Abstract
Allergic pulmonary inflammation is driven by allergen-specific Th2/Th17 responses counterregulated by tolerogenic dendritic cells (tDCs) and regulatory T cells (Tregs). While early-life colonization with commensal Clostridia induces the generation of colonic Tregs, the therapeutic potential of Clostridium leptum (CL) monotherapy to treat asthma patients with comorbid gut dysbiosis remains unexplored. Therefore, this study aimed to investigate whether 14 days of oral CL administration restores gut–lung immune homeostasis and suppresses allergic inflammation via tDC/Treg induction in a dysbiosis-asthma comorbidity model. To this end, adult BALB/c mice were subjected to ovalbumin (OVA) sensitization/challenge and antibiotic-induced gut dysbiosis. Interventions included 14 days of oral CL (5 × 109 CFUs/day) and/or 5 days of budesonide (BUD) nebulization. The gut microbiota (16 S rRNA sequencing), metabolome (untargeted LC‒MS), and immune parameters were assessed via histopathology, flow cytometry (tDCs: CD11c+CD80−CD86−; Tregs: CD4+CD25+FoxP3+), cytokine profiling (ELISA), and analyses of the TLR/NF-κB pathway (IHC/qPCR/WB). As a result, CL monotherapy restored gut microbial α diversity and enriched tryptophan-derived indole derivatives. These microbial metabolites suppressed TLR2/4-NF-κB activation in pulmonary tDCs, driving Treg differentiation while inhibiting Th2 and Th17 responses. Similar to budesonide (BUD), CL significantly reduced airway hyperresponsiveness and eosinophilic infiltration, with a synergistic effect observed for the combination therapy. In conclusion, oral CL alleviates allergic asthma by the microbiota-mediated production of immunoregulatory indoles that activate the TLR2/4-tDC-Treg axis, suggesting a microbiota-targeted strategy for treating steroid-resistant asthma.
Similar content being viewed by others
Introduction
The pathogenesis of asthma is driven by the complex interplay of Th2/Th17-mediated inflammation and deficient regulatory T (Treg) cell responses, culminating in airway hyperreactivity, eosinophilic infiltration, and mucus hypersecretion1,2. While inhaled corticosteroid (ICS) remain first-line therapies, their limited efficacy and side effects in individuals with ICS-insensitive phenotypes and failure to address underlying immune dysregulation underscore the need for novel strategies targeting upstream immunomodulatory pathways3,4. Accumulative evidence implicates gut dysbiosis as a critical contributor to asthma development, with antibiotic-induced microbial depletion exacerbating Th2 polarization and airway inflammation5—a phenomenon mitigated by commensal Clostridia species6,7. Among these, Clostridium leptum (CL), a dominant member of Clostridium cluster IV, has emerged as a potent immunoregulator capable of restoring gut homeostasis and inducing systemic tolerance through metabolite-dependent mechanisms8.
The gut–lung axis operates via microbial metabolites that condition dendritic cells (DCs) and T-cell subsets. While short-chain fatty acids (SCFAs) are well characterized in this crosstalk, recent work highlights tryptophan catabolites—particularly indole derivatives—as pivotal mediators of mucosal and systemic immunity9,10,11. Indole-3-propionic acid (IPA) and related tryptophan metabolites promote the differentiation of tolerant dendritic cells (tDCs), the expansion of Foxp3 + Tregs, and the inhibition of Th2/Th17 cells through certain signaling pathways12,13,14. CL, a prolific producer of tryptophan metabolites, is uniquely positioned at this interface, yet its capacity to leverage these compounds for asthma mitigation remains unexplored.
The clinical outcomes of current probiotic interventions, although promising, are inconsistent due to limited metabolic versatility and transient colonization. In contrast, the robust induction of migratory Tregs by CL and the sustained modulation of tryptophan metabolism offer distinct therapeutic advantages. The results of our preliminary studies indicated that CL supplementation reverses gut dysbiosis in asthmatic mice, which is correlated with reduced lung inflammation and AHR. Therefore, we hypothesized that CL alleviates asthma through two mechanisms: (1) restoring microbial tryptophan metabolism to generate immunoregulatory indole derivatives and (2) programming tDCs to drive Treg-mediated suppression of Th2/Th17 responses.
Here, using a murine model of ovalbumin (OVA)-induced allergic asthma, we establish CL as a master regulator of gut–pulmonary immunity via tryptophan metabolite signaling. Metabolomic profiling revealed the CL-dependent enrichment of indole-3-lactic acid, indoleacetic acid, and IPA, compounds that suppressed TLR2/4-NF-κB activation in pulmonary tDCs, thereby inhibiting Th2/Th17 differentiation while increasing Treg responses. Our findings position CL at the nexus of microbial metabolism and immune tolerance, providing a mechanistically grounded strategy to recalibrate inflammatory cascades in patients with steroid-refractory asthma.
Materials and methods
Clostridium leptum preparation
Clostridium leptum (XuanKe, Shanghai, China), which was cryopreserved at -80 °C, was anaerobically cultured in chopped meat broth for 24 h. The bacterial cells were harvested by centrifugation (3,000 × g, 10 min), washed twice with PBS, and resuspended in fresh PBS for oral gavage administration.
Animals
Female BALB/c mice (6–8 weeks, 18–22 g) from Nanjing Medical University’s Laboratory Animal Center were housed under specific pathogen-free conditions with 12-h light/dark cycles and provided food and water ad libitum. Mice were anesthetized with 2,2,2-tribromoethanol (T48402, Sigma-Aldrich) dissolved in tert-amyl alcohol (721123, Sigma-Aldrich) and diluted in PBS to 20 mg/mL, administered intraperitoneally at 300–400 µL per mouse. All procedures complied with the NIH animal care guidelines and the ARRIVE guidelines and were approved by the Institutional Animal Care and Use Committee of Nanjing Medical University (Approval No: IACUC-2504001).
Model establishment and grouping
Female BALB/c mice (6–8 weeks) were subjected to a modified intestinal disordered flora complicated by allergic asthma (IDFAA) model in which intestinal dysbiosis and allergic asthma induction are combined (Fig. 1A). Gut microbiota depletion was achieved via the oral administration of cefoperazone–sulbactam (1 g/L in drinking water) for 7 days, concurrent with systemic sensitization via intraperitoneal (i.p.) injections of 200 µL of ovalbumin (OVA, 200 µg/mL; Sigma‒Aldrich, S7951) on days 0, 7, and 14. Airway challenge commenced 24 h after the final sensitization using aerosolized 2% OVA (30 min/day, 7 days), whereas control mice received PBS on the same schedule. On day 14, sensitized mice were randomized into five groups (n = 5 mice/group): CON, untreated controls; IDFAA, dysbiosis-asthma model (no treatment); IDFAA_CL, oral gavage with Clostridium leptum (CL, 5 × 109 CFU/day in 200 µL of PBS) for 14 days; IDFAA_BUD, budesonide (BUD) nebulization (500 µg/kg, 40 min/day) from day 22; and IDFAA_CL_BUD, combined CL and BUD therapy. On day 29, the mice were euthanized for bronchoalveolar lavage (BALF), the collection of serum, and the harvesting of tissues harvesting (lung, spleen, and colon). BALF was analyzed via ELISA and flow cytometry; tissues were subjected to histopathological and molecular profiling. The groups of IDFAA, IDFAA_CL, IDFAA_BUD, and IDFAA_CL_BUD underwent the same antibiotic treatment regimen followed by an identical natural recovery period prior to OVA challenge, which ensured that any residual effects from antibiotic withdrawal were uniformly distributed across all groups, thereby minimizing confounding influences. This design ensured that any residual effects from antibiotic withdrawal were uniformly distributed across all groups, thereby minimizing confounding influences. Budesonide (Pulmicort Suspension, AstraZeneca) was used as the inhaled corticosteroid in this study. The drug was prepared as a nebulized suspension at a concentration of 0.5 mg/mL. Aerosol generation was performed using a Medicair nebulizer (UK) with an output rate of approximately 0.3 mL/min. The aerosol was delivered into a whole-body inhalation exposure system, consisting of a plexiglass chamber (20 × 20 × 40 cm) designed to accommodate animal cages, with a total chamber airflow of approximately 3 L/min15,16,17,18.
