Abstract
Despite extensive research, spinal cord injuries (SCI), which could cause severe sensory, motor and autonomic dysfunction, remain largely incurable. Oligodendrocytes and oligodendrocyte precursor cells (ODC/OPC) play a crucial role in neural morphological repair and functional recovery following SCI. We performed single-cell sequencing (scRNA-seq) on 59,558 cells from 39 mouse samples, combined with microarray data from 164 SCI samples and 3 uninjured samples. We further validated our findings using a large clinical cohort consisting of 38 SCI patients, 10 healthy controls, and 10 trauma controls, assessed with the American Spinal Cord Injury Association (ASIA) scale. We proposed a novel SCI classification model based on the expression of prognostic differentially expressed ODC/OPC differentiation-related genes (PDEODGs). This model includes three types: Low ODC/OPC Score Classification (LOSC), Median ODC/OPC Score Classification (MOSC), and High ODC/OPC Score Classification (HOSC). Considering the relationship between these subtypes and prognosis, we speculated that enhancing ODC/OPC differentiation and inhibiting inflammatory infiltration may improve outcomes. Additionally, we identified potential treatments for SCI that target key genes within these subtypes, offering promising implications for therapy.
Similar content being viewed by others
Introduction
SCI is a severe neurological condition associated with high morbidity and mortality rate. Annually, approximately 250,000 to 500,000 new SCI cases are reported globally1. Treatment options for SCI primarily involve surgical decompression, spinal stabilization, and drug therapies targeting neurotrophic factors2. However, the clinical effectiveness of regenerative and neuroprotective treatments remains inadequate. Potential cellular mechanisms include prolonged inflammation at lesion sites, which can exacerbate nervous tissue damage. Balancing these mechanisms is crucial; excessive inflammation can damage tissue, while insufficient inflammation may result in abnormal cytotoxin accumulation3. Circulating microbial signaling molecules, such as lipopolysaccharides (LPS), can affect glial cells post-SCI, significantly altering their transcriptional profiles toward neurotoxic phenotypes4,5. Furthermore, peripheral infections and inflammatory responses can generate blood-borne signaling molecules, such as cytokines or LPS, activating immune cells that prolong inflammation at lesion sites and exacerbate tissue damage post-SCI6. The weeks following SCI, during which various cellular subtypes interact and organize into mature lesion sites with distinct compartment7, offer significant opportunities for protective interventions. Inflammatory and immune cell populations are also critical for nerve regrowth after SCI. Activated macrophages contribute to significant axonal retraction and dieback following SCI8. Conversely, neutrophils around neuronal cell bodies promote nerve regrowth. Despite substantial efforts in traditional research areas, including inflammatory responses, axon regeneration, and cell transplantation, these approaches have not achieved satisfactory results in neural regeneration and circuit reconstruction following SCI9,10. Recent immunofluorescence and transcriptome analyses have been essential for elucidating the spatial distribution of multicellular populations and the temporal dynamics of gene expression. Several studies suggest that high-throughput therapies using extensive transcriptome sequencing may identify candidate drugs for SCI, providing various molecular and cellular targets11. Oligodendrocytes and oligodendrocyte precursor cells (ODC/OPC) are vital in the pathophysiology of SCI; their differentiation stage significantly influences the regeneration of myelin sheaths and axons12. Genes that regulate differentiation influence and determine cell fate, potentially limiting the efficacy of single-target clinical approaches.
In this study, we performed a comprehensive bioinformatics analysis of single-cell RNA sequencing (scRNA-seq), RNA sequencing (RNA-Seq), microarray data, and integrated multi-omics data to reconstruct the differentiation trajectory of ODC/OPC in SCI. We then developed an ODC/OPC differentiation-based SCI classification (ODBSC) system via multidimensional analysis and external validation. We also constructed SCI subtype-specific regulatory networks to clarify the continuity of ODC/OPC differentiation. This approach enabled us to identify rare and critical differentiation transitions, along with both classical and novel candidate transcriptional regulators linked to ODC/OPC differentiation13. Finally, we identified a panel of small molecule drugs targeting differentially expressed transcription factors (DETFs) linked to each SCI subtype marker. In summary, we developed a more precise molecular classification of SCI that incorporates the heterogeneity of ODC/OPC differentiation more comprehensively.
Materials and methods
Data collection
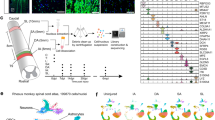
We collected 59,558 single cells from uninjured tissue and a mouse model of incomplete SCI using the 10X Chromium technology platform. The datasets are accessible via the identifier (https://doi.org/10.6084/m9.figshare.17702045)14. We utilized microarray data for a rat model of incomplete SCI. Gene expression was measured using the Affymetrix Rat Genome 230 2.0 Array (Rat230_2) platform, sourced from the Gene Expression Omnibus (GEO) database (accession number: GSE69334)15.Notably, our study’s reliance on the GSE69334 database, which only includes samples from female rats, is a significant limitation. This is especially important considering the known neuroprotective effects of female hormones on SCI. Future research should use databases with samples from both male and female subjects to gain a more complete understanding of the molecular mechanisms of SCI. We analyzed peripheral blood mononuclear cells (PBMCs) from SCI patients using RNA-seq data generated on an Illumina HiSeq 4000 sequencer. The study included 58 samples: 10 healthy individuals without central nervous system (CNS) pathology (healthy control, HC), 10 patients with non-CNS trauma (trauma control, TC), and 38 patients with acute traumatic SCI. These samples were sourced from the GEO database (accession number: GSE151371). We processed the raw microarray data using the affy package, which included robust multi-array average (RMA) background correction, normalization, probe-specific background correction, and summarization of probe set values into a single expression measure15.
Single cell RNA-seq data processing and RNA-seq data processing
Sample preprocessing and scRNA-seq data were performed according to the 10x Genomics Chromium protocol16. The sequencing results were divided into two paired-end FASTQ files, which were then trimmed to remove the template switch oligo (TSO) sequence and poly(A) tail. These files were quantified using Cell Ranger Software (Version 1.0.0)17. Cells expressing more than 100,000 transcripts and with less than 10% of mapped mitochondrial genes were selected for subsequent analysis. Additionally, genes expressed in more than two single cells were included in further analyses. First, FastQC and Trimmomatic software were employed to assess and control the quality of the raw FASTQ files. The data were mapped to the human reference genome using HISAT2 (Version: Ensembl GRCh38), generating output alignments in Sequence Alignment/Map (SAM) format. Subsequently, the SAM files were converted to binary alignment/map (BAM) format using SAMtools (Version: 1.8) and analyzed with FeatureCounts to estimate transcript counts18. The transcript count was expressed as fragments per kilobase of mapped reads per million (FPKM), which was subsequently converted to transcripts per million (TPM). Finally, a gene expression matrix was created with gene symbols as rows and sample IDs as columns. All subsequent analyses were based on this matrix19.
