Abstract
Remote ischemic preconditioning (RIPC) is a non-invasive strategy to protect tissues from ischemia-reperfusion injury (IRI), yet its systemic molecular mediators remain incompletely understood. Extracellular vesicles (EVs), particularly their microRNA (miRNA) cargo, have emerged as potential conveyors of RIPC-induced protection. In this translational study, we performed paired EV-miRNA profiling in five human subjects undergoing free flap surgery, with plasma collected before and 30 min after a standardized RIPC protocol. EVs were isolated, RNA was extracted and profiled using a 384-plex qPCR array. Differential expression was assessed using paired statistics and false discovery correction. Functional impact was inferred via validated target mapping, gene set enrichment analysis (GSEA), and miRNA-gene-pathway network modeling. RIPC induced a rapid, systemic shift in the circulating EV-miRNA profile. Principal component analysis revealed consistent directional changes post-RIPC. Four miRNAs, hsa-miR-505-3p (log2FC = -3.30, p = 0.025), hsa-miR-374a-5p (log2FC = -4.52, p = 0.028), hsa-miR-200a-3p (log2FC = -7.27, p = 0.044), and hsa-miR-181b-5p (log2FC = -3.94, p = 0.045), were significantly downregulated across the cohort. No miRNAs were consistently upregulated. GSEA of validated targets showed positive enrichment of stress-responsive and pro-survival pathways, including TNF-alpha signaling, TGF-beta, Notch, hypoxia, angiogenesis, and apoptosis. Network analysis identified miR-181b-5p and miR-374a-5p as central regulators affecting multiple interconnected pathways. These results suggest that RIPC elicits systemic protection not by introducing new molecular signals but by selectively removing suppressive EV-bound miRNAs. This may reflect a subtractive regulatory mechanism and enable rapid derepression of protective gene programs, offering a plausible explanation for the early benefits of RIPC. These findings should be validated in larger, multi-timepoint cohorts.
Similar content being viewed by others
Introduction
Ischemia reperfusion injury (IRI) is a pathological process that drives tissue damage in acute conditions such as myocardial infarction, stroke, organ transplantation and free flap surgery, ultimately contributing to morbidity and mortality1. In these settings, timely reperfusion is essential to salvage ischemic tissue; however, paradoxically, the restoration of blood flow can itself exacerbate cell death and inflammation2. This phenomenon is called IRI. Despite advances in revascularization therapies, there are currently no broadly effective pharmacological interventions to prevent IRI, underscoring an urgent need for novel cytoprotective strategies3. One promising approach is remote ischemic preconditioning (RIPC), which involves applying brief, non-lethal cycles of ischemia and reperfusion to a limb to protect distant organs from subsequent IRI4,5,6. This simple, non-invasive procedure, often accomplished by inflating a blood-pressure cuff on the arm or leg for a few minutes at a time, utilizes an innate protective mechanism of the body. RIPC has demonstrated robust protective effects across a range of clinical settings, including reducing myocardial injury, preserving neurological function, and improving outcomes in major cardiovascular and surgical interventions4,7,8,9. However, the central mechanism by which RIPC confers systemic protection remains only partially understood and the biological pathways conveying the “preconditioning” signal from the limb to vital organs are not yet fully elucidated.
Current evidence suggests that the underlying mechanisms of RIPC involve a complex interplay of neural and humoral pathways10,11,12,13,14,15,16. While neural circuits, such as the vago-splenic axis, play a crucial role in signal transduction, the humoral pathway is characterized by the release of protective factors into the circulation. Among the leading candidates for these humoral mediators are extracellular vesicles (EVs) – nanoscale, membrane-bound vesicles released into the bloodstream17,18. Indeed, recent studies indicate that EVs released during RIPC are both necessary and sufficient for its remote benefits, as EVs isolated after a preconditioning stimulus have been shown to carry protective properties to distant cells17,19. EVs, which include exosomes (typically 30–150 nm in size) and slightly larger microvesicles, serve as intercellular messengers, carrying cargos of proteins, lipids, and nucleic acids between cells. Notably, they can circulate through the blood and deliver their cargo to specific target cells, thereby triggering signaling cascades in tissues far from their cell of origin. This has led to intense interest in EVs as the humoral factors of RIPC.