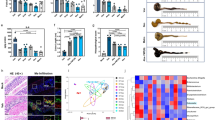
CL relieved antibiotic-induced colon tissue damage. (A)Grouping and modeling of mice. (B) Body weight; (C) DAI; (D,E) Length of colon; (F) H&E sections of colon(All tissue images were obtained at 40× magnification and the scale bar shows 2000 μm); (G) Histological score; (H) Relative mRNA levels of Zo-1.; (I–L) Relative protein levels of IL-6, occludin, and ZO-1, as detected by western blotting(Each bar represents the densitometric value from the respective lane of the blot (lane 1 = CON, lane 2 = IDFAA, lane 3 = IDFAA_BUD, lane 4 = IDFAA_CL, lane 5 = IDFAA_CL_BUD)). The immunohistochemistry staining and the relative optical density of ZO-1 (M-N) in the colon tissue (All tissue images were obtained at 100× magnification and the scale bar shows 800 μm). Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA followed by Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
16 S rRNA sequencing of gut microbiota
Fecal DNA was extracted (E.Z.N.A.® Soil DNA Kit), quantified (NanoDrop 2000/Qubit 4.0), and PCR-amplified using primers targeting the 16 S V3-V4 regions (primers 338 F/806R; 27 cycles, 55 °C annealing). Triplicate amplicons were pooled, gel-purified, and sequenced on the Illumina MiSeq PE300 platform. The raw reads were quality-filtered (fastp v0.21.0; Q20), merged (FLASH v1.2.7; ≥10 bp overlap), and analyzed via dual pipelines: (1) OTUs clustered at 97% similarity (UPARSE, SILVA 138 database) and (2) denoised ASVs (DADA2/QIIME2 v2022.8). Taxonomy was assigned using the RDP Classifier (confidence ≥ 0.7), with rarefaction to 16,000 reads/sample. Alpha/beta diversity (vegan v2.6-4) and differentially abundant taxa (LEfSe: LDA > 2.0, FDR < 0.05) were assessed in R v4.1.3.
Analysis of metabolomic profiles
Metabolomic profiling was conducted using a Nexera UHPLC LC-30 A system (Shimadzu) coupled to a Q Exactive HF-X mass spectrometer (Thermo Fisher). Chromatographic separation was achieved on an ACQUITY UPLC BEH amide column (1.7 μm, 2.1 × 100 mm; Waters) with mobile phases A (10 mM ammonium acetate in acetonitrile/water [95:5, 0.1% formic acid, pH 8.0 for negative mode]) and B (10 mM ammonium acetate in acetonitrile/water [50:50, 0.1% formic acid, pH 8.0 for negative mode]). The gradient elution (0.3 mL/min) involved 2% B (0–0.5 min), 2–98% B (0.5–12.0 min), 98% B (12.0–16.0 min), and reequilibration with 2% B (16.0–18.0 min).
Full MS scans (70–1050 m/z) were acquired at 120,000 resolution (AGC 3 × 106, max IT 100 ms) in both polarities, with data-dependent MS² scans (200–2000 m/z) at 7,500 resolution (AGC 2 × 105, max IT 50 ms) using stepped collision energies (20, 40, 60 eV). The raw data were processed via Progenesis QI (v3.0) for peak alignment, metabolite annotation (HMDB/KEGG), and z score normalization. Hierarchical clustering (Euclidean distance, average linkage) and differential expression analyses [logFC > 1.2, -log10 (FDR) > 2] were performed in R (v4.2.1). System suitability was validated with pooled QC samples (retention time RSD < 2%, intensity RSD < 15%).
Airway hyperresponsiveness
Airway hyperresponsiveness (AHR) was assessed using whole-body plethysmography (Buxco® FinePointe System, Data Sciences International) under controlled environmental conditions (22–24 °C, 50–60% humidity). The mice were acclimatized in the chamber for 15 min before baseline measurements were recorded with aerosolized PBS. Sequential challenges with increasing methacholine (MCh) concentrations (3.12, 6.25, 12.5, 25, and 50 mg/mL) were administered via nebulization (1.5 L/min airflow, 10 s/dose), with enhanced pause (Penh) values recorded 5 min after exposure. Penh was calculated as follows: Penh=(Peak expiratory flow/Peak inspiratory flow)×Pause duration. Measurements were recorded 5 min after nebulization. The results were normalized to the baseline values and are reported as the percentage change in Penh relative to the PBS control.
Histopathological analysis
The mice were euthanized by cervical dislocation. For colonic pathology, the distal colon was excised, fixed in 4% paraformaldehyde (PFA) for 48 h, embedded in paraffin, and sectioned at a thickness of 5 μm. The sections were stained with hematoxylin and eosin (H&E), and two blinded independent pathologists used a semiquantitative scoring system to evaluate the following: inflammatory cell infiltration (0: none; 5: transmural), crypt damage (0: intact; 4: complete loss), ulceration (0: absent; 3: extensive), and edema (0: absent; 1: present). For the pulmonary assessment, the right middle lung lobe was fixed in 4% PFA, processed identically, and stained with H&E. A blinded histopathological analysis was performed using an Olympus IX71 microscope (400× magnification). Peribronchial and perivascular inflammatory cell infiltration was scored as follows: 0 (none), 1 (scattered inflammatory cells), 2 (1 cell layer cuffing), 3 (2–4 cell layers), or 4 (> 4 cell layers). Digital images were acquired and analyzed using RS image analysis software (v3.0, Runcim Technology), with the interrater reliability validated (Cohen’s κ > 0.80).
Immunohistochemistry (IHC) analysis
Paraffin-embedded tissue sections were deparaffinized in xylene and rehydrated by means of a graded ethanol series. Antigen retrieval was performed by microwaving the sections in 0.01 M sodium citrate buffer (pH 6.0) for 15 min, followed by cooling at room temperature for 30 min and two washes with phosphate-buffered saline (PBS). Endogenous peroxidase activity was quenched with 3% H2O2 (10 min, RT), and nonspecific binding was blocked with 5% bovine serum albumin (BSA) in PBS (37 °C, 30 min). The relevant primary antibodies were applied, and the sections were incubated overnight at 4 °C. After three washes with PBS, the sections were incubated with HRP-conjugated secondary antibodies (37 °C, 30 min), developed with 3,3′-diaminobenzidine (DAB) substrate (5 min, RT), and counterstained with hematoxylin. All the antibodies were validated for specificity using isotype and negative controls.
Bronchoalveolar lavage
Bronchoalveolar lavage (BAL) was performed 48 h after the final ovalbumin (OVA) challenge. Following cardiac blood collection, the mice were euthanized by cervical dislocation, and the thoracic cavity was exposed. The right main bronchus was ligated, and the left lung was lavaged three times with 0.5 mL of sterile PBS following tracheal cannulation. The BAL fluid (BALF) was centrifuged at 600 × g for 10 min at 4 °C, and the supernatants were stored at -80 °C for cytokine analyses.
The cell pellets were resuspended in PBS, and viability was assessed via trypan blue exclusion (> 95% viability threshold). For differential counts, cells (1 × 105 cells/slide) were cytocentrifuged (Cytospin™ 4, Thermo Fisher Scientific) and stained with Wright‒Giemsa (Sigma‒Aldrich). Two blinded investigators counted the number of macrophages, eosinophils, lymphocytes, and neutrophils by examining 200 cells per sample under a light microscope (400×, Olympus BX53), with the interrater concordance validated (Cohen’s κ > 0.85).
Flow cytometry analysis
Single-cell suspensions from lung and spleen tissues were prepared by mechanical dissociation through 70-µm cell strainers, followed by erythrocyte lysis using ACK buffer (Thermo Fisher Scientific). The cells were resuspended in PBS (1 × 106 cells/mL) and stained with the following fluorochrome-conjugated antibodies: anti-CD3ε-FITC (Clone 145-2C11, Cat# 11-0031-82), anti-CD4-PE (Clone GK1.5, Cat# 12-0041-82), anti-CD25-APC (Clone PC61.5, Cat# 17-0251-82), anti-CD11c-BV421 (Clone N418, Cat# 117343), anti-CD80-PerCP-Cy5.5 (Clone 16-10A1, Cat# 45-0801-82), and anti-CD86-PE-Cy7 (Clone GL1, Cat# 25-0862-82). Surface staining was performed for 30 min at 4 °C in PBS supplemented with 2% FBS. For intracellular FoxP3 detection, the cells were fixed and permeabilized using the Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher, Cat# 00-5523-00) for 1 h, followed by an incubation with PE-Cy5-conjugated anti-FoxP3 (Clone FJK-16s, Cat# 15-5773-82) for 45 min at 4 °C. Flow cytometry data were acquired on a BD FACSAria™ III (BD Biosciences) using FACSDiva™ software (v8.0.1), with compensation adjustments verified daily using UltraComp eBeads™ (Thermo Fisher, Cat# 01-2222-42).