Assigning cell types to single cell clusters
Biomarkers obtained from the Cell Marker database were used for comprehensive annotation of the cell types in each unsupervised cluster20. First, known cell surface biomarkers were utilized to annotate various cell types, including astrocytes (NFIA, GFAP, S100B), endothelial cells (EGFL7, FLT1, ITM2A), ependymal cells (CCDC153, CLU, MIA), erythrocytes (AIDH1A1, CAR2, CITED2), granulocytes (CD81, CSF3R, CD63), fibroblasts (ABI3, MGP, CD47), lymphocytes (CD2, CD3D, NKG7), microglia (AIF1, CD68, ITGAM), myeloid cells (CD33, LYZ2, CD14), neurons (RTN1, MAP2, MEG3), and oligodendrocyte/oligodendrocyte precursor cells (CSPG5, LHFPL3, OLIG1).Additionally, cellular feature plots, violin plots, dot plots, and heat maps were generated to display the marker genes for each cell type using SCANPY (Version: 1.7.1) and the Seurat R package (Version: 3.2.2) with Python 3.621.
Trajectory analysis and Cellular communication analysis
To simulate the gene expression dynamics of ODC/OPC during the continuous differentiation trajectory following SCI, we reconstructed the ODC/OPC developmental trajectory using the “Monocle2” package in R. To analyze the pseudo-temporal trajectory of the SCI ODC/OPC lineage, we extracted previously annotated ODC/OPC cells and integrated them into a Seurat object. Using the Monocle R package (Version: 2.18.0), we delineated the transcriptional relationships among ODC/OPC clusters22. Initially, we merged the preprocessed Seurat object into Monocle 2 using the “newCellDataSet” function. Subsequently, genes associated with ODC/OPC differentiation (ODGs) were identified through functional assessment with the “differential Gene Test.” The data was then reduced to two dimensions using the discriminative dimensionality reduction with trees (DDR Trees) method within the “reduce Dimension” function. Pseudo-time was computed for each ODC/OPC cell, and the ODC/OPC clusters from the Seurat object were mapped onto a minimum spanning tree based on cell differentiation order using the “plot cell trajectory” function. To analyze essential cellular communication characteristics and ligand-receptor (LR) interactions among different ODC/OPC clusters in SCI tissues, the iTALK R package (Version: 0.1.0) was utilized. As there is a scarcity of Rattus norvegicus data in prevalent cellular communication algorithms, the bionaRt package (version: 2.46.0) was employed to convert the top 2,000 variable genes into their human equivalents using the “getLDS” function23. Subsequently, the normalized expression matrix of these genes was integrated into an iTALK object via the “rawParse” function. Finally, the top 200 LR plots were generated and visualized using the LR plots and iTALK networks supported by the edge bundle R package.
Cell cycle analysis and principal component analysis (PCA)
Potential novel clusters of differentially expressed ODGs (genes associated with ODC/OPC differentiation) were identified through consensus cluster analysis using the “ConsensusClusterPlus” R software24. This process entailed subsampling features from state-related gene sets and partitioning them into a maximum of k clusters using an agglomerative hierarchical clustering algorithm. Subsequently, agglomerative hierarchical consensus clustering based on 1 - consensus values distance was performed for each k value, defining consensus clusters. Molecular subtypes of SCI were characterized based on the expression levels of 34 PDEODGs. Utilizing the PAM algorithm with the “1-Pearson correlation coefficient” as the distance metric, 500 bootstraps were conducted, each containing 80% of SCI samples. The identified PDEODGs were clustered consistently, and the PDEODG modules were derived using the same parameters and settings as previously described. We used PCA analysis to further characterize the molecular pathological features of ODC/OPC differentiation in each SCI molecular subtype and determine the optimal number of consensus clusters (K) for the ODC/OPC score based on PDEODG expression levels. Following this, to elucidate the mechanisms of PDEODGs and showcase their clinical relevance, we conducted an extensive 90-day transcriptome investigation of spinal cord segments at the injury site post-SCI (evaluated at 1, 3, 10, 20, 30, 60, and 90 dpi). Additionally, we examined the correlation between PCA-derived ODC/OPC scores and other multi-omics data, such as transcriptomics and immune cell profiles, using single-sample gene set enrichment analysis (ssGSEA)25.
Immune cell Infiltration patterns in SCI
The gene expression matrix data of ODC/OPC clusters was uploaded to the CIBERSORT database to extract cell subtype-specific gene expression26. Only immune cells and patient genes with P < 0.05 were deemed statistically significant for further analysis. A nonparametric test was employed to assess correlations between SCI molecular subtypes and 22 types of immune cells. Additionally, the Single-Sample Gene Set Enrichment Analysis (ssGSEA) algorithm was used to gauge the level of immune cell infiltration in the microenvironment of the injury site. Moreover, Pearson correlation analysis was conducted to unveil associations between DEGs and immune-related signaling pathways or genes, with statistical significance set at P < 0.05.
Construction regulatory network and identification of potential drugs
Co-expression Pearson correlation analysis was conducted involving DETFs, PDEODGs, immune cells, and hallmark pathways. The “igraph” R package was employed to build a regulatory network. The pRRophetic package was used to identify potential agents targeting marker PDEODGs and DETFs specific to each SCI subtype within their respective networks27. Subsequently, a selection of drugs targeting PDEODGs and DETFs in each SCI subtype was identified, with a significance level set at P < 0.05.
Results
The gene expression landscapes of cells revealed by scRNA-Seq analysis
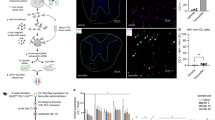
After rigorous filtration and quality control, scRNA-seq data for 59,558 single cells from uninjured tissue and a mouse model of incomplete SCI were gathered for further examination. Subsequently, tSNE analysis was employed for dimensionality reduction, leading to the clear identification of 21 clusters and 11 cell types utilizing the specified cell-type markers as shown in Fig. 1A–C. Additionally, a bar plot was utilized to display the average count and proportion of these cell clusters (Fig. 1D). Notably, the absolute number percentage of various inflammatory cells (lymphocytes, granulocytes, and microglia) and ODC/OPC in SCI samples markedly exceeded those in uninjured samples. Feature plots exhibiting the expression levels and spatial distribution of cell type-specific markers sourced from the Cell Marker database were visualized (Fig. 2). Subsequent to this, 1930 ODC/OPC were identified based on canonical markers and clustering, selected for all ensuing analyses.
