Abstract
The spread of antibiotic resistance through horizontal gene transfer (HGT) in food-associated bacteria represents an emerging public health concern. Staphylococcus equorum strain KS1030, isolated from a high-salt fermented food, carries plasmids encoding the lincomycin resistance gene lnuA and the relaxase gene rlx, both of which contribute to resistance dissemination. Previous studies have shown that strain KS1030 can transfer the lnuA gene both within and across subspecies when exposed to lincomycin. To investigate the transcriptional basis of this phenomenon, we performed RNA sequencing (RNA-Seq) to analyze the global gene expression profile of KS1030 under lincomycin stress (30 mg/L). Transcriptome analysis revealed more differentially expressed genes (DEGs) at 2 h than at 4 h, with enriched categories including amino acid transport and metabolism (22.9%), transcription (19.3%), and inorganic ion transport and metabolism (14.7%). Genes involved in ornithine, Fe³⁺, siderophore, and tryptophan metabolism, as well as stress regulators such as sigB, dcuSR, and helix-turn-helix transcriptional regulators, were strongly induced. Genome analysis further identified the competence (Com) operon and DNA translocase (ftsK) as potential transport systems, with comGC classified as a DEG. To capture short-term dynamics not resolved by RNA-Seq, quantitative real-time PCR was performed at 30-min intervals. Several genes, including comC, comEC, comFA, and ftsK, peaked at 1.5 h, while lnuA and rlx peaked at 1 h. Although the roles of the Com and FtsK systems in HGT remain unresolved, their induction under lincomycin stress suggests a potential contribution to plasmid transfer, offering new insight into the adaptive and gene transfer responses of S. equorum. However, as this study relies solely on transcriptional data from a single strain and antibiotic condition, functional validation—such as targeted gene disruption—will be required to confirm the involvement of these candidate HGT-related genes.
Similar content being viewed by others
Introduction
The extensive use of antibiotics for infection control and disease treatment has accelerated the emergence of antibiotic-resistant microorganisms1. Resistance is further exacerbated when antibiotic resistance genes are horizontally transferred within microbial communities, thereby increasing the prevalence of resistant strains2. This phenomenon has resulted in clinical challenges where conventional antibiotics become ineffective during patient treatment or surgical procedures3. Recognizing this threat, the U.S. Centers for Disease Control and Prevention (CDC) issued a landmark report in 2013 highlighting the risks posed by the horizontal transfer of antibiotic resistance genes4. The report emphasized that resistance determinants could potentially be transmitted not only between humans but also across food-to-human and animal-to-human interfaces. Subsequently, the World Health Organization published a global priority list of antibiotic-resistant bacteria in 20175, and the CDC identified 18 antibiotic-resistant bacterial and fungal pathogens4, both underscoring the urgent need for global intervention strategies.
Given this context, increasing attention has been directed toward the safety of microorganisms employed in food and feed applications. The European Union has established guidelines prohibiting the use of strains harboring acquired antibiotic resistance genes6. Similarly, microorganisms registered under the Generally Recognized As Safe system in the U.S. must demonstrate antibiotic susceptibility7. In Korea, candidate strains for provisional approval as food sources are also required to be confirmed as non-resistant7. Despite these regulatory measures, reports of horizontal transfer of antibiotic resistance genes by food-associated microorganisms in fermented foods remain scarce. Moreover, experimental evidence confirming horizontal transfer from foodborne microorganisms to the human gut microbiota is limited.
In previous work, we analyzed microbial communities in high-salt fermented foods and identified coagulase-negative staphylococci as dominant members8. To investigate their functional potential, candidate starter strains were screened for antibiotic susceptibility and enzymatic activities that could enhance the quality of fermented foods9,10. Several candidates were shown to contribute to flavor compound development during fermentation11,12,13. Although most food-derived strains were antibiotic-sensitive, four isolates displaying acquired resistance to lincomycin were identified14. Resistance was attributed to the lincosamide O-nucleotidyltransferase gene (lnuA), located on plasmids capable of intra- and interspecific transfer14,15. Strain KS1030, unlike strain KM1031, exhibited horizontal transfer activity, and genomic analysis revealed that only strain KS1030 carries the plasmid pKS1030-3, which contains the relaxase gene14,16. This finding suggests that the horizontal transfer of antibiotic resistance requires not only resistance genes themselves but also mobile genetic elements such as relaxase16. Typically, relaxase-mediated relaxasome complexes are transferred though a Type IV secretion system (T4SS); however, interestingly, no genes associated with a T4SS were identified in the genome of S. equorum including strain KS103016,17. Therefore, the objective of this study was to identify candidate genetic systems that may contribute to antibiotic resistance gene transfer in strain KS1030 and to establish a basis for further functional studies. By analyzing gene expression profiles in response to lincomycin as an inducer, we sought to generate hypothesis-driven insights into genes associated with secretion and to characterize transcriptional responses potentially linked to horizontal transfer under antibiotic exposure.