Among the diverse bioactive cargo carried by EVs, microRNAs (miRNAs) have emerged as particularly intriguing mediators of RIPC’s effects18,20,21. MiRNAs are small (~ 22 nucleotide) non-coding RNAs that regulate gene expression post-transcriptionally, typically by binding to target mRNAs and inhibiting their translation or stability22. These molecules have been identified in EVs and are remarkably stable in circulation, protected from enzymatic degradation within the vesicle lumen. It is well established that miRNAs can orchestrate cytoprotective or injury-response pathways in cells including pathways involved in apoptosis, inflammation, and metabolism18,20,21,23. RIPC has been hypothesized to provoke the release of specific EV-associated miRNAs into the bloodstream, which then home to vulnerable organs (e.g., heart or brain) and preemptively activate cytoprotective gene programs19,20,21,24. However, while a number of individual miRNAs have been postulated to play such roles, a comprehensive profile of the EV-bound miRNAs altered by RIPC is lacking. The present study addresses this gap by characterizing the circulating EV-miRNA signature before and after a standardized RIPC stimulus in humans. The aim of this study was to identify key miRNAs modulated by an RIPC stimulus by performing a direct, high-resolution comparative analysis of circulating EV-associated miRNA profiles immediately before vs. immediately after the RIPC intervention in human subjects undergoing free flap surgery. We hypothesized that RIPC would induce a rapid and significant alteration in circulating EV-miRNAs, resulting in the release of specific miRNAs that target pathways involved in cytoprotection.
Materials and methods
Study population and sample origin
The samples analyzed in this study were selected from the intervention (RIPC) arm of a larger clinical trial investigating the effects of RIPC on IRI in free flap surgery25. The present sub-study focuses on a representative cohort of five patients from this intervention group. Free flap reconstructions in this cohort were all performed because of a defect caused by cancer (e.g., breast reconstruction following mastectomy). All patients were cancer-free at the time of blood drawing and prior to their reconstructive surgery. RIPC was performed using a tourniquet applied to the patient’s arm, inflated to 250 mmHg for three cycles of ten-minute occlusion followed by ten-minute reperfusion. For these patients, plasma samples were collected at two time points: immediately before the RIPC procedure (pre-RIPC) and within 30 min post-intervention (post-RIPC). Blood was collected from the contralateral arm using a 14G peripheral intravenous access established without tourniquet application. Baseline blood sample collection, the RIPC maneuver, and the post-intervention blood sample collection were completed prior to the induction of anesthesia. Patient characteristics can be found in Table 1.
Ethics declarations
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Institutional Ethics Committee of Ruhr University Bochum (Approval Number: 19–6752, 08 January 2020). All participants gave written informed consent prior to enrolment. In line with the General Data Protection Regulation (GDPR) and institutional policies, all samples were anonymized before analysis. The present sub-study analyzed only samples obtained from participants in the RIPC arms of the parent randomized trial.
EV Isolation, MiRNA Extraction, and cDNA synthesis
Extracellular vesicles (EVs) were isolated from EDTA plasma using the ExoQuick (EQ) method as previously described26. Briefly, EVs were precipitated from 500 µl EDTA plasma using ExoQuick Exosome Precipitation (System Biosciences, Palo Alto, USA) as recommended by the manufacturer with minor modifications. After precipitation of the EVs from the plasma, we carried out a secondary purification step. Using PD Spin Traps G25 (Cytiva, Marlborough, USA) the EVs were filtered through sepharose to remove any remaining EQ from the extracted EVs. EV isolation efficiency and marker profile (e.g., Western Blot for Alix, CD63) using this specific ExoQuick protocol have been published by our group previously26,27. An aliquot of the isolated EVs was used for quantification and size determination via Nanoparticle Tracking Analysis (NTA) with a ZetaView® (Particle Metrix, GmbH, Meerbusch, Germany) instrument as previously described. In short, the particle concentration was measured after concentration correction to 50–200 particles/position. We included five cycles and 11 positions. The “region of interest” (ROI) was set to 50–150 nm. NTA quantification data comparing pre- and post-RIPC samples is provided in Supple Figure S1 and Supplementary Table S1. For miRNA analysis, the remaining EV fraction was treated with RNase A (Thermo Fisher Scientific, Waltham, MA, USA) to remove miRNA or any other extra-vesicular RNA, ensuring that only intra-vesicular cargo was analyzed. Subsequent microRNA extraction was performed with the mirVANA™ miRNA Isolation Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and the addition of a synthetic cel-mir-39-3p spike-in control (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) to monitor extraction efficiency. The extracted miRNAs were stored at −80 °C until they were reverse-transcribed into cDNA using the TaqMan™ Advanced miRNA-cDNA Synthesis Kit according to the manufacturer’s instructions with a slight modification to the number of cycles in the mirAMP amplification step.
