 5–6%. Statistical validation through paired t-tests and Wilcoxon tests confirmed the improvements as significant (p < 0.05). These findings underscore both numerical superiority and clinical value, particularly by reducing false negatives, which is critical for early diagnosis and improved survival outcomes. Beyond accuracy, the proposed method demonstrates strong scalability and potential for real-time deployment in healthcare workflows. Its design supports integration into automated decision-support systems, reducing manual interpretation errors and expediting diagnosis. Furthermore, incorporating explainable AI (XAI) can enhance transparency and clinician trust, enabling smoother adoption in medical practice.
Data availability
Data will be available on request to the corresponding Author.
References
Sharif, M. et al. Recognition of different types of leukocytes using YOLOv2 and optimized bag-of-features. IEEE Access. 8, 167448–167459. https://doi.org/10.1109/ACCESS.2020.3021660 (2020).
Winter, G. et al. Clinical and genetic characteristics of children with acute lymphoblastic leukemia and Li-Fraumeni syndrome. Leukemia 35, 1475–1479. https://doi.org/10.1038/s41375-021-01163-y (2021).
Abir, W. H. et al. Explainable AI in diagnosing and anticipating leukemia using transfer learning method. Computational Intelligence and Neuroscience, 2022, 5140148. (2022). https://doi.org/10.1155/2022/5140148
Hegde, R. B., Prasad, K., Hebbar, H. & Singh, B. M. K. Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybernetics Biomedical Eng. 39, 382–392. https://doi.org/10.1016/j.bbe.2019.01.005 (2019).
Ghanem, P. et al. The vitamin E derivative gamma Tocotrienol promotes anti-tumor effects in acute myeloid leukemia cell lines. Nutrients 11, 2808. https://doi.org/10.3390/nu11112808 (2019).
Alfayez, M. et al. The clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia 35, 691–700. https://doi.org/10.1038/s41375-020-0920-z (2021).
Pan, L. et al. Machine learning applications for prediction of relapse in childhood acute lymphoblastic leukemia. Sci. Rep. 7, 7408. https://doi.org/10.1038/s41598-017-07408-0 (2017).
Godoy, P. B. G., Simionato, N. M., de Mello, C. B. & Suchecki, D. Assessment of executive functions after treatment of childhood acute lymphoid leukemia: A systematic review. Neuropsychol. Rev. 30, 386–406. https://doi.org/10.1007/s11065-020-09446-4 (2020).
Fujita, T. C., Sousa-Pereira, N., Amarante, M. K. & Watanabe, M. A. E. Acute lymphoid leukemia etiopathogenesis. Mol. Biol. Rep. 48, 817–822. https://doi.org/10.1007/s11033-020-06073-3 (2021).
Kumar, V. et al. Addressing binary classification over class imbalanced clinical datasets using computationally intelligent techniques. Healthcare 10, 1293. https://doi.org/10.3390/healthcare10071293 (2022).
Alsalem, M. et al. Systematic review of an automated multiclass detection and classification system for acute leukaemia in terms of evaluation and benchmarking, open challenges, issues and methodological aspects. J. Med. Syst. 42, 1064. https://doi.org/10.1007/s10916-018-1064-9 (2018).
Lalotra, G. S., Kumar, V. & Rajput, D. S. Predictive performance analysis of ensemble learners on BCD dataset. In 2021 IEEE International Conference on Technology, Research, and Innovation for Betterment of Society (TRIBES) 1–6 (IEEE, 2021). https://doi.org/10.1109/TRIBES52498.2021.9751648.
Kumar, V., Lalotra, G. S. & Kumar, R. K. Improving performance of classifiers for diagnosis of critical diseases to prevent COVID risk. Comput. Electr. Eng. 102, 108236. https://doi.org/10.1016/j.compeleceng.2022.108236 (2022).
Daqqa, K. A. A., Maghari, A. Y. & Al Sarraj, W. F. Prediction and diagnosis of leukemia using classification algorithms. In 2017 8th International Conference on Information Technology (ICIT) 638–643 (IEEE, 2017). https://doi.org/10.1109/ICITECH.2017.8079919.