Materials and methods
Bacterial strains and culture conditions
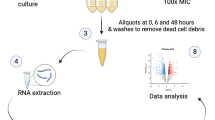
Staphylococcus equorum strain KS1030, which isolated from a high-salt fermented seafood and possessed lincomycin resistance gene, lnuA, and mobile element relaxase gene, rlx, in plasmids8,14,16,18, was subjected to genomic and transcriptomic analyses. S. equorum strains were cultured in tryptic soy broth (TSB; Becton, Dickinson and Co., Franklin Lakes, NJ, USA) supplemented with 30 mg/L lincomycin—a sub-inhibitory concentration previously shown to maintain normal cell growth while inducing antibiotic-responsive gene expression19—at 30 °C for 24 h.
Extraction and purification of RNA
S. equorum strain KS1030 was initially cultured overnight in TSB supplemented with 30 mg/L lincomycin. The overnight culture was then inoculated into fresh TSB medium at 1% (v/v) and incubated at 30 °C with shaking. When the culture reached an optical density of 0.5 at 600 nm, lincomycin was added again at a final concentration of 30 mg/L. The cultures were subsequently incubated for an additional 2 and 4 h at 30 °C. A control group was prepared under identical conditions without the addition of the antibiotic. Cells were harvested by centrifugation, treated with lysostaphin (1 mg/mL) at 37 °C for 20 min, and total RNA was extracted using TRIzol™ reagent (Invitrogen, Carlsbad, CA, USA). Total RNA from each sample was subjected to rRNA removal using the NEBNext rRNA Depletion Kit (New England BioLabs, Wrexham, UK) according to manufacturer instructions.
RNA sequencing analysis
The purified RNA was then used for mRNA-Seq library construction with the Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA, USA). RNA sequencing was performed on the Illumina NovaSeq 6000 platform using paired-end 150 bp reads. All RNA-Seq datasets generated and analyzed in this study, including whole transcriptome profiles (Supplementary Table 1), have been deposited in the Sequence Read Archive (accession numbers SRR28760434-SRR28760435 for 2 h-exposure and SRR32872724-SRR32872725 for 4 h-exposure). The reference-genome sequence was obtained from the NCBI database (http://ncbi.nlm.nih.gov/genomes), and quality-filtered reads were aligned to this reference using Bowtie220. Sequencing reads were mapped to the S. equorum KS1030 genome and normalized using the CLRNASeq program (version 1.00.06; CJ Bioscience, Inc., Seoul, Republic of Korea) based statistical packages like edgeR and DEseq2. Gene expression levels were quantified using the RPKM (Reads Per Kilobase of transcript, per Million mapped reads) method. Statistical significance (p-values) was calculated using edgeR, and subsequently, the FDR correction was applied to these p-values. Fold changes were determined as the ratio of RPKM values under antibiotic treatment to those under control conditions (RPKM_antibiotic/RPKM_control). Differentially expressed genes (DEGs) were defined as those with FDR-corrected p < 0.05 and an absolute fold change greater than 2. These DEGs were visualized using the CLRNASeq program. Functional classification of all strain KS1030 genes was performed using the evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) database (version 5.0)21, and the proportion of DEGs in each functional category was subsequently calculated. Functional enrichment analysis including Kyonto Encyclopedia of Genes and Genomes was performed22.
Genomic analyses
The complete genome sequence of S. equorum strain KS1030 was published previously (GenBank accession: CP068576–CP068580). CLgenomics™ software (version 1.55; CJ Bioscience, Inc) and the web-hosted BLAST programs of NCBI were used to find genes and gene products with sequence identity. Gene functions were analyzed by a search against the COG database.
Quantitative real-time PCR (qRT-PCR)
The expression levels of selected genes predicted to be involved in DNA uptake or export were validated using qRT-PCR. Reactions were performed with gene-specific primer sets (Supplementary Table 2) designed based on the strain KS1030 genome sequence, using the iQ™ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) on a C1000 Thermal Cycler (Bio-Rad). Thermal cycling conditions consisted of an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The 16 S rRNA gene served as the reference gene for normalization23. RNA samples used for qRT-PCR were extracted and reverse-transcribed to cDNA using the same procedures as those employed for transcriptome analysis. To precisely evaluate time-dependent gene expression under short-term antibiotic exposure, samples were collected not only at 2 h and 4 h but also at 0.5 h, 1 h, and 1.5 h following lincomycin treatment. Untreated cultures grown under identical conditions served as controls. Gene expression levels were quantified from three independent biological replicates, and normalized fold changes were calculated using the 2^(-ΔΔCt) method24.
