Abstract
Hepatocellular carcinoma (HCC) is a globally prevalent and aggressive form of hepatic cancer. Both N6-methyladenosine (m6A) and long non-coding RNAs (lncRNAs) modification are identified as key mediators of HCC progression. However, comprehensive genome-wide studies and functional annotations of m6A-modified lncRNAs in HCC are still limited. In this study, LINC01315 demonstrated a significant increase in HCC and was closely associated with patient survival. METTL3 was shown to increase LINC01315 expression by acting as its m6A methyltransferase, while YTHDF1 served as the respective m6A reader. LINC01315 promoted the growth and metastatic ability of HCC. LINC01315 acts as a competing endogenous RNA (ceRNA) for miR-185-5p, which elevated β-catenin levels and activated the WNT signaling cascade in HCC cells. These findings highlight that METTL3-driven LINC01315 accelerates HCC cell propagation, invasion, and migration via the LINC01315/miR-185-5p/β-catenin/WNT signaling axis. LINC01315 could serve as a potent prognostic marker and treatment option in HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC), a type of hepatic cancer, is the common cancer type globally and has high rates of morbidity and deaths1. Although there have been significant advances in previous treatments, the overall prognosis for HCC patients remains poor2. This highlights the need to better understand the underlying causes and molecular processes that drive HCC, as well as find new and effective treatments that can be used in a clinical setting3,4,5,6.
N6-methyladenosine (m6A) modification, the predominant and extensively studied epitranscriptomic mark in eukaryotic RNA, has emerged as a key regulator of gene expression7. METTL3, a well-known methyltransferase, serves as a core component of the m6A writer complex, which is responsible for catalyzing this modification8. While initial studies primarily focused on the m6A regulation of mRNAs, growing evidence shows that non-coding RNAs, i.e., long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs), are also regulated by m6A modification9. Uncontrolled m6A levels have been associated with HCC progression through their impact on various aspects of lncRNA biology, such as secondary structure, maturation, splicing, stability, nuclear export, degradation, and translation10,11,12,13.
LncRNAs, a class of transcripts > 200 nucleotides that lack protein-coding potential, have been shown to exert various regulatory functions in oncogenesis14. Distinct expression patterns of lncRNAs are closely associated with HCC initiation and progression15. Its dysregulation has been reported to contribute to key oncogenic processes, such as cellular growth, penetration, metastasis, and resistance to apoptosis, in HCC cells16. This study identified LINC01315 as a downstream target of METTL3, which promotes its expression through an m6A-dependent mechanism. Upregulation of LINC01315 is closely associated with aggressive clinicopathological traits and poor prognosis in HCC. Functional studies further showed that LINC01315 promotes cellular growth, migration, and invasive potential by modulating the β-catenin/WNT signaling cascade through the sequestration of miR-185-5p. These findings demonstrate that the METTL3/LINC01315/miR-185-5p/β-catenin/WNT axis forms a key regulatory pathway in HCC, emphasizing the broader significance of m6A modification in the post-transcriptional regulation of lncRNA activity.
Materials and methods
Cell culture conditions
Human HCC cell lines (HepG2 and Huh7) were supplied by the Cell Bank of the Chinese Academy of Sciences. Both cell lines were cultured under cell culture conditions (5% CO2, 37 °C) and allowed to grow in DMEM (Gibco, Grand Island, NY, USA) enriched with 10% FBS (Gibco, Grand Island, NY, USA) and 1% penicillin-streptomycin (Invitrogen, CA, USA).
MeRIP assay
The Magna MeRIP™ m6A Kit (Millipore, USA) was used to perform MeRIP as per the provided guidelines. Briefly, total RNA was first fragmented using the supplied fragmentation buffer. The fragmented RNA was then subjected to incubation with an m6A-specific antibody and Magna ChIP Protein A/G magnetic beads with gentle rotation at ambient temperature for a duration equal to 30 min. The bead–antibody–RNA complexes were incubated in RNase and IP buffers at 4 °C for 2 h. Bound RNA was eluted from the beads while employing the elution buffer with constant shaking for 60 min at 4 °C. The purification of the resulting RNA was then carried out, and immunoprecipitated transcripts were quantified by qRT-PCR.
