Abstract
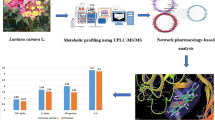
This study investigated the anti-inflammatory properties of Lonicera macranthoides and its active component isochlorogenic acid C (ILAC) through an integrated approach combining spectrum-effect relationship analysis, network pharmacology, and molecular docking. Five extracts (S1-S5) were evaluated in LPS-stimulated RAW 264.7 macrophages, with S4 demonstrating the strongest inhibition (45.53 ± 0.23%). HPLC fingerprinting identified 12 characteristic peaks, including ILAC and chlorogenic acid. PLS regression analysis revealed these two compounds were most positively correlated with the observed anti-inflammatory activity. Network pharmacology predicted 113 potential anti-inflammatory targets for ILAC, with PPI network analysis identifying 10 core targets (e.g., CASP3, HIF1A, NF-κB1, TLR4). Molecular docking studies suggested ILAC’s potential high binding affinity to these targets (<-5 kcal/mol). Together, these in vitro and in silico analyses indicated that ILAC is a key anti-inflammatory constituent in L. macranthoides, likely acting via multi-target interactions with critical inflammatory mediators. The study provided preliminary molecular-level insights into the traditional use of L. macranthoides for inflammatory conditions and suggested ILAC’s potential as a candidate for further anti-inflammatory research. Further in vivo studies are required to substantiate its therapeutic potential and mechanism of action.
Similar content being viewed by others
Introduction
The Lonicera macrantha complex (Lonicera sect. Nintooa), is one of the most complicated groups in the genus Lonicera. It includes five species: L. macrantha (D. Don) Spreng, L. macranthoides, L. similis Hemsl., L.hypoglauca Miq. and L. ferruginea Re- hd., which share similar floral structures, various leaf shapes, habitats, and sympatric distributions. These species are found in southern China and adjacent areas, typically growing at the edges of forests or roadsides1. Basing the data of Flora Reipublicae popularis Sinicae(https://www.iplant.cn/frps2019), an authoritative website for Chinese natural plants, these five types of Lonicera are distinct. Chen2 noted that these species were traditionally used in “da hua ren dong”. L. macranthoides is an economically and medicinally valuable traditional Chinese medicine (TCM) that has been utilized for centuries. Its medicinal value is nearly as high as that of Honeysuckle (Lonicera japonica Thunb), a plant highly valued for its strong anti-inflammatory effects1. L. macranthoides is most widely distributed in Guizhou province in southeastern China, where it is traditionally used to clinically treat various ailments, from ulcers and abscesses to arthralgia3. Contemporary pharmacological studies also reported that the extract of L. macranthoides possesses antibacterial, anti-inflammatory, and antioxidant effects4, which further validate its traditional uses.
In the last several years, the integration of modern scientific methodology and TCM research has accelerated significantly. Spectrum-effect relationship research has come to the forefront, connecting the chemical fingerprints with the pharmacological effects. Not only is this beneficial for the establishment of quality control specifications, but it also illuminates the therapeutic basis of TCM preparations5. For example, Wubuli et al.6 identified quality markers for the anti-inflammatory and antioxidant activities of Artemisia absinthium L. based on a spectrum-effect relationship. Another agent, Cnidii Fructus, derived from the dried ripe fruit of Cnidium monnieri (L.) Cuss, has been demonstrated to possess anti-hepatoma effects using this method7.
Network pharmacology has emerged as an increasingly significant methodology in TCM research, yielding new avenues for investigating complex interactions in biological systems, the safety research of TCM, and network toxicology research8. Specifically, Song’s team elucidated the anti-hypoxia mechanism of sesamoside, an active component of Phlomis younghusbandii Mukerjee, through network pharmacology9. Additionally, the anti-inflammatory potency of triterpenoids from SG and their potential mechanisms of action were explored using this approach10 These methodologies reinforce the modernization and internationalization of TCM, offering key support for drug discovery and precision medicine11.