Statistical analysis
Duncan’s multiple range test following one-way analysis of variance was applied to evaluate significant differences between the average values obtained in qRT-PCR. All statistical analysis was performed using the SPSS software package (version 29.0; IBM SPSS Statistics, Armonk, NY, USA).
Results
Comprehensive transcriptome analysis under Lincomycin stress
S. equorum strain KS1030 harbors the lnuA on plasmid pSELNU1 and the relaxase gene on plasmid pKS1030-3, both involved in the horizontal transfer of resistance determinants14,16. Previous studies demonstrated that strain KS1030 is capable of transferring the lincomycin resistance gene between strains and even across subspecies boundaries14. Based on these findings, we hypothesized that exposure to lincomycin would induce the expression of resistance- and transfer-associated genes. To evaluate this, transcriptome profiles were generated from RNA samples collected after 2 h and 4 h of lincomycin (30 µg/mL) exposure, time points selected based on a previous study to capture both early (short-term) and sustained (long-term) transcriptional responses19. RNA-Seq data were processed as described in the methods (Supplementary Table S1).
The strain KS1030 genome comprises 2,948 predicted genes, of which 2,788 and 2,781 were expressed following 2 h and 4 h lincomycin exposure, respectively (Supplementary Table S3). Compared with the control, 352 genes were DEGs after 2 h, whereas only 119 DEGs were detected after 4 h (log2 fold-change value; Fig. 1A and Supplementary Table S3).
Classification of differentially expressed genes (DEGs) from RNA-Seq analysis. (A) DEGs in S. equorum strain KS1030 after 2 h and 4 h of lincomycin treatment compared with untreated controls. The x-axis represents log-scaled Reads Per Kilobase of transcript per Million mapped reads (RPKM) values under untreated conditions, while the y-axis represents log-scaled RPKM values following lincomycin exposure. (B) Functional categorization of DEGs based on the Clusters of Orthologous Groups (COG) database. Genes were considered significantly differentially expressed with a P-value ≤ 0.05 and a fold-change ≥ 2.
At 2 h, excluding genes of unknown function, 101 were upregulated and 8 downregulated. The most affected functional category was amino acid transport and metabolism (22.9%; 25/109), followed by transcription (19.3%; 21/109) and inorganic ion transport and metabolism (14.7%; 16/109) (Fig. 1B). At 4 h, DEG numbers decreased markedly to 16 upregulated and 6 downregulated genes, with the largest decreases observed in transcription (31.8%; 7/22) and inorganic ion transport and metabolism (27.3%; 6/22). Overall, lincomycin exposure significantly altered genes related to amino acid metabolism, transcriptional regulation, and ion transport.
Lincomycin and differentially expressed genes (DEGs)
Lincomycin exposure differentially modulated gene expression over time. Notably, the number of upregulated DEGs declined from 315 at 2 h to 37 at 4 h, while downregulated DEGs slightly increased from 37 to 39 (Fig. 1B). We hypothesized that resistance- and transfer-related genes would be selectively upregulated under lincomycin stress. Therefore, subsequent analyses focused on the upregulated DEGs (Table 1).
Effects of Lincomycin on amino acid transport and metabolism
Among the 315 DEGs upregulated after 2 h of lincomycin exposure, excluding those of unknown function, the most enriched COG category was “amino acid transport and metabolism” (Table 1). Genes involved in ornithine biosynthesis from glutamate were enriched (Fig. 2A and Supplementary Fig. S1). Ornithine has been reported to confer resistance or tolerance to antibiotic stress25. Consistent with this, our results suggest that lincomycin exposure induces the expression of genes responsible for ornithine biosynthesis from glutamate, thereby contributing to resistance or tolerance against the antibiotic.
Metabolic pathways associated with DEGs of S. equorum KS1030 under lincomycin exposure. (A) Ornithine biosynthesis, (B) tryptophan biosynthesis, and (C) staphyloferrin B biosynthesis pathways are shown. Gene names are labeled in blue, and DEG locus tags are highlighted in red. Biosynthetic compounds are represented in light purple boxes, and related amino acids are shown in light pink boxes. Expression levels at 2 h and 4 h after lincomycin exposure are indicated adjacent to each locus tag on log₂ fold-change values.
In addition, genes responsible for tryptophan biosynthesis from chorismate were detected in an operon structure and identified as DEGs after 2 h of lincomycin exposure (Fig. 2B and Supplementary Fig. 1). As tryptophan is a precursor of indole, which promotes horizontal gene transfer (HGT) and antibiotic resistance26,27. This finding suggests an adaptive role for tryptophan metabolism. Interestingly, genes encoding biosynthesis of the siderophore staphyloferrin B were also upregulated at 2 h (Fig. 2C and Supplementary Fig. S1). Siderophores play a key role in iron acquisition under iron-limiting conditions28, and reduced intracellular Fe³⁺ availability during antibiotic stress may drive siderophore-mediated uptake29.