RNA Immunoprecipitation (RIP)
The assay of RNA immunoprecipitation was carried out utilizing the EZ-Magna RIP kit (Millipore) based on the manual’s instructions. Lysis of the cells was carried out while employing a lysis buffer, and the lysates were then subjected to pre-clearing with recombinant protein A/G agarose beads (Thermo Fisher Scientific) at 4 °C for 30 min to minimize nonspecific interactions. A fraction (1–10%) of each lysate was kept as input. This was followed by subjecting equal volumes of lysates to overnight incubation with antibodies against YTHDF1 or IgG (negative control) at 4 °C. The capturing of the RNA-protein complexes was achieved while using recombinant protein A/G agarose, and RNA was then eluted, reverse-transcribed into cDNA, and measured by RT-qPCR.
RNA stability (half-life) assay
HCC cell subclones were exposed to 5 µg/mL of actinomycin D (Merck-Millipore, Darmstadt, Germany) to inhibit de novo RNA transcription. Total RNA was isolated at 0, 12, 24, 36, and 48 h after treatment, and transcript levels were analyzed by RT-qPCR to determine RNA stability.
Statement
All methods were performed in accordance with the relevant guidelines and regulations.
Statistical analysis
Data were statistically examined using GraphPad Prism 6.0 (San Diego) and SPSS 20.0 software (SPSS, Inc.). The results are illustrated as mean ± standard deviation. Statistical analyses included the Chi-square test, Student’s t-test, Kaplan-Meier (KM) survival analysis, log-rank test, one-way analysis of variance (ANOVA), and Pearson’s correlation analysis (PCA). A p-value < 0.05 was depicted as a significant cutoff value.
More detail materials and methods were listed in supplement information.
Results
Overexpression of LINC01315 and its prognostic association in HCC patients
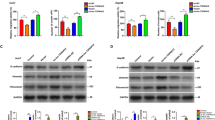
In this study, the TCGA dataset was analyzed, showing that LINC01315 expression was substantially higher in various cancers, including HCC, suggesting its potential relevance in cancer progression (Fig. 1A, B). A stage-dependent increase was observed, with significantly higher expression in advanced tumor grades (T2-T4) compared to early-stage disease (T1) (Fig. 1C). The analysis of receiver operating characteristic (ROC) curve indicated 0.824 as the value of area under the curve, suggesting the potential diagnostic value of LINC01315 in distinguishing HCC from normal tissue (Fig. 1D). To explore its clinical significance, patients were distributed into two groups (low- and high-expression) based on median LINC01315 levels. Higher expression was strongly correlated with reduced overall survival (OS) (Fig. 1E). These findings indicated that LINC01315 is substantially overexpressed in HCC, and its overexpression is associated with an unfavorable prognosis.
Overexpression of LINC01315 and its prognostic association in HCC patients. (A). Analysis of TCGA RNA-sequencing reveals the expression patterns of LINC01315 in different tumor types using the Xiantao platform. (B). Comparison of LINC01315 expression in HCC tissues vs. respective healthy tissues from TCGA datasets. (C). LINC01315 levels in HCC tissues are stratified by pathological stages T1 vs. T2–T4 based on TCGA data. (>D). A ROC curve showing LINC01315 for differentiating HCC from healthy tissues. False positive rate (X-axis); true positive rate (Y-axis). (E). KM survival analysis shows the association between LINC01315 level and OS in HCC patients. The obtained data have been reported as the mean accompanied by the standard deviation (SD) (n = 3). p < 0.001 (***), p < 0.01 (**), p < 0.05 (*).
