Abstract
To thoroughly analyze the characteristics of the chloroplast genome in the genus Agropyron Gaertn and provide a theoretical basis for its evolutionary biology and the molecular marker-assisted taxonomic identification of the broad-spike and narrow spike clades. This study conducted a systematic analysis of the chloroplast genomes from seven Agropyron species and two closely related species (Elymus trachycaulus and Elytrigia elongata). The chloroplast genome of Agropyron Gaertn exhibits a typical quadripartite structure, ranging in size from 135 to 137 kb and containing 131 genes. The rps12 and ycf3 genes show high variability, while the intergenic regions (IR) exhibit high GC content of 43.91% to 44.01%. SSR and scattered repetitive sequences exhibit significant interspecific differences among the Agropyron Gaertn and the closely related species. For instance, Elymus trachycaulus and Elytrigia elongata share the locus (CCATA)3, while simultaneously retaining genus-specific markers. For instance, locus (ATATA)3 is unique to Elytrigia elongata, and Elymus trachycaulus lacks the rps12 gene intron variation, forming a sequence profile that combines both conservation and differentiation. Based on the distribution patterns of SSR loci and nucleotide diversity analysis in this study, the combination patterns of SSR loci in the chloroplast genome can serve as candidate basis for the molecular-assisted taxonomic identification of the broad-spike/narrow-spike clades. Phylogenetic analysis revealed that most species within the genus Agropyron form monophyletic clades, while Elymus trachycaulus clusters closely with Elytrigia elongata due to shared characteristics such as high GC content (44.01%) in the intercalary region of the chloroplast genome and with LSC length (80,642 bp). Combined with its narrow-spike morphology, this result supports the molecular marker-assisted taxonomic identification to identify Elymus trachycaulus into the narrow-spike clade, demonstrating the synergy between morphological identification and molecular evidence in auxiliary taxonomy. This study lays the foundation for characterizing the chloroplast genomes of Agropyron species and developing molecular markers for the identification of the broad-spike/narrow-spike clades. Further multidisciplinary research is needed to explore the potential applications of these molecular markers and the mechanisms underlying the species’ adaption.
Similar content being viewed by others
Introduction
Agropyron, a genus within the tribe Triticeae of the family Poaceae, is classified as a perennial grass species widely distributed across arid and semi-arid regions of Eurasia. As an important forage grass and ecological restoration plant, Agropyron plays an important role in grassland improvement, soil and water conservation and biodiversity conservation1,2. These ecological and agronomic roles underscore the importance of further deep study especially unraveling its genetic architecture to advance both theoretical understanding and practical applications.
The taxonomy of Agropyron Gaertn has been progressively refined through decades of research, integrating morphological, cytological, and genomic data. In 1933, the Soviet botanist S. A. Nevski proposed a narrow definition of the genus based on morphological traits, which delineated its core species. Later, in the late twentieth century, Dewey3 developed a modern classification system centered on the P chromosome group, which is shared by all species in the genus. Subsequent studies incorporated ploidy levels, clearly distinguishing diploids (2n = 14) and tetraploids (2n = 28) species4. This chromosome-based system has been continually refined and now recognizes three main species within Agropyron Gaertn., a framework that remains standard today5.
The classification of subordinate taxa also relied on differences between broad-spike and narrow-spike forms. For instance, in 1960, British cytotaxonomist Keith Jones categorized populations of flat-spiked wheatgrass into western broad-spike, eastern broad-spike, and narrow-spike types based on spike morphology6. In China, the mainstream classification has long followed the concept of flat-spiked wheatgrass established by Professor Geng Yili7 which is based on Nevski system, and has remained relatively stable. However, significant challenges persist in precisely defining broad-spike and narrow-spike clades: on one hand, morphological traits are highly influenced by environmental conditions, which often leads to subjective judgements when identifying transitional forms or closely related species. On the other hand, while chromosome-based classification is accurate, its complex experimental procedures and long timelines make it impractical for rapidly screening large germplasm collections. Therefore, developing molecular markers to efficiently and accurately distinguish broad-spike and narrow-spike clades is crucial. Such techniques would resolve current ambiguities in classification and advance the precise identification and efficient use of Agropyron germplasm resources.
Chloroplasts are vital organelles in plant cells responsible for photosynthesis and various metabolic processes. They not only carry out photosynthesis but also contain abundant genetic information. Their genome exhibits characteristics such as maternal inheritance, structural conservation, and moderate evolutionary rates, making it a crucial sequence feature for plant phylogenetics, species identification, and genetic improvement research8. Since the first tobacco chloroplast genome was sequenced in the 1980s, significant progress has been made in chloroplast genomics research9,10. In recent years, with the rapid advancement of sequencing technology, an increasing number of plant chloroplast genomes have been successfully sequenced and analyzed, providing rich data support for revealing the genetic code and evolutionary patterns of plant chloroplast genomes. Within the grass family, chloroplast genome research has yielded substantial results. For example, Luo et al.11 conducted a population genetic study of Agropyron Gaertn across four populations from the Qinghai-Tibet Plateau, Central Asia, East Asia, and Europe using Accl and GBSSI gene sequences. The results revealed that Agropyron Gaertn exhibits rich genetic diversity at the population level, with the Central Asian population potentially serving as the center of differentiation for Agropyron Gaertn. This finding indicates Central Asia as the origin center for Agropyron Gaertn, providing crucial theoretical support for the conservation and utilization of its genetic diversity. These studies not only reveal the structural characteristics and evolutionary patterns of grass chloroplast genomes but also provide an important molecular foundation for crop improvement and germplasm resource conservation12. However, research on complete chloroplast genome sequences for additional species within the genus Agropyron Gaertn remains relatively scarce and lacking systematic analysis and comparison. Therefore, the identification of complete chloroplast genome sequences for 7 species of the genus Agropyron Gaertn and 2 closely related species is of great significance for comprehensively revealing the genetic characteristics, evolutionary patterns, and potential functions of the chloroplast genomes within the genus Agropyron Gaertn.