While the medicinal effectiveness of L. macranthoides has been acknowledged, studies on the anti-inflammatory active components of it are focused primarily on chlorogenic acid, with few reports on other potential active components12,13. This leaves room to examine the full spectrum of bioactive compounds in L. macranthoides. The aim of this study is to identify and characterize more anti-inflammatory compounds in L. macranthoides by combining spectrum-effect relationship analysis, network pharmacology, and molecular docking technology. By leveraging these advanced technologies, we expect to have a full understanding of the anti-inflammatory mechanism of L. macranthoides and uncover new bioactive compounds that can account for its therapeutic efficacy.
The results of this study are anticipated to provide a scientific basis for the standardized production of L. macranthoides and its application in anti-inflammatory fields. Furthermore, the identification of novel active ingredients and their mechanisms of action may facilitate the development of new therapeutic agents derived from L. macranthoides. This research not only supports the scientific validation of traditional medicinal practices but also promotes the modernization and internationalization of TCM, ultimately contributing to global health through the development of effective treatments.
Materials and methods
Materials
Full confluence of RAW 264.7 macrophage cells was achieved by culture in a 5% CO2 humidified environment at 37 °C using Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum. The cells were obtained from the Culture Collection of Kunming Institute of Zoology at the Chinese Academy of Sciences in Kunming, China.
Reagents
The plant material used in this study was the flower of L. macranthoides (Lonicera family), which was commercially sourced from Zunyi Jinlei Yinhua Agricultural Industry Development Co., Ltd. (Batch number: 20250720). The botanical origin of the material was authenticated by Professor Wei Shenghua of Guizhou University of Traditional Chinese Medicine.
The medicinal plant specimens were packaged in vacuum-sealed bags and stored in the herbarium of Guizhou University of Traditional Chinese Medicine under standardized conditions (temperature: 22 °C; relative humidity: 40%; with proper ventilation). Analytical standards for neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, caffeic acid, isochlorogenic acid B, isochlorogenic acid A, and isochlorogenic acid C were sourced from Tanmo Quality Inspection Standard Material Center (Sichuan, China). The demethylbellidifolin standard was obtained from Toronto Research Chemicals Company (Toronto, Canada). Analytical grade methanol, xylene, and n-butanol were sourced from Nanjing Reagent Co., Ltd. (Nanjing, China). Distilled water was produced using the experimental distilled water apparatus.
Instrument
Instruments included an Electronic Balance (Precision: 0.0001, Model: FA1004, Liangping Instruments Co., Ltd., Shanghai, China), Rotary Evaporator (Model: RE-52AA, Shanghai Yarong Biochemical Instruments Co., Ltd., Shanghai, China), Vacuum Drying Oven (Model: BZF-50, Nanjing Wanzhong Technology Co., Ltd., Nanjing, China), Recirculating Water Multipurpose Vacuum Pump (Model: GZX-9146MBE, Shanghai Linke Instruments Co., Ltd., Shanghai, China), Powder Grinder (Model: 800 A, Yongkang Aizera Electric Appliance Co., Ltd., Jinhua, Zhejiang), and High-Performance Liquid Chromatography System (Model: 1260, Agilent Technologies, Santa Clara, California, USA).
Sample preparation
Preparation of extracts and resin treatment
20 g of L. macranthoides powder were separately extracted with 200 mL of methanol and distilled water, respectively, through two rounds of extraction, each lasting 2 h. The residues were filtered out, and the methanol extracts combined, as were the aqueous extracts. The combined methanol extract was concentrated to a paste using a rotary evaporator and subsequently lyophilized to yield a methanol extract powder, designated as S1. Similarly, the combined aqueous extract was concentrated and lyophilized to produce a water extract powder, designated as S2. Additionally, another 20 g of L. macranthoides were extracted twice with 200 mL of methanol, each extraction lasting 2 h. The residues were filtered out, and the methanol extracts were combined and concentrated to a paste, then reconstituted with water to achieve a solution with a crude drug concentration of 2 mg/mL. This solution was partitioned using two solvent systems: (a) n-butanol saturated with water and (b) water saturated with n-butanol, resulting in two partitioned extracts, designated as S3 and S4, respectively14.