Functional role of LINC01315 in HCC cell propagation, migration, and invasion in vitro
The functional role of LINC01315 in tumor cell behavior was further examined in HepG2 and Huh7 cells by targeted silencing using siRNAs (si-LINC01315). The efficiency of knockdown was confirmed by RT-qPCR (Fig. 2A). Cell Counting Kit-8 (CCK-8) assays showed a considerable reduction in cell viability after LINC01315 silencing (Fig. 2B). Similarly, colony formation assays showed a substantial reduction in the clonogenic ability of both cell lines after depletion of LINC01315 (Fig. 2C). Invasion assays and transwell migration were conducted to evaluate effects on metastatic potential. The findings indicated that the number of HepG2 and Huh7 cells crossing through the membrane or Matrigel was significantly lower in LINC01315-silenced cells relative to controls (Fig. 2D, E). These results collectively depict that LINC01315 stimulates the proliferation, as well as the migratory and invasive behaviors, of HCC cells, indicating a pro-tumorigenic role in the progression of HCC.
Functional role of LINC01315 in HCC cell proliferation, migration, and invasion in vitro. (A). RT-PCR quantitatively validates the LINC01315 level in HCC cells lines after LINC01315 knockdown. (B). CCK-8 assay showing decreased proliferation of both HCC cell lines after LINC01315 silencing. C. Colony formation assay indicating reduced clonogenic capacity of both HCC cell lines after LINC01315 knockdown. D-E. Microscopic images and quantification of (D) Transwell migration and (E) invasion assays of both HCC cell lines. Scale = 200x. The obtained data have been reported as the mean accompanied by the standard deviation (SD) (n = 3). p < 0.001 (***), p < 0.01 (**), p < 0.05 (*).
METTL3 enhances LINC01315 expression via m6A modification in HCC
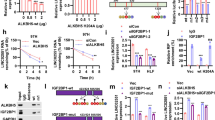
The m6A modification has a significant effect on the advancement of HCC, with METTL3 serving as a key regulator17. In this study, METTL3, WTAP, YTHDF3, YTHDC1, YTHDF1, YTHDF2, IGF2BP1, and IGFB2P2 were found to be positively correlated with LINC01315 in HCC tissues, as determined by TCGA sequencing data (Fig. 3A and B). Moreover, silencing METTL3 in HCC cell lines resulted in a significant decrease in LINC01315 level (Fig. 3C, Fig. S1A). The SRAMP algorithm (http://www.cuilab.cn/sramp) predicted 4 possible m6A modification sites within the LINC01315 transcript (Fig. 3D). Plasmids with A-to-G substitutions at these sites were constructed. MeRIP-qPCR showed a substantial reduction in m6A enrichment in HepG2 and Huh7 cells transfected with the mutant (MT) LINC01315 in comparison to cells with the wild-type (WT) construct (Fig. 3E). These m6A readers are essential in mediating m6A modification.
METTL3 enhances LINC01315 level via m6A modification in HCC. (A). Forest plot illustrating correlations between LINC01315 expression and multiple m6A regulators (METTL3, METTL14, WTAP, YTHDF1/2/3, ALKBH5, YTHDC1/2, FTO, IGF2BP1/2/3) in HCC tissues. (B). Spearman correlation analysis of LINC01315 with selected regulators (METTL3, WTAP, YTHDF1/2/3, YTHDC1, IGF2BP1, IGFB2P2). (C). RT-PCR showing LINC01315 levels in HepG2 and Huh7 cells after siRNA-mediated knockdown of METTL3, METTL14, WTAP, FTO, or ALKBH5. (D). Predicted m6A sites in LINC01315 identified using the SRAMP database. (E). MeRIP-qPCR showing reduced m6A enrichment of LINC01315 after METTL3 knockdown in HepG2 and Huh7 cells. The obtained data have been reported as the mean accompanied by the standard deviation (SD) (n = 3).p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
Considering the key role of m6A reader proteins in regulating m6A-modified transcripts, the contribution of YTHDF1 was examined. Silencing of YTHDF1 resulted in a considerable reduction in LINC01315 expression (Fig. 4A, Fig. S1B). RIP-qPCR confirmed that YTHDF1 had the strongest binding affinity for LINC01315 among the tested readers in HepG2 and Huh7 cells (Fig. 4B). Treatment with 10 µg/mL actinomycin D, an antagonist of RNA polymerase II elongation, showed that LINC01315 transcripts decayed faster after YTHDF1 silencing (Fig. 4C). These results found that METTL3 improves LINC01315 expression in an m6A-dependent mechansim, mainly through YTHDF1 in HCC cells.