This study employed high-throughput sequencing technology to analyze the chloroplast genomes of seven Agropyron Gaertn species and two closely related species (Elymus trachycaulus and Elytrigia elongata), elucidating their genetic characteristics and evolutionary patterns. As a representative of the closely related genus within the Triticeae13, the 2 closely related species (Elymus trachycaulus and Elytrigia elongata) can be used in comparative genomic analysis to screen for specific molecular markers distinguishing the broad-spike and narrow spike clades within the genus Agropyron Gaertn, thereby providing a reference supporting taxonomic identification. This study aims to provide theoretical support for the precise utilization of Agropyron Gaertn germplasm resources and the establishment of an efficient identification system.
Materials and methods
Plant materials
This study screened seven species of the genus Agropyron Gaertn and two closely related species: ACPE(Agropyron cristatum var. pectiniforme)14,15, ACPL(Agropyron cristatum var. pluriflorum)14,15 AD(Agropyron dasystachyum var. subvillosum)16,17, ADP(Agropyron desertorun var. pilosiusculum)14,15, AS(Agropyron sibiricum f. sibiricum)14,15, AMV (Agropyron mongolicum var. villosum) 14,15, ASP(Agropyron sibiricum f. pubiflorum)14,15, ET(Elymus trachycaulus) 18 and EE(Elytrigia elongata)14,15,18,19,20. The complete chloroplast genomes were characterized and de novo assembled (Table 1). Species identification was primarily based on the identification keys in Tomus 9 (Part 2) of the Chinese Academy of Sciences15 and Flora Intramongolica (Editio Tertia) Tomus 614. The morphological identification reference by Yan Weihong21 employs a chromosome-based identification system to categorize Agropyron Gaertn into broad-spike and narrow-spike clades. This identification has long adhered to the concept of narrow-spike narrow-spike agropyron wheatgrass proposed by Professor Geng Yili 22 based on the Nevski identification system. This study does not validate this morphological identification through molecular data but instead focuses on screening chloroplast molecular markers that can assist in distinguishing the two clades. All species samples were collected from the Shalqin Experimental Station of the Grassland Research Institute, Chinese Academy of Agricultural Sciences, located at N 40°35’, E 111°47’. Genomic DNA was extracted using a modified CTAB method. All samples are stored at the National Pasture Germplasm Resource Intermediate Repository (Hohhot, Inner Mongolia).
cpDNA sequencing and de novo assembly
Raw data were filtered using fastp v0.20.0 (https://github.com/OpenGene/fastp) to obtain clean data23. Bowtie2 v2.2.4 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) was employed in very-sensitive-local mode to align against the chloroplast genome database, reducing assembly complexity. Sequences aligned were designated as chloroplast genome sequences (cpDNA sequences) for the project samples. The core assembly module employed SPAdes v3.10.1 (http://cab.spbu.ru/software/spades/) to assemble chloroplast genomes, utilizing kmer sizes of 55, 87, and 121, with assembly performed independently of reference genomes. The complete chloroplast genomes of seven Agropyron species and two closely related species (E. trachycaulus, E. elongata) have been deposited in the NCBI database under the following accession numbers: SAMN47853882 (E. elongata), SAMN47853883 (A. sibiricum), SAMN47853884 (E. trachycaulus), SAMN47853885 (A. cristatum var. pectiniforme), SAMN47853886 (A. cristatum var. pluriflorum), SAMN47853887 (A. mongolicum var. villosum), SAMN47853888 (A. desertorun var. pilosiusculum), SAMN47853889 (A. sibiricum f. pubiflorum) and SAMN47853890 (A. dasystachyum).
Chloroplast gene annotation
Two methods were employed to annotate the chloroplast genome, enhancing annotation accuracy. First, Prodigal v2.6.3 (https://www.github.com/hyattpd/Prodigal) was used to annotate chloroplast CDSs24, predicted rRNA using hmmmer v3.1b2 (http://www.hmmer.org/)25, and predicted tRNA using aragorn v1.2.38 (http://www.ansikte.se/ARAGORN/)26. Next, gene sequences were extracted from published relatives in NCBI, then aligned against the assembled sequences using blast v2.6 (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to obtain a second annotation result27. Finally, manually examine genes with discrepancies between the two annotation sets, remove erroneous or redundant annotations, and define multi-exon boundaries to obtain the final annotation.
Chloroplast genome map
Nine chloroplast genomes of the genus Cynodon were assembled using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html)28. MISA v1.0 (MIcroSAtellite identification tool, https://webblast.ipk-gatersleben.de/misa/) was used to identify chloroplast SSRs ranging from single-nucleotide to octanucleotide repeats29. RSCU values were analyzed using MEGA 7. Gene sequences were aligned using MAFFT v7.427 (https://mafft.cbrc.jp/alignment/software)30, and synonymous and non-synonymous substitution rates were calculated with KaKs_Calculator v2.0 (https://sourceforge.net/projects/kakscalculator2/)31. Global alignment of homologous gene sequences across different species was performed using MAFFT. Nucleotide diversity (Pi)32 for each gene was calculated using DNASP v5 (http://www.ub.edu/dnasp/).