MiRNA expression profiling and data processing
miRNA expression was profiled using quantitative real-time PCR and the Human Advanced miRNA Array Card A (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions, without modification. Data acquisition was performed on a QuantStudio 7 Flex Real-Time PCR System. Raw amplification data (CT values) were processed using a custom Python pipeline. CT values reported as “undetermined” were conservatively imputed with a value of 40 to represent the limit of detection. A quality control filtering step was then applied: miRNAs were retained for analysis only if they were detected (CT < 40) in at least two of the five samples in either the pre-RIPC or post-RIPC group. This filtering removes uninformative assays while retaining miRNAs that may be specifically activated or repressed by the I/R event. For the retained miRNAs, CT values were normalized to the cel-mir-39 spike-in control to compute ΔCT values (ΔCT = CTmiRNA−CTspike − in). The final log2-transformed relative expression values were calculated as − ΔCT and used for all downstream statistical analyses.
Paired differential expression analysis
To identify miRNAs that were differentially expressed following RIPC, a paired analysis of the log2-transformed miRNA expression profiles was performed. For each miRNA, a paired, two-tailed Student’s t-test was used to compare the pre- and post-RIPC expression values within each of the five patients. The log2 fold change (log2FC) was calculated as the difference between the mean post-RIPC expression and the mean pre-RIPC expression. To account for multiple hypothesis testing, the resulting p-values were adjusted using the Benjamini-Hochberg method to generate a False Discovery Rate (FDR) q-value28. A miRNA was considered statistically significant if its FDR q-value was less than 0.2. To further increase confidence, “consistently regulated” miRNAs were identified as those that changed expression in the same direction (up or down) in at least three of the five patients.
Bioinformatic pathway enrichment analysis
To infer the biological impact of the observed miRNA changes, we performed Gene Set Enrichment Analysis (GSEA). This was achieved by first translating the list of differentially expressed miRNAs into a ranked list of their validated gene targets. Raw miRNA identifiers were systematically cleaned and normalized to ensure compatibility with annotation databases. The normalized identifiers were mapped to their experimentally validated gene targets using the miRTarBase database, accessed via the gseapy (v1.1.0) Python library29. A ranked list of all unique target genes was generated. The ranking metric for each gene was set to the negative log2FC of its most strongly regulated targeting miRNA, reflecting the canonical inverse relationship between miRNA and target gene expression. Preranked GSEA was performed on this gene list using gseapy.prerank with 1,000 permutations. Enrichment was tested against the Human MSigDB Hallmark gene set30,31. A pathway was considered significantly enriched at an FDR q-value < 0.2. Results were visualized using dot plots and volcano plots.
Network visualization
To synthesize these findings, a miRNA-gene-pathway interaction network was constructed. The network included miRNAs with a nominal p-value < 0.05, Hallmark pathways with an FDR q-value < 0.2, and the genes that were both validated targets of the included miRNAs and members of a leading-edge enrichment subset. The network was visualized using the networkx library with a Kamada-Kawai layout to illustrate the relationships between these components32.
Results
A distinct EV-miRNA signature is induced by remote ischemic preconditioning
To characterize the circulating EV-miRNA profile in response to RIPC, we performed high-throughput qPCR analysis on plasma samples from five patients taken before and after the intervention. Our data processing and filtering cascade is summarized in Fig. 1. Of the 384 target miRNA assays on the array, 203 were robustly detected in at least one sample in either the pre- or post-RIPC group. Applying our pre-defined filtering criterion of retaining miRNAs detected in at least two samples per group resulted in a final set of 134 high-confidence miRNAs that were used for subsequent analyses.
miRNA Data Filtering Cascade. The bar chart illustrates the number of miRNAs retained at each stage of the data filtering process. Starting from the 384 target miRNA assays on the array, 203 were detected in at least one sample in either the pre- or post-RIPC group. The final set of 134 miRNAs used for differential expression analysis was obtained by requiring detection in at least two samples in either group.
RIPC induces a systemic and asymmetric change in the Circulating EV-miRNome
To assess the global impact of the RIPC procedure on this miRNA profile, we first performed a Principal Component Analysis (PCA). The PCA plot revealed a clear and consistent trend, with post-RIPC samples shifting away from their pre-RIPC counterparts for each patient, indicating a distinct, systemic effect of the intervention on the circulating miRNome (Fig. 2A). To identify the specific miRNAs driving this change, we conducted a paired differential expression analysis. The volcano plot visualizes both the statistical significance and the magnitude of change for all 134 miRNAs (Fig. 2B). Several miRNAs clustered around a log2 fold change of zero, with no significant change in expression. However, a clear asymmetric response was observed: four miRNAs were significantly downregulated (p < 0.05), all with a strong magnitude of change (log2FC < −2.5), while no miRNAs were found to be significantly upregulated at this threshold. This suggests that the primary response to RIPC is a consistent downregulation of specific miRNAs.
Global miRNA Profile Changes Following RIPC. (A) Principal Component Analysis of the 134 miRNA profiles. Each point represents a sample, with lines connecting the pre- and post-RIPC samples for each patient. The plot shows a consistent directional shift post-intervention. (B) Volcano plot illustrating the log2 fold change and statistical significance (-log10 p-value) for each miRNA. The four significantly downregulated miRNAs (p < 0.05) are highlighted in red.