YTHDF1 promotes the stability of LINC01315 in HCC cells. (A). RT-PCR analysis of LINC01315 expression in both HCC cell lines after knockdown of YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, IGF2BP1, IGF2BP2, or IGF2BP3 (si-NC as control). (B). RIP-qPCR assessing the binding of LINC01315 with YTHDC1/2, YTHDF1/2/3, and IGF2BP1/2/3 in both HCC cell lines (anti-IgG as control). (C). RNA degradation assay showing decay curves and half-life of LINC01315 in both HCC cell lines after YTHDF1 knockdown (si-NC vs. si-YTHDF1). The obtained data have been reported as the mean accompanied by the standard deviation (SD) (n = 3). p < 0.001 (***), p < 0.01 (**), p < 0.05 (*).
LINC01315 drives HCC progression via β-catenin/WNT signaling
The potential mechanism by which LINC01315 regulates the propagation and metastatic potential of HCC cells was further examined. The WNT signaling pathway, known to play a key role in HCC progression18, was evaluated. Knockdown of LINC01315 reduced TOP flash luciferase activity in both HepG2 and Huh7 cells, indicating suppressed WNT/β-catenin signaling (Fig. 5A). Similarly, β-catenin (CTNNB1) mRNA levels were decreased under the same conditions (Fig. 5B). TCGA sequencing data depicted a positive relationship between LINC01315 and β-catenin, as well as several WNT-associated target genes, including c-MYC, c-JUN, AXIN2, MMP7, and MMP9 (Fig. 5C, D). Moreover, qRT-PCR confirmed that knockdown of LINC01315 resulted in reduced expression of these target genes in HCC cells (Fig. 5E). Functional rescue experiments further established the involvement of the LINC01315/β-catenin/WNT signaling axis. In HepG2 and Huh7 cells, silencing LINC01315 suppressed the proliferative, migratory, and invasive potential, as demonstrated by CCK-8, wound healing, and Transwell assays. These inhibitory effects were effectively reversed by co-transfection with pcDNA-CTNNB1 (Fig. 5F-H, Fig. S1C). These results reveal that LINC01315 stimulates HCC cell growth, migration, and invasion by activating the β-catenin/WNT signaling pathway.
LINC01315 drives HCC progression via β-catenin/WNT signaling. (A) β-catenin luciferase reporter assay (Tcf4 transcriptional activity) in both HCC cell lines transfected with si-NC or si-LINC01315 (#1 or #2). (B) RT-PCR analysis of CTNNB1 mRNA levels in both HCC cell lines after LINC01315 knockdown. (C-D). Correlation analysis between LINC01315 and CTNNB1, MMP7, MMP9, JUN, MYC, and AXIN2 based on TCGA RNA sequencing data (xiantao platform). (E). RT-PCR quantification of MMP7, MMP9, JUN, MYC, and AXIN2 mRNA levels in both HCC cell lines after silencing LINC01315. (F). CCK-8 assay showing cell proliferation in the indicated groups of both HCC cell lines. G-H. Microscopic images and quantification of (G) Transwell migration and (H) invasion assays in both HCC cell lines. Scale = 200x. The obtained data have been reported as the mean accompanied by the standard deviation (SD) (n = 3). p < 0.001 (***), p < 0.01 (**), p < 0.05 (*).