Nazari, E. et al. Deep learning for acute myeloid leukemia diagnosis. J. Med. Life. 13, 382 (2020).
Hossain, M. A. et al. An effective leukemia prediction technique using supervised machine learning classification algorithm. In Proceedings of International Conference on Trends in Computational and Cognitive Engineering 219–229 (Springer, 2021). https://doi.org/10.1007/978-981-33-4673-4_19.
Bodzas, A. Diagnosis of malignant haematopoietic diseases based on the automation of blood microscopic image analysis (Master’s thesis). Technical University of Ostrava, Ostrava, CZ (2019).
Aljaboriy, S., Sjarif, N. & Chuprat, S. Segmentation and detection of acute leukemia using image processing and machine learning techniques: A review. AUS 26, 511–531. https://doi.org/10.4206/aus.2019.n26.2.60 (2019).
Putzu, L., Caocci, G. & Di Ruberto, C. Leucocyte classification for leukaemia detection using image processing techniques. Artif. Intell. Med. 62, 179–191. https://doi.org/10.1016/j.artmed.2014.09.002 (2014).
Hariprasath, S., Dharani, T. & Santh, M. Detection of acute lymphocytic leukemia using statistical features. In Proceedings of the 4th International Conference on Current Research in Engineering Science and Technology (ICCREST), Trichy (2019). http://www.internationaljournalssrg.org/uploads/specialissuepdf/ICCREST/2019/ECE/IJECE-ICCREST-P102-JRCE1119.pdf.
Moradiamin, M., Samadzadehaghdam, N., Kermani, S. & Talebi, A. Enhanced recognition of acute lymphoblastic leukemia cells in microscopic images based on feature reduction using principal component analysis. Front. Biomedical Technol. 2, 128–136 (2015).
Joshi, M. D., Karode, A. H. & Suralkar, S. R. White blood cells segmentation and classification to detect acute leukemia. Int. J. Emerg. Trends Technol. Comput. Sci. 2, 147–151 (2013).
Kazemi, F., Najafabadi, T. & Araabi, B. Automatic recognition of acute myelogenous leukemia in blood microscopic images using K-means clustering and support vector machine. J. Med. Signals Sens. 6, 183–193 (2016).
Neoh, S. et al. An intelligent decision support system for leukaemia diagnosis using microscopic blood images. Sci. Rep. 5, 14938. https://doi.org/10.1038/srep14938 (2015).
Eckardt, J. N., Bornhäuser, M., Wendt, K. & Middeke, J. M. Application of machine learning in the management of acute myeloid leukemia: current practice and future prospects. Blood Adv. 4, 6077–6085. https://doi.org/10.1182/bloodadvances.2020002997 (2020).
Alsuwaidi, L., Hachim, M. & Senok, A. Novel markers in pediatric acute lymphoid leukemia: the role of ADAM6 in B cell leukemia. Front. Cell. Dev. Biol. 9, 1680. https://doi.org/10.3389/fcell.2021.706129 (2021).
Hossain, M. A., Islam, A. M., Islam, S., Shatabda, S. & Ahmed, A. Symptom-based explainable artificial intelligence model for leukemia detection. IEEE Access. 10, 3176274. https://doi.org/10.1109/ACCESS.2022.3176274 (2022).
Clinton, L. P. Jr., Somes, K. M., Chu, Y. & Javed, F. Acute lymphoblastic leukemia detection using depthwise separable convolutional neural networks. SMU Data Sci. Rev. 3, 4 (2020).
Chatila, R. et al. Epidemiology and survival of colorectal cancer in lebanon: A sub-national retrospective analysis. Cancer Control. 28, 10732748211041221. https://doi.org/10.1177/10732748211041221 (2021).
Sleiman, Z. et al. Laparoscopic management of uncommon benign uterine tumors: a systematic review. Updates Surg. 71, 637–643. https://doi.org/10.1007/s13304-019-00651-2 (2019).