Enzyme-linked immunosorbent assay (ELISA)
Cytokine concentrations in serum and bronchoalveolar lavage fluid (BALF) were quantified using commercial sandwich ELISA kits (Jining, Shanghai, China) according to the manufacturer’s protocols. Precoated 96-well plates immobilized with capture antibodies were sequentially incubated with samples, standards, and horseradish peroxidase (HRP)-conjugated detection antibodies (1 h, 37 °C), followed by five washes with PBS-Tween 20 (0.05%). Colorimetric detection was performed using 3,3’,5,5’-tetramethylbenzidine (TMB) as the substrate (15 min, dark), and the reactions were terminated with 2 M H2SO4. The absorbance at 450 nm was measured using a Synergy H1 microplate reader (BioTek, USA), and the concentrations were interpolated from standard curves (4-parameter logistic fit, 0–1000 pg/mL).
The assayed cytokines included IFN-γ (Cat: JN17030), TNF-α (Cat: TNF-α), IL-2 (Cat: JN16860), IL-4 (Cat: JN16952), IL-5 (Cat: JN16894), IL-13 (Cat: IL-13), IL-17 (Cat: JN16874), TGF-β (Cat: JN7509), IL-10 (Cat: IL-10), and IL-12p40 (Cat: JN16857). For absolute quantification of microbial indole metabolites, serum were analyzed using commercial ELISA kits for indole-3-propionic acid (IPA, Cat. CB11098-Mu), indole-3-acetic acid (IAA, Cat. CB11069-Mu), and indole-3-lactic acid (ILA, Cat. MM-46497M1) following the manufacturers’ protocols. The intra- and interassay coefficients of variation were maintained at < 10% and < 15%, respectively, and were validated through triplicate measurements of pooled controls.
Western blot (WB)
Proteins were extracted by homogenizing tissues or cell pellets in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.2 mM PMSF), followed by an incubation on ice for 30 min and centrifugation at 12,000 × g (4 °C, 15 min). The supernatants were quantified using a BCA protein assay kit (Beyotime, Shanghai, China), denatured in 5× SDS loading buffer (100 °C, 5 min), and stored at -80 °C.
For SDS‒PAGE, 100 µg of total protein per lane was resolved on 10–12% Tris‒glycine gels (Bio‒Rad) at 70 V (stacking gel) and 110 V (separating gel) until the bromophenol blue reached the bottom of the gel. Proteins were transferred to PVDF membranes (0.45 μm, Millipore) via wet transfer (100 V, 60–120 min, 4 °C) in Tris-glycine buffer containing 20% methanol. The membranes were blocked with 5% nonfat milk/TBST (1 h, RT) and then incubated with primary antibodies (1:1000 dilution in blocking buffer, 4 °C overnight). After washes with TBST (3 × 10 min), the membranes were probed with HRP-conjugated secondary antibodies (goat anti-mouse 1:4000, goat anti-rabbit 1:2000; Abcam, 1 h, RT) and washed again.
The signals were developed using ECL Prime (Cytiva, Shanghai, China) and captured on a ChemiDoc™ XRS + System (Bio-Rad, USA). Band intensities were quantified via Image Lab™ Software (v6.1), normalized to β-actin and analyzed across three biological replicates.
Quantitative reverse transcription PCR (qRT–PCR)
Total RNA was isolated from lung and colon tissues using TRIzol® Reagent (Thermo Fisher Scientific) and reverse-transcribed into cDNA with PrimeScript™ RT Master Mix (Takara Bio, Japan). Target mRNA expression was quantified using TB Green® Premix Ex Taq™ II (Takara Bio, Japan) on a 7500 Real-Time PCR System (Applied Biosystems, USA). The data were normalized to β-actin expression and analyzed via the 2−ΔΔCt method.
Primer Sequences.
Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
ZO-1 | GAGCAGGCTTTGGAGGAGAC | TGGGACAAAAGTCCGGGAAG |
T-bet | AGCAAGGACGGCGAATGTT | GTGGACATATAAGCGGTTCCC |
GATA-3 | AAGCTCAGTATCCGCTGACG | GTTTCCGTAGTAGGACGGGAC |
RORγt | TCCACTACGGGGTTATCACCT | AGTAGGCCACATTACACTGCT |
FoxP3 | CACCTATGCCACCCTTATCCG | CATGCGAGTAAACCAATGGTAGA |
TLR2 | CTCTTCAGCAAACGCTGTTCT | GGCGTCTCCCTCTATTGTATTG |
TLR4 | ATGGCATGGCTTACACCACC | GAGGCCAATTTTGTCTCCACA |
MyD88 | CATACGCAACCTTAATGTGGGG | GGAACTGATTGTATCTGTCGTCG |
IRAK4 | CATACGCAACCTTAATGTGGGG | GGAACTGATTGTATCTGTCGTCG |
TRAF6 | TACGATGTGGAGTTTGACCCA | CACTGCTTCCCGTAAAGCCAT |
IKKα | GGTTTCGGGAACGTCAGTCTG | GCACCATCGCTCTCTGTTTTT |
β-actin | AGGGAAAATCGTCGTGACAT | GAACCGGCTCGTTGCCGATAG |
Generation and treatment of bone marrow-derived dendritic cells (BMDCs)
Bone marrow cells were isolated from the femurs and tibias of mice under sterile conditions. After carefully removing the surrounding muscle tissues, the bones were disinfected in 70% ethanol for 1 min and washed twice with sterile PBS. Both ends of the bones were cut, and the bone marrow was flushed out with PBS using a syringe. The cell suspension was filtered through a 70 μm cell strainer and centrifuged at 1000 rpm for 5 min. Red blood cells were lysed with RBC lysis buffer for 5 min, followed by washing and resuspension in complete RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 2 mM L-glutamine.
Cells were seeded at a density of 1 × 10⁶ cells/mL and cultured in the presence of 20 ng/mL GM-CSF (Cat. No. CK02, Novoprotein) and 20 ng/mL IL-4 (Cat. No. CK15, Novoprotein) at 37 °C in 5% CO₂. Half of the medium was replaced on days 2 and 4 with fresh cytokine-supplemented medium. On days 6–8, non-adherent and loosely adherent cells were collected as immature BMDCs for subsequent experiments.
For in vitro stimulation, BMDCs were seeded in 6-well plates at 1 × 10⁶ cells/mL and divided into five groups: CON group: untreated cells; TNF-α group: treated with 50 ng/mL TNF-α; IPA group: treated with 500 µg/mL indole-3-propionic acid (IPA; Cat. No. 220027, Sigma-Aldrich) dissolved in DMSO (final DMSO concentration 0.1%); IPA_LPS group: treated with 500 µg/mL IPA and 1 µg/mL lipopolysaccharide (LPS; Cat. No. L2654, Sigma-Aldrich); IPA_CH223191 group: treated with 500 µg/mL IPA and 5 µg/mL CH223191 (Cat. No. M7727, AbMole) dissolved in DMSO (final DMSO concentration 0.1%).
All groups were incubated for an additional 48 h under standard culture conditions (37 °C, 5% CO₂). Cells were subsequently harvested and analyzed by flow cytometry to assess dendritic cell phenotype and activation status.
Statistical analysis
Statistical analyses were performed using one-way ANOVA followed by Tukey’s post hoc test in GraphPad Prism (v8.0.2, GraphPad Software). The data are presented as the means ± standard deviations (SDs), with statistical significance defined as p < 0.05. All graphical representations (bar graphs and scatter plots) were generated in GraphPad Prism, with figure panels assembled and annotated in Adobe Illustrator (v26.0.1). For multiple comparisons, p values were adjusted using the Benjamini‒Hochberg false discovery rate (FDR) correction to control for type I errors.
Results
Oral administration of CL mitigates colonic tissue damage
Antibiotic-treated mice exhibited weight loss starting on day 3 (Fig. 1B). The CL intervention effectively alleviated weight loss, with the rebound beginning on day 17, and this was more pronounced when CL was combined with BUD. Additionally, the mice developed soft stools, diarrhea, and occult blood in feces following oral antibiotic treatment. These symptoms were largely resolved after 7 days of CL administration, as reflected by the disease activity index (DAI) score, indicating improved intestinal health (Fig. 1C). Furthermore, both CL and BUD restored the shortened colon length (Fig. 1D,E).
Hematoxylin and eosin (H&E) staining (Fig. 1F) revealed significant morphological abnormalities in the IDFAA group, including mucosal damage, a loss of the normal villous architecture, extensive epithelial cell shedding, marked inflammatory cell infiltration, and villous shortening. However, CL administration reversed these pathological changes, with the IDFAA_CL and IDFAA_BUD_CL groups exhibiting a largely intact crypt architecture, preserved epithelial structure, and elongated villi. Moreover, the IDFAA_CL, IDFAA_BUD, and IDFAA_BUD_CL groups exhibited a superior recovery of colonic epithelial and mucosal integrity. The histological score (Fig. 1G) also revealed that CL could reduce colon tissue damage.