LINC01315 functions as a molecular sponge for miR-185-5p to regulate CTNNB1 in HCC cells
To elucidate the regulatory mechanism underlying LINC01315-mediated upregulation of CTNNB1, nuclear-cytoplasmic fractionation was performed to examine the subcellular localization of LINC01315. The transcript was mainly localized within the cytoplasm of HepG2 and Huh7 cells (Fig. 6A), suggesting a potential post-transcriptional regulatory role. Since lncRNAs can act as ceRNAs by sequestering miRNAs from their mRNA targets, bioinformatic prediction using the miRanda and miRDB databases (https://mirdb.org/) was conducted. Four candidate miRNAs, miR-185-5p, miR-4708-5p, miR-3162-3p, and miR-6511a-5p, were identified as potential common regulators of LINC01315 and CTNNB1 (Fig. 6B). RNA pull-down assays were then used to validate the correlation between LINC01315 and these candidates, showing a strong enrichment of miR-185-5p with biotin-labeled LINC01315 compared to other miRNAs (Fig. S2A). Sequence alignment confirmed complementary binding sites between LINC01315 and miR-185-5p (Fig. 6C). Dual-luciferase reporter assays demonstrated that overexpression of miR-185-5p via mimics (Fig. S2B) substantially lower the luciferase activity of LINC01315-WT but not the mutant construct, further confirming the direct binding relationship (Fig. 6D). Similarly, database analysis revealed that the 3′UTR of CTNNB1 also harbored binding sites for miR-185-5p (Fig. 6E). Dual-luciferase reporter assays confirmed this interaction, showing suppressed reporter activity after miR-185-5p overexpression (Fig. 6F). Meanwhile, CTNNB1 mRNA expression was decrewased in HCC with miR-185-5p mimic(Fig. 6G). Functional rescue assays indicated that LINC01315 silencing reduced CTNNB1 mRNA expression, whereas this suppressing effect was reversed by co-transfection with a miR-185-5p inhibitor (Fig. 6H, Fig. S2C). These findings demonstrate that LINC01315 acts as a ceRNA by sponging miR-185-5p, thereby promoting CTNNB1 upregulation in HCC cells.
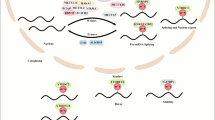
LINC01315 functions as a molecular sponge for miR-185-5p to regulate CTNNB1 in HCC cells. (A). Subcellular localization of LINC01315 in both HCC cell lines by nuclear–cytoplasmic fractionation and RT-qPCR. (B). Venn diagram identifying candidate miRNAs predicted to be co-regulated by LINC01315 and CTNNB1. (C). Predicted binding sequences between miR-185-5p and WT LINC01315, along with the respective MT construct. (D). Dual-luciferase reporter assay evaluating the interaction between miR-185-5p and WT or MT LINC01315 in both HCC cell lines, transfected with mimic NC or miR-185-5p mimic. (E). Predicted binding sequences between miR-185-5p and the 3′UTR of CTNNB1, and the respective MT construct with a mutated binding site. (F). Dual-luciferase reporter assay confirming the interaction between miR-185-5p and WT or MT CTNNB1 3′UTR in both HCC cell lines. (G) RT-PCR analysis of CTNNB1 mRNA levels in both HCC cell lines, transfected with mimic NC or miR-185-5p mimic. (H). RT-PCR analysis of CTNNB1 mRNA level in both HCC cell lines, transfected with si-NC + inhibitor NC, si-LINC01315#1 + inhibitor NC, si-NC + miR-185-5p inhibitor, or si-LINC01315#1 + miR-185-5p inhibitor. (I). Diagram of METTL3 indcued N6-methyladenosine (m6A) of LINC01315 promotes hepatocellular carcinoma progression by activating β-Catentin/WNT pathway. The obtained data have been reported as the mean accompanied by the standard deviation (SD) (n = 3). p < 0.001 (***), p < 0.01 (**), p < 0.05 (*).