Top candidate MiRNAs exhibit a uniform downregulation pattern
To further characterize the RIPC-induced EV-miRNA signature, we visualized the top 25 differentially expressed miRNAs (ranked by p-value) using a heatmap (Fig. 3A). This analysis revealed a clear separation between pre- and post-RIPC samples, primarily driven by a prominent cluster of downregulated miRNAs. Examination of individual miRNA changes across all five patients demonstrated a highly uniform downregulation pattern for the most significantly altered candidates. The top four (down-)regulated miRNAs were hsa-miR-505-3p (log2FC = −3.30, p = 0.025), hsa-miR-374a-5p (log2FC = −4.52, p = 0.028), hsa-miR-200a-3p (log2FC = −7.27, p = 0.044), and hsa-miR-181b-5p (log2FC = −3.94, p = 0.045), all of which decreased consistently in patients following RIPC (Fig. 3B; Supplementary Figure S2). In contrast, several miRNAs with a positive overall fold change, such as hsa-miR-501-3p (log2FC = 10.56, p = 0.137), exhibited heterogeneous expression profiles. These upregulation patterns were typically driven by one or two outlier patients, while the remaining individuals showed minimal change (Fig. 3C). This asymmetry was further supported by a consistency analysis across patients: miRNAs downregulated in ≥ 3 of 5 individuals (Supplementary Table S3) displayed substantially lower p-values compared to those in the upregulated group (Supplementary Table S4).
Characterization of Top Differentially Expressed miRNAs. (A) Heatmap of the top 25 miRNAs ranked by p-value. The columns represent individual samples, and rows represent miRNAs. The clear separation between pre- and post-RIPC samples is evident. (B) Variability plot for the top significant candidate, hsa-miR-505-3p, showing a consistent decrease in expression across all five patients. (C) Variability plot for hsa-miR-501-3p, illustrating an outlier upregulation pattern driven by a strong response in only two patients.
Functional analysis predicts modulation of key protective and signaling pathways
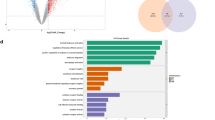
To elucidate the potential biological function of the observed miRNA signature, we performed Gene Set Enrichment Analysis (GSEA). Given the canonical inverse relationship between miRNAs and their targets, a ranked list of validated target genes was generated based on the negative log2 fold change of their targeting miRNAs, using a comprehensive miRNA-target interaction map derived from the miRTarBase database (Supplementary Table S5). This analysis, focused on the MSigDB Hallmark collection, revealed a significant enrichment of several pathways critical to cellular signaling and stress responses (Fig. 4; Supplementary Figure S3).
Notably, the analysis showed a strong and consistent positive enrichment for pathways central to inflammation, cell fate, and tissue remodeling. The top five enriched pathways were TNF-alpha Signaling Via NF-kB (NES = 2.02, FDR q-val = 0.001), UV Response Down (NES = 2.02, FDR q-val = 0.0005), TGF-beta Signaling (NES = 1.87, FDR q-val = 0.002), Notch Signaling (NES = 1.77, FDR q-val = 0.008), and Angiogenesis (NES = 1.76, FDR q-val = 0.006). The uniformly positive Normalized Enrichment Scores (NES) indicate that the leading-edge genes of these pathways are concentrated among those whose targeting miRNAs were downregulated by RIPC. This suggests a functional derepression of these programs, consistent with a cellular “priming” mechanism. Furthermore, pathways directly relevant to ischemia-reperfusion injury, such as Hypoxia (NES = 1.69, FDR q-val = 0.003) and Apoptosis (NES = 1.52, FDR q-val = 0.005), were also significantly enriched. The coordinated regulation of these programs implies that the systemic EV-miRNA signal may prepare distant tissues for subsequent ischemic stress by modulating fundamental inflammatory, cell-survival and tissue-remodeling cascades.
Functional Enrichment of the RIPC-Induced miRNA Signature. Dot plot of Gene Set Enrichment Analysis (GSEA) results against the Hallmark pathway database. The x-axis shows the Normalized Enrichment Score (NES). The size of each dot corresponds to the number of leading-edge genes in the pathway, and the color represents the FDR q-value. All significantly enriched pathways (FDR < 0.2) show a positive NES, indicating that their member genes are targets of the downregulated miRNAs.