(A) The tSNE dimension reduction analysis of unsupervised clustering identified 21 clusters based on 59,558 single cells. (B) The dimension reduction analysis annotated 11 main cell types. Notably, The (A) and (B) are using the same icon. (C) The tSNE scatter plot showed the distribution of all the cells in samples. (D) The bar plot showed the cell number and proportion of each cell type in all samples.
Integrated analyses of DEG and cell cycle
DEGs were extracted from the top 2,000 variably expressed genes based on the gene expression profiles of 11 cell types. A dot plot following Cleveland’s design was utilized to illustrate the expression levels of 3 classical marker genes: AIF1, MBP, and COI1A2 (Supplementary Fig. S1A). The heat map displayed the expression levels of the top 5 marker DEGs across the 11 cell types (Supplementary Fig. S1B). Moreover, cell cycle analysis indicated that a majority of ODC/OPC and inflammatory cells were predominantly in the G2M and S phases, significantly influencing immune and inflammation-related processes (Supplementary Fig. S1C,D).
Identification of ODC/OPC lineages’ characteristics
These ODC/OPC were integrated into a single Seurat object, where was performed and segregated into 8 major cell subpopulations (ODC/OPC 0-7). In this section, we extracted 1930 ODC/OPC annotated previously from Seurat for further analysis of scRNA-seq preprocessing and cell clustering was conducted by utilizing tSNE analysis (Fig. 3A,B). Importantly, the 8 ODC/OPC clusters showed significantly strong intercellular communication, where ODC/OPC3 occupied the cellular communication center. By comparing LR with the cell type-specific markers described above, hypothetical pairs of LR interactions among each cell subpopulation were identified. (Fig. 3C,D). It showed that high-level expression of OLIG2 was significantly apparent in all ODC/OPC clusters. The heat map showed the expression levels of the top 5 marker DEGs in 8 ODC/OPC clusters (Fig. 4A). Cleveland’s dot plot was used to indicate the expression levels of key markers genes (MBP, OLIG1, OLIG2 and TOP2A) in 8 ODC/OPC clusters (Fig. 4B).
(A) The tSNE analysis of unsupervised clustering identified 8 ODC/OPC clusters (ODC/OPC 0–7). (B) The tSNE plot showed the distribution of ODC/OPCs in different samples. (C) The ligand-receptor plot showed the specific cellular communication mechanisms of 8 ODC/OPC clusters, as well as the key genes that were involved in the cellular communication. (D) The iTalk network showed the intersected cellular communication among 8 ODC/OPC clusters.
Trajectory analyses recapitulated ODC/OPC differentiation
Subsequently, all cells from the eight ODC/OPC clusters were individually selected for trajectory analysis (Fig. 5A). Notably, ODC/OPC exhibited significantly different gene expression states at each time point, with the cell trajectory distinctly separating into six future cellular states at branch points 1, 2, and 3 (Fig. 5B). The trajectory plots were employed to illustrate the specific locations of all ODC/OPC clusters along the differentiation trajectory (Fig. 5C). Importantly, ODC/OPC clusters 5 and 6 predominantly occupied starting states 1 and 7 of the progression trajectory, which were designated as the least mature branches in pseudo-time. Clearly, state-5 and state-7 cells were primarily composed of ODC/OPC5, while state-1 cells were primarily composed of ODC/OPC1. Notably, states 1 and 7 were enriched for genes that were highly expressed early but not sustained over pseudo-time. In contrast, gene expression levels in clusters 2, 3, 5, and 6 were highest at later time points (Fig. 5D).
(A) The pseudo-time plot showed the differentiation sequence of ODC/OPCs. The darker the color, the more immature the ODC/OPCs might be. (B) The cell trajectory state plot illustrated the developmental trajectory of ODC/OPCs with 6 cell states shown in different colors. (C) The cluster plot illustrated the developmental trajectory of ODC/OPCs and all cell sub populations were illustrated in the trajectory. (D) The cell trajectory state plot showed the 6 different cell states of ODC/OPC, respectively.
Identification of key ODGs
As discussed, we have successfully delineated the transcriptomic roadmap of ODC/OPC differentiation states following SCI. By analyzing the intersection of the Venn diagram, we defined key ODGs, or PDEODGs, based on the following conditions, which were retained for constructing the ODBSC. (1) ODGs with P < 0.05, representing a total of 12,939 genes used to rank the differentiation trajectory; (2) ODGs whose gene expression significantly correlated with the ASIA grade in PMBC of SCI patients (non-parametric test, P < 0.05), totaling 40 genes; (3) ODGs with significant expression differences in PMBC among healthy controls, trauma controls, and SCI patients, totaling 34 genes; The intersection of the Venn diagram yielded 34 genes, which will be used for subsequent consistent clustering analysis to construct the PDEODG set (Supplementary Fig. S2A,B).
Unsupervised clustering to identify SCI molecular subtypes
The PDEODG sets may elucidate additional functions of biological programs that influence clinical outcomes in both uninjured and SCI samples. Based on their consistency, these genes were divided into k clusters, as shown in the corresponding consensus matrix plot (Supplementary Fig. S3A). Additionally, varying the parameters to different k values allowed the delta area plot to display changes in the area under the cumulative distribution function (CDF) curve (Supplementary Fig. S3B,C). Consequently, we identified three SCI molecular subtypes, leading to the creation of a novel ODC/OPC Differentiation-Based SCI Classification (ODBSC). The heat map illustrated the expression levels of 34 PDEODGs across various clusters, including consensus clusters (SCI subtypes versus ODC/OPC state clusters) and different time and treatment groups (Supplementary Fig. S3D). Notably, strong correlations were identified between the ODC/OPC differentiation states and the consensus clustering. Samples in consensus cluster 1 (ODBSC subtype 1) exhibited high expression of PDEODGs that dominated the terminal states of the progression trajectory, representing the most mature branches of ODC/OPC in pseudo-time. Additionally, samples in consensus cluster 2 (ODBSC subtype 2) exhibited high expression of PDEODGs that dominated the initial states of the progression trajectory. Furthermore, chi-square tests confirmed significant differences among the three SCI molecular subtypes at different time points (P < 0.001) (Supplementary Fig. S3E). Significant differences in treatment strategies were observed among the three SCI molecular subtypes (P < 0.001) (Supplementary Fig. S3F).