Analysis and identification of cpSSR and scattered repeat sequences
Analysis of cpSSR was performed using MISA v1.0 (MIcroSAtellite identification tool, https://webblast.ipk-gatersleben.de/misa/) with parameters 1–8 (single-base repeats occurring 8 or more times), 2–5, 3–3, 4–3, 5–3, 6–3. Repeat sequences were identified using vmatch v2.3.0 (http://www.vmatch.de/) combined with a Perl script33. This included forward, reverse, complementary, and palindromic tandem repeats with a minimum length of 30 bp and an edit distance less than 3 bp.
Chloroplast sequence homology analysis, collinearity identification, and phylogenetic tree construction
Visualize the boundary information between IR and LSC/SSC using CPJSdraw (http://cloud.genepioneer.com:9929/#/tool/alltool/detail/296), expressed as LSC-IRb, IRb-SSC, SSC-IRa, and IRa-LSC34. Perform phylogenetic analysis using the entire genome. Set the same starting point for circular sequences. Perform multiple sequence alignment of interspecies sequences using MAFFT v7.427 software (–auto mode). Process the aligned data using MrBayes v3.2.7a (http://nbisweden.github.io/MrBayes/) software. using the GTR + I + G model. Ngammacat was set to 5, with statefreqpr, revmat, pinvar, and shapepr configured according to the optimal model identified by jModelTest software. Other parameters remained at default values to construct Bayesian phylogenetic trees35. Genome alignment was performed using Mauve software with default parameters36.
Results
Basic traits of the chloroplast genome
Sequencing of the chloroplast genomes from seven Agropyron Gaertn species and two closely related species (E. trachycaulus, E. elongata) yielded 17,707,085 to 20,670,776 clean paired-end reads, respectively (Table 2). Among these, E. trachycaulus (ET) yielded 19,727,229 clean reads, with a complete cp genome length of 135,037 bp. This length is significantly shorter than that of the seven Agropyron Gaertn-clade species (135,448–135,483 bp) but comparable to that of the closely related species E. elongata (EE, 135,067 bp) (Table 2 and Fig. 1).
In terms of genetic composition, E. trachycaulus exhibits unique intermediate characteristics: its chloroplast genome contains 131 genes, specifically 40 tRNA genes, 8 rRNA genes, and 83 mRNA genes. This numerical profile is identical to that of seven Agropyron Gaertn species within the broad-spike clades; However, it differs significantly from E. elongata (129 genes, 38 tRNA genes), sharing only the numbers of rRNA (8) and mRNA (83) genes with E. elongata. This characteristic—where the total gene count aligns with the broad-spike clades of the genus Agropyron Gaertn while the genome length approximates that of E. elongata—provides crucial chloroplast genomic evidence for E. trachycaulus taxonomic identification. It not only reflects its phylogenetic relationship with Agropyron Gaertn but also reveals genomic structural signals indicating its differentiation toward the narrow-spike clades (Table 2).
The chloroplast genomes of all species exhibit a single-circular quadripartite structure: a large single-copy region (LSC), a small single-copy region (SSC), and two inverted repeat regions (IRa and IRb) (Fig. 1). This architecture is similar to chloroplast genomes in various plant species37. Within the Agropyron Gaertn chloroplast genome, the IR regions harbor 8 rRNA genes, 16 tRNA genes, and 14 mRNA genes; the SSC region contains 1 tRNA gene and 10 mRNA genes, while the LSC region contains 23 tRNA genes and 59 mRNA genes (Table 3 and Fig. 1). Although the overall GC content of chloroplast genomes across different Agropyron Gaertn species is relatively similar (38.32%–38.34%), the GC content in the IR region (43.91%–44.01%) is significantly higher than that in the LSC region (36.28%–36.38%) and SSC region (32.21%–32.26%). This difference is closely related to the enrichment of high-GC-content rRNA genes in the inverted repeat region (Table 2).
Among these genes, 14 genes (atpF, rpl2, rpl16, rps16, ndhA, ndhB, petB, petD, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) all contained one intron, while the remaining three genes (ycf3 and rps12) contained two introns (Table 4). Among these, the rps12 gene exhibited the highest nucleotide diversity (Pi = 0.05198). Its high variability makes it a potential candidate marker for molecular-assisted taxonomic identification of the broad-spike/narrow-spike clades. rps12 is located in the IR region, while ycf3 is situated in the LSC region, while ndhA, containing only one intron, is localized in the SSC region. The remaining related genes are distributed in the LSC and IR regions (Table 4).