Discussion
Globally, HCC is a prevalent and lethal malignancy, representing a major health burden due to its highly aggressive biological behavior, metastatic potential, and rapid proliferation19. Despite advances in diagnostic modalities, the overall prognosis for patients with HCC remains unsatisfactory, highlighting the urgent need to elucidate the basic mechanistic role and to find prognostic indicators and therapeutic options. RNA epigenetic modifications, particularly m6A, have a substantial impact on tumor development and progression20. One key regulatory protein, METTL3, in association with METTL14, forms the catalytic core of the methyltransferase complex that adds methyl groups to RNA transcripts21. This study identified LINC01315 as a novel downstream effector of METTL3 in HCC, with its expression being positively regulated in an m6A-dependent way. This m6A reader protein YTHDF1 was shown to stabilize methylated LINC01315 transcripts, which in turn increases their oncogenic activity. This highlights the key role of the METTL3-YTHDF1 axis in regulating the expression and stability of lncRNA within the HCC cellular environment. LINC01315 has been previously associated with the pathogenesis of several cancers, including thyroid, breast, and gastric cancers22,23,24,25. In colorectal cancer, LINC01315, which is enriched in exosomes from cancer stem cells, has been reported to increase cell viability, propagation, stemness, and dissemination26,27. Based on these findings, this study showed that LINC01315 levels were considerably higher in HCC tissues relative to normal tissues and were associated with poor clinical outcomes. This suggests that LINC01315 may act as an oncogenic driver in HCC and can serve as a prognostic biomarker for the disease.
Functional analyses further confirmed that LINC01315 promotes tumor growth. Loss-of-function experiments showed that silencing of LINC01315 substantially suppresses HCC cellular growth, migration, and invasion, highlighting its role in malignant progression. Mechanistic studies demonstrated that the oncogenic function of LINC01315 is partly driven by stimulation of the WNT/β-catenin signaling cascade. This pathway is conserved throughout evolution and regulates vital biological processes. Its abnormal activation is widely associated with tumor growth and metastasis28,29,30,31. LINC01315 was found to positively regulate CTNNB1 expression, and rescue experiments showed that restoring CTNNB1 expression reversed the suppressive action of LINC01315 knockdown on tumor growth and invasion.
Along with direct pathway modulation, the current results show that LINC01315 acts as a ceRNA. The ceRNA regulatory interaction, in which lncRNAs act as sponges for microRNAs to regulate mRNA targets, has emerged as a key post-transcriptional mechanism in cancer biology32,33,34. Bioinformatic prediction, supported by luciferase reporter and RNA pull-down assays, identified miR-185-5p as a direct interactor of LINC01315. Among the four candidate miRNAs, only miR-185-5p showed a direct association with LINC01315 in HCC cells. Further gene analysis revealed that LINC01315 negatively regulates miR-185-5p levels, indicating that it acts as a molecular sponge for this microRNA. MicroRNAs are widely recognized for their post-transcriptional regulatory roles, primarily by suppressing gene translation or promoting the degradation of target mRNA35,36. In this case, β-catenin (CTNNB1) was recognized as a direct target of miR-185-5p.These findings support a mechanistic model in which LINC01315 sequesters miR-185-5p, alleviating its inhibitory effect on β-catenin and increasing β-catenin levels in HCC cells.
The current analysis was mainly restricted to in vitro studies, and additional validation in vivo using animal models and larger clinical samples with larger cohorts is necessary to confirm the oncogenic role of LINC01315 in HCC.Additional rescue experiments-such as restoring METTL3 or YTHDF1 in knockdown backgrounds-would help establish causality more firmly. FISH staining could provide higher resolution and confirm cytoplasmic enrichment. Additionally, the study focuses heavily on two cell lines (HepG2 and Huh7); including additional HCC cell lines or primary cells would improve the generality of the conclusions. Further study would supplement LINC01315 overexpression experiments to confirm its oncogenic role.Besides, further study would also need to directly compare the half-life of wild-type and m6A-site-mutated LINC01315 to verify that m6A modification mediates its stability via YTHDF1. Moreover, the possibility that LINC01315 may interact with other microRNAs or regulatory proteins beyond the miR-185-5p/CTNNB1 axis cannot be excluded, indicating the need for more comprehensive transcriptomic and proteomic studies. Lastly, therapeutic approaches targeting LINC01315 or its related molecular partners require further exploration to examine their efficacy and safety in translational applications.