Similarly, we constructed the ODBSC based on the expression of 34 key PDEODGs from the RNA-seq data of PMBC in SCI patients (Supplementary Fig. S4A–D). Diagnosing SCI severity is crucial for the emergency treatment of patients, particularly using the ASIA grading system. The earliest ASIA Impairment Scale (AIS) grades were obtained from patients between days 3 and 10 post-SCI to avoid unstable AIS scores caused by factors such as strong sedation and spinal shock during the first two days28. To evaluate the correlations between SCI severity and our previous findings, we divided SCI patients based on assigned AIS grades obtained from neurological examinations conducted between days 3 and 10 in the hospital. The chi-square test results indicated that the ODBSC, which comprised three categorical variables, was significantly correlated with patients AIS grades (Supplementary Fig. S4E). Furthermore, significant differences were observed among healthy control, trauma control, and SCI groups (Supplementary Fig. S4F). We assessed that the prognosis for ODBSC subtype 1 was the most favorable, while the prognoses for samples in ODBSC subtype 2 and subtype 3 were relatively poorer.
Clinical correlation analysis and principal component analysis
A co-expression regulatory network was constructed using non-parametric tests and Pearson correlation (Fig. 6A). In this regulation network, genes determining 6 directions of cellular differentiation were illustrated using labels in different colors. All genes were significantly co-expressed in the SCI region during ODC/OPC cellular differentiation (P < 0.001). The identified gene set was significantly associated with clinical outcomes in patients with SCI. The box plot revealed significant differences in overall survival (OS) among the three SCI molecular subtypes (P < 0.001). Specifically, SCI subtype 2 exhibited the highest OS, while SCI subtype 1 exhibited the lowest OS (Fig. 6B). In conclusion, we identified three new molecular subtypes of SCI based on ODC/OPC differentiation status and corresponding biological characteristics: Low ODC/OPC Score Classification types (LOSC), including SPARCL129and SOX1030; Median ODC/OPC Score Classification types (MOSC), including CISD231 and CUL3; and High ODC/OPC Score Classification types (HOSC), including PPBP32 and CTSZ33.
(A) A regulatory network for 34 ODGs. The circles indicated genes, the color of which corresponded to ODC/OPC differentiation states. Lines indicated statistically significant correlations. Pink represented the positive correlation while blue represented the negative correlation. Circle size was inversely proportional to the P-value (non-parametric test). (B) PCA score plot showed the PCA ODC/OPC score of samples in 3 consensus clusters, which indicated significant differences among these clusters.
Immune infiltration patterns of ODBSC
The box plots indicated that inflammation-promoting immune cells (activated CD4(+) T cells and neutrophils) exhibited a lower degree of infiltration in the Low ODC/OPC Score Classification (LOSC) group (Supplementary Fig. S5A). Subsequently, we observed significant positive correlations between PCA ODC/OPC and the infiltration of various inflammatory cells in SCI tissues, as indicated by the correlation matrix (P < 0.05) (Supplementary Fig. S5B). Furthermore, we identified that the known ODC/OPC markers (MBP, CNP, OLIG1) were significantly overexpressed in the low PCA ODC/OPC group. Therefore, PCA ODC/OPC was significantly negatively correlated with the degree of infiltration of ODC/OPC and inflammatory cells (Supplementary Fig. S5C). Considering the relationship between SCI subtypes in ODBSC and patient prognosis, we speculated that the Low ODC/OPC Score Classification is favorable for improving SCI prognosis.
Construction of the prognosis prediction model and regulatory network
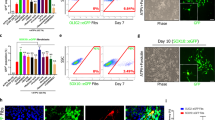
To investigate the functional significance of 34 fate-related genes, we conducted over-representation analysis (ORA) of PDEODGs in gene ontology (GO) terms and pathways (Fig. 7A). To prevent overfitting of the prognostic prediction model, we performed LASSO regression analysis and PCA based on the 34 PDEODGs (Fig. 7B). 164 SCI samples were randomly divided into training and testing sets. The training set (100 SCI samples) included 44 samples treated with a 5-mm chitosan tube loaded with NT3 and 56 untreated samples (LC and ET). The testing set (64 SCI samples) comprised 16 samples treated with NT3 and 48 untreated samples (LC and ET). The prognostic value of the signature was further validated using the receiver operating characteristic (ROC) curve in the training set (AUC = 0.886), testing set (AUC = 0.738), and overall set (AUC = 0.837) (Fig. 7C,D).
(A) The bar plot showed the enrichment degree of PDEODGs in GO biological processes by over-representation analysis. (B) The cvfit plot showed the variation of binomial deviance with log(λ) by Lasso regression analysis. (C) The box plots showed the differential expression levels of 34 PDEODGs between the low-risk and high-risk groups in all, train, and test sets. (D) The ROC curve of the total, train, and test sets indicated excellent predictive value of the PCA model for SCI (total set AUC = 0.837, train set AUC = 0.886, test set AUC = 0.738).
Construction of regulatory network and identification of drugs
The state transition of ODC/OPC during differentiation is regulated by PDEODGs, DETFs, and related co-factors, which collaboratively mediate target expression. Thus, we constructed a regulatory network centered on key PDEODGs for each subtype to elucidate the regulatory links governing state transitions (Fig. 8A). To quantify the interaction coefficients of key biomarkers in each SCI subtype (PDEODGs, DETFs, hallmark pathways, immune gene sets, immune cells), we conducted Pearson correlation analysis at the transcriptional level, as illustrated in the heat maps (Fig. 8B). Given the important roles of PDEODGs and DETFs in the pathological processes following SCI, there is an urgent need to identify novel potential drugs targeting these factors to optimize treatment. Consequently, we performed PRRophetic analysis based on the correlation results to identify small molecule drugs that have been shown to be target-specific for SCI (Fig. 8C).
(A) Regulatory network of each subtype showed tight gene linkage of cooperative expression regulation. Blue circles indicated PDEODGs, yellow circles indicated DETFs, red circles indicated pathways, brown circles indicated immune gene sets identified by ssGSEA, and green circles indicated immune cells. (B) Co-expression heat map of each subtype identified co-expression patterns and correlations among all components within the regulatory network. (C) The Violin plot showed subtype-specific drugs identified by the PRRophetic package that may target key PDEODGs and/or DETFs.