Codon usage bias analysis
The relative synonymy codon usage (RSCU) quantifies codon usage frequency, revealing codon preferences within the chloroplast genome of Agropyron Gaertn. These preferences reflect characteristics of natural selection, species mutation, and genetic variation. When RSCU > 1, it indicates that the codon is used more frequently and exhibits strong preference; when RSCU < 1, it indicates that the codon is used less frequently and exhibits weak preference; when RSCU = 1, it indicates that the codon shows no preference38. Among the chloroplast genomes of seven species of Agropyron Gaertn and two closely related species (E. trachycaulus and E. elongata), 33 codons exhibit an RSCU value greater than 1. The codon with the highest RSCU value is AUG for methionine (Met), at 6.97; followed by UUA for leucine (Leu), at 2.074; while the lowest was GUG for methionine (Met) at 0.03. Amino acid specificity analysis revealed that methionine (Met), arginine (Arg), leucine (Leu), and serine (Ser) exhibited the highest occurrence frequencies. Tryptophan (Trp) was the only codon showing no preference (RSCU = 1.00), potentially related to its strict monocodonic coding nature in the chloroplast genome. Among codons with RSCU > 1, 29 codons (96.67%) terminated with A or U, while only 3 codons (3.33%) terminated with G or C. This pattern aligns strongly with the AU-enriched nature of the chloroplast genome and transcription optimization mechanisms (Table 5 and Fig. 2A). This preference pattern provides a foundational background for developing molecular markers for identifying clades, aiding in the selection of specific markers at the codon level.
Analysis of interspersed repeats
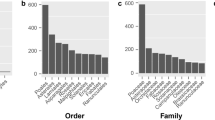
Analysis of repetitive sequences in seven species of the Agropyron Gaertn and two closely related species (E. trachycaulus, E. elongata) revealed 29 to 39 forward repeats and 13 to 18 palindromic repeats. No reverse or complementary repeat sequences were detected, reflecting an evolutionary strategy for maintaining core functional stability in the chloroplast genome. Notably, E. trachycaulus (ET) and E. elongata (EE) exhibited significantly higher total repeat sequences than the other seven Agropyron Gaertn species. This clades-specific repeat sequence pattern can serve as a reference indicator for molecular marker-assisted taxonomic identification of the broad-spike/narrow-spike clades. (Fig. 2B).
cpSSR analysis
SSR loci were most densely distributed in the LSC region, with total numbers varying among species (Table 6). All species contained SSRs ranging from mononucleotides to hexanucleotides, and clade-specific loci were identified: E. elongata possesses a unique (ATATA)3 pentanucleotide locus (exclusive to the narrow-spike clades); A. desertorun var. pilosiusculum possesses a unique (TC)5 locus (exclusive to the broad-spike clades A. cristatum var. pluriflorum and A. desertorun var. pilosiusculum); A. sibiricum f. pubiflorum possesses a unique (TAAA)4 locus (exclusive to the broad-spike clades A. sibiricum and A. sibiricum f. pubiflorum); E. trachycaulus shares the (CCATA)3 locus with E. elongata (common to the narrow-spike clades), but lacks the (ATATA)3 locus, serving as a marker to distinguish E. trachycaulus from E. elongata. SSR loci in the broad-spike clades are highly conserved, yet exhibit single-nucleotide repeat differences. For example, A. mongolicum var. villosum lacks the (A)11 site, which can serve as a specific auxiliary marker for A. mongolicum var. villosum. The clades specificity and shared patterns of these SSR sites provide a candidate marker library for molecular-assisted taxonomic identification of the broad-spike/narrow-spike clades (Fig. 3).
Analysis of chloroplast nucleotide diversity
Nucleotide diversity (Pi value) serves as a crucial indicator for measuring the degree of nucleic acid sequence variation among different species, with highly variable regions potentially serving as sequence features for population genetics research. Global homology analysis using Mafft software revealed that the rps12 gene within the large single-copy region (LSC) exhibited the highest genetic diversity, with a Pi value peak of 0.05198. Its high variability makes it a core candidate marker for molecular-assisted taxonomic identification of the broad-spike/narrow-spike clades. Further comparisons revealed that genetic variation in single-copy regions (LSC and SSC) significantly exceeded that in inverted repeat (IR) regions. This difference is closely related to the high conservation of IR regions maintained through gene conversion mechanisms during evolution (Fig. 4).
Chloroplast boundary analysis
The chloroplast genome adopts a circular structure, with the intercalary region (IR) sharing four boundaries with the left supercalicinal region (LSC) and the right supercalicinal region (SSC): LSC-IRb, IRb-SSC, SSC-IRa, and IRa-LSC. During genomic evolution, IR boundaries undergo expansion and contraction, causing certain genes to enter the IR region or the single-copy region. Therefore, CPJSdraw was employed to visualize this boundary information. By comparing critical boundary connections within the chloroplast genomes of 10 Agropyron species, 1 Elymus species, 1 Elytrigia species, 1 Australopyrum species, and 1 Psathyrostachys species, the study focuses on linkages between the inverted repeat region (IR) and the large single-copy region (LSC) as well as the small single-copy region (SSC). Results revealed that across all examined species:—The rpl22 gene resides within the LSC, spanning 450 bp;—The rps19 and rps15 genes are located within IRb, with rps19 adjacent to the LSC region and rps15 adjacent to the SSC region; The ndhF gene was located within the SSC region; the ndhH gene was situated at the SSC/IRa boundary; the rps19 and rps15 genes were within IRa; and the psbA gene was located within the LSC region. The boundary genes and their connecting lengths showed consistency across seven Agropyron species and two closely related species (Fig. 5).
Chloroplast sequence homology analysis
A collinearity analysis was performed on the chloroplast genomes of 10 Agropyron Gaertn, 1 Elytrigia species, 1 Elymus species, 1 Australopyrum species, and 1 Psathyrostachys species (Table S3). Results revealed homology across all genome sequences, with no significant insertions or deletions detected. The 14 chloroplast genomes were connected by a single red line, indicating highly conserved chloroplast genome structures without gene rearrangements (Table S2 and Fig. 6).