Oral CL administration increases tight junction protein transcription and expression in intestinal epithelial cells
Compared with that in the CON group, Zo-1 mRNA expression in the IDFAA group was significantly reduced, and protein both the ZO-1 and occludin protein levels were significantly decreased in colonic tissues, indicating impaired intestinal barrier integrity (Fig. 1H–L). In the IHC analysis, ZO-1 levels in the colonic epithelium were further confirmed to be markedly downregulated in the IDFAA group (Fig. 1M–N). Notably, the oral administration of CL significantly restored Zo-1 expression at both the mRNA and protein levels, whereas the effect of nebulized budesonide (BUD) on Zo-1 expression in colonic tissues was minimal. The combined intervention of CL and BUD was synergistic, leading to the most robust restoration of tight junction protein expression.
With respect to colonic epithelial inflammation, IL-6 expression was markedly increased in the IDFAA group, reflecting a heightened inflammatory state. Oral CL treatment significantly decreased IL-6 levels, surpassing the anti-inflammatory efficacy of BUD monotherapy. Importantly, the effect of cotreatment with CL and BUD was additive, resulting in a more substantial reduction in the expression of inflammatory markers and further alleviation of intestinal inflammation.
Oral CL administration modulates the gut microbiota
As shown in the Venn diagram in Fig. 2A, there were differences in operational taxonomic units (OTUs) across groups, with the hierarchical clustering analysis (Fig. 2B) indicating distinct microbial community structures. The IDFAA_CL group exhibited greater divergence from the IDFAA group, and the IDFAA_BUD_CL group presented the most pronounced intergroup dissimilarity. The α diversity analysis (Fig. 2C) revealed a significant reduction in microbial richness in the IDFAA and IDFAA_BUD groups, whereas CL increased the richness but decreased the overall diversity, potentially due to the selective enrichment of beneficial bacteria. The β diversity analysis (Fig. 2D), as assessed via principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS), indicated a significant shift in the microbial composition following IDFAA induction. The CL intervention induced further alterations, whereas BUD had a moderate effect.
CL regulated the gut microbiota of antibiotic-induced mice. (A) Venn diagram of the average numbers of operational taxonomic units (OTUs) and overlapping OTUs in different mouse groups; (B) Sample-level clustering in different mouse groups. The length of the branch represents the distance between the samples. (C) Observed index, Chao 1index, ACE index, Shannon index, Simpson index, and Pielou index; (D) Principal coordinate analysis (PCoA) of each group; Nonmetric multidimensional scaling (NMDS) analysis of each group. (E) At the phylum level, the top 8 with the highest abundance; At the class level, the top 8 with the highest abundance; At the order level, the top 12 with the highest abundance; At the family level, the top 19 with the highest abundance. (F,G) Effect Size (LEfSe) analysis of the microbiota in different mouse groups. (H) PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) based on KEGG gene family database between IDFAA_CL group and IDFAA group, (I) PICRUSt2 based on KEGG gene family database in different mouse groups. Significant differences in LDA scores > 2.0 and FDR < 0.05 are shown in Biomakers, i.e., Biomakers with statistical differences.
Taxonomic alterations in the gut microbiota (Fig. 2E) suggested that, at the phylum level, IDFAA induction resulted in an increased Firmicutes/Bacteroidetes (F/B) ratio, which was reversed by CL treatment. Notably, the Verrucomicrobia abundance increased in the IDFAA_BUD_CL group, with an enrichment of Akkermansia muciniphila. At the family level, the abundance of Prevotellaceae decreased, whereas that of Lachnospiraceae increased in the IDFAA group. CL treatment restored the abundance of Prevotellaceae, and LEfSe analysis (LDA score > 4, p < 0.05) revealed 41 differentially abundant taxa across groups (Fig. 2F,G). The CL-treated group exhibited an enrichment of anti-inflammatory commensals, including Erysipelotrichaceae (family), Eubacterium (genus), and Clostridium sp. Culture-27 (species), and the IDFAA group demonstrated enrichment in pathogenic bacteria Saccharimonadia, Patescibacteria, Odoribacter, Rikenella microfusus DSM 15,922, and Peptococcaceae, taxa previously associated with colitis progression and pyogenic infections. Notably, Odoribacter and Patescibacteria dominated the IDFAA microbiota, suggesting their roles in sustaining gut–lung inflammatory crosstalk.
The PICRUSt2 functional prediction (Fig. 2H-I) revealed that gut microbiota functions were significantly enriched in tryptophan metabolism in the IDFAA_CL group compared with the IDFAA group. Similarly, tryptophan metabolism was the most enriched pathway in the IDFAA_BUD_CL group across all comparisons19,20,21.
Oral administration of CL modulates gut metabolites in mice
The untargeted metabolomics analysis revealed that prolonged antibiotic exposure disrupted the gut microbial metabolite profile, significantly altering the abundance of multiple metabolites (Fig. 3A–D). We focused on metabolites whose levels were reduced under IDFAA conditions and identified significantly differentially abundant metabolites with thresholds of logFC > 1.2 and -log10(FDR) > 2. Among these differentially abundant metabolites, the levels of three indole derivatives—indole-3-propionic acid (IPA), indole-3-lactic acid (ILA), and indole-3-acetic acid (IAA)—were consistently significantly reduced in IDFAA mice, whereas the CL intervention markedly restored their levels, with IPA exhibiting the most pronounced fold change (Fig. 4A). This trend strongly correlated with CL administration, which is consistent with previous findings linking Clostridium species to the tryptophan–indole metabolic pathway.
Volcano plot and Heatmap illustrating the intestinal microbiota metabolomic differences between (A) CON and IDFAA mice; (B) IDFAA_BUD and IDFAA mice; (C) IDFAA_CL_BUD and IDFAA mice; (D) IDFAA_CL_BUD and IDFAA mice. The relative abundances of metabolites were normalized to the internal standard and expressed as fold change compared with the control group. Significant differences in LDA scores > 2.0 and FDR < 0.05 are shown in Biomakers, i.e., Biomakers with statistical differences.
Quantitative analysis of indole-3-propionic acid (IPA), indole-3-lactic acid (ILA), and indole-3-acetic acid (IAA) in different treatment groups. (A) Volcano plot illustrating the intestinal microbiota metabolomic differences in different mouse groups. Untargeted metabolomic quantification of IPA, ILA, and IAA was performed using liquid chromatography–mass spectrometry (LC–MS) analysis. The relative abundances of metabolites were normalized to the internal standard and expressed as fold change compared with the control group. Significant differences in LDA scores > 2.0 and FDR < 0.05 are shown in Biomakers, i.e., Biomakers with statistical differences. (B–D) Absolute quantification of IPA, ILA, and IAA concentrations was determined using enzyme-linked immunosorbent assay (ELISA). Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA followed by Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
To further confirm the role of indole derivatives in the gut–lung immune axis, we performed targeted quantification of serum IPA, ILA, and IAA levels using ELISA (Fig. 4B–D). Compared with the IDFAA group, CL supplementation markedly increased serum IPA and ILA concentrations (p < 0.01), whereas the combined treatment with CL and budesonide (IDFAA_CL_BUD) resulted in the highest levels of all three metabolites. These findings were consistent with the untargeted LC–MS profiling, thereby validating the reliability of indole derivative identification and quantification.
Oral administration of CL mitigates allergic airway hyperresponsiveness and inflammation in a mouse model of asthma
Airway hyperresponsiveness (AHR) was assessed via whole-body plethysmography in response to increasing concentrations of MCh (Fig. 5A). Compared with CON mice, IDFAA mice presented a nearly fourfold increase in airway reactivity. In contrast, within the 50 mg/mL range, CL-treated mice presented airway reactivity similar to that of CON-treated mice. Compared with IDFAA-treated mice, CL-treated mice presented significantly reduced AHR, with no significant differences from the IDFAA_BUD and CON groups upon exposure to 50 mg/mL MCh. These findings suggest that oral CL administration alleviates allergic AHR in asthmatic mice, showing efficacy comparable to that of BUD, with a potential synergistic effect when CL is combined with BUD.