Effects of Lincomycin on transcription
Transcription-related genes represented the second largest category of DEGs at 2 h (Table 1). Approximately 10% (20/196) of transcription-associated genes in the genome were differentially expressed, including six helix-turn-helix transcriptional regulators, which are known to contribute to antibiotic resistance30. These findings suggest that transcriptional regulators involved in antibiotic resistance were upregulated in response to lincomycin stress. Interestingly, the expression of a metalloregulator belonging to the ArsR/SmtB family, which was low at 2 h, was significantly increased at 4 h of exposure. Members of this family are well known as stress-inducible regulators31. Thus, it can be inferred that strain KS1030 recognized lincomycin as an external stressor, resulting in delayed induction of this transcription factor relative to other regulators. It is expected that exposure to lincomycin would require transcriptional factors to activate genes involved in environmental adaptation, including antibiotic resistance genes. Consistent with this notion, S. equorum KS1030 showed a high number of differentially expressed genes related to transcription, including transcriptional regulators—the second most enriched functional category among the DEGs. However, the specific relationship between these transcriptional regulators and HGT remains to be elucidated in future studies.
Effects of Lincomycin on inorganic ion transport and metabolism
The third most enriched COG category among the DEGs upregulated after 2 h of lincomycin exposure was “inorganic ion transport and metabolism” (Table 1). At 4 h, this category contained the largest number of DEGs overall. As shown in Table 1, no clear trend was observed across all transport genes; however, specific patterns were evident. For example, genes encoding transporters of betaine and choline (JL104_RS01920 and JL104_RS03745), which function as osmoprotectants and contribute to salt tolerance, were highly expressed at 2 h. In contrast, genes associated with arsenite efflux showed elevated expression at 4 h. Betaine and choline uptake promotes the intracellular accumulation of osmoprotectants, thereby protecting cells from environmental stress19, whereas arsenite efflux systems remove toxic compounds such as arsenite from the cell32. Accordingly, the observed transcriptional changes suggest that strain KS1030 responds to lincomycin-induced stress by enhancing osmoprotectant accumulation at early time points and activating efflux systems to eliminate harmful compounds during prolonged exposure.
Notably, five of the 14 DEGs identified in the COG category “inorganic ion transport and metabolism” after 2 h of lincomycin exposure were associated with Fe³⁺ uptake (Table 1). Previous studies have reported that the intracellular accumulation of exogenous Fe³⁺ enhances bacterial resistance to antibiotics33. These findings therefore suggest that, in addition to activating lincomycin resistance genes, strain KS1030 may increase Fe³⁺ uptake as a complementary mechanism to strengthen its tolerance against lincomycin stress.
Effects of Lincomycin on the regulator genes
In addition to the genes described above, increased expression was also observed for several genes involved in signal transduction. In particular, the DcuSR two-component system (TCS), known to regulate genes in response to citrate and to control both virulence factors and antibiotic resistance determinants34,35,36, showed elevated expression (Table 1 and Supplementary Fig. 1). Genes encoding universal stress proteins, which play protective roles against antibiotics and other environmental challenges37, were also upregulated at 2 h, and their expression remained high at 4 h (Table 1). Furthermore, the expression of the stress-responsive sigma factor sigB was significantly increased at 2 h, along with its regulatory partners rsbV, rsbU, and rsbW, which are required to maintain sigB activation (Table 1 and Supplementary Fig. 1).
Changes of DEGs during exposure time of Lincomycin
Analysis of the transcriptional profiles of strain KS1030 after 2 h and 4 h of lincomycin exposure revealed increased expression of transport- and biosynthesis-related genes associated with ornithine, Fe³⁺, siderophores, and tryptophan, all of which are linked to cellular protection against environmental stress (Table 1; Fig. 2)25,26,27,28,29,33. In addition, regulatory genes required for adaptation to external stressors, including sigB, dcuSR TCS, and several helix-turn-helix transcriptional regulators, were also highly expressed (Table 1)30,31,32,34,35,36,37. To further assess the temporal differences in gene expression, a Venn diagram analysis was conducted (Fig. 3). While DEGs analysis excluded genes classified as “function unknown”, the Venn diagram analysis included these genes to provide a more comprehensive view of transcriptional changes under lincomycin stress (Fig. 3 and Supplementary Table 4).