Network analysis reveals a coordinated regulatory hub
To synthesize our pathway enrichment and target prediction results into an integrative framework, we constructed a miRNA-gene-pathway interaction network using the Hallmark gene sets (Fig. 5). This network connects the four significantly downregulated EV-miRNAs (p < 0.05) to their predicted target genes within the leading-edge subsets of enriched pathways. The resulting topology reveals a highly coordinated regulatory hub in which a small set of miRNAs exerts broad influence across multiple interrelated stress-response pathways. Within this network, hsa-miR-181b-5p and hsa-miR-374a-5p emerge as central regulators, targeting 68 and 53 genes respectively, and dominating the interaction space compared to hsa-miR-200a-3p (35 genes) and hsa-miR-505-3p (25 genes). Notably, many of these target genes are not limited to a single function; they are shared components across several critical Hallmark pathways, including TNF-alpha/NF-κB signaling, hypoxia response, TGF-beta signaling, and apoptosis. This high degree of interconnectivity suggests a concerted, systems-level response, where RIPC-triggered EV-miRNA modulation reprograms stress-adaptive gene networks across tissues. Furthermore, the central positioning of these miRNAs suggests a model in which systemic EV-miRNA downregulation coordinates tissue-wide priming for ischemic tolerance. By targeting genes with such pleiotropic effects, the miRNA response to RIPC appears to induce a multi-layered protective phenotype that enhances resilience against subsequent IRI.
Predicted miRNA-Gene-Pathway Interaction Network. The network visualizes the connections between significantly downregulated miRNAs (p < 0.05, blue nodes), their validated gene targets that are part of a leading-edge subset (grey nodes), and the significantly enriched Hallmark pathways (FDR < 0.2, green nodes). The layout highlights the central role of the downregulated miRNAs in regulating a large number of genes across multiple protective pathways.
Discussion
In this study, we demonstrate that a brief RIPC stimulus in humans provokes a rapid, systemic shift in the circulating EV-associated microRNA profile. Notably, this shift is defined by a coordinated downregulation of a small subset of miRNAs, with no miRNAs showing consistent or significant upregulation. All five patients exhibited a similar directional change post-RIPC, as evidenced by principal component analysis and paired comparisons. Four miRNAs (hsa-miR-181b-5p, hsa-miR-374a-5p, hsa-miR-200a-3p, and hsa-miR-505-3p) were significantly decreased in EVs after RIPC, with consistent trends across the cohort. In contrast, miRNAs with nominal positive fold-changes varied considerably between individuals and did not reach significance. This asymmetric pattern suggests that the primary early response to RIPC is not the addition of new protective signals, but the selective removal of key regulatory miRNAs from the EV compartment, potentially to relieve suppression of downstream stress-adaptive pathways.
These findings both complement and differ from earlier work on RIPC-induced molecular effects18,20,33,34. Several preclinical studies have described upregulation of specific cardioprotective miRNAs in EVs following RIPC, such as miR-2418, miR-126a-3p35, and miR-14420, which contribute to anti-apoptotic signaling or kinase activation in target tissues. In human cardiac surgery settings, RIPC has been associated with increased EV concentrations and altered EV miRNA content, including elevation of canonical cytoprotective miRNAs like miR-2121. However, our study did not identify consistent upregulation of any of these miRNAs in the acute phase following RIPC. Instead, the dominant response was a uniform reduction in a small set of EV-bound miRNAs. These differences likely reflect variations in timing and physiology. Our study captured the acute humoral response within 30 min of RIPC, whereas many prior findings pertain to later stages of conditioning or require subsequent ischemic insults to manifest fully36,37. It is therefore plausible that the early systemic response to RIPC in humans relies more on the withdrawal of inhibitory miRNAs than on the deployment of new effectors.
The striking downregulation of these miRNAs suggests that RIPC initiates a systemic release of constraints on multiple stress-responsive and pro-survival pathways. Gene set enrichment analysis revealed that the validated targets of these miRNAs cluster within Hallmark pathways involved in inflammation, cellular adaptation, and tissue remodeling, including TNFα and NF-κB signaling, TGF-β and Notch signaling, angiogenesis, hypoxia responses, and apoptosis regulation. These pathways exhibited positively skewed enrichment scores, indicating predicted upregulation of their member genes following miRNA downregulation. In functional terms, this suggests that RIPC removes miRNA-mediated repression from critical survival and repair pathways, transiently priming endothelial, immune, and parenchymal compartments for future ischemic stress.
The individual miRNAs reinforce this mechanistic interpretation: miR-181b-5p is a known repressor of NF-κB signaling through direct inhibition of importin-α3, a nuclear transport protein essential for NF-κB translocation38,39. Its reduction is expected to facilitate controlled NF-κB activation, a recognized early component of ischemic preconditioning. Similarly, miR-374a-5p is a known suppressor of chemokines such as MCP-1 and cytokines including IL-6 and TNF-α40,41. Its downregulation may allow a transient rise in inflammatory signaling that contributes to immune system priming. MiR-200a-3p targets components of the TGF-β/Smad and Notch pathways; its reduction is predicted to enhance anti-apoptotic and regenerative signaling42,43,44. Finally, miR-505-3p acts as a negative regulator of inflammation through pathways involving HMGB1 and Uromodulin45,46,47. Lowering miR-505-3p levels may permit a controlled increase in pro-inflammatory signaling that initiates protective cascades, such as M2 macrophage recruitment and ischemic tolerance.