CL suppresses lung airway inflammation. (A) The AHR was evaluated by methacholine administration after 24 h of the last BUD atomization. The graph indicates Penh ratio. (B) Total inflammatory cells, eosinophils and lymphocytes in the BAL fluid were counted using trypan blue stain or Giemsa stain, respectively. (C,D) The lung sections were stained with hematoxylin and eosin (All tissue images were obtained at 100× magnification and the scale bar shows 400 μm). The inflammatory score was assessed by measuring the infiltration of inflammatory cells into the peribronchial and perivascular tissues. (E) Relative mRNA levels of Gata-3 (Th2) in different mouse groups; Th2-related cytokine levels in different mouse groups. (F) Relative mRNA levels of T-bet (Th1) in different mouse groups. Th1-related cytokine levels in different mouse groups. (G) Relative mRNA levels of Rorγt (Th17) in different mouse groups; Th17-related cytokine levels in different mouse groups. Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA followed by Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Moreover, compared with the IDFAA controls, CL-treated mice presented significant reductions in the total inflammatory cells, and eosinophil counts. Notably, the combined administration of CL and BUD further reduced inflammatory cell infiltration (Fig. 5B). At one week after OVA challenge, hematoxylin‒eosin (H&E) staining of lung sections revealed airway epithelial congestion, edema, and increased goblet cell numbers in the IDFAA mice. Compared with the IDFAA mice, the IDFAA_CL and IDFAA_BUD mice had markedly fewer goblet cells and less inflammatory cell infiltration, along with attenuated epithelial congestion and edema. Macroscopically, the IDFAA_CL, IDFAA_BUD, and IDFAA_CL_BUD groups were not significantly different from the CON group (Fig. 5C). Inflammation scoring indicated significantly increased inflammation in the IDFAA group compared with the CON group, whereas IDFAA_CL, IDFAA_BUD, and IDFAA_CL_BUD mice exhibited significantly reduced pulmonary inflammation compared with IDFAA mice (Fig. 5D). These results indicate that oral CL administration alleviates airway and lung inflammation, exhibiting comparable efficacy to BUD.
Oral administration of CL modulates T-Cell responses by suppressing Th2/Th17 cells and enhancing Th1 cells in an allergic airway inflammation model
Compared with that in the IDFAA control mice, the expression of the Th2-differentiation transcription factor Gata-3 and the levels of Th2-associated cytokines IL-4, IL-5, and IL-13 were significantly lower in the CL-treated mice, indicating a marked reduction in the Th2 cell population (Fig. 5E). Similarly, in the BUD-treated mice, the expression of Gata-3 and Th2 cytokines was reduced. Additionally, in the CL-treated mice, the expression of the Th17-differentiation transcription factor Rorγt and the Th17-associated cytokine IL-17 A was decreased, reflecting a significant decrease in the Th17 cell population (Fig. 5G).
Conversely, treatment with oral CL administration upregulated the expression of IL-2, TNF-α, and IFN-γ in CD4⁺ T cells and increased the level of the Th1 differentiation transcription factor T-bet, indicating a shift toward Th1 dominance (Fig. 5F), an effect comparable to that of BUD treatment. Notably, the combination of CL and BUD further suppressed Gata-3, Rorγt, IL-4, IL-5, IL-13, and IL-17 A expression, resulting in a more pronounced inhibition of the Th2 and Th17 responses. Concurrently, the expression of T-bet, IL-2, TNF-α, and IFN-γ was further upregulated, reinforcing the Th1-promoting effect.
Oral CL administration induces tolerogenic dendritic cells in asthmatic mice
Compared with that in the CON group, CD80⁺ and CD86⁺ expression in both the lung (Fig. 6A-B) and spleen (Fig. 6C-D) in the IDFAA group was significantly increased, indicating the greater proportion of mDCs and stronger inflammatory response. In contrast, in CL-treated mice, CD80 expression on DCs was significantly reduced in both the lung and spleen, and CD86 expression was significantly decreased on splenic DCs, with a downward trend in pulmonary DCs (although not significant). These results suggest that CL treatment reduces DC maturation, thereby increasing the proportion of tDCs.
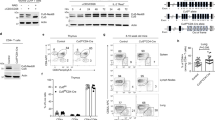
CL suppresses lung airway inflammation. (A,B) The proportion of tDCs in the lung was determined by flow cytometry. (C,D) The proportion of tDCs in the spleen was determined by flow cytometry. (E,F) The proportion of Treg cell in the lung was determined by flow cytometry. (G,H) The proportion of Treg cell in the spleen was determined by flow cytometry; (I) Relative mRNA levels of FoxP3 in the lung of different mouse groups. Cytokine production in serum of different mouse groups during allergic airway inflammation was measured by ELISA. (J) IL-10; (K) IL-12p40; (L) TGF-β. Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA followed by Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Similarly, in BUD-treated mice, there was a significant reduction in CD80 expression on DCs in both the lungs and spleen, along with significantly decreased CD86 expression in pulmonary DCs and a decreasing trend in splenic DCs (not significant). Notably, in IDFAA_CL_BUD mice, there was a greater reduction in CD80 expression in both the lung and spleen, and a reduction in CD86 expression in the lung, than in the CON mice did, indicating the more pronounced induction of tDCs with combined CL and BUD treatment.
Oral CL administration increases Treg responses in asthmatic mice
Flow cytometry revealed a significant reduction in the number of Tregs in the lungs (Fig. 6E-F) and spleens (Fig. 6G-H) of IDFAA mice compared with those in the CON group, indicating compromised immune tolerance in asthmatic mice. However, CL-treated mice presented a marked increase in the number of CD4⁺CD25⁺Foxp3⁺ Tregs in both the lungs and spleen, similar to the findings in BUD-treated mice and the combination (CL + BUD) treatment group.
Moreover, the expression of Foxp3 (Fig. 6I), a key transcription factor regulating Treg function, was significantly lower in the lungs of IDFAA mice than in those of CON mice. Conversely, Foxp3 expression was significantly upregulated in IDFAA_CL, IDFAA_BUD, and IDFAA_CL_BUD mice, with the combination treatment resulting in the most pronounced increase.
Oral CL administration enhances Treg differentiation via IL-10 and TGF-β induction by tDCs in asthmatic mice
ELISAs of serum cytokine levels (Fig. 6J-L) revealed a significant reduction in the TGF-β level in the IDFAA group compared with the CON group, whereas the IL-10 level showed a decreasing trend that was not significant. These findings suggest that reduced Treg proliferation in the IDFAA model may be attributed to decreases in IL-10 and TGF-β levels.
Compared with IDFAA treatment, CL or BUD treatment significantly increased serum IL-10 levels, whereas TGF-β levels were increased but did not reach significance. However, the combination treatment (CL + BUD) resulted in significant increases in both IL-10 and TGF-β levels, suggesting a synergistic effect on promoting Treg differentiation through cytokine regulation. Additionally, the level of mDC-secreted IL-12p40 was significantly increased in IDFAA mice compared with CON mice, indicating a heightened proinflammatory DC response. CL, BUD, and their combination treatment significantly reduced IL-12p40 levels, indirectly indicating a decrease in the proportion of mDCs and an increase in the number of tDCs in asthmatic mice.
Oral CL administration suppresses the transcription and expression of TLR2- and TLR4-associated proteins
IHC results revealed markedly increased TLR2 (Fig. 7A) and TLR4 (Fig. 7B) expression in the lung tissues of IDFAA mice compared with those of CON mice. Interestingly, BUD treatment further increased TLR2 and TLR4 expression beyond the levels observed in the IDFAA group. In contrast, the CL intervention significantly reduced the expression of both receptors.
CL inhibits the transcription and expression of downstream signaling pathways associated with TLR2 and TLR4. The immunohistochemistry staining and the relative optical density of TLR2 (A) and TLR4 (B) in the lung tissue(All tissue images were obtained at 100× magnification and the scale bar shows 400 μm). (C) Relative mRNA levels of Tlr2, Tlr4, MyD88, Irak4, Traf6, IκK-α were measured by PCR. (D,E) Relative protein levels of TLR2&TLR4 and the associated proteins were measured by WB. Relative protein levels of downstream signaling pathways associated with TLR2 and TLR4 were measured by WB: (F,G) TLR2&4-AKT, (H,I) TLR2&4-IRF3, (J,K) TLR2&4-NF-κB, (L,M)TLR2&4-MAPKs. Each bar represents the densitometric value from the respective lane of the blot (lane 1 = CON, lane 2 = IDFAA, lane 3 = IDFAA_BUD, lane 4 = IDFAA_CL, lane 5 = IDFAA_CL_BUD). Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA followed by Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Original blots/gels are presented in Supplementary Information file.