Discussion
Currently, specific and effective treatment targets for SCI are lacking. As a result, treatment strategies primarily involve surgical interventions for stabilization and rehabilitative care. Cell transplantation has emerged as a novel approach to promote neurological recovery in early-phase clinical trials; however, its safety and long-term efficacy remain unclear. However, progress in treatment options to improve clinical outcomes has been limited, despite efforts to decode the dynamic gene expression changes of single OPCs along their differentiation trajectories. Consequently, over the past few decades, significant efforts have been made to elucidate the pathophysiology of SCI and to reveal the mechanisms of tissue regeneration and nerve repair at the molecular and cellular levels. Despite extensive research, spinal cord injuries, which could cause severe sensory, motor and autonomic dysfunction, remain largely incurable34. These defects might be due to loss of ODC/OPC and demyelination of retained axons30,35. Despite intense research efforts on these different cell types and their related biomarkers, the regenerative potential of axons is likely conserved in the SCI, whereas it is blocked by multiple inhibitory factors generated at the lesion sites. Immediately post SCI, oligodendrocyte/myelin-derived axon regrowth inhibitors, including Nogo and MAG are generated OPCs proliferated and then differentiated into new ODC and Schwann cells to remyelinate axons, which ultimately leaded to efficient myelin regeneration after SCI. ODC/OPCs contributed to scar formation after SCI and limited regeneration of injured axons. In addition, ODC/OPCs altered gene expression after demyelination, expressed cytokines and maintained immune responses36. However, little is known about the mechanisms by which individual cell types facilitate tissue repair and functional improvements. Differentiation trajectories and genealogical hierarchies play an increasingly important role in exploring specific mechanisms. In our study, scRNA-seq was used to define the transcriptional status of mouse and human spinal cord ODC/OPC. In mice, 21 clusters and 11 cell types were identified by analyzing a large number (59,558) of single cells, of which 1,930 ODC/OPC were further extracted using typical marker genes and classified into 8 subgroups (DOC/OPC 0–7). In this study, intercellular communication between neurons and other cells was identified in scRNA-seq data using iTalk analysis. In addition, ODC/OPC subpopulations exhibit the most extensive intercellular contacts with each other. Based on previous studies, these cellular and cellular communication targets have been reported as potential therapeutic targets for SCI. However, how to regulate immune responses and communications between the nervous and immune systems at the appropriate intensity and duration still needs to be further explored. As an active and key regulator of the immune microenvironment, ODC/OPC influence immune infiltration, myelin regeneration and motor recovery through intercellular factors. Considering the diversity driven by the combined effects of ODC/OPC differentiation and ODGs, we reconstructed the ODC/OPC differentiation trajectory using Monocle2. The results suggest that cellular communication, as a dynamic mechanism, plays an important role in determining the differentiation fate and plasticity of ODC/OPC37. In this study, we conducted an integrated multi-omics analysis using large sample microarray data from an SCI animal model, along with RNA-seq data from PBMCs collected from SCI patients. This analysis revealed the transcriptomic landscape along the ODC/OPC differentiation trajectory, identifying 34 PDEODGs to evaluate treatment efficacy and predict prognosis.
Most previous research has targeted microglia, neural stem cells, and astrocytes for SCI treatment, focusing on resolving and regulating cell differentiation using various molecular methods, such as repopulating astrocyte transformation38,39,40. ODC/OPCs are well recognized as myelin-forming cells in the CNS41.The SRY-box transcription factor 10 (SOX10) is a high-mobility group transcription regulator that is essential for myelin gene expression. In the CNS, Sox10 expression increases during the development of glial progenitor cells into OPCs, and it persists throughout the differentiation and maturation of ODCs. Numerous studies have shown that SOX10 binds to the promoters of myelin-related genes, enhances gene expression, and recruits chromatin remodeling and histone-modifying enzymes42. Sox10 is essential for OPC proliferation and ODC regeneration following SCI in adults. It is temporarily upregulated during ODC lineage progression and maturation, as well as in newly regenerated ODCs during myelin regeneration. Therefore, we propose that SOX10, as a marker of the LOSC subtype, may activate transcriptional regulation and participate in intercellular communication among oligodendrocytes. It may also promote local myelin regeneration and contribute to the recovery of neurological function in patients.
SPARCL1 (hevin) is a protein secreted by astrocytes that regulates excitatory synaptogenesis both in vitro and in vivo. It positively influences synaptic formation, and by modulating hevin levels, astrocytes can control synaptic formation, maturation, and plasticity in vivo. As a synaptogenic protein, SPARCL1 is formed by linking neuroprotein 1α and neuroprotein 1b, which are two non-interacting subtypes. These findings suggest that astrocytes promote synaptic formation through the secretion of hevin, a regulatory neurotrophic factor29. In this study, the expression level of SPARCL1 (hevin) in the LOSC subtype was significantly elevated and correlated with a favorable prognosis for patients. Previous studies have demonstrated that OPCs can differentiate into ODCs, as well as into astrocytes and neurons43,44. We speculate that the increased expression of SPARCL1 during SCI may stimulate the proliferation of astrocytes, thereby promoting synaptic formation and nerve regeneration at the SCI site14.
CDGSH iron-sulfur domain 2 (CISD2), encoded by the CISD2 gene located at position 24 on the long arm of human chromosome 4 (4q24), is one of the three members (CISD1–3) of the human NEET family. The structure of the CISD2 protein consists of homodimers, with each monomer containing a CDGSH domain, classifying it as a class I NEET protein. CISD2 exerts anti-inflammatory effects by modulating the upstream components of the peroxisome proliferator-activated receptor-β (PPAR-β)/IκB/NF-κB signaling pathway43. This modulation is expected to directly inhibit the inflammatory cascade in the CNS and alleviate inflammation-induced neurological sequelae in patients with CNS injuries. It may also prevent astrocytic activation, astrocyte-mediated neuroinflammation, apoptosis, and neuronal loss45,46.
4-phenyl-1-(4-phenylbutyl) piperidine (PPBP) is a sigma receptor agonist. Previous studies have demonstrated that PPBP protects neurons in animal models47, and this protective effect is associated with reduced coupling of nNOS to PSD-95. Additionally, sigma receptor agonism has been shown to enhance nerve growth factor (NGF)-induced neurite outgrowth. We hypothesize that PPBP protects neurons by activating the transcription factor cyclic adenosine monophosphate response element binding protein and involves the antiapoptotic protein bcl-248.
Currently, specific and effective treatment targets for SCI are lacking. As a result, treatment strategies primarily involve surgical interventions for stabilization and rehabilitative care. Cell transplantation has emerged as a novel approach to promote neurological recovery in early-phase clinical trials; however, its safety and long-term efficacy remain unclear48. However, progress in treatment options to improve clinical outcomes has been limited, despite efforts to decode the dynamic gene expression changes of single OPCs along their differentiation trajectories. Consequently, over the past few decades, significant efforts have been made to elucidate the pathophysiology of SCI and to reveal the mechanisms of tissue regeneration and nerve repair at the molecular and cellular levels. ODBSC is a proposed molecular classification for the clinical treatment of SCI. It is significantly associated with patient prognosis and enhances understanding of the molecular pathology of OPCs in SCI. Importantly, bioinformatics analyses are widely employed to explore and validate pharmacological hypotheses. They are often used to identify and optimize novel small molecules with a unique affinity for specific targets49. Our proposed ODBSC and potential targeting agents refine the concept of precision therapy for SCI, providing a significant theoretical basis for such treatments.