Our findings support a framework in which RIPC induces a rapid, coherent reprogramming of the circulating EV miRNA landscape. Rather than broadcasting a single protective signal, RIPC modifies the composition of circulating EVs in a targeted manner, removing specific regulatory miRNAs that collectively govern survival, hypoxia, and immune-related gene networks. This mechanism may complement other forms of RIPC signal transmission, including neural and cytokine-mediated pathways, but is uniquely well-suited for targeted intercellular communication. EVs can home to specific organs and deliver cargo directly into recipient cells, and the absence of particular miRNAs from these vesicles may act as a functional cue48. Our network analysis further highlights miR-181b-5p and miR-374a-5p as central regulatory nodes with broad influence over multiple interconnected pathways. These miRNA changes likely contribute to the systems-level shift that prepares tissues for future ischemic insult. That such changes are detectable within the short time frame investigated in this study underscores the speed and coordination of this protective signaling mechanism.
This study has several limitations. First, the small sample size (n = 5) and the specific demographic distribution (predominance of female patients with prior breast cancer) limits generalizability, though the paired design provides internal consistency. These factors caution against extrapolating the findings to other populations without further validation. Second, our analysis was limited to a single early time point; dynamic changes occurring before or after 30 min may have been missed. Future studies should incorporate detailed time-course analyses to capture transient or delayed miRNA responses. Third, we relied on a predefined panel of 384 miRNAs using qPCR-based profiling. Unbiased small RNA sequencing may identify additional RIPC-responsive species, including rare or unannotated miRNAs. Fourth, we could not determine the cellular origin of the EV-bound miRNAs. RIPC likely affects multiple donor cell types, including endothelium, circulating immune cells, and limb muscle, and further studies using EV surface markers or tissue-specific profiling are warranted. Lastly, our study was observational in nature. Although bioinformatic analysis predicts derepression of protective pathways, these in silico predictions are hypothetical. Functional experiments are needed to establish causality and confirm that removal of these miRNAs contributes directly to ischemic protection.
Future research should aim to validate these findings in larger, independent cohorts with appropriate control groups. Time-course profiling of EV-miRNA content will be essential to map the kinetics of the response and identify optimal therapeutic windows. Functional studies using gain- and loss-of-function approaches are required to determine whether specific miRNAs mediate RIPC-induced protection. Investigating the upstream cues that lead to miRNA downregulation in EVs such as shear stress, hypoxia, or adrenergic signaling may provide further insight into how RIPC modifies vesicle content. From a translational perspective, targeted inhibition of the downregulated miRNAs using antagomirs could potentially mimic the effects of RIPC without requiring ischemic stimuli. Such a strategy would need to be carefully evaluated for safety and specificity but may open new options for prophylactic organ protection in high-risk surgical or ischemic settings.
Taken together, this study provides novel evidence that RIPC in humans rapidly alters the circulating EV miRNA landscape through the selective downregulation of key regulatory miRNAs. These changes appear to relieve repression on stress-responsive pathways, enabling broad activation of pro-survival, inflammatory, angiogenic, and anti-apoptotic gene networks. Rather than introducing new signals, RIPC may act primarily by withdrawing suppressive ones. This subtractive molecular editing, detectable within minutes of the stimulus, offers a compelling mechanistic explanation for the systemic benefits of RIPC. We emphasize, however, that given the sample size and statistical design, these results are exploratory in nature and should be viewed as hypothesis-generating. Nevertheless, our findings suggest that EV-associated miRNAs are not only markers of conditioning but may also serve as functional mediators, with potential implications for biomarker development and therapeutic innovation in IRI.
Data availability
All processed data supporting the findings of this study are included in this published article and its supplementary files. The raw qPCR data and code used for analysis are available from the corresponding author upon reasonable request.
References
Tang, L. et al. Autophagy: a double-edged sword in ischemia–reperfusion injury. Cell. Mol. Biol. Lett. 30, 42 (2025).
Wu, M. Y. et al. Current mechanistic concepts in ischemia and reperfusion injury. Cell. Physiol. Biochem. 46, 1650–1667 (2018).
Davidson, S. M. et al. Multitarget strategies to reduce myocardial Ischemia/Reperfusion injury: JACC review topic of the week. J. Am. Coll. Cardiol. 73, 89–99 (2019).
Lv, J. et al. RIPC provides neuroprotection against ischemic stroke by suppressing apoptosis via the mitochondrial pathway. Sci. Rep. 10, 1–13 (2020).