PCR (Fig. 7C) and Western blot (WB) analyses (Fig. 7D-E) revealed the significant upregulation of TLR2 and TLR4 expression in IDFAA mice compared with the CON group. Notably, CL treatment effectively reduced the elevated TLR2 and TLR4 levels in IDFAA mice (IDFAA_CL group). In contrast, BUD treatment (IDFAA_BUD group) significantly increased TLR2 and TLR4 expression. This observation aligns with Rozkova’s study, suggesting that glucocorticoids upregulate TLR2 and TLR4 expression while simultaneously blocking downstream TLR signaling, thereby preventing DC maturation22. The results of PCR (Fig. 7C) of lung tissue revealed significant increases in Tlr2, Tlr4, MyD88, Irak4, Traf6, IκK-α, and Trif expression in the IDFAA group compared with the CON group. CL treatment significantly reduced the expression of these molecules. WB (Fig. 7D-E) analysis further confirmed that, compared with the levels in CON mice, TLR2, TLR4, and MyD88 expression in IDFAA mice increased by 2.00-, 1.29-, and 1.66-fold, respectively. The IRAK4, TRAF6, IκK-α, and TRIF levels increased by 1.33-, 1.57-, 1.95-, and 1.60-fold, respectively. Compared with IDFAA treatment, CL treatment suppressed TLR2- and TLR4-mediated MyD88 expression, reducing TLR2, TLR4, and MyD88 levels by 55.4%, 85.8%, and 66.0%, respectively. Additionally, CL inhibited TLR2- and TLR4-mediated IRAK4, TRAF6, IκK-α, and TRIF expression, with reductions of 72.8%, 27.7%, 75.2%, and 74.7%, respectively.
Oral CL administration suppresses the TLR2- and TLR4-associated downstream signaling pathways
An investigation of the downstream signaling pathways of TLR2 and TLR4 revealed that CL inhibited the activation of the MAPK pathway (Fig. 7L-M), leading to reductions in p-ERK, p-JNK, and p-p38 levels of 40.7%, 51.4%, and 52.3%, respectively. Similarly, BUD treatment suppressed MAPK activation and downregulated p-ERK, p-JNK, and p-p38 levels by 41.5%, 65.4%, and 63.1%, respectively. Notably, the combined CL and BUD treatment had a more pronounced inhibitory effect, reducing p-ERK, p-JNK, and p-p38 levels by 64.2%, 91.6%, and 88.8%, respectively.
Regarding the NF-κB pathway (Fig. 7J-K), BUD significantly inhibited the increases in p-IκBα and p-p65 levels by 70.5% and 68.5%, respectively, compared with those in the IDFAA group. CL had a weaker inhibitory effect on p-IκBα and p-p65 levels (13.6% and 18.2%, respectively). However, CL primarily suppressed NF-κB signaling by inhibiting p-p65 nuclear translocation by 59.8%. The combination of CL and BUD further enhanced the inhibition of p-IκBα and p-p65 levels (74.6% and 59.8%, respectively) and significantly suppressed p-p65 nuclear translocation by 88.5%.
With respect to the AKT signaling pathway (Fig. 7F-G), CL had minimal inhibitory effects, reducing p-AKT levels by only 8.7%, whereas BUD significantly suppressed p-AKT levels by 68.3%. The combined treatment further reduced p-AKT levels by 70.1%.
In the IRF3 pathway (Fig. 7H-I), CL decreased p-IRF3 levels by 62.8%, whereas BUD significantly reduced p-IRF3 levels by 75.1%. The combination therapy further enhanced this effect, reducing p-IRF3 levels by 81.5%.
IPA mediates the tolerogenic effect of clostridium leptum through TLR4 rather than AhR signaling
To determine whether indole-3-propionic acid (IPA) mediates the tolerogenic effect of Clostridium leptum (CL) via TLR4 or AhR signaling, in vitro rescue and blocking experiments were performed using exogenous IPA supplementation and the AhR antagonist CH223191. Flow cytometric analysis revealed that IPA markedly increased the proportion of tolerogenic dendritic cells (tDCs; CD11c⁺CD80⁻CD86⁻) while reducing mature DCs compared with TNF-α stimulation alone (p < 0.001). However, co-treatment with LPS reversed the IPA-induced tolerogenic phenotype (p < 0.01; Fig. 8A, B), whereas CH223191 failed to abolish the effects of IPA, showing no significant difference from IPA treatment alone. These findings indicate that the immunomodulatory and anti-inflammatory effects of CL-derived IPA are primarily mediated via TLR4-dependent signaling rather than AhR activation.
Rescue and blocking experiments in vitro demonstrated that Clostridium leptum (CL) suppresses airway inflammation through tolerogenic dendritic cells (tDCs). (A,B) The proportion of tolerogenic dendritic cells (tDCs; CD11c⁺CD80⁻CD86⁻) in lung tissue was determined by flow cytometry Data are means ± standard deviation (SD). Data are presented as mean ± SD. Statistical significance was evaluated using one-way ANOVA followed by Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Discussion
Our study establishes Clostridium leptum (CL) as a pivotal modulator of gut–lung immunity, attenuating allergic airway inflammation through a triad of mechanisms: gut microbiota restoration, indole metabolite signaling, and systemic tDC–Treg axis activation. CL supplementation reversed antibiotic-induced dysbiosis, notably restoring the Firmicutes/Bacteroidetes ratio—a hallmark of microbial equilibrium23—while enriching Akkermansia muciniphila, a mucin-degrading bacterium associated with intestinal barrier fortification and IAA production24. At the family level, the abundance of Prevotellaceae decreased, and that of Lachnospiraceae increased in the IDFAA group, indicating a dysbiotic profile linked to inflammation. CL treatment significantly restored the abundance of Prevotellaceae, a key family involved in mucosal immunity and regulatory T-cell induction25,26 and decreased the abundance of Lachnospiraceae, which has been implicated in pediatric asthma and gut barrier disruption27 and is negatively correlated with serum IPA levels28. Notably, the IDFAA_BUD_CL group presented the highest A. muciniphila abundance, suggesting a synergistic effect of CL and BUD on gut microbiota modulation and barrier enhancement.
Crucially, the CL-driven enrichment of Erysipelotrichaceae29, Eubacterium30, and Clostridium sp. Culture-27 11 (a C. sporogenes strain) amplified microbial tryptophan catabolism, yielding a spectrum of immunomodulatory indole derivatives, including IPA, ILA, and IAA. Previous studies have shown that these taxa regulate host immunity via tryptophan metabolism, and our findings are consistent with these reports. These metabolites have emerged as central immunoregulators that suppress TLR2/4-NF-κB signaling in pulmonary and splenic tolerogenic dendritic cells (tDCs; CD11c⁺CD80−CD86−), thereby promoting Foxp3⁺ Treg polarization while suppressing Th2/Th17 effector responses (IL-4, IL-13, and IL-17 A) (Fig. 6).
Recent evidence indicates that asthma, although classically characterized as an airway-centered disorder, also involves systemic immune and metabolic dysregulation. Circulating mediators serve as critical intermediates linking pulmonary inflammation with systemic immune, reflecting a broader systemic pathophysiological process31. Indole derivatives have been implicated in obesity, lipid metabolism, allergic diseases, and inflammation. These metabolites exert systemic anti-inflammatory effects and may contribute to the therapeutic modulation of asthma by attenuating both airway and systemic inflammatory responses.The systemic immunomodulatory effects of indole derivatives distinguish CL from conventional probiotics reliant on short-chain fatty acid (SCFA)-mediated mechanisms32,33. While SCFAs primarily modulate colonic immunity, CL-generated indoles display pleiotropic anti-inflammatory activity across distal tissues.
IPA inhibited NF-κB in macrophages, reducing IL-1β and IL-6 production13 and inflammatory cell infiltration in the lung tissue and significantly (p < 0.05) downregulating inflammatory cytokine levels11,12. ILA suppressed Th17 differentiation via RORγt antagonism14. Furthermore, ILA enhances immune regulation in early infancy by upregulating galectin-1 expression in Th2 and Th17 cells, linking a beneficial microbiota to neonatal immune development34. Notably, IAA enhanced gut barrier integrity12 and attenuated inflammatory cell infiltration in lung tissue and inflammatory cytokine levels, synergizing with CL-induced Treg expansion to resolve airway hyperreactivity. Indole derivatives are microbial-dependent deamination products of tryptophan in both humans and mice and are enriched primarily in the indole metabolic pathway. This finding aligns with our 16 S rRNA gut microbiota analysis and PICRUSt2 functional predictions, which indicated that the CL intervention enriched gut microbial metabolites in the tryptophan metabolism pathway. This multiorgan efficacy positions indole derivatives as “pharmacomicrobiotics” capable of bridging microbial metabolism and host immunity—a paradigm shift from species-centric to metabolite-driven probiotic therapies.