A total of 37 genes were identified as common DEGs after both 2 h and 4 h of lincomycin exposure (Fig. 3 and Supplementary Table 4). Excluding hypothetical proteins, 22 genes remained, including two helix-turn-helix domain-containing proteins, as well as universal and general stress protein genes. These findings suggest that certain regulatory and protective genes are consistently expressed throughout lincomycin exposure to facilitate bacterial adaptation. By contrast, genes upregulated at 2 h—such as those involved in ornithine, Fe³⁺, siderophore, and tryptophan transport and biosynthesis, as well as sigB and dcuSR TCS—were no longer identified as DEGs at 4 h. Instead, the DEGs at 4 h included three arsenite efflux genes (JL104_RS09790, JL104_RS14300, and JL104_RS14320), which are known to mediate the export of intracellular arsenic acid but can also contribute to antibiotic efflux38,39. This suggests that, during prolonged exposure, strain KS1030 activates efflux systems to expel accumulated lincomycin.
Interestingly, the sigma-70 family RNA polymerase sigma factor, a general transcriptional regulator under normal conditions, was also detected as a DEG at 4 h. Moreover, the lnuA gene (JL104_RS14695), which confers lincomycin resistance, was strongly expressed at 2 h but markedly reduced at 4 h. Although this gene is classified as “function unknown” in COG analysis and thus not highlighted in DEG categorization, its induction at 2 h under lincomycin exposure is consistent with its known role in resistance. Taken together, these results indicate that strain KS1030 mounts a rapid adaptive response within the first 2 h of lincomycin exposure, upregulating metabolic, stress-response, and resistance genes. By 4 h, however, expression largely returns toward baseline, with efflux systems becoming the predominant mechanism of defense.
Putative horizontal gene transfer related genes
We hypothesized that exposure of strain KS1030 to lincomycin for 2 h and 4 h would induce expression of lincomycin resistance genes and genes required for their horizontal transfer across species or genera. Transcriptome analysis at the DEG level confirmed that the lincomycin resistance gene lnuA was differentially expressed. However, neither the relaxase gene (JL104_RS14645) nor the mobilization relaxasome protein gene mobC (JL104_RS14640)—both previously proposed to contribute to horizontal transfer16—showed significant changes in expression (Table 2). Nevertheless, despite the lack of strong induction, we reasoned that genes supporting horizontal transfer of antibiotic resistance must exist in strain KS1030 and therefore extended our analysis to membrane-associated proteins potentially involved in DNA transfer. Notably, genome analysis did not identify a T4SS, the classical mechanism implicated in horizontal transfer of resistance genes, in strain KS1030 or in other S. equorum strains examined.
Based on these findings, we next searched the genome of strain KS1030 for genes associated with “secretion,” “translocase,” and “DNA export/DNA uptake” (Table 3). Genes related to the general secretion system (Sec-SRP) and secretion accessory proteins were identified under the “secretion” category. Although the Sec-SRP system is primarily involved in protein secretion40, genome analysis confirmed that strain KS1030 possesses the complete set of Sec-SRP components (Fig. 4). Transcriptome analysis showed that only secE (JL104_RS12770) was classified as a DEG at 4 h, while expression changes in other genes were minimal.
When searched under the “translocase” keyword, two DNA translocase ftsK genes were detected, in addition to the Sec-SRP components (Table 3; Fig. 4). FtsK is known to function in chromosome segregation and cell division, but has also been implicated in HGT41. Although not classified as DEGs, both ftsK genes showed increased expression at 2 h.
Finally, no genes associated with “DNA export” were identified, whereas genes related to “DNA uptake” were detected in the competence (Com) operon (Table 3; Fig. 4). The Com operon is reported to mediate HGT through the uptake of extracellular DNA42, and all required competence-associated genes were present in the strain KS1030 genome. Transcriptome data revealed that comGC (JL104_RS06960), encoding the type IV pilus major pilin, was a DEG at 2 h, whereas the other Com operon genes did not show significant differential expression.
Taken together, genome analysis indicated that strain KS1030 harbors several membrane-associated transport systems, including the Sec-SRP system, the Com operon, and DNA translocase FtsK (Fig. 4). Since our aim was to identify potential secretion or transport systems involved in HGT under lincomycin stress, we cautiously propose that the Com operon and FtsK may contribute to gene transfer in this strain. However, as their expression was generally low, further research is required to determine their functional relevance. The transient upregulation of comGC at 2 h suggests that competence-related genes may be induced early, followed by rapid decline, highlighting the need for finer time-point analyses. Consequently, we performed qRT-PCR at 30-min intervals to more precisely assess temporal expression dynamics.