Preparation and activation of NKA-9 macroporous resin
30 g of NKA-9 macroporous resin was weighed and immersed in 95% ethanol solution for 24 h. The resin was filtered and washed multiple times with distilled water until the washing solution did not have the smell of ethanol and was clear and not turbid. Then, the resin was immersed in 5% HCl solution for 5 h and filtered and washed multiple times with distilled water until the washing solution was neutral on pH paper. Last, the resin was immersed in 5% NaOH solution for 5 h and washed multiple times with distilled water until the washing solution was neutral on pH paper, indicating the resin was fully activated. Activated resin was kept in distilled water for later use.
Adsorption and elution of extracts using NKA-9 resin
Methanol-extracted L. macranthoides powder was concentrated to a paste and dissolved to a 0.1 g/mL concentration. A precise amount of activated NKA-9 resin was prepared, dried, and placed in a conical flask with an equal volume of L. macranthoides solution for static adsorption. After 12 h, the resin was washed until colorless, and the adsorbed compounds were eluted with 70% methanol15. The eluate was collected and designated as sample S5.
Concentration and drying of samples
All samples were concentrated using rotary evaporation and freeze-dried to obtain sample powders.
Nitric Oxide(NO) determination
The cells were treated with 100 µg/mL of each L. macranthoides extract (S1-S5) and 10 µg/mL of LPS (Sigma, USA) for 24 h, referred to as the Sample group; cells treated with 10 µg/mL of LPS for 24 h were termed the Model group; and cells treated with culture medium for the same duration were designated the Blank group. Each group comprised 5 replicates. The subsequent accumulation of NO in the supernatant was quantified using the NO Assay Kit (Nanjing Jiancheng Bioengineering Research Institute). Briefly, 50 µL of cell supernatant were collected and mixed with 50 µL of Griess Reagent I per well, followed by the addition of an equal volume of Griess Reagent II. The mixture was thoroughly mixed, incubated at room temperature, and the absorbance at 540 nm was measured16. The anti-inflammatory inhibitory rate was calculated according to Eq. (1), where a higher rate indicates superior anti-inflammatory activity. Each sample was assayed in triplicate to ensure reproducibility and accuracy.
Note:
Amodel is the absorbance of model group; Asample is the absorbance of sample group; Ablank is the absorbance of blank group.
HPLC conditions
The chromatographic separation was successfully conducted utilizing an XTerra MS-C18 column (Waters Corporation, Milford, USA). The column was maintained at a temperature of 30 °C, with a flow rate established at 0.8 mL/min. The volume of the injection was measured at 2 µL, while the wavelength for detection was set at 327 nm. The composition of the mobile phase is presented in Table 1.
Collection of ILAC targets
The structural formula and SMILES notation of ILAC were obtained by querying “Isochlorogenic acid C” in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Using this data, potential targets for ILAC were retrieved from the Swiss Target Prediction (http://swisstargetprediction.ch/) and SuperPred database (http://bioinformatics.charite.de/superpred), employing the keyword “Isochlorogenic acid C” and restricting the search to “Homo sapiens”. The structural integrity and consistency of the search results were verified. Duplication was eliminated, and target names were standardized using the UniProt database (https://www.uniprot.org/). A comprehensive target database for ILAC was constructed.
Selection of target networks related to anti-inflammatory effects
Key targets associated with anti-inflammatory activity were identified utilizing the GeneCards (https://www.genecards.org/) and OMIM (https://www.omim.org/) databases. Venn diagram analysis was subsequently applied to discern common targets between ILAC and known anti-inflammatory agents, thereby isolating the specific anti-inflammatory targets of ILAC.
Protein interaction network construction and core target screening
To find possible anti-inflammatory targets of ILAC, the STRING database was queried using the species “Homo sapiens.” Next, the data was imported into a tool for network biology visualization, and Cytoscape software (version 3.10.1) was used to assess the node properties and display the molecular connections. Central targets were identified by employing Centiscape (version 2.2) via visual network examination. Furthermore, the MCODE plugin performed module analysis, identifying ten crucial genes within the most significant module.