Targeted quantitative analysis confirmed that CL treatment significantly elevated circulating levels of IPA, ILA, and IAA, in agreement with our untargeted metabolomic results. Moreover, rescue and blockade experiments provided mechanistic evidence linking the CL–indole–TLR axis to immune modulation. Exogenous IPA administration reprogrammed dendritic cells toward a tolerogenic phenotype even under inflammatory conditions, whereas LPS (a TLR4 agonist) abolished this effect, confirming its TLR4 dependency. In contrast, the AhR antagonist CH223191 failed to reverse IPA-induced tolerance, suggesting that AhR signaling is not the primary mediator. Although prior studies have implicated AhR activation in mucosal homeostasis, our findings reveal a distinct TLR4-dependent mechanism through which IPA modulates DC maturation and function. Together, these data identify microbial tryptophan catabolites as central intermediates linking microbial metabolism to systemic immune tolerance, thereby expanding the conceptual framework of microbiota–host interactions.
Despite the comprehensive mechanistic framework established in this study, several limitations should be acknowledged to ensure a balanced interpretation of our findings.
First, airway hyperresponsiveness (AHR) was assessed using the noninvasive plethysmographic parameter Penh, which allows longitudinal monitoring in conscious mice and avoids confounding effects of anesthesia or tracheotomy. Several studies have validated the methodological reliability of Penh in murine asthma models. Hamelmann et al.35 first established its strong correlation with airway eosinophilia, while Dohi et al.36 demonstrated that Penh accurately tracks allergen-specific bronchial responses and the degree of eosinophilic inflammation. Consistent with these findings, Verheijden et al.37confirmed its sensitivity under severe inflammatory conditions, and Dijoux et al.38 reported comparable trends between Penh and the forced oscillation technique (FOT) in HDM-induced models. However, Penh is an indirect surrogate of airway resistance and can be influenced by nonrespiratory factors. We therefore acknowledge its limitations and have standardized measurement conditions to minimize variability. In future animal experiments, we plan to apply the FlexiVent forced oscillation technique to directly assess invasive lung mechanics. This method will provide precise measurements of airway resistance and compliance, thereby serving as a robust validation and extension of our current findings.
Second, the phenotypic definition of tolerogenic dendritic cells (tDCs) in this work primarily relied on CD11c⁺CD80⁻CD86⁻ gating combined with functional readouts such as Treg expansion and Th2/Th17 suppression. While this minimal marker panel has been widely adopted in prior reports39,40,41,42,43,44,45, we recognize that additional markers—including MHC-IIlow, PD-L1high, IL-10⁺, or Aldh1a2—would provide a more refined definition. Due to antibody availability and throughput constraints, these markers were not systematically assessed in all samples. Future work will expand the flow-cytometric panel and include functional co-culture assays to directly confirm the tolerogenic capacity of CL-primed DCs.
Third, although our targeted and untargeted metabolomics consistently demonstrated increased levels of indole-3-propionic acid (IPA), indole-3-lactic acid (ILA), and indole-3-acetic acid (IAA) in serum and lung tissue, the causal role of the TLR4 pathway remains to be fully validated in vivo. The current evidence, derived from rescue and blockade experiments in cell culture and mouse models, supports a TLR4-dependent rather than AhR-mediated mechanism. Nevertheless, definitive proof will require genetic or pharmacologic loss-of-function approaches—such as TLR4-knockout animals, receptor antagonism, or metabolite-tracing studies—to delineate the tissue specificity and necessity of this signaling axis.
Fourth, the present findings primarily reflect the regulatory effects of Clostridium leptum on airway inflammation under conditions of attenuated responsiveness to inhaled corticosteroids (ICS), rather than representing generalized corticosteroid resistance. Future investigations are warranted to determine whether C. leptum can modulate systemic glucocorticoid responsiveness or serve as an adjunctive therapeutic approach in more clinically relevant models of severe asthma.
Finally, this study was conducted in adult mice, which may underestimate the prophylactic potential of Clostridium leptum, as early-life microbial interventions often exert stronger immunoregulatory effects. Future comparative studies in neonatal versus adult models could clarify age-dependent therapeutic windows. The absence of human data also limits the direct clinical extrapolation of our findings. Therefore, we emphasize that this work represents a mechanistic “proof-of-concept” study. Subsequent research should extend to ex vivo human immune-cell assays, cohort-based metabolomic correlations, and early-phase clinical trials to evaluate safety, dosing, and biomarker responses (e.g., circulating IPA/ILA/IAA levels and peripheral tDC/Treg indices).
In light of these limitations, we have carefully moderated our conclusions to avoid overinterpretation. Collectively, our findings delineate a CL-driven microbiota–metabolite–immune axis, wherein microbial tryptophan catabolites orchestrate Tregs through TLR4-dependent reprogramming of dendritic cells(Fig. 9). The identification of tryptophan catabolites as central mediators not only advances our understanding of microbiota‒host crosstalk but also opens new avenues for developing metabolite-based therapeutics.
CL inhibits the inflammation in asthmatic mice. (A) In the IDFAA group, gut barrier dysfunction allows LPS and PGN to enter circulation, activating DCs via TLR2/TLR4, promoting Th2/Th17 responses, inhibiting Th1/Treg differentiation, and exacerbating lung inflammation. CL intervention enhances tryptophan metabolism, generating indole derivatives that restore gut integrity, induce tolerogenic DCs, suppress Th2/Th17, and promote Th1/Treg differentiation. Reduced pro-inflammatory cytokines and increased anti-inflammatory factors foster immune tolerance, reducing pulmonary inflammation in asthmatic mice. (B) CL inhibits the expression of downstream signaling pathways associated with TLR2 and TLR4. BUD can upregulate the expression of TLR2 and TLR4, while significantly inhibiting the activation of NF-κB, IκK-α, p38, ERK, JNK, AKT, and IRF3. In contrast, CL downregulates the expression of TLR2 and TLR4, inhibits the activation of p38, ERK, JNK, and IRF3, but has no significant inhibitory effect on AKT. Additionally, CL exerts a strong inhibitory effect on the nuclear translocation of NF-κB.
Data availability
The data that support the findings of this study are openly available in Mendeley Data at https://doi.org/10.17632/7wyykpktfz.1. The sequencing data have been deposited in the NCBI Sequence Read Achieve(SRA) database (http://www.ncbi.nlm.nih.gov/sra) with BioProject accession number PRJNA1365055.
References
Peters, M. C. & Wenzel, S. E. Intersection of biology and therapeutics: type 2 targeted therapeutics for adult asthma. Lancet 395, 371–383. https://doi.org/10.1016/s0140-6736(19)33005-3 (2020).
Hammad, H. & Lambrecht, B. N. The basic immunology of asthma. Cell 184, 1469–1485. https://doi.org/10.1016/j.cell.2021.02.016 (2021).
Wang, H. et al. Gut microbiota-derived tryptophan metabolites alleviate allergic asthma inflammation in ovalbumin-induced mice. Foods 13 https://doi.org/10.3390/foods13091336 (2024).
Gross, N. J. & Barnes, P. J. New therapies for asthma and chronic obstructive pulmonary disease. Am. J. Respir Crit. Care Med. 195, 159–166. https://doi.org/10.1164/rccm.201610-2074PP (2017).
Li, J. et al. Causal associations between gut microbiota, metabolites and asthma: a two-sample Mendelian randomization study. BMC Pulm Med. 24, 72. https://doi.org/10.1186/s12890-024-02898-x (2024).
Cait, A. et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11, 785–795. https://doi.org/10.1038/mi.2017.75 (2018).
Xiong, Y. et al. High-throughput 16S rDNA sequencing of the pulmonary Microbiome of rats with allergic asthma. Genes Dis. 7, 272–282. https://doi.org/10.1016/j.gendis.2019.03.006 (2020).
Atarashi, K. et al. Induction of colonic regulatory T cells by Indigenous clostridium species. Science 331, 337–341. https://doi.org/10.1126/science.1198469 (2011).
Chajadine, M. et al. Harnessing intestinal Tryptophan catabolism to relieve atherosclerosis in mice. Nat. Commun. 15, 6390. https://doi.org/10.1038/s41467-024-50807-x (2024).
Zheng, K. Y. et al. Melatonin ameliorates Depressive-Like behaviors in ovariectomized mice by improving Tryptophan metabolism via Inhibition of gut microbe alistipes Inops. Adv. Sci. (Weinh). 11, e2309473. https://doi.org/10.1002/advs.202309473 (2024).
Wang, Z. et al. The gut microbiota-derived metabolite indole-3-propionic acid enhances leptin sensitivity by targeting STAT3 against diet-induced obesity. Clin. Transl Med. 14, e70053. https://doi.org/10.1002/ctm2.70053 (2024).