Conclusion
This study concluded that LINC01315 acts as an oncogenic factor in HCC, with its upregulation driven by METTL3-mediated m6A modification under the regulation of YTHDF1. LINC01315 overexpression was correlated with an unfavorable clinical prognosis. Functional analyses revealed that LINC01315 increases the migratory, invasive, and proliferative capabilities of HCC cells, thus promoting tumor progression. Mechanistic studies further elucidate the role of the LINC01315/miR-185-5p/β-catenin regulatory axis in HCC pathogenesis. METTL3 was shown to stabilize LINC01315 levels by increasing its m6A modification(Fig. 6I). These findings identify LINC01315 as a promising biomarker for the possible prognosis and diagnosis, as well as an effective therapeutic option in HCC.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Zheng, J. et al. Hepatocellular carcinoma: signaling pathways and therapeutic advances. Signal. Transduct. Target. Ther. 10 (1), 35 (2025).
Renne, S. L. et al. Hepatocellular carcinoma: a clinical and pathological overview. Pathologica 113 (3), 203–217 (2021).
Ye, W. et al. Association between higher expression of Vav1 in hepatocellular carcinoma and unfavourable clinicopathological features and prognosis. Protein Pept. Lett. 31 (9), 706–713 (2024).
Gudivada, I. P. et al. Integrative bioinformatics analysis for targeting hub genes in hepatocellular carcinoma treatment. Curr. Genomics. 26 (1), 48–80 (2025).
Qi, L. et al. Proteogenomic identification and analysis of KIF5B as a prognostic signature for hepatocellular carcinoma. Curr. Gene Ther. 25 (4), 532–545 (2025).
Liu, C. et al. SHR6390 combined with Cabozantinib inhibits tumor progression in the hepatocellular carcinoma mouse model. Curr. Gene Ther. 24 (5), 453–464 (2024).
Wei, G. RNA m6A modification, signals for degradation or stabilisation? Biochem. Soc. Trans. 52 (2), 707–717 (2024).
Liu, W. W. et al. RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy. Signal. Transduct. Target. Ther. 9 (1), 70 (2024).
Li, Y. et al. The role of RNA methylation in tumor immunity and its potential in immunotherapy. Mol. Cancer. 23 (1), 130 (2024).
Shi, J. X. et al. RNA m6A modification in ferroptosis: implications for advancing tumor immunotherapy. Mol. Cancer. 23 (1), 213 (2024).
Tang, F., Xiao, D., Li, X. & Qiao, L. The roles of lactate and the interplay with m6A modification in diseases. Cell. Biol. Toxicol. 40 (1), 107 (2024).
Chen, M., Chen, Y., Wang, K., Deng, X. & Chen, J. Non-m6A RNA modifications in haematological malignancies. Clin. Transl Med. 14 (6), e1666 (2024).
Bu, F. T. et al. The role of m6A-associated membraneless organelles in the RNA metabolism processes and human diseases. Theranostics 14(12), 4683–4700 (2024).
Liu, F., Gu, W. & Shao, Y. Cross-talk between circrnas and m6A modifications in solid tumors. J. Transl Med. 22 (1), 694 (2024).
Qu, Y. et al. Role of N6-methyladenosine RNA modification in cancer. MedComm 5(9), e715 (2020).
Ahi, E. P. Regulation of skeletogenic pathways by m6A RNA modification: A comprehensive review. Calcif Tissue Int. 116 (1), 58 (2025).
Xiang, Z. et al. Cell differentiation-related signaling pathways in hepatocellular carcinoma metastasis. Cancer Lett. 627, 217846 (2025).
Amin, N., Anwar, J., Sulaiman, A., Naumova, N. N. & Anwar, N. Hepatocellular Carcinoma: A Comprehensive Review. Diseases 13(7), 207 (2025).