Aoun, R., Kaul, M. & Sahni, A. Excessive daytime sleepiness due to brain tumor. J. Clin. Sleep Med. 16, 2117–2119. https://doi.org/10.5664/jcsm.8788 (2020).
Ali, T. et al. A sequential machine learning-cum-attention mechanism for effective segmentation of brain tumor. Front. Oncol. 12, 873268. https://doi.org/10.3389/fonc.2022.873268 (2022).
Rizwan, M. et al. Brain tumor and glioma grade classification using Gaussian convolutional neural network. IEEE Access. 10, 29731–29740. https://doi.org/10.1109/ACCESS.2022.3153108 (2022).
Genovese, A., Hosseini, M. S., Piuri, V., Plataniotis, K. N. & Scotti, F. Acute lymphoblastic leukemia detection based on adaptive unsharpening and deep learning. In ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 1205–1209 (IEEE, 2021). https://doi.org/10.1109/ICASSP39728.2021.9414362.
de Oliveira, J. E. M. & Dantas, D. O. Classification of normal versus leukemic cells with data augmentation and convolutional neural networks. In Proceedings of the 16th International Conference on Computer Vision Theory and Applications (VISIGRAPP 2021) 685–692 (2021).
Bibi, N., Sikandar, M., Ud Din, I., Almogren, A. & Ali, S. IoMT-based automated detection and classification of leukemia using deep learning. J. Healthcare Eng. 2020, 6648574. https://doi.org/10.1155/2020/6648574 (2020).
Tanwar, V., Acharya, U. R. & Mandal, D. An explainable vision transformer model for white blood cells classification and localization. Diagnostics 13 (14), 2459. https://doi.org/10.3390/diagnostics13142459 (2023).
Raghaw, C. S., Sharma, A., Bansal, S., Rehman, M. Z. U. & Kumar, N. CoTCoNet: an optimized coupled transformer-convolutional network with an adaptive graph reconstruction for leukemia detection. Comput. Biol. Med. 193, 108821. https://doi.org/10.1016/j.compbiomed.2023.108821 (2024).
Katar, O. & Yildirim, Ö. An explainable vision transformer model based on white blood cells classification and localization. Diagnostics 13 (14), 2459. https://doi.org/10.3390/diagnostics13142459 (2023).
Guan, H., Yap, P. T., Bozoki, A. & Liu, M. Federated learning for medical image analysis: a survey. Pattern Recogn. 146, 110424. https://doi.org/10.1016/j.patcog.2024.110424 (2024).
Zhang, Y., Xu, H. & Chen, Z. When federated learning meets medical image analysis. In Lecture Notes in Computer Science 101–116 (Springer, 2024). https://doi.org/10.1007/978-3-031-71602-7_10.
Zhou, Z., Luo, G., Chen, M., Weng, Z. & Zhu, Y. Federated learning for medical image classification: a comprehensive benchmark. arXiv preprint, arXiv:2504.05238 (2025). https://arxiv.org/abs/2504.05238.
Dahiwade, D., Patle, G. & Meshram, E. Designing disease prediction model using machine learning approach. In 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC) 1211–1215 (IEEE, 2019). https://doi.org/10.1109/ICCMC.2019.8819782.
Díaz, G. & Manzanera, A. Automatic analysis of microscopic images in hematological cytology applications. In Biomedical Image Analysis and Machine Learning Technologies: Applications and Techniques (eds. González, F. A. & Romero, E.) 167–196 (Yurchak Printing Inc., 2009).
Funding
No Funding has been received.
Author information
Authors and Affiliations
Contributions
M.S Written the manuscript G. S Analysis & ResearchR. Written and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mohan, S.B., Sathya, S., Rajalakshmi, S. et al. Efficient convolutional neural networks for acute lymphoblastic leukaemia prediction in computer vision. Sci Rep 16, 1135 (2026). https://doi.org/10.1038/s41598-025-30777-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30777-w