Phylogenetic tree analysis
Fourteen species from different genera within the wheat tribe were selected for phylogenetic tree analysis, including 10 Agropyron species, 1 Elymus species, 1 Elytrigia species, 1 Australopyrum species, 1 Psathyrostachys species, and 1 cultivar from Psathyrostachys. Results indicate that the 14 species are divided into two clades. Clade I comprised 10 Agropyron species whose chloroplast genomic characteristics (e.g., codon third position A/U preference > 87%, forward repeat sequence enrichment) strongly aligned with the morphological criteria for broad-spike clades (spike width > 5 mm, lanceolate glumes), supporting the broad-spike clades feature in the North American taxonomic system39. Clade II comprises four species from Elymus species, Campeiostachys species, Elytrigia species, and Australopyrum species, indicating that E. trachycaulus and E. elongata are more closely related to species of the Campeiostachys species and Australopyrum species. These phylogenetic branch results provide molecular evidence at the evolutionary level for molecular marker-assisted identification of the broad-spike/narrow-spike clades, corroborating findings from SSR and IR length markers (Table 1 and Fig. 7).
Discussion
Structural variation in chloroplast genomes and its value as molecular markers
Agropyron Gaertn, an important perennial forage resource within the Triticeae tribe of the gramineae family, has garnered significant attention due to its strong stress tolerance and rich genetic diversity. Research indicates that the chloroplast genome of Agropyron Gaertn exhibits a typical quadripartite structure (LSC-IR-SSC-IR), ranging in size from 135 to 137 kb. It contains 130 to 134 annotated functional genes, including 89 to 91 protein-coding genes, 37 to 39 tRNAs, and 8 rRNAs, consistent with the chloroplast genome characteristics of most higher plants40. However, variations with taxonomic and phylogenetic significance were also identified within the conserved framework.
Coding gene variants: identification of core candidate gene markers
Nucleotide diversity (Pi) analysis revealed that the genes rps12 and ycf3 exhibited significantly high variability: The Pi value of the rps12 gene in the LSC region reached 0.05198, the highest among all tested genes. This result strongly aligns with findings from studies on Triticeae relatives (such as Setaria and Hordeum) that “high sequence polymorphism in the rps12 gene can serve as a taxonomic marker”41,42. Its sequence variation effectively distinguishes broad-spike/narrow-spike clades, making it a core candidate gene for molecular marker-assisted identification. Additionally, the ycf3 gene in the LSC region exhibits potential for clade differentiation due to its structural characteristics—containing two introns—and sequence length polymorphism. This finding corroborates the conclusion by Xie et al.43 that “intron features of the ycf3 gene can serve as markers for species and clades identification” in the gramineae, further enhancing the reliability of this gene as an auxiliary taxonomic marker.
Variation in scattered repeat sequences: identification of clade-specific structural markers
Scattered repetitive sequences comprise only forward and palindromic types, reflecting genomic structural stability. Heidari et al.44 noted that forward repeats can promote local sequence amplification through sliding mismatches, while palindromic repeats participate in transcription termination or RNA editing by forming stem-loop structures. The synergistic interaction between these two types may regulate genomic functional diversity. Regarding clades differences, the closely related species E. elongata exhibits a total of 57 repetitive sequences, significantly higher than the Agropyron Gaertn’s broad-spike clades. Similar phenomena have been applied in interspecific hybrid identification within the Triticeae, where differences in repetitive sequence numbers have been confirmed as a key indicator for distinguishing hybrids from their parents45. Wicher et al.46 further confirmed that repeat sequence expansion in gramineae often accompanies fine-tuning of genomic architecture, potentially linked to adaptive potential. Concurrently, the distribution characteristics of repetitive sequences in this study, coupled with codon bias (96.67% of highly biased codons terminate with A/U), collectively reflect the AU enrichment feature of the chloroplast genome in response to mutational pressure and natural selection47. This coevolutionary pattern was also observed in the genus Leymus48 within the gramineae, revealing a common evolutionary principle in the chloroplast genomes of Triticeae plants and providing supplementary evidence for resolving phylogenetic relationships among clades.
SSR and IR region variation: screening of target marker for precise identification
The clade specificity of SSR loci provides direct clues for the precise identification of the broad-spike/narrow-spike clades: E. elongata of the narrow-spike clade possesses a unique (ATATA)3 pentanucleotide locus, while A. desertorun var. pilosiusculum possesses a unique (TC)5 site, while A. sibiricum f. pubiflorum exhibits a distinctive (TAAA)4 site. These sites serve as specific markers at the taxon and species levels, aligning with Deng et al.49 ‘s conclusion in their Triticeae SSR study that “site combination patterns support auxiliary identification.” Furthermore, IR region length variation also holds clear taxonomic value: the IRb length in narrow-spike clades (E. trachycaulus, E. elongata) is uniformly 20,813 bp, significantly shorter than that in broad-spike clades (21,530–21,547 bp). This structural difference can serve as an auxiliary indicator for rapid differentiation between broad-spike and narrow-spike clades.
Molecular marker-assisted taxonomic identification systems and application
Phylogenetic analysis based on chloroplast genomes provides core support for defining the broad-spike/narrow-spike clades within Agropyron species: the maximum likelihood phylogenetic tree reveals two highly supported monophyletic clades, with branch clustering perfectly matching broad-spike/narrow-spike phenotypic traits. The integration of molecular markers—including SSR loci, intergenic region (IR) length, and highly variable genes (rps12, ycf3)—established a multidimensional molecular marker-assisted identification system for the broad-spike/narrow-spike clades.