Gene function and pathway enrichment analysis of target protein
Biological functions of ILAC’s potential targets, particularly in relation to anti-inflammatory effects, were further elucidated through data integration from the Metascape and DAVID databases. The species was set to ”Homo sapiens”, and the screening criterion was set at a significance level of P < 0.01. This process involved a detailed GO analysis, which included evaluations of BP, CC, and MF. This analysis provided detailed insights into the primary BP of these targets. The top 10 items were selected based on P-values for visualization analysis. Further, the main pathways linked to ILAC’s anti-inflammatory properties were identified by a KEGG analysis17,18,19, with an emphasis on therapeutic and possible harmful pathways. The top 20 items were selected based on P-values for visualization analysis. Our goal in doing these investigations was to gain a better grasp of the molecular mechanisms underlying the anti-inflammatory benefits of ILAC.
Molecular docking of ILAC with core targets
Protein information was retrieved from the UniProt database (https://www.uniprot.org/uniprotkb), and the protein structure was obtained from the SCSB PDB database (https://www.rcsb.org/). The acquired structure was imported into Discovery Studio 2019 for protein structure optimization. This process primarily included removing water molecules, adding hydrogen atoms, completing charges, filling in missing amino acids, and reconstructing side chains, ultimately yielding an optimized protein structure. The optimized structure was then exported as a PDB file. Small molecule structures were acquired from the PubChem database and subjected to energy minimization using Discovery Studio 2019, after which they were saved as PDB files.
A grid box was defined to encompass the binding pocket of the receptor, with detailed parameters provided in Table 2 to ensure it accommodated all extended conformations of the ligands. Protein and small molecule PDBQT files were prepared using AutoDock 1.5.6, and molecular docking was performed using AutoDock Vina (version 1.2.6.) Finally, the results were visualized and analyzed using PyMOL (version 3.1) and Discovery Studio 2019.
Statistical analysis
Data from this investigation were processed using GraphPad Prism (version 10)for t-tests (two-sided) on two groups and one-way ANOVA on three or more groups. P < 0.05 or lower is considered statistically significant.
Results
Anti-inflammatory results
As depicted in Table 3, each sample exhibited potent anti-inflammatory activity. The highest inhibition rate recorded was 45.53 ± 0.23%, while the lowest was 33.02 ± 0.23%, indicating that the anti-inflammatory activity of S4 is the most potent and that of S1 is the least.
HPLC fingerprint profiles
HPLC fingerprints of the five samples showed 12 common peaks, eight of which were identified by comparison with reference standards (Table 4, chemical structures in Fig. 1). Notably, the eight identified compounds showed excellent agreement with the findings reported by Yan et al.20., further validating the reliability of our analytical approach.
Spectral effect relationship of anti-inflammatory experiments
Based on the anti-inflammatory data in Table 3 and peak areas from Table 5, PLS analysis was performed using Minitab (version 6.4.1), yielding Eq. (2). The magnitude of the correlation coefficient indicated the level of contribution to anti-inflammatory activity: higher absolute values denoted substantial contributions, while lower values indicated minor contributions. The sign of the correlation coefficient determined the nature of the relationship with anti-inflammatory activity, where a negative coefficient indicated an inverse relationship and a positive coefficient suggested a direct relationship. Peaks showing a positive correlation with the pharmacological effect are termed pharmacologically effective peaks. This analysis identified peaks 5, 6, 9, 11, and 12 as efficacy peaks, with peak 6 contributing most significantly, followed by peak 5, and then peak 12. These peaks are significantly associated with anti-inflammatory activity. Peak 6, identified as chlorogenic acid3, is noted for its anti-inflammatory properties. Peak 5 remains unidentified, while peak 12, corresponding to ILAC, is highlighted as a promising candidate for further investigation. Consequently, ILAC will be included in subsequent network pharmacology studies.
Identification of targets of ILAC affecting anti-inflammatory effects
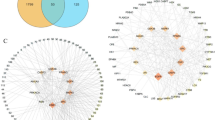
Initially, this research identified 145 targets associated with ILAC from the Swiss Target Prediction and SuperPred databases. Subsequent analyses using the GeneCards and OMIM databases expanded this to include 3,724 targets with significant links to anti-inflammatory effects. After rigorous integration and deduplication, a refined set of 113 intersecting targets was determined, posited to influence ILAC’s impact on anti-inflammatory mechanisms. These targets are illustrated in Fig. 2.
