Abstract
Various factors have been implicated in the pathogenesis of ulcerative colitis (UC), with immune system failure being the most important one. Calcitonin gene-related peptide (CGRP), a neuropeptide with two isoforms, CGRPα and CGRPβ, has been reported to regulate the immune system. In this study, we investigated the role of CGRP isoforms in UC pathogenesis. We induced UC-like symptoms in CGRPα and CGRPβ knockout (KO) mice using dextran sulphate sodium. Compared to wild-type and CGRPα KO mice, CGRPβ-deficient mice exhibited severe symptoms with increased blood in the stool and diarrhoea. Proteome analysis revealed significant up-regulation of immune-related proteins and immunoproteasome components in CGRPβ-deficient mice, suggesting that an enhanced immune response contributes to the severity of this disease. Treatment with ONX-0914, an immunoproteasome inhibitor, markedly improved these symptoms, highlighting the role of the immunoproteasome in exacerbating UC. This study provides the first evidence that CGRPβ protects against UC by modulating immune responses, particularly those mediated by the immunoproteasome. Our findings suggest that functional differences in CGRP isoforms may influence the severity and management of UC. This insight into the neuro-immune mechanism of UC opens avenues for novel therapies that address both the neural and immune aspects of this disease.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD) is a chronic condition that causes diarrhoea, bloody stools, and abdominal pain, among other symptoms. Ulcerative colitis (UC) and Crohn’s disease are the two major types of IBD1. UC is a chronic inflammatory disease confined to the colon and is characterized by erosion and ulceration of the colon mucosa. Patients with UC often experience severe bloody stools, diarrhoea, fever, and abdominal pain, and tend to go through periods of exacerbation and remission1,2. Current UC treatments, which include 5-aminosalicylic acid, immunomodulatory agents, and surgery, focus on maintaining remission1,3. Various factors, such as genetic factors4,5,6, psychological stress7,8, and dysbiosis of the intestinal microbiota, all of which cause abnormalities in the innate and acquired immune systems, have been investigated for their role in the pathogenesis of UC9,10,11. UC is accompanied with neutrophil accumulation and sustained inflammation mediated by inflammatory cytokines, such as TNF-α and interleukin 1β12. In addition, a shift to a less permissive subset of dendritic cells has been suggested to reduce their ability to produce regulatory T cells13,14. The disruption of homeostasis of effector T cells, Th1, Th2, and Th17 cells, enhances the production of inflammatory cytokines, including interleukin-1315,16,17. This induces apoptosis of epithelial cells and affects the formation of tight junction, resulting in tissue damage16. Considering these factors, regulation of the immune system and inhibition of inflammation are promising therapeutic strategies. Realisation of such strategies requires a detailed elucidation of the mechanisms that cause immune system disruption in UC.
Calcitonin gene-related peptide (CGRP) is a 37-amino acid-long neuropeptide derived from alternative splicing of the calcitonin gene. It exists in two isoforms, namely CGRPα and CGRPβ, which are encoded by different genes. CGRPα is encoded by the Calca gene, located on Chr7:114,225,223 − 114,236,145 (GRCm39), while CGRPβ is encoded by the Calcb gene, located on Chr7:114,315,790 − 114,322,603 (GRCm39). CGRP is expressed in both the central and enteric nervous systems, where it induces vasodilation, nociception, and neurogenic inflammation18,19. Only CGRPα functions in the central nervous system and is believed to play a role in trigeminal afferent sensory nerves and trigeminal pain-mediating regions18,20. CGRP acts as a mediator between the nervous and immune systems21. It inhibits the maturation of dendritic cells22, promotes differentiation of Th2 cells23,24, and affects the production of anti-inflammatory cytokines in macrophages25,26. Therefore, CGRP might be involved in some immune-related diseases.
A link between CGRP and UC pathology has previously been noted—significant down-regulation of CGRP at both mRNA and protein levels in serum and lesion sites of patients with UC is negatively correlated with disease severity27. Furthermore, CGRP induces anti-inflammatory macrophages and aids in the recovery from colitis28. In addition, CGRP derived from nociceptor neurons may contribute to the suppression of UC by stimulating goblet cells and, thereby, inducing mucus secretion29. UC model mice deficient in RAMP1, a receptor for CGRP, exhibit exacerbated disease symptoms30. The possibility that CGRP contributes to the suppression of UC has been discussed in several papers. However, differences in the roles of the two CGRP isoforms have not been fully investigated. Moreover, the pathological pathways at the core of CGRP-mediated pathogenesis of UC, especially those involved in the exacerbation phase, are unknown. In this study, we investigated the functional differences between the CGRP isoforms by evaluating their anti-inflammatory effects in UC using CGRPα and CGRPβ knockout (KO) mice.