Brevoord, D. et al. Remote ischemic conditioning to protect against ischemia-reperfusion injury: A systematic review and meta-analysis. PLoS ONE 7, e42179 (2012).
Rossaint, J. et al. Remote ischemic preconditioning causes transient cell cycle arrest and renal protection by a NF-κB-dependent Sema5B pathway. JCI Insight 7, e158523 (2022).
Billah, M. et al. Egr-1 functions as a master switch regulator of remote ischemic preconditioning-induced cardioprotection. Basic Res. Cardiol 115, 3 (2020).
Yang, L., Wang, G., Du, Y., Ji, B. & Zheng, Z. Remote ischemic preconditioning reduces cardiac troponin I release in cardiac surgery: a meta-analysis. J. Cardiothorac. Vasc Anesth. 28, 682–689 (2014).
Chen, G., Thakkar, M., Robinson, C. & Doré, S. Limb remote ischemic conditioning: Mechanisms, Anesthetics, and the potential for expanding therapeutic options. Front. Neurol. 9, 40 (2018).
Heinen, A. et al. The release of cardioprotective humoral factors after remote ischemic preconditioning in humans is age- and sex-dependent. J. Transl Med. 16, 112 (2018).
Sawashita, Y. et al. Remote ischemic preconditioning reduces myocardial ischemia–reperfusion injury through unacylated ghrelin-induced activation of the JAK/STAT pathway. Basic. Res. Cardiol. 115, 50 (2020).
Hildebrandt, H. A. et al. Kinetics and signal activation properties of Circulating Factor(s) from healthy volunteers undergoing remote ischemic Pre-Conditioning. JACC Basic. Transl Sci. 1, 3–13 (2016).
Lieder, H. R. et al. Platelet-Mediated transfer of cardioprotection by remote ischemic conditioning and its abrogation by aspirin but not by Ticagrelor. Cardiovasc. Drugs Ther. 37, 865–876 (2023).
Lieder, H. R. et al. Vago-splenic signal transduction of cardioprotection in humans. Eur. Heart J. 45, 3164–3177 (2024).
Lieder, H. R. et al. Bioassays of humoral cardioprotective factors released by remote ischemic conditioning in patients undergoing coronary artery bypass surgery. J. Cardiovasc. Pharmacol. Ther. 27, 10742484221097273 (2022).
Heusch, G. & Kleinbongard, P. The spleen in ischaemic heart disease. Nat. Rev. Cardiol. 22, 497–509 (2025).
Giricz, Z. et al. Cardioprotection by remote ischemic preconditioning of the rat heart is mediated by extracellular vesicles. J. Mol. Cell. Cardiol. 68, 75–78 (2014).
Minghua, W. et al. Plasma exosomes induced by remote ischaemic preconditioning attenuate myocardial ischaemia/reperfusion injury by transferring miR-24. Cell. Death Dis. 9, 320 (2018).
Frey, U. H. et al. Remote ischaemic preconditioning increases serum extracellular vesicle concentrations with altered micro-RNA signature in CABG patients. Acta Anaesthesiol. Scand. 63, 483–492 (2019).
Li, J. et al. MicroRNA-144 is a Circulating effector of remote ischemic preconditioning. Basic. Res. Cardiol. 109, 423 (2014).
Pan, T. et al. Delayed remote ischemic preconditioning confers renoprotection against septic acute kidney injury via Exosomal miR-21. Theranostics 9, 405–423 (2019).
Cannell, I. G., Kong, Y. W. & Bushell, M. How do MicroRNAs regulate gene expression? Biochem. Soc. Trans. 36, 1224–1231 (2008).
Jiang, J. et al. MiR-499/PRDM16 axis modulates the adipogenic differentiation of mouse skeletal muscle satellite cells. Hum. Cell. 31, 282–291 (2018).
Zhu, W. et al. miR-142-5p encapsulated by Serum-Derived extracellular vesicles protects against acute lung injury in septic rats following remote ischemic preconditioning via the PTEN/PI3K/Akt axis. J. Innate Immun. 14, 532–542 (2022).
Drysch, M. et al. Ischemia–Reperfusion injury in free flaps: molecular mechanisms and protective effects of remote ischemic preconditioning. J. Cell. Mol. Med. 29, e70739 (2025).
Gluth, L. et al. Influence of the anesthetic technique on Circulating extracellular vesicles in bladder cancer patients undergoing radical cystectomy: A Prospective, randomized trial. Cells 12, 2503 (2023).
Pham, P. N. V. et al. Influence of anesthetic regimes on extracellular vesicles following remote ischemic preconditioning in coronary artery disease. Int. J. Mol. Sci. 25, 9304 (2024).
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 (2001).