Ganesan, P. & Kulik, L. M. Hepatocellular carcinoma: new developments. Clin. Liver Dis. 27 (1), 85–102 (2023).
Wang, S. et al. The emerging importance role of m6A modification in liver disease. Biomed. Pharmacother. 162, 114669 (2023).
Shi, Q. et al. Non-coding RNA methylation modifications in hepatocellular carcinoma: interactions and potential implications. Cell. Commun. Signal. 21 (1), 359 (2023).
Ren, J., Zhang, F. J., Wang, J. H. & Tang, J. D. LINC01315 promotes the aggressive phenotypes of papillary thyroid cancer cells by sponging miR-497-5p. Kaohsiung J. Med. Sci. 37, 459–467 (2021).
Xue, J. et al. Overexpression of long Non-Coding RNA Linc01315 predicts poor prognosis in breast cancer. Front. Oncol. 11, 562378 (2021).
Jiang, M., Fang, C. & Ma, Y. Prognosis risk model based on Pyroptosis-Related LncRNAs for gastric cancer. Biomolecules 13, 469 (2023).
Han, C., Zhang, C., Wang, H., Li, K. & Zhao, L. Angiogenesis-related LncRNAs predict the prognosis signature of stomach adenocarcinoma. BMC Cancer. 21, 1312 (2021).
Li, Y. et al. Colorectal cancer stem cell-derived Exosomal long intergenic noncoding RNA 01315 (LINC01315) promotes proliferation, migration, and stemness of colorectal cancer cells. Bioengineered 13, 10827–10842 (2022).
Li, Y. et al. LncRNA LINC01315 Silencing modulates cancer stem cell properties and epithelial-to-mesenchymal transition in colorectal cancer via miR-484/DLK1 axis. Cell. Cycle. 21, 851–873 (2022).
Clevers, H. & Nusse, R. Wnt/beta-catenin signaling and disease. Cell 149 (6), 1192–1205 (2012).
Zou, Y., Salinas, P. & Introduction Wnt signaling mechanisms in development and disease. Dev. Neurobiol. 74 (8), 757–758 (2014).
Wang, X. et al. LncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathway. J. Toxicol. Environ. Health Part. A. 79 (9–10), 407–418 (2016).
Wang, Y. et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell. Stem Cell. 16 (4), 413–425 (2015).
Chodurska, B. & Kunej, T. Long non-coding RNAs in humans: Classification, genomic organization and function. Noncoding RNA Res. 11, 313–327 (2025).
Zhang, J., Xiong, C., Wei, X., Yang, H. & Zhao, C. Modeling NcRNA synergistic regulation in cancer. Methods Mol. Biol. 2883, 377–402 (2025).
Virciglio, C., Abel, Y. & Rederstorff, M. Regulatory Non-Coding rnas: an overview. Methods Mol. Biol. 2300, 3–9 (2021).
Hill, M. & Tran, N. MiRNA interplay: mechanisms and consequences in cancer. Dis. Model. Mech. 14 (4), dmm047662 (2021).
Hussen, B. M. et al. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 138, 111528 (2021).
Author information
Authors and Affiliations
Contributions
Wen-xiao Li were responsible for conception and design. Ming-xuan Xing, Bao-zhuang Sun, and Teng-yue Zou provided statistical analysis. Ren-zheng Liu assisted in statistical analysis. Ming-xuan Xing, Bao-zhuang Sun, Teng-yue Zou, and Guang-jin Li were involved in drafting the manuscript. All authors were involved in reviewing the manuscript. All authors verified the underlying data. All authors approved the final manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xing, Mx., Sun, Bz., Zou, Ty. et al. N6-methyladenosine (m6A) of LINC01315 promotes hepatocellular carcinoma progression by activating β-Catentin/WNT pathway. Sci Rep 16, 2346 (2026). https://doi.org/10.1038/s41598-025-32019-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-32019-5