Additionally, using the PRRophetic package, we identified potential SCI subtype-specific PDEODGs/DETF targets for LOSC, MOSC, and HOSC: metformin, epothilone, and rapamycin, respectively. Metformin has been shown to reduce neuronal apoptosis, promote axon regeneration, and significantly facilitate the removal of myelin debris while preserving myelin sheaths after SCI. Treatment of SCI with metformin can increase the number of OPCs, enhance the reactivity of aging OPCs to pro-differentiation factors, and facilitate myelin regeneration. Metformin has the potential to promote functional recovery after SCI during both the acute and subacute phases, increasing neural stem cell (NSPC) and OPC activation, as well as enhancing OPCs proliferation49,50. Huang51 et al. demonstrated that metformin treatment after SCI enhances functional recovery by reducing white matter loss and promoting myelin regeneration in Schwann cells. Metformin also facilitates oligodendrocyte-mediated myelin regeneration by activating the Nrg1/ErbB (Neuregulin-1(Nrg1) binds to the ErbB receptor family, leading to downstream signaling events that regulate cell growth, differentiation, and survival) signaling pathway. Additionally, a study by Zhang52 et al. indicated that metformin may protect neural tissues after injury by downregulating inflammatory factors such as TNF-α and IL-1β during the acute inflammatory phase. Recent research further suggests that metformin may enhance neural stem cell proliferation and inhibit neural cell ferroptosis by activating the AMPK (regulates cellular energy balance and metabolism by phosphorylating target proteins) signaling pathway53.
Studies indicate that axon growth is partially dependent on the mammalian target of rapamycin (mTOR): a key regulator of cell growth, proliferation, and survival. This mechanism may involve rapamycin enhancing autophagy, which increases Beclin-1 and LC3 expression, decreases p62 expression, and significantly improves motor function after SCI. Rapamycin can reduce inflammation, provide neuroprotection, decrease glial scar formation, promote motor function recovery, enhance OPCs recruitment, and facilitate OPC-mediated myelin regeneration. Targeting the differentiation of OPCs to form a new myelin sheath may represent a potential therapeutic strategy for treating SCI54,55. mTOR signaling plays a crucial role in regulating actin polymerization, cell branching, and axon sheath formation during early oligodendrocyte differentiation. It also governs two stages of cytoskeletal reorganization essential for the formation of both axon and myelin sheaths56,57. High levels of reactive oxygen species during SCI can have detrimental effects, including lipid peroxidation. Rapamycin enhances the antioxidant stress response by modulating the antioxidant response element and the transcription factor Nrf259. Furthermore, the inflammatory response following SCI is influenced by the mTOR regulatory pathway, which exerts neuroprotective effects by promoting autophagy and reducing inflammation59. Rapamycin also protects neurons after SCI by decreasing the number of microglia and neutrophils, minimizing the activation of inflammatory cells, and downregulating the expression of inflammatory factors60. In summary, rapamycin provides neuroprotective activity in SCI through multiple mechanisms.
Epothilone disrupts fibroblast polarization and enhances axon growth. Furthermore, when these neurons were exposed to high levels of inhibitory molecules, such as Nogo-A, CSPGs, and semaphorin 3 A at the SCI site, Epothilone facilitated axon growth. Epothilone promotes microtubule protrusion and axon elongation in neurons, modulating microtubule dynamics at the cell periphery. This modulation is essential for structural polarization and directed cell migration61. Epothilone improved hindlimb function after spinal cord contusion injury in rats. To inhibit secondary injury following acute SCI, epothilone enhances mitochondrial function, reduces oxidative stress, and prevents apoptosis through various mechanisms. These include increasing ATP levels in mitochondria, boosting glutathione content, and enhancing the activities of antioxidant enzymes such as catalase. Additionally, epothilone regulates the expression of Bax and Bcl-2 proteins62. Furthermore, epothilone modulates the inflammatory response after SCI by inhibiting scar formation, which subsequently affects immune cell function. This process may involve the IL6-JAK-STAT3 (Interleukin-6(IL6) binds to its receptor, activating Janus Kinase (JAK)which in turn phosphorylates Signal Transducer and Activator of Transcription 3(STAT3), leading to its translocation to the nucleus and regulation of gene expression) signaling pathway. Simultaneously, epothilone decreases the expression of inflammatory factors, providing a neuroprotective effect63. Therefore, our work provides insights into small molecule drugs associated with OPCs differentiation, proliferation, and myelin regeneration, potentially leading to new treatments for SCI64.
Conclusion
In conclusion, we identify key fate-related mechanisms involved in the differentiation of OPCs within the pathophysiology of SCI at various time points. This study reveals new diagnostic biomarkers and therapeutic targets, and predicts potential drugs for LOSC, MOSC, and HOSC. Although these drugs are FDA-approved for human use, they have not been thoroughly investigated in the context of SCI. Therefore, we will conduct systematic cellular, animal, and pharmacological experiments to determine whether these drugs can effectively target key molecular markers associated with oligodendrocyte-derived spinal cord (ODBSC)-related subtypes. Additionally, clinical trials will be initiated to evaluate the efficacy of these drugs in appropriate SCI patients, ultimately aiming for precise treatment of SCI.
Data availability
Data is provided within the manuscript or supplementary information files. The datasets for this study can be found in the GEO data repository and included the accession numbers as follows: GSE69334, GSE151371 (Suppl Tables 1–2). The results of sequencing data sets can be found in the figshare, identifier(https://doi.org/10.6084/m9.figshare.17702045).
References
Courtine, G. & Sofroniew, M. V. Spinal cord repair: advances in biology and technology. Nat. Med. 25, 898–908. https://doi.org/10.1038/s41591-019-0475-6 (2019).
Karsy, M. & Hawryluk, G. Modern medical management of spinal cord Injury. Curr. Neurol. Neurosci. 19, 65. https://doi.org/10.1007/s11910-019-0984-1 (2019).
Burda, J. E. & Sofroniew, M. V. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–248. https://doi.org/10.1016/j.neuron.2013.12.034 (2014).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. https://doi.org/10.1038/nature21029 (2017).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410. https://doi.org/10.1523/jneurosci.6221-11.2012 (2012).
Russo, M. V. & McGavern, D. B. Inflammatory neuroprotection following traumatic brain injury. Science 353, 783–785. https://doi.org/10.1126/science.aaf6260 (2016).