Interaction network of potential targets and acquisition of core genes
The PPI network comprised 110 targets with 583 interactions (Fig. 3a). Topological analysis identified ten core targets, among which CASP3 exhibited the highest connectivity (Fig. 3b). Module analysis further highlighted a densely connected cluster containing these key targets (Fig. 3c).
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG)analysis of potential targets
The GO analysis showed that the BP were primarily involved in cellular responses to nitrogen compounds, inflammatory responses, and positive regulation of response to external stimuli; CC included the perinuclear region of the cytoplasm, the apical part of the cell, and the cell body; MF related to peptidase activity, histone modifying activity, and kinase binding. The KEGG pathways focused on processes such as apoptosis, neutrophil extracellular trap formation, serotonergic synapse, and calcium signaling pathway. Employing the Metascape database, GO and KEGG enrichment analyses of the 113 potential targets were performed. The GO analysis identified significant terms related to inflammatory response, response to external stimuli, and peptidase activity (top terms shown in Fig. 4). KEGG pathway analysis revealed 172 significantly enriched pathways. To precisely delineate the anti-inflammatory mechanism, we focused our interpretation on those pathways most directly associated with inflammation and immune regulation. As illustrated in Fig. 5, the key inflammation-related pathways included NF-kappa B signaling, TNF signaling, Apoptosis, Toll-like receptor signaling, and NOD-like receptor signaling. These pathways strongly suggest that ILAC may exert its effects by modulating core inflammatory signaling networks and immune cell fate.
GO analysis of potential targets (top 10). (a) Each bubble’s size corresponds to gene expressions in a particular pathway, with enrichment significance indicated by color saturation. (b) This histogram presents the top 10 enriched entries for each GO category (BP, CC, MF), where smaller P-values signify higher statistical importance and the bar height reflects the level of enrichment.
KEGG analysis of potential targets. (a) The bubble diagram illustrates the top 20 enriched KEGG pathways. In this representation, the size of each bubble corresponds to the quantity of enriched genes, while the intensity of the color reflects the level of enrichment significance. (b) The histogram depicts the frequency and importance of enrichment for each pathway, where the length of each bar represents gene counts and levels of enrichment.
Pathway enrichment analysis of core targets
The top 10 proteins ranked by their degree values are considered as the ten primary targets. These ten primary targets of ILAC were examined utilizing the DAVID database (https://david.ncifcrf.gov/). A detailed pathway analysis, incorporating KEGG enrichment, was represented in a horizontal gradient bar chart (Fig. 6), showcasing the top 20 enrichment pathways characterized by the lowest FDR values. The apoptosis pathway emerged as a crucial mechanism in ILAC’s anti-inflammatory effects, with CASP8, CASP3, NF-κB1, and CTSB assuming essential roles, highlighting their important contributions to the regulatory process. This emphasizes the unique importance and potential therapeutic applications of the apoptosis pathway in mitigating the anti-inflammatory effect, opening new possibilities for future research and intervention strategies.
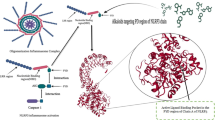
Molecular docking of ILAC and anti-inflammatory effect core target protein
The interactions between ILAC and ten crucial target genes were examined using molecular docking analysis. Utilizing AutoDock software, the simulations indicated remarkably low binding energies(Table 6), all beneath − 5 kcal/mol, showcasing the spontaneous and strong binding ability of ILAC to these essential proteins, emphasizing its significant affinity and vital function in the molecular processes underlying its anti-inflammatory effects. PyMOL was subsequently used to visualize and detail the lowest energy binding conformations (Fig. 7.), clearly depicting the intimate relationship between ILAC and its key targets, offering invaluable insights into the structural basis of their interactions and reaffirming the significance of ILAC in modulating the anti-inflammatory effect at the molecular level.