Highly variable gene markers form the core of the system. The rps12 gene which has the highest nucleotide diversity (Pi = 0.05198) is particularly informative. Its unique structure and copy number variation in the IR region, combined with the length polymorphism of the ycf3 gene intron in the LSC region, provide reliable sequence-level evidence for broad-spike/narrow-spike clades discrimination. This supports findings bu Wu et al.50 in Setaria and Han et al.51 in Agropyron Geartn, confirming the universality of these genes in distinguishing closely related species.
Simple sequence repeat (SSR) loci offer a high-resolution identification tool. The study identified a (CCATA)₃ locus, shared by all narrow-spike clades, that serves as a reliable clade-specific marker. Furthermore, an (ATATA)₃ locus unique to E. elongata, cleanly distinguishes it from E. trachycaulus. This multi-locus identification strategy significantly improves identification accuracy and specificity, an approach widely validated in wheat tribe genomic research49.
Genomic structural variation enable rapid initial screening. We confirmed that the length of the IR region is stably 20,813 bp in the narrow-spike clades, which is significantly shorter than the 21,530–21,547 bp range in the broad-spike clades. This macrostructural difference is easily detectable via conventional PCR and electrophoresis, making it an ideal screening marker for large-scale germplasm resources. This aligns with reports by Qin et al.52 in legumes and Jiang et al.53 in Setaria who also found that IR region variation correlates with clades differentiation.
Scattered repetitive sequences provide supplementary corroborating evidence. The total number of repetitive sequences in the closely related species E. elongata and E. trachycaulus is significantly higher than that in the broad-spike Agropyron clades. This genomic structural difference offers further support for the classification.
In summary, the molecular markers identified in this study provide a practical tool for efficient and precise classification and identification of the broad-spike/narrow-spike Agropyron clades. They also serve as a reference for developing molecular markers in other wheat tribe species, underscoring broad value of chloroplast genomes in plant phylogenetics and taxonomy.
Conclusions
This study conducted an in-depth analysis of the chloroplast genomes of seven species of Agropyron Gaertn and two closely related species, revealing their evolutionary characteristics and taxonomic value. Analysis of chloroplast genome characteristics indicates that Agropyron Gaertn species exhibit a typical quadripartite structure (LSC-IR-SSC-IR), with genome sizes ranging from 135 to 137 kb and containing 131 genes. Among these, rps12 (Pi = 0.05198) and ycf3 were screened out as core candidate genes for molecular marker-assisted taxonomic identification of the broad-spike/narrow-spike clades. The high GC content (43.91%–44.01%) in the IR region correlates with gene conversion mechanisms, while the trnK-UUU intron length variation (2487–2504 bp) in the LSC region serves as a supplementary marker. Specific combination patterns of chloroplast genome SSR loci (e.g., the (CCATA)3 locus present in all narrow-spike clades) aid in distinguishing between wide-spike and narrow-spike clades. Combined features in Elymus trachycaulus which are highly similar with features of Elytrigia elongata such as the high GC content (44.01%) in the IR region and the length (80,642 bp) of the LSC region, supports the traditional identification of Elymus trachycaulus into the narrow-spike clades at the molecular level. Phylogenetic analysis further confirms the evolutionary validity of the identification system for the broad-spike and narrow-spike clades within Agropyron Gaertn. This study identifies chloroplast molecular markers to aid in the taxonomic identification within Agropyron Gaertn. These markers provide a tool for the precise identification, utilization, and conservation of Agropyron germplasm. Future work should focus on validating marker stability across larger populations and integrating gene markers to establish a more robust identification system.
Data availability
The raw sequencing data from the Illumina NovaSeq 6000 platform and the chloroplast genome sequences have been deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/) with the accession numbers SAMN47853882, SAMN47853883, SAMN47853884, SAMN47853885, SAMN47853886, SAMN47853887, SAMN47853888, SAMN47853889, and SAMN47853890.
Abbreviations
- rRNA:
-
Ribosoma RNA
- tRNA:
-
Transfer RNA
- mRNA:
-
Messenger RNA
- LSC:
-
Large single-copy region
- SSC:
-
Small single-copy region
- IRA:
-
Inverted repeat region A
- IRB:
-
Inverted repeat region B
- RSCU:
-
Relative synonymous codon usage
- SSR:
-
Simple sequence repeats
- cpSSR:
-
Chloroplast simple sequence repeats
- Pi:
-
Nucleotide diversity
- LCBs:
-
Locally collinear blocks
References
Gao, H. J. et al. Research advance on genetic diversity of Agropyrong Gaertn plants. J. Grassl. Forage Sci. 06, 31–35 (2011).
Zhao, Y. et al. Functional Identification of MwMYB4 Gene from Agropyron mongolicum Keng. Acta Botan. Boreali-Occiden. Sin. 40(04), 565–571 (2020).
Dewey, D.R. The Genomic System of Classification as a Guide to Intergeneric Hybridization with the Perennial Triticeae. In: Gustafson, J.P. (eds) Gene Manipulation in Plant Improvement. Stadler Genetics Symposia Series. Springer, Boston, MA. https://doi.org/10.1007/978-1-4613-2429-4_9 (1984).
Gao, H. J. et al. Study on the morphology and cytology of Agropyron cristatum. Pratacultural Sci. 27(10), 54–59 (2010).