Ransohoff, R. M. How neuroinflammation contributes to neurodegeneration. Science 353, 777–783. https://doi.org/10.1126/science.aag2590 (2016).
Horn, K. P., Busch, S. A., Hawthorne, A. L., van Rooijen, N. & Silver, J. Another barrier to regeneration in the CNS: activated macrophages induce extensive retraction of dystrophic axons through direct physical interactions. J. Neurosci. 28, 9330–9341. https://doi.org/10.1523/jneurosci.2488-08.2008 (2008).
Curtis, E. et al. A first-in-Human, phase I study of neural stem cell transplantation for chronic spinal cord Injury. Cell. Stem Cell. 22, 941–950e946. https://doi.org/10.1016/j.stem.2018.05.014 (2018).
Khaing, Z. Z. et al. Clinical trials targeting secondary damage after traumatic spinal cord Injury. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms24043824 (2023).
Hu, X. et al. Spinal cord injury: molecular mechanisms and therapeutic interventions. Signal. Transduct. Target. Ther. 8, 245. https://doi.org/10.1038/s41392-023-01477-6 (2023).
Yang, J. et al. Oligodendrocyte precursor cell transplantation promotes functional recovery following contusive spinal cord injury in rats and is associated with altered microRNA expression. Mol. Med. Rep. 17, 771–782. https://doi.org/10.3892/mmr.2017.7957 (2018).
Huang, R. et al. Identification of key eRNAs for spinal cord injury by integrated multinomial bioinformatics analysis. Front. Cell. Dev. Biol. 9 https://doi.org/10.3389/fcell.2021.728242 (2021).
Li, C. et al. Temporal and spatial cellular and molecular pathological alterations with single-cell resolution in the adult spinal cord after injury. Signal. Transduct. Target. Ther. https://doi.org/10.1038/s41392-022-00885-4 (2022). 7.
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. https://doi.org/10.1093/bioinformatics/btg405 (2004).
Chen, W. T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991e919. https://doi.org/10.1016/j.cell.2020.06.038 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. https://doi.org/10.1038/nbt.4096 (2018).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. https://doi.org/10.1093/bioinformatics/btp352 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinf. (Oxford England). 30, 923–930. https://doi.org/10.1093/bioinformatics/btt656 (2014).
Zhang, X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728. https://doi.org/10.1093/nar/gky900 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19. (2018). https://doi.org/10.1186/s13059-017-1382-0
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 14, 979–982. https://doi.org/10.1038/nmeth.4402 (2017).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. https://doi.org/10.1038/nprot.2009.97 (2009).
Ringnér, M. What is principal component analysis? Nat. Biotechnol. 26, 303–304. https://doi.org/10.1038/nbt0308-303 (2008).
Yi, M., Nissley, D. V., McCormick, F. & Stephens, R. M. ssGSEA score-based Ras dependency indexes derived from gene expression data reveal potential Ras addiction mechanisms with possible clinical implications. Sci. Rep. 10. https://doi.org/10.1038/s41598-020-66986-8 (2020).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Barbour, J. D., Geeleher, P., Cox, N. & Huang, R. S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 9 https://doi.org/10.1371/journal.pone.0107468 (2014).
Ahuja, C. S. et al. Traumatic spinal cord injury. Nat. Rev. Dis. Primers 3. https://doi.org/10.1038/nrdp.2017.18 (2017).
Singh, S. K. et al. Astrocytes assemble thalamocortical synapses by bridging NRX1α and NL1 via Hevin. Cell 164, 183–196. https://doi.org/10.1016/j.cell.2015.11.034 (2016).
Kuhn, S., Gritti, L., Crooks, D. & Dombrowski, Y. Oligodendrocytes in development, myelin Generation and Beyond. Cells 8 https://doi.org/10.3390/cells8111424 (2019).
Lin, M. S. CISD2 attenuates inflammation and regulates Microglia polarization in EOC microglial Cells-As a potential therapeutic target for neurodegenerative dementia. Front. Aging Neurosci. 12, 260. https://doi.org/10.3389/fnagi.2020.00260 (2020).
Yang, S., Alkayed, N. J., Hurn, P. D. & Kirsch, J. R. Cyclic adenosine monophosphate response element-binding protein phosphorylation and neuroprotection by 4-phenyl-1-(4-phenylbutyl) piperidine (PPBP). Anesth. Analg. 108, 964–970. https://doi.org/10.1213/ane.0b013e318192442c (2009).
Tica, J., Bradbury, E. & Didangelos, A. Combined transcriptomics, proteomics and Bioinformatics identify drug targets in spinal cord Injury. Int. J. Mol. Sci. 19. https://doi.org/10.3390/ijms19051461 (2018).
Hu, X. et al. Spinal cord injury: molecular mechanisms and therapeutic interventions. Signal. Transduct. Target. Ther. 8. https://doi.org/10.1038/s41392-023-01477-6 (2023).
Elbaz, B. & Popko, B. Molecular control of oligodendrocyte development. Trends Neurosci. 42, 263–277. https://doi.org/10.1016/j.tins.2019.01.002 (2019).
Clayton, B. L. L. & Tesar, P. J. Oligodendrocyte progenitor cell fate and function in development and disease. Curr. Opin. Cell. Biol. 73, 35–40. https://doi.org/10.1016/j.ceb.2021.05.003 (2021).
Wu, Y., Tang, Z., Zhang, J., Wang, Y. & Liu, S. Restoration of spinal cord injury: from endogenous repairing process to cellular therapy. Front. Cell. Neurosci. 16. https://doi.org/10.3389/fncel.2022.1077441 (2022).
Ma, D. et al. A novel hydrogel-based treatment for complete transection spinal cord injury repair is driven by microglia/macrophages repopulation. Biomaterials 237, 119830. https://doi.org/10.1016/j.biomaterials.2020.119830 (2020).
Hara, M. et al. Interaction of reactive astrocytes with type I collagen induces astrocytic scar formation through the integrin-N-cadherin pathway after spinal cord injury. Nat. Med. 23, 818–828. https://doi.org/10.1038/nm.4354 (2017).
Willis, E. F. et al. Repopulating microglia promote brain repair in an IL-6-dependent manner. Cell 180, 833–846e816. https://doi.org/10.1016/j.cell.2020.02.013 (2020).
Sankavaram, S. R. et al. Adult neural progenitor cells transplanted into spinal cord injury differentiate into oligodendrocytes, enhance myelination, and contribute to recovery. Stem Cell. Rep. 12, 950–966. https://doi.org/10.1016/j.stemcr.2019.03.013 (2019).