Li, J. et al. Indole-3-propionic acid improved the intestinal barrier by enhancing epithelial barrier and mucus barrier. J. Agric. Food Chem. 69, 1487–1495. https://doi.org/10.1021/acs.jafc.0c05205 (2021).
Han, Z., Fu, J., Gong, A. & Ren, W. Bacterial indole-3-propionic acid inhibits macrophage IL-1β production through targeting methionine metabolism. Sci. China Life Sci. https://doi.org/10.1007/s11427-024-2789-1 (2025).
Han, J. X. et al. Microbiota-derived Tryptophan catabolites mediate the chemopreventive effects of Statins on colorectal cancer. Nat. Microbiol. 8, 919–933. https://doi.org/10.1038/s41564-023-01363-5 (2023).
Fu, H. et al. Chemoprevention of lung carcinogenesis by the combination of aerosolized Budesonide and oral Pioglitazone in A/J mice. Mol. Carcinog. 50, 913–921. https://doi.org/10.1002/mc.20751 (2011).
Sagar, S. et al. Bifidobacterium Breve and Lactobacillus rhamnosus treatment is as effective as Budesonide at reducing inflammation in a murine model for chronic asthma. Respir Res. 15, 46. https://doi.org/10.1186/1465-9921-15-46 (2014).
Phalen, R. F. & Mendez, L. B. Dosimetry considerations for animal aerosol inhalation studies. Biomarkers 14(Suppl 1), 63–66. https://doi.org/10.1080/13547500902965468 (2009).
Dabisch, P. A., Kline, J., Lewis, C., Yeager, J. & Pitt, M. L. Characterization of a head-only aerosol exposure system for nonhuman primates. Inhal Toxicol. 22, 224–233. https://doi.org/10.3109/08958370903191023 (2010).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–d677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Rozkova, D., Horvath, R., Bartunkova, J. & Spisek, R. Glucocorticoids severely impair differentiation and antigen presenting function of dendritic cells despite upregulation of Toll-like receptors. Clin. Immunol. 120, 260–271. https://doi.org/10.1016/j.clim.2006.04.567 (2006).
Stojanov, S., Berlec, A. & Štrukelj, B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 8 https://doi.org/10.3390/microorganisms8111715 (2020).
Gu, Z. et al. Akkermansia muciniphila and its outer protein Amuc_1100 regulates Tryptophan metabolism in colitis. Food Funct. 12, 10184–10195. https://doi.org/10.1039/d1fo02172a (2021).
Chen, Y. et al. Prevotellaceae produces butyrate to alleviate PD-1/PD-L1 inhibitor-related cardiotoxicity via PPARα-CYP4X1 axis in colonic macrophages. J. Exp. Clin. Cancer Res. 41, 1. https://doi.org/10.1186/s13046-021-02201-4 (2022).
Ye, S. et al. Diversity analysis of gut microbiota between healthy controls and those with atopic dermatitis in a Chinese population. J. Dermatol. 48, 158–167. https://doi.org/10.1111/1346-8138.15530 (2021).
Zhang, J. et al. Beneficial effect of butyrate-producing lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroenterol. Hepatol. 34, 1368–1376. https://doi.org/10.1111/jgh.14536 (2019).
Menni, C. et al. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut Microbiome diversity. Gut Microbes. 10, 688–695. https://doi.org/10.1080/19490976.2019.1586038 (2019).
Singh, S. et al. Distinct intestinal microbial signatures linked to accelerated systemic and intestinal biological aging. Microbiome 12, 31. https://doi.org/10.1186/s40168-024-01758-4 (2024).
Hao, Z. et al. Positive mood-related gut microbiota in a long-term closed environment: a multiomics study based on the lunar palace 365 experiment. Microbiome 11, 88. https://doi.org/10.1186/s40168-023-01506-0 (2023).
Hirano, T. et al. A novel role of growth differentiation factor (GDF)-15 in overlap with sedentary lifestyle and cognitive risk in COPD. J. Clin. Med. https://doi.org/10.3390/jcm9092737 (2020).
Nastasi, C. et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci. Rep. 5, 16148. https://doi.org/10.1038/srep16148 (2015).
Mann, E. R., Lam, Y. K. & Uhlig, H. H. Short-chain fatty acids: linking diet, the Microbiome and immunity. Nat. Rev. Immunol. https://doi.org/10.1038/s41577-024-01014-8 (2024).
Henrick, B. M. et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 3884–3898e3811. https://doi.org/10.1016/j.cell.2021.05.030 (2021).
Hamelmann, E. et al. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Am. J. Respir Crit. Care Med. 156, 766–775. https://doi.org/10.1164/ajrccm.156.3.9606031 (1997).
Dohi, M. et al. Noninvasive system for evaluating the allergen-specific airway response in a murine model of asthma. Lab. Invest. 79, 1559–1571 (1999).
Verheijden, K. A., Henricks, P. A., Redegeld, F. A., Garssen, J. & Folkerts, G. Measurement of airway function using invasive and non-invasive methods in mild and severe models for allergic airway inflammation in mice. Front. Pharmacol. 5, 190. https://doi.org/10.3389/fphar.2014.00190 (2014).
Dijoux, E. et al. Allergic sensitization driving immune phenotyping and disease severity in a mouse model of asthma. Allergy Asthma Immunol. Res. 15, 246–261. https://doi.org/10.4168/aair.2023.15.2.246 (2023).
Bauer, C. A. et al. Dynamic Treg interactions with intratumoral apcs promote local CTL dysfunction. J. Clin. Invest. 124, 2425–2440. https://doi.org/10.1172/jci66375 (2014).
Mo, C. et al. Indoleamine 2,3-dioxygenase 1 limits hepatic inflammatory cells recruitment and promotes bile duct ligation-induced liver fibrosis. Cell. Death Dis. 12, 16. https://doi.org/10.1038/s41419-020-03277-0 (2021).
Schmidt, S. V., Nino-Castro, A. C. & Schultze, J. L. Regulatory dendritic cells: there is more than just immune activation. Front. Immunol. 3, 274. https://doi.org/10.3389/fimmu.2012.00274 (2012).
Tian, X. et al. Bifidobacterium animalis KV9 and Lactobacillus vaginalis FN3 alleviated β-lactoglobulin-induced allergy by modulating dendritic cells in mice. Front. Immunol. 13, 992605. https://doi.org/10.3389/fimmu.2022.992605 (2022).
Xu, Y. et al. Exopolysaccharide from trichoderma Pseudokoningii promotes maturation of murine dendritic cells. Int. J. Biol. Macromol. 92, 1155–1161. https://doi.org/10.1016/j.ijbiomac.2016.06.064 (2016).
Zhang, B. et al. Ephedrae herba polysaccharides inhibit the inflammation of ovalbumin induced asthma by regulating Th1/Th2 and Th17/Treg cell immune imbalance. Mol. Immunol. 152, 14–26. https://doi.org/10.1016/j.molimm.2022.09.009 (2022).
Zhuang, Q. et al. Tolerogenic dendritic cells: the Pearl of immunotherapy in organ transplantation. Front. Immunol. 11, 552988. https://doi.org/10.3389/fimmu.2020.552988 (2020).
Funding
This study was sponsored by grants from the Top Talent of Changzhou “The 14th Five-Year Plan” High-Level Health Talents Training Project (2022CZBJ006; 2024BJHB013), The Jiangsu Provincial Health Commission Medical Research Youth Project (MQ2024001), The National Natural Science Foundation of China (No. 82400025), Youth Science and Technology Project of Changzhou Municipal Health Commission (QN202349; QN202348; QN202316), the Research Project of Changzhou Medical Center, Nanjing Medical University (CMCB202301; CMCB202423 ), Changzhou Municipal Science and Technology Bureau Applied Basic Research Program (CJ20230067), Natural Science Foundation of Xinjiang Uygur Autonomous Region (2022D01A307).
Author information
Authors and Affiliations
Contributions
Heng Ding, Liwen Zhang and Zhiying Huang conceived the idea and designed this study. Heng Ding, Yu Wan, Shiting Su, Yidong Zhao, Wenjie Mao acquired experimental data. Heng Ding and Liang Ma performed the analysis, visualized the data and drafted the manuscript. Liwen Zhang and Zhiying Huang provided material support and obtaining funding. All authors read and provided edits to the paper and contributed to the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ding, H., Wan, Y., Ma, L. et al. Clostridium leptum attenuates allergic airway inflammation via tDC‒Treg axis activation in a murine model of gut dysbiosis-associated asthma. Sci Rep 16, 2259 (2026). https://doi.org/10.1038/s41598-025-31935-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-31935-w