Discussion
The extracts of L. macranthoides (Sample S1-S5) exhibited dose-dependent anti-inflammatory activity in vitro, with the Sample S4 extract showing the highest potency (45.53% inhibition). This aligns with established reports on the anti-inflammatory properties of this species3,4,6. HPLC fingerprinting identified 12 common peaks, eight of which were characterized, including chlorogenic acid and ILAC. These results are in agreement with those reported by other researchers3, who observed similar chemical profiles in their studies of L. macranthoides.
PLS analysis revealed that peaks 6 (chlorogenic acid), 5 (unknown), and 12 (ILAC) were positively correlated with anti-inflammatory activity. Numerous studies have demonstrated the anti-inflammatory properties of chlorogenic acid and have further elucidated its mechanisms of action. However, research on ILAC is relatively limited, particularly regarding its anti-inflammatory mechanisms, which remain unclear. Therefore, we focused our subsequent analysis on ILAC to provide a more comprehensive understanding of its potential therapeutic applications.
Using network pharmacology, 113 potential targets of ILAC related to anti-inflammatory effects were identified. A protein-protein interaction network highlighted ten core targets (e.g., CASP3, NF-κB1, TLR4). KEGG pathway analysis enriched these targets in inflammation-related pathways such as NF-κB signaling and apoptosis, suggesting ILAC may act through a multi-target mechanism centered on these core nodes.
Our pathway analysis suggested ILAC’s action may involve apoptosis, a process known to interplay with inflammation21. This aligns with the established roles of CASP3 and NF-κB1 in regulating inflammatory responses22,23 and is supported by a recent report that ILAC can inhibit the NF-κB pathway in mammary cells24.
Molecular docking studies were performed on ten key targets, and it was found that the binding energies of ILAC to these targets were all below − 5 kcal/mol. Extensive literature underscores the crucial roles of CASP3, HIF1A, NF-κB1, TLR4, and APP in anti-inflammatory mechanisms25,26,27,28,29,30. The interplay of these factors plays a crucial role in the regulation of inflammatory responses, influencing the expression of inflammatory cytokines, the recruitment of inflammatory cells, and the synthesis of anti-inflammatory mediators. As potential targets for anti-inflammatory therapy, they offer a theoretical foundation for the development of innovative anti-inflammatory medications.
Compared with existing studies, the innovations of this study are highlighted by the comprehensive evaluation of the chemical constituents of L. macranthoides extracts through HPLC fingerprint analysis, the systematic identification and analysis of the potential targets of ILAC, and the construction of a PPI network elucidating its multi-target mechanism in anti-inflammatory effects. Molecular docking experiments validated the binding affinity of ILAC to core target proteins, providing direct molecular evidence for its anti-inflammatory activity. This research represents a pioneering effort to integrate network pharmacology with molecular docking, facilitating an in-depth examination of the anti-inflammatory mechanisms associated with ILAC.
Despite the achievements of this study, several limitations must be acknowledged. First, our anti-inflammatory experiments were conducted in vitro, lacking in vivo validation. Future research should involve verifying the anti-inflammatory effects of L. macranthoides extracts in animal models to confirm their activity in vivo. Additionally, the anti-inflammatory role of ILAC remains at the analytical stage. Further investigations are required to validate and explore the anti-inflammatory mechanisms of ILAC both in vitro and in vivo, involving cellular experiments and animal models. This will also require further confirmation of the relevant targets, with the ultimate goal of providing more effective therapeutic options for inflammatory diseases.