Validation of RNA-Seq data by qRT-PCR
To validate the transcriptional profiles of S. equorum strain KS1030 obtained by RNA-Seq under lincomycin exposure, qRT-PCR was performed. While RNA-Seq was conducted at 2 h and 4 h post-exposure, qRT-PCR included these time points as well as additional earlier time points (0.5 h, 1 h, and 1.5 h) to capture short-term transcriptional dynamics. Target genes included those predicted to be involved in DNA uptake or export and located at the membrane, specifically the Com operon genes (comC, comEC, comFA, comGC) and two DNA translocase genes (ftsK JL104_RS06005 and JL104_RS14955). In addition, the lincomycin resistance gene lnuA and the transfer-related genes rlx and mobC were analyzed.
qRT-PCR results at 2 h and 4 h were consistent with RNA-Seq data, showing relatively modest expression changes (Fig. 5). However, as expected, several genes showed earlier peaks in expression. At 1.5 h, five genes (comC, comEC, comFA, ftsK JL104_RS06005, and ftsK JL104_RS14955) exhibited their highest expression levels. The resistance gene lnuA and rlx peaked at 1 h before declining, while comGC and mobC showed maximal expression at 2 h. Although fold change values were not large enough to classify these genes as DEGs, their consistent upregulation compared with untreated controls confirms their induction by lincomycin. Thus, RNA-Seq results were validated by qRT-PCR, and additional short-term analysis revealed transient induction of several transport- and transfer-associated genes.
Relative expression levels of selected genes predicted to be involved in DNA uptake or export in S. equorum KS1030 upon lincomycin exposure. Gene expression was measured at 0.5, 1, 1.5, 2, and 4 h using quantitative real-time PCR (qRT-PCR). Expression levels were normalized against the 16 S rRNA gene, and fold changes were calculated using the 2^(-ΔΔCt) method. Data represent the mean ± standard deviation of three independent experiments. Different letters above bars indicate statistically significant differences between time points (p < 0.05, Duncan’s multiple range test).
Discussion
S. equorum strain KS1030 harbors the lincomycin resistance gene lnuA on plasmid pSELNU1, as well as a relaxase gene on plasmid pKS1030-314,16,18. Previous studies demonstrated that strain KS1030 is capable of horizontally transferring the resistance plasmid both within and across subspecies boundaries when exposed to lincomycin14,15. Furthermore, resistance gene transfer was observed not only under in vitro conditions but also when strain KS1030 was used as a starter culture in fermented foods or administered orally, followed by antibiotic exposure15. These findings strongly suggest that strain KS1030 possesses the genetic potential to mediate plasmid transfer. However, despite the presence of a transferable plasmid and relaxase, the specific secretion system required for gene transfer has remained unclear. Therefore, we hypothesized that lincomycin exposure would induce not only lincomycin resistance genes but also genes required for horizontal gene transfer, and we used RNA-Seq to identify candidate secretion systems potentially involved in this process. Transcriptomic analysis confirmed that the lincomycin resistance gene lnuA was differentially expressed; however, the relaxase gene and the mobilization relaxosome gene mobC, both previously implicated in horizontal transfer, showed no significant transcriptional changes. This outcome was unexpected, given earlier findings that plasmids carrying a relaxase gene mediate the horizontal transfer of lnuA. One possibility is that the conditions used in this study were not optimal to induce their expression. Alternatively, these genes may function without requiring substantial transcriptional upregulation, as some resistance-associated genes are known to exert effects even at low expression levels19.
Contrary to our initial expectation that lincomycin exposure would trigger marked expression of secretion system components facilitating plasmid transfer, the major DEGs were instead associated with stress adaptation. Genes involved in osmoprotection (e.g., opuD encoding glycine-betaine uptake), environmental adaptation (e.g., sigB), and iron acquisition (e.g., staphyloferrin B biosynthesis) were significantly upregulated (Table 1). These results indicate that strain KS1030 primarily mounts a physiological response to survive antibiotic stress rather than directly inducing secretion machinery. Interestingly, among the DEGs, we detected comGC, a structural component of the type IV pilus encoded by the Com operon, which is known to mediate DNA uptake. Genomic analysis further confirmed that strain KS1030 contains the full set of Com operon genes (Fig. 4). Although expression changes were modest, qRT-PCR revealed that comGC expression peaked at 1.5 h, earlier than the 2 h and 4 h RNA-Seq time points (Fig. 5). This observation is consistent with prior studies reporting that transcriptional responses to antibiotic stress often occur within the first hour of exposure43,44,45, suggesting that the sampling times used for RNA-Seq in this study may have been too long to capture peak activity. Taken together, the transcriptomic and qRT-PCR data indicate that early (≤ 1.5 h) time points are optimal for capturing lincomycin-induced expression dynamics.
Our initial hypothesis posited the presence of a T4SS or a related DNA export pathway in strain KS1030. However, transcriptomic data did not reveal clear evidence of such a system. Instead, we hypothesize that weak activation of the Com operon and FtsK may transiently promote DNA mobilization capacity, thereby enhancing plasmid dissemination under antibiotic stress. This hypothesis could explain why plasmid transfer occurs upon lincomycin exposure even when classical secretion systems such as the T4SS are not transcriptionally induced. The Com operon, which encodes pilins structurally analogous to type IV secretion components46,47,48,49, may represent a functional alternative mechanism contributing to horizontal transfer. The DNA translocase FtsK is known to be involved in chromosome segregation and cell division, but it has also been suggested to contribute to HGT41. However, the functional validation of candidate genes presumed to participate in HGT—such as the com operon and ftsK—has not yet been conducted. Moreover, the RNA-Seq analysis used in this study detects transcriptional changes but cannot directly measure the extent of plasmid transfer. To overcome these limitations, additional experimental validation—including transcriptomic analysis with earlier sampling and functional characterization of the corresponding genes—is required.