Hsu, S. D. et al. MiRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39, D163–169 (2011).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005).
Hagberg, A., Swart, P. J. & Schult, D. A. Exploring Network Structure, Dynamics, and Function Using NetworkX. (2008). https://www.osti.gov/biblio/960616
Li, S. et al. Limb remote ischemic conditioning promotes neurogenesis after cerebral ischemia by modulating miR-449b/Notch1 pathway in mice. Biomolecules 12, 1137 (2022).
Kohns, M., Huhn, R., Bauer, I. & Brandenburger, T. MiRNA-Mediated mechanisms of cardiac protection in ischemic and remote ischemic Preconditioning—A qualitative systematic review. Shock 51, 44 (2019).
Li, D. et al. Plasma exosomes at the late phase of remote ischemic Pre-conditioning attenuate myocardial Ischemia-Reperfusion injury through transferring miR-126a-3p. Front. Cardiovasc. Med. 8, 736226 (2021).
Hummitzsch, L. et al. Effects of remote ischemic preconditioning (RIPC) and chronic remote ischemic preconditioning (cRIPC) on levels of plasma cytokines, cell surface characteristics of monocytes and in-vitro angiogenesis: a pilot study. Basic. Res. Cardiol. 116, 60 (2021).
Gedik, N. et al. Potential humoral mediators of remote ischemic preconditioning in patients undergoing surgical coronary revascularization. Sci. Rep. 7, 12660 (2017).
Florio, T. J. et al. Differential recognition of canonical NF-κB dimers by importin α3. Nat. Commun. 13, 1207 (2022).
Sun, X. et al. MicroRNA-181b regulates NF-κB–mediated vascular inflammation. J. Clin. Invest. 122, 1973–1990 (2012).
Chen, Z., Hu, Y., Lu, R., Ge, M. & Zhang, L. MicroRNA-374a-5p inhibits neuroinflammation in neonatal hypoxic-ischemic encephalopathy via regulating NLRP3 inflammasome targeted Smad6. Life Sci. 252, 117664 (2020).
Perez-Sanchez, C. et al. miR-374a-5p regulates inflammatory genes and monocyte function in patients with inflammatory bowel disease. J. Exp. Med. 219, e20211366 (2022).
Xue, L. et al. miR-200-3p suppresses cell proliferation and reduces apoptosis in diabetic retinopathy via blocking the TGF-β2/Smad pathway. Biosci. Rep. 40, BSR20201545 (2020).
Wang, B. et al. miR-200a prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes 60, 280–287 (2010).
Brabletz, S. et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 30, 770–782 (2011).
He, D. et al. The Effect of miR-505-5p on Inhibition of Serum Uromodulin Ameliorates Myocardial Inflammation and Apoptosis Induced by Ischemia-Reperfusion. Oxid. Med. Cell. Longev. 3521971 (2022).
Wang, S. & Zhang, Y. HMGB1 in inflammation and cancer. J. Hematol. Oncol. J. Hematol. Oncol. 13, 116 (2020).
Liu, J. et al. MiR-505 as an anti-inflammatory regulator suppresses HMGB1/NF-κB pathway in lipopolysaccharide-mediated endometritis by targeting HMGB1. Int. Immunopharmacol. 88, 106912 (2020).
Zhou, X. et al. Tumour-derived extracellular vesicle membrane hybrid lipid nanovesicles enhance SiRNA delivery by tumour-homing and intracellular freeway transportation. J. Extracell. Vesicles. 11, e12198 (2022).
Funding
Open Access funding enabled and organized by Projekt DEAL. The senior author A.S. received a grant (K128-19) from the “Junior Clinician Scientist” program by the FoRUM-program of the medical faculty of Ruhr-University Bochum.
Author information
Authors and Affiliations
Contributions
M.D. contributed to conceptualization, formal analysis, data curation, visualization, and writing of the original draft as well as review and editing; A.F., S.V.S., F.R., and F.P. contributed to investigation and manuscript review and editing; T.K. contributed resources and manuscript review and editing; U.H.F. contributed methodology, supervision, resources, and manuscript review and editing; C.O. contributed to methodology, investigation and manuscript review and editing; M.L. contributed supervision, resources, and manuscript review and editing; C.W. and A.S. contributed to conceptualization, funding acquisition, and manuscript review and editing; A.S. also contributed to project administration and supervision. C.W. and A.S. contributed equally to this work.https://submission.springernature.com/new/submission/1aea15b5-0092-494c-8831-06ce35377cc0/declarations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Drysch, M., Fiedler, A., Schmidt, S.V. et al. Systemic downregulation of EV-associated MiRNAs following remote ischemic preconditioning. Sci Rep 15, 43657 (2025). https://doi.org/10.1038/s41598-025-31356-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-31356-9