Conclusions
In this study, spectrum-effect relationship analysis, network pharmacology, and molecular docking techniques were employed to preliminarily investigate the principal active constituents, potential targets, and molecular interactions underlying the anti-inflammatory effects of L. macranthoides. Our in vitro assays and chemical analysis identified chlorogenic acid and ILAC as key components positively correlated with the observed anti-inflammatory activity in macrophages. Through network pharmacology prediction, we pinpointed 113 potential targets linked to the anti-inflammatory properties of ILAC. A predicted interaction network of these targets was developed, highlighting ten central nodes, such as CASP3 and NF-κB1. Pathway analysis suggested significant links to the apoptosis pathway, with CASP3 and NF-κB1 emerging as potential key mediators in modulating inflammatory responses. Furthermore, molecular docking simulations indicated that the binding energies between ILAC and these predicted targets were favorable (below − 5 kcal/mol). Collectively, these integrated in vitro and in silico findings propose that ILAC serves as a key anti-inflammatory constituent in L. macranthoides, acting potentially through multi-target interactions. This work provided a preliminary mechanistic hypothesis and a computational foundation for further exploration of ILAC. However, it is crucial to emphasize that these conclusions are primarily derived from cell-based and computational models. Therefore, additional in vivo pharmacological studies and, ultimately, clinical validations are indispensable to substantiate its actual therapeutic potential and mechanism of action.
Data availability
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to institutional requests.
Abbreviations
- PPI:
-
protein-protein interaction
- BP:
-
biological processes
- CC:
-
cellular components
- GO:
-
Gene Ontology
- HPLC:
-
High-Performance Liquid Chromatography
- ILAC:
-
Isochlorogenic acid C
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- L. macranthoides :
-
Lonicera macranthoides Hand.-Mazz
- MF:
-
molecular functions
- PLS:
-
Partial Least Squares
- TCM:
-
Traditional Chinese medicine
References
Lin, A. T. et al. Network Pharmacology and data analysis method to explore the traditional Chinese medicine regulates ferroptosis key genes in the occurrence and prognosis of lupus nephritis. TMR Pharmacol. Res. 3 (3), 25–32 (2023).
Chen, Z. H. et al. New records of lonicera plants in Zhejiang Province. Bull. Bot. Res. 42 (1), 1–11 (2022).
Qi, D. M. et al. Differentiation between lonicera Japonica and lonicera hypoglauca based on fingerprint analysis and quality evaluation of different varieties of lonicera Japonica. Chin. Tradit Herb. Drugs. 54 (6), 1946–1952 (2023).
Zeng, A. Q. et al. Anti-inflammatory Pharmacological effects of lonicera Japonica and lonicera hypoglauca. Chin. J. Chin. Mater. Med. 45 (16), 3938–3944 (2020).
Luorong, Q. et al. Comprehensive quality evaluation of lysimachia Christinae hance via Fingerprint, Spectrum–Effect Relationship, and quantitative analyses of multiple components by single marker. Phytochem Anal. 35(6). (2024).
Wubuli, A. et al. Exploring Anti-inflammatory and Antioxidant‐related quality markers of Artemisia absinthium L. Based on Spectrum–Effect relationship. Phytochem Anal. 35 (5), 1152–1173 (2024).
Lv, Z. et al. Spectrum–Effect Relationship Study Between Ultra-high‐performance Liquid Chromatography Fingerprints and Anti‐hepatoma Effect in Vitro of Cnidii Fructus. Biomed. Chromatogr. 38(5), e5847 (2024).
Qian, Z. et al. Chemical Composition, Pharmacological effects and clinical applications of cinobufacini. Chin. J. Integr. Med. 30 (4), 366–378 (2024).
Song, D. et al. The Anti-hypoxic mechanism of Sesamoside determined using network Pharmacology. Dose-Response 22 (3), 15593258241282574 (2024).
Wei, Y. et al. Network Pharmacology and experimental verification to explore the Anti-inflammatory activities of triterpenoids from Siraitia grosvenorii. Nat. Prod. Res. 1–5. (2024).
Yang, J. et al. Exploring the mechanism of TCM formulae in the treatment of different types of coronary heart disease by network Pharmacology and machining learning. Pharmacol. Res. 159, 105034 (2020).
Wang, Y. Y. et al. Simultaneous determination of chlorogenic acid and luteoloside in lonicera macranthoides by HPLC. J. Chin. Med. Mater. 32 (11), 1705–1707 (2009).
Chen, Y. C. Mechanism of Light Affecting Chlorogenic Acid Accumulation in Lonicera macranthoides. Ph.D. Thesis, Hunan University, (2023).
Bai, Z. J. Study on the Spectrum-Effect Relationship of Lonicera hypoglauca. Master’s Thesis, Guizhou University, (2015).