This study presents the first integrated genomic and transcriptomic analysis of S. equorum KS1030 under lincomycin stress, revealing that the strain primarily activates metabolic and stress-adaptation pathways rather than classical secretion systems. Although lnuA was strongly upregulated, the relaxase and mobC genes showed no significant induction, while competence-associated components and the DNA translocase ftsK exhibited only modest, early transcriptional responses. These findings suggest that KS1030 may rely on alternative or condition-dependent mechanisms for plasmid transfer. By delineating a focused set of candidate genes, this work provides a hypothesis-generating foundation for targeted functional studies—including gene disruption, complementation, protein-level assays, and earlier time-point transcriptomics—to clarify their roles in horizontal gene transfer. Ultimately, the results offer valuable insights into resistance dissemination in food-derived S. equorum and support the development of safer fermented-food starter cultures through improved understanding of HGT-associated genetic factors.
Data availability
RNA-Seq data analyzed in this study were deposited in the Sequence Read Archive (SRR28760434-SRR28760435 and SRR32872724-SRR32872725). The data presented in this study are available in the article/Supplementary information file; further inquiries can be directed to the corresponding author.
References
Ventola, C. L. The antibiotic resistance crisis: part 1: causes and threats. Pharm. Ther. 40, 277–283 (2015).
von Wintersdorff, C. J. et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173 (2016).
Sengupta, S., Chattopadhyay, M. K. & Grossart, H. P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4, 47 (2013).
Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States (U.S. Departement of Health and Human Services, CDC, 2013).
Tacconelli, E. & Magrini, N. Global priority list of anitbiotic-resistant bacteria to guide research, discovery, and development of new antibiotics (World Health Organization, 2017).
EFSA. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 10, 2740–2749 (2012).
Heo, S. et al. Safety assessment systems for microbial starters derived from fermented foods. J. Microbiol. Biotechnol. 32, 1219–1225 (2022).
Guan, L., Cho, K. H. & Lee, J. H. Analysis of the cultivable bacterial community in jeotgal, a Korean salted and fermented seafood, and identification of its dominant bacteria. Food Microbiol. 28, 101–113 (2011).
Jeong, D. W., Lee, B., Her, J. Y., Lee, K. G. & Lee, J. H. Safety and technological characterization of coagulase-negative Staphylococci isolates from traditional Korean fermented soybean foods for starter development. Int. J. Food Microbiol. 236, 9–16 (2016).
Jeong, D. W., Han, S. & Lee, J. H. Safety and technological characterization of Staphylococcus equorum isolates from jeotgal, a Korean high-salt-fermented seafood, for starter development. Int. J. Food Microbiol. 188, 108–115 (2014).
Jeong, D. W. et al. Effects of starter candidates and NaCl on the production of volatile compounds during soybean fermentation. J. Microbiol. Biotechnol. 29, 191–199 (2019).
Jeong, D. W. et al. Effects of the predominant bacteria from Meju and Doenjang on the production of volatile compounds during soybean fermentation. Int. J. Food Microbiol. 262, 8–13 (2017).
Jeong, D. W. et al. Effects of Enterococcus faecium and Staphylococcus succinus starters on the production of volatile compounds during Doenjang fermentation. LWT-Food Sci. Technol. 122, 108996 (2020).
Lee, J. H. & Jeong, D. W. Characterization of mobile Staphylococcus equorum plasmids isolated from fermented seafood that confer Lincomycin resistance. PLoS One. 10, e0140190 (2015).
Heo, S., Bae, T., Lee, J. H. & Jeong, D. W. Transfer of a lincomycin-resistant plasmid between coagulase-negative Staphylococci during soybean fermentation and mouse intestine passage. FEMS Microbiol. Lett. 366, fnz113 (2019).
Heo, S. et al. Staphylococcus equorum plasmid pKS1030-3 encodes auxiliary biofilm formation and trans-acting gene mobilization systems. Sci. Rep. 13, 11108 (2023).
Jeong, D. W., Heo, S., Ryu, S., Blom, J. & Lee, J. H. Genomic insights into the virulence and salt tolerance of Staphylococcus equorum. Sci. Rep. 7, 5383 (2017).
Kim, T., Heo, S., Lee, J. H. & Jeong, D. W. Complete genome sequence of Staphylococcus equorum KS1030 exhibiting acquired Lincomycin resistance. Korean J. Microbiol. 57, 210–212 (2021).