Li J. X. Genetic diversity of germplasm resources of Agropyron cristatum in Inner Mongolia. https://kns.cnki.net/kcms2/article/abstract?v=vK4oI_BKqSVzjRVqfq55625CRxaejeNWXWBg3loN4n-8f7HO6Ktk_N9M7BE5EmDehpAMALx-_FKPuB3mkWuFGXXANTOSXU-mMr6rvkOBUr06rXbcXVnB4LuvEZ4m7ac3ZimQ8zr1zwCTWaHJCNAg41KqShao9SWfVYVB3ybkew5ZO8wpamZPTQ==&uniplatform=NZKPT&language=CHS (2005).
Yun, J. F. & Mi, F. G. A historical review of taxonomic research on the genus Agropyron Gaertn. Chin. J. Grassl. 02, 3–7 (1989).
Geng, Y. L. Illustrated Guide to Major Plants of China—Gramineae (Volumes 1–2) (Science Press, 1959).
Cay, S. B. et al. Genome skimming approach reveals the gene arrangements in the chloroplast genomes of the highly endangered Crocus L species: Crocus istanbulensis (B.Mathew) Rukšāns. PLoS ONE 17(6), e0269747. https://doi.org/10.1371/journal.pone.0269747 (2022).
Zhang, J. F. et al. Recent advances in tobacco chloroplast genetic engineering. Tobacco Sci. Technol. 50(06), 88–98. https://doi.org/10.16135/j.issn1002-0861.2017.0035 (2017).
Chen, N. et al. Complete structure and variation of the chloroplast genome of Agropyron cristatum (L.) Gaertn. Gene 640, 86–96. https://doi.org/10.1016/j.gene.2017.10.009 (2018).
Luo Y. 2019 The study genetic differentiation of Agropyron cristatum populations based on genomic size, phenotypic traits and chlorophyll content. Doctoral Dissertation, Sichuan Agricultural University. https://doi.org/10.27345/d.cnki.gsnyu.2019.000307
Yang, T. et al. A comprehensive analysis of chloroplast genome provides new insights into the evolution of the genus Chrysosplenium. Int. J. Mol. Sci. 24(19), 14735. https://doi.org/10.3390/ijms241914735 (2023).
Meng L. et al. Study on germplasm resources of Elytrigia species.Beijing: Science press, 2020: 1–9.
Zhao Y. Z. et al. Flora Intramongolica (Editio Tertia) Tomus 6. Huhhot: Typis intramongolicae popularis, 2020: 139–145.
Editorial Committee of Flora of China. Chinese Academy of Sciences. Tomus 9. Flora Reipublicae Popularis Sinicae. Beijing: Science Press, 2002.
Clayton, W. D., Harman, K.T. & Williamson, H. World Grass Species - Synonymy database The Board of Trustees of the Royal Botanic Gardens, Kew. (2006).
Lavin, M. et al. Physical disturbance shapes vascular plant diversity more profoundly than fire in the sagebrush steppe of southeastern Idaho USA. Ecol. Evol. 3(6), 1626–1641. https://doi.org/10.1002/ece3.574 (2013).
Liu J. et al. Chloroplast genome characteristies and phylogenetic analysis of Elytrigia elongata and Elymus trachycaulus. Pratacultural Science, 1–26[2025–11–23]. https://link.cnki.net/urlid/62.1069.S.20250904.1434.007 (2025).
Zhang, Y. et al. Domestic research progress on Agropyron elongatum in our country. Mod. Agric. Sci. Technol. 11, 162–166 (2022).
Zhang, Y. et al. Study on carbon, nitrogen and phosphorus contents and stoichiometric characteristics of Agropyron cristatum in different phenological periods. Agric. Technol. 42(13), 22–25. https://doi.org/10.19754/j.nyyjs.20220715006 (2022).
Yan W. H. Genetic Analysis of Some Germplasm Resources of Agropyron Gaertn. and Roegneria C. Koch. Dissertation, Chinese Academy of Agricultural Sciences, 2010.
Geng, K. X. et al. Geng Yili: The founder of Gramineae taxonomy in China. Zhongshan Fengyu 6, 21–24 (2002).
Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2(2), e107–e107. https://doi.org/10.1002/imt2.107 (2023).
Doug, H. et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11(1), 119. https://doi.org/10.1186/1471-2105-11-119 (2010).
Ananth, P. et al. The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinform. 60(1), 3–15. https://doi.org/10.1002/cpbi.40 (2017).
Laslett, D. & Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1), 11–16. https://doi.org/10.1093/nar/gkh152 (2004).
Christiam, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10(1), 421. https://doi.org/10.1186/1471-2105-10-421 (2009).
Lohse, M., Drechsel, O. & Bock, R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52(5–6), 267–274. https://doi.org/10.1007/s00294-007-0161-y (2007).
Thiel, T., Michalek, W., Varshney, R. K. & Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L) TAG. Theor. Appl. Gen. Theoretische und angewandte Genetik 106(3), 411–422. https://doi.org/10.1007/s00122-002-1031-0 (2003).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30(4), 772–780. https://doi.org/10.1093/molbev/mst010 (2013).
Wang, D., Zhang, Y., Zhang, Z., Zhu, J. & Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinform. 8(1), 77–80. https://doi.org/10.1016/S1672-0229(10)60008-3 (2010).
Librado, P. & Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11), 1451–1452. https://doi.org/10.1093/bioinformatics/btp187 (2009).
Breitenbach, J. E., Shelby, K. S. & Popham, H. J. Baculovirus induced transcripts in hemocytes from the larvae of Heliothis virescens. Viruses 3(11), 2047–2064. https://doi.org/10.3390/v3112047 (2011).