Zhang, S. et al. Sox2 is essential for oligodendroglial proliferation and differentiation during postnatal brain myelination and CNS remyelination. J. Neurosci. 38, 1802–1820. https://doi.org/10.1523/JNEUROSCI.1291-17.2018 (2018).
Kung, W. M. et al. Anti-inflammatory CDGSH iron-sulfur domain 2: A Biomarker of central nervous system insult in cellular, animal models and patients. Biomedicines 10. https://doi.org/10.3390/biomedicines10040777 (2022).
Llorens-Bobadilla, E. et al. A latent lineage potential in resident neural stem cells enables spinal cord repair. Sci. (New York N Y). 370, eabb8795. https://doi.org/10.1126/science.abb8795 (2020).
Lin, C. C. et al. CISD2 serves a novel role as a suppressor of nitric oxide signalling and curcumin increases CISD2 expression in spinal cord injuries. Injury 46, 2341–2350. https://doi.org/10.1016/j.injury.2015.07.040 (2015).
Kung, W. M., Chang, C. J., Chen, T. Y. & Lin, M. S. Cryogen spray cooling mitigates inflammation and injury-induced CISD2 decline in rat spinal cord hemisection model. J. Neurosci. 19, 619–628. https://doi.org/10.31083/j.jin.2020.04.255 (2020).
Goyagi, T. et al. Neuroprotective effect of sigma(1)-receptor ligand 4-phenyl-1-(4-phenylbutyl) piperidine (PPBP) is linked to reduced neuronal nitric oxide production. Stroke 32, 1613–1620. https://doi.org/10.1161/01.str.32.7.1613 (2001).
Assinck, P., Duncan, G. J., Hilton, B. J., Plemel, J. R. & Tetzlaff, W. Cell transplantation therapy for spinal cord injury. Nat. Neurosci. 20, 637–647. https://doi.org/10.1038/nn.4541 (2017).
Gilbert, E. A. B., Livingston, J., Garcia-Flores, E., Kehtari, T. & Morshead, C. M. Metformin improves functional outcomes, activates neural precursor cells, and modulates Microglia in a sex-dependent Manner after spinal cord Injury. Stem Cells Transl. Med. 12, 415–428. https://doi.org/10.1093/stcltm/szad030 (2023).
Wu, Y. et al. Metformin promotes microglial cells to facilitate myelin debris clearance and accelerate nerve repairment after spinal cord injury. Acta Pharmacol. Sin. 43, 1360–1371. https://doi.org/10.1038/s41401-021-00759-5 (2021).
Huang, Z. et al. Metformin promotes Schwann cell remyelination, preserves neural tissue and improves functional recovery after spinal cord injury. Neuropeptides 100, 102348. https://doi.org/10.1016/j.npep.2023.102348 (2023).
Zhang, T. et al. Therapeutic effect of metformin on inflammation and apoptosis after spinal cord injury in rats through the Wnt/β-catenin signaling pathway. Neurosci. Lett. 739, 135440. https://doi.org/10.1016/j.neulet.2020.135440 (2020).
Wang, Z., Zhou, W., Zhang, Z., Zhang, L. & Li, M. Metformin alleviates spinal cord injury by inhibiting nerve cell ferroptosis through upregulation of heme oxygenase-1 expression. Neural Regen. Res. 19, 2041–2049. https://doi.org/10.4103/1673-5374.390960 (2024).
Lu, P. et al. Long-distance growth and connectivity of neural stem cells after severe spinal cord Injury. Cell 150, 1264–1273. https://doi.org/10.1016/j.cell.2012.08.020 (2012).
Liu, J. et al. Rapamycin preserves neural tissue, promotes Schwann cell myelination and reduces glial scar formation after hemi-contusion spinal cord Injury in mice. Front. Mol. Neurosci. 13. https://doi.org/10.3389/fnmol.2020.574041 (2021).
Musah, A. S. et al. Mechanistic target of rapamycin regulates the oligodendrocyte cytoskeleton during myelination. J. Neurosci. 40, 2993–3007. https://doi.org/10.1523/jneurosci.1434-18.2020 (2020).
Zhang, X. et al. Therapeutic effects of rapamycin and surgical decompression in a rabbit spinal cord injury model. Cell. Death Dis. 11 https://doi.org/10.1038/s41419-020-02767-5 (2020).
Jia, Z. et al. Oxidative stress in spinal cord injury and antioxidant-based intervention. Spinal Cord. 50, 264–274. https://doi.org/10.1038/sc.2011.111 (2012).
Li, X. G., Du, J. H., Lu, Y. & Lin, X. J. Neuroprotective effects of rapamycin on spinal cord injury in rats by increasing autophagy and Akt signaling. Neural Regen. Res. 14, 721–727. https://doi.org/10.4103/1673-5374.247476 (2019).
Song, Y., Xue, H., Liu, T. T., Liu, J. M. & Chen, D. Rapamycin plays a neuroprotective effect after spinal cord injury via anti-inflammatory effects. J. Biochem. Mol. Toxicol. 29, 29–34. https://doi.org/10.1002/jbt.21603 (2015).
Ruschel, J. et al. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science 348, 347–352. https://doi.org/10.1126/science.aaa2958 (2015).
Jia, Z. Q. et al. Ebselen protects mitochondrial function and oxidative stress while inhibiting the mitochondrial apoptosis pathway after acute spinal cord injury. Neurosci. Lett. 678, 110–117. https://doi.org/10.1016/j.neulet.2018.05.007 (2018).
Zhao, W. et al. Mechanisms responsible for the inhibitory effects of epothilone B on scar formation after spinal cord injury. Neural Regen. Res. 12, 478–485. https://doi.org/10.4103/1673-5374.202921 (2017).
Tica, J., Bradbury, E. J., Didangelos, A. . Combined transcriptomics: Proteomics and Bioinformatics identify drug targets in spinal cord Injury. Int. J. Mol. Sci. 19, 1461. https://doi.org/10.3390/ijms19051461 (2018).
Funding
This work was supported by Henan Province Medical Science and Technology Research Project Joint Construction Project (LHGJ20200304).
Author information
Authors and Affiliations
Contributions
X.Y. and X.C. designed and conceived the study. Y.Z. and J.W. analyzed data. G.C. completed, drafted and revised the manuscript. H.T. and K.Y. provided advice and technical assistance. All authors have contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
All data are sourced from public databases, with ethics approval obtained by the data providers.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yue, X., Chen, X., Zang, Y. et al. Bioinformatics analysis reveals key mechanisms of oligodendrocytes and oligodendrocyte precursor cells regulation in spinal cord Injury. Sci Rep 15, 6400 (2025). https://doi.org/10.1038/s41598-025-90489-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-90489-z