Gao, W. et al. Establishment of HPLC Fingerprint, cluster analysis, and principal component analysis of clerodendrum Cyrtophyllum. China Pharm. 29 (16), 2215–2219 (2018).
Zhou, M. J. et al. Chemical constituents and Anti-neuroinflammatory activity of Piper sarmentosum. Chin. Tradit Herb. Drugs. 56 (1), 44–51 (2025).
Kanehisa, M. et al. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Yan, W. N. Study on Fingerprint of Lonicera Japonica and Lonicera Hypoglauca in Guizhou (Guizhou University, 2018).
Wang, Y. et al. Dietary Chlorogenic Acid Supplementation Protects Against Lipopolysaccharide-Induced Oxidative Stress, Inflammation and Apoptosis in Intestine of Amur Ide (Leuciscus waleckii). Aquat. Toxicol. 279, (2025).
Yan, J. W. et al. Quality evaluation of Runbi Tongqiao drops based on HPLC fingerprint and determination of seven components. Chin. J. Pharm. Anal. 44 (12), 2064–2071 (2024).
Huang, T. L. et al. Anethole mitigates H₂O₂-Induced inflammation in HIG-82 synoviocytes by suppressing the Aquaporin 1 expression and activating the protein kinase A pathway. Environ. Toxicol. 39(1/2). (2024).
Lossi, L. et al. Ex vivo imaging of active caspase 3 by a FRET-Based molecular probe demonstrates the cellular dynamics and localization of the protease in cerebellar granule cells and its regulation by the Apoptosis-Inhibiting protein survivin. Mol. Neurodegener. 11 (1), 1–20 (2016).
Chen, X. et al. PIM1/NF-κB/CCL2 Blockade enhances Anti-PD-1 therapy response by modulating macrophage infiltration and polarization in tumor microenvironment of NSCLC. Oncogene 43 (33), 1–14 (2024).
Chen, X. H. et al. Analysis of the inhibitory effect of isochlorogenic acid C on mammary inflammation via the NF-κB signaling pathway using bovine mammary cells and mouse mammary tissue. Acta Vet. Zootech Sin. 54 (9), 3931–3940 (2023).
Dhahbi, J. et al. Circulating blood leukocyte gene expression profiles: effects of the Ames Dwarf mutation on pathways related to immunity and inflammation. Exp. Gerontol. 42 (8), 772–788 (2007).
Feng, F. et al. DendroX: Multi-level Multi-cluster selection in dendrograms. BMC Genom. 25(1). (2024).
Nasr, S. et al. Anti-inflammatory potential of Aspergillus Unguis SP51-EGY: TLR4-Dependent effects & chemical diversity via Q-TOF LC-HRMS. BMC Biotechnol. 24(1). (2024).
Huang, P. et al. Glucocorticoid activates STAT3 and NF-κB synergistically with inflammatory cytokines to enhance the Anti-inflammatory factor TSG6 expression in mesenchymal Stem/Stromal cells. Cell. Death Dis. 15(1). (2024).
Funding
This research was funded by Science and Technology Fund of Guizhou Health Commission(No. gzwkj2024-230), Key Advantageous Discipline Construction Project of Guizhou Provincial Health Commission in 2025, Guizhou CDC Doctoral Studio Project(No. bsgzs2025-01) and Guizhou Provincial Center for Disease Prevention and Control Youth Science Fund Project (No. 2024-E-6).
Author information
Authors and Affiliations
Contributions
Conceptualization, Z.W. and Z.L.; investigation, Z.L., H.J. Z.J, L.Q. and C.Y.; resources, P.S.; writing —original draft preparation, Z.W., Z.L.; writing—review and editing, Q.X., P.S. and Z.L.; funding acquisition, Z.J., L.L., S.L. and Z. L. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, Z., Junfei, H., Runsang, P. et al. Anti-inflammatory effects of Lonicera macranthoides Hand.-Mazz based on Spectrum-effect relationship, network pharmacology and molecular docking technology. Sci Rep 16, 3396 (2026). https://doi.org/10.1038/s41598-025-33416-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-33416-6