Heo, S. et al. Transcriptomic analysis of Staphylococcus equorum KM1031 from the high-salt fermented seafood Jeotgal under chloramphenicol, erythromycin and Lincomycin stresses. Sci. Rep. 12, 15541 (2022).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 9, 357–359 (2012).
Huerta-Cepas, J. et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Clifford, R. J. et al. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. 7, e48558 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods 25, 402–408 (2001).
Kim, S. K. et al. Bacterial ornithine lipid, a surrogate membrane lipid under phosphate-limiting conditions, plays important roles in bacterial persistence and interaction with host. Environ. Microbiol. 20, 3992–4008 (2018).
Dong, W. et al. Indole may help the horizontal transmission of antibiotic resistance genes in E. coli under subinhibitory concentrations of cefotaxime stress. Cellular Microbiology 9018205 (2024). (2024).
Lee, J. H. & Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34, 426–444 (2010).
Kramer, J., Ozkaya, O. & Kummerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163 (2020).
Ezraty, B. & Barras, F. The ‘liaisons dangereuses’ between iron and antibiotics. FEMS Microbiol. Rev. 40, 418–435 (2016).
Huffman, J. L. & Brennan, R. G. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12, 98–106 (2002).
Busenlehner, L. S., Pennella, M. A. & Giedroc, D. P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143 (2003).
Shi, K. et al. Efflux transporter ArsK is responsible for bacterial resistance to arsenite, antimonite, trivalent roxarsone, and Methylarsenite. Appl. Environ. Microbiol. 84, e01842–e01818 (2018).
Huang, Y. F. et al. Promoting effect of Fe(3+) on gentamicin resistance in Escherichia coli. Biochem. Biophys. Res. Commun. 625, 134–139 (2022).
Zheng, Z., Deng, C., He, W., Qian, W. & Li, A. The two-component system DcuS-DcuR is involved in virulence and stress tolerance in the Poplar canker bacterium Lonsdalea Populi. Phytopathology 110, 1763–1772 (2020).
Abo-Amer, A. E. et al. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J. Bacteriol. 186, 1879–1889 (2004).
Zientz, E., Bongaerts, J. & Unden, G. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180, 5421–5425 (1998).
Luo, D. et al. Universal stress proteins: from gene to function. Int. J. Mol. Sci. 24, 4725 (2023).
Zhang, M. et al. Co-selection and stability of bacterial antibiotic resistance by arsenic pollution accidents in source water. Environ. Int. 135, 105351 (2020).
Murray, L. M. et al. Co-selection for antibiotic resistance by environmental contaminants. NPJ Antimicrob. Resist. 2, 9 (2024).
Freudl, R. Signal peptides for Recombinant protein secretion in bacterial expression systems. Microb. Cell. Fact. 17, 52 (2018).
Vogelmann, J. et al. Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J. 30, 2246–2254 (2011).
Mell, J. C. & Redfield, R. J. Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483 (2014).
O’Rourke, A. et al. Mechanism-of-action classification of antibiotics by global transcriptome profiling. Antimicrob. Agents Chemother. 64, e01207–e01219 (2020).
Molina-Santiago, C. et al. Differential transcriptional response to antibiotics by Pseudomonas Putida DOT-T1E. Environ. Microbiol. 17, 3251–3262 (2015).
Howden, B. P. et al. Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure. Antimicrob. Agents Chemother. 57, 3864–3874 (2013).
Piepenbrink, K. H. DNA uptake by type IV filaments. Front. Mol. Biosci. 6, 1 (2019).
Wallden, K., Rivera-Calzada, A. & Waksman, G. Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12, 1203–1212 (2010).
Fischer, W., Tegtmeyer, N., Stingl, K. & Backert, S. Four chromosomal type IV secretion systems in Helicobacter pylori: Composition, structure and function. Front. Microbiol. 11, 1592 (2020).
Cascales, E. & Christie, P. J. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149 (2003).
Funding
This work was supported by the National Research Foundation of Korea (NRF) [NRF-RS-2024-00334769].
Author information
Authors and Affiliations
Contributions
**Y.M.** Data curation, Formal analysis, Investigation, Methodology, Writing-original draft, Writing - review & editing. **S.H.** Data curation, Formal analysis, Investigation, Methodology. **M.K.** Investigation, Validation. **G.L.** Investigation, Validation. **J.H.L.** Conceptualization, Investigation, Methodology. **D.W.J.** Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, and Writing - review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Moon, Y., Heo, S., Kim, M. et al. The transcriptomic response of Staphylococcus equorum KS1030 to Lincomycin stress reveals transporters associated with horizontal gene transfer. Sci Rep 16, 2492 (2026). https://doi.org/10.1038/s41598-025-32198-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-32198-1