Li, H. et al. CPJSdraw: analysis and visualization of junction sites of chloroplast genomes. PeerJ 11, e15326. https://doi.org/10.7717/peerj.15326 (2023).
Ronquist, F. et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61(3), 539–542. https://doi.org/10.1093/sysbio/sys029 (2012).
Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7), 1394–1403. https://doi.org/10.1101/gr.2289704 (2004).
Wang, P., Guo, J., Zhou, J. & Wang, Y. Characterization of the complete chloroplast genome and development of molecular markers of Salix. Sci. Rep. 14(1), 28528. https://doi.org/10.1038/s41598-024-79604-8 (2024).
Wang, J.-H., Moore, M. J., Wang, H., Zhu, Z.-X. & Wang, H.-F. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies. Gene 765, 145103–145103. https://doi.org/10.1016/j.gene.2020.145103 (2021).
Rabeler R K, Hartman R L. Flora of North America. From Internet, 2007, 767.
Sathyamurthy, D. et al. Comparative chloroplast genome analysis of six millet species along with related Poaceae family members. Nucleus 68, 13–24. https://doi.org/10.1007/s13237-023-00464-0 (2025).
Su, Q. et al. The complete chloroplast genomes of seventeen Aegilops tauschii: genome comparative analysis and phylogenetic inference. PeerJ 8, e8678. https://doi.org/10.7717/peerj.8678 (2020).
Ou, T., Wu, Z., Tian, C., Yang, Y. & Li, Z. Complete mitochondrial genome of Agropyron cristatum reveals gene transfer and RNA editing events. BMC Plant Biol. 24(1), 830. https://doi.org/10.1186/s12870-024-05558-8 (2024).
Xie, W. et al. LpY3IP1 Enhances the drought and salt tolerance of perennial ryegrass by protecting the photosynthetic apparatus. Sci. Hortic. 2024, 338113645–339113645. https://doi.org/10.1016/J.SCIENTA.2024.113645 (2024).
Heidari, B. & Shariatipour, N. Meta-analysis of expression of the stress tolerance associated genes and uncover their -regulatory elements in rice (L.). Open Bioinform. J. 13, 39–49 (2020).
Feng, H. et al. Hordeum I genome unlocks adaptive evolution and genetic potential for crop improvement. Nat. Plants 11(3), 438–452. https://doi.org/10.1038/s41477-025-01942-w (2025).
Wicker, T. et al. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 19(1), 103. https://doi.org/10.1186/s13059-018-1479-0 (2018).
Zhang, Y. R. et al. Analysis of codon usage patterns of the chloroplast genomes in the Poaceae family. Aust. J. Bot. 60(5), 461–470 (2012).
Li, Y. Y. et al. Phylogenetic analysis of Leymus (Poaceae: Triticeae) based on random amplified polymorphic DNA. Biologia 71, 396–401. https://doi.org/10.1515/biolog-2016-0048 (2016).
Deng, P. et al. Genome-wide characterization of microsatellites in Triticeae species: Abundance, distribution and evolution. Sci. Rep. 6, 32224. https://doi.org/10.1038/srep32224 (2016).
Wu, Z. N. et al. Comparative and phylogenetic analysis of complete chloroplast genomes in Leymus (Triticodae, Poaceae). Genes 13(8), 1425. https://doi.org/10.3390/genes13081425 (2022).
Han, H. et al. Analysis of chloroplast genomes provides insights into the evolution of Agropyron. Front. Genet. 13, 832809. https://doi.org/10.3389/fgene.2022.832809 (2022).
Qin, Q. et al. Comparative analysis of chloroplast genomes reveals molecular evolution and phylogenetic relationships within the Papilionoideae of Fabaceae. BMC Plant. Biol. 25, 157. https://doi.org/10.1186/s12870-025-06138-0 (2025).
Jiang, L. et al. The chloroplast genome of Elytrigia repens (L.) Nevski (Poaceae: Triticeae), a widespread perennial grass from Qinghai-Tibetan Plateau China. Mitochondrial DNA B Resour. 6(12), 3490–3492. https://doi.org/10.1080/23802359.2021.1993098 (2021).
Funding
This study was supported by the Inner Mongolia Key Science and Technology Project “Key Technology Research on Germplasm Innovation and Application of Fine Native Grasses” (2021ZD0031); the General Project of the Inner Mongolia Autonomous Region Natural Science Foundation “Development of Salt-Tolerant Related Sequence Features for Agropyron cristatum and Creation of New Germplasm with Marker Assistance” (2023MS03033); and the Inner Mongolia Autonomous Region Science and Technology Plan Project “Integration of High-Efficiency Breeding, Processing, and Utilization Technologies for Multi-Functional Native Grasses in Xilingol League” (2022YFDZ0083).
Author information
Authors and Affiliations
Contributions
WXP prepared figures and tables and wrote the first draft of the manuscript. WHY designed the experiments, performed data analyses, and revised the manuscript. ZYL and ZNW collected plant materials and extracted DNA. QST and YD conceived and designed the experiments. CZW and WHY contributed to the analysis of results and revision of the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was conducted the plant material that complies with relevant institutional, national, and international guidelines and legislation.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pan, W., Yan, W., Li, Z. et al. Analysis of chloroplast genomes and SSR classification and identification of Agropyron species and closely related species. Sci Rep 16, 3579 (2026). https://doi.org/10.1038/s41598-025-33570-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-33570-x