Results
CGRPβ deficiency exacerbates DSS-induced colitis in mice
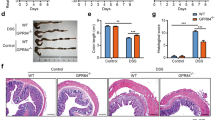
To elucidate the functional involvement of CGRP in UC, we employed KO mice deficient in either CGRPα or CGRPβ (Supplementary Fig. 1). Wild-type (WT), CGRPα KO, and CGRPβ KO mice were allowed to consume 3% dextran sulphate sodium (DSS) ad libitum to induce UC-like symptoms (Fig. 1a). The severity of UC symptoms was evaluated using the disease activity index (DAI) score. The symptoms of enteritis were significantly more severe in CGRPβ KO mice than in the WT and CGRPα KO groups (Fig. 1b). Although no difference in weight loss was observed among the UC model mice (Fig. 1c), typical UC symptoms, such as haematochezia and diarrhoea were notably exacerbated in CGRPβ KO mice (Fig. 1d, e). Morphological evaluation of the colon using haematoxylin and eosin (HE) staining revealed well-defined crypts in the colonic mucosa of mice in the control group without DSS treatment. In contrast, mice with DSS-induced enteritis exhibited characteristic features of UC, including loss of goblet cells, crypt damage, and progressive sub-mucosal dilation. However, no significant difference in the colon morphology was observed among the groups (Fig. 1f, g). These results indicate that CGRPβ deficiency exacerbates UC-like symptoms, such as bloody stools and diarrhoea, and alters the inflammatory profile of the disease but only to the extent that it does not significantly affect the tissue morphology.
Exacerbation of dextran sodium sulphate (DSS)-induced ulcerative colitis (UC) symptoms in CGRPβ knockout (KO) mice. (a) Schematic diagram of the experimental model: the UC model was prepared in wild-type (WT), CGRPα KO, and CGRPβ KO mice by allowing them to consume 3% DSS through drinking water for 5 days. (b) Disease activity index (DAI), scored and integrated for rate of weight loss, faecal blood, and diarrhoea, over 5 days. For each experiment, n = 5, for all the mice groups. Error bars represent standard deviation (SD). Weight loss rate (c), blood stool score (d), and diarrhoea score (e) over 5 days of DSS consumption. (f) Images of haematoxylin and eosin (HE)-stained colon tissue sections from WT, CGRPα KO, and CGRPβ KO mice with and without DSS treatment. The black scale bar represents 100 μm. (g) Pathology scores were calculated based on HE-stained images performed on colon tissue after 5 days of DSS treatment. The experiment was performed using the same number of mice for which DAI scores were obtained, and representative HE staining results are shown. Error bars represent SD. p-values were determined using one-way ANOVA with Tukey’s post hoc correction. *p < 0.05 for WT vs. CGRPβ KO, **p < 0.01 for WT vs. CGRPβ KO, ***p < 0.0001 for WT vs. CGRPβ KO, #p < 0.05 for CGRPα KO vs. CGRPβ KO, ##p < 0.01 for CGRPα KO vs. CGRPβ KO, ###p < 0.0001 for CGRPα KO vs. CGRPβ KO.
Proteome analysis at the end of the experiment indicates differences in the blood coagulation-fibrinolysis system in the different mice groups
To identify the factors contributing to the exacerbation of UC symptoms in CGRPβ KO mice, we performed proteome analysis of the colon tissue. Principal component analysis (PCA) of the proteome data sampled at the endpoint of the experiment revealed only slight differences among WT, CGRPα KO, and CGRPβ KO mice (Fig. 2a). Protein expression between WT and CGRPα KO mice, between WT and CGRPβ KO mice and between CGRPα KO and CGRPβ KO mice was compared in volcano plots (Fig. 2b, c, d). As suggested by the PCA, the number and extent of altered proteins were greater in CGRPβ KO mice than in WT or CGRPα KO mice. Furthermore, gene set enrichment analysis (GSEA) of these proteins was performed to identify altered associated pathways (Fig. 2e, f, g). Interestingly, it was observed that the expression of proteins associated with the coagulation and fibrinolysis pathways was reversed in CGRPα KO and CGRPβ KO mice when compared to WT mice. These changes may contribute to the development of thrombocytopenia in CGRPβ KO mice. Comparison of CGRPα KO and CGRPβ KO mice also suggests that the manners of response with regards to immune responses tend to differ between CGRP isoforms. On the contrary, comparison of both WT mice vs. CGRPα KO mice and WT mice vs. CGRPβ KO mice unexpectedly showed similar results as well as changes associated with various immune responses. Considering the volcano plot results, the magnitude of changes in CGRPβ KO mice might contribute to the severity of the disease; however, the proteomic data at the endpoint did not allow us to define the underlying cause of UC exacerbation in CGRPβ KO mice.
Analysis of the colon proteome at the experimental endpoint indicated changes in the blood coagulation-fibrinolytic system. (a) Results of the principal component analysis based on the colon proteome analysis of samples prepared after 5 days of DSS consumption. For proteomic analysis, four WT mice, three CGRPα KO mice and four CGRPβ KO mice were used. (b) Volcano plot showing comparison of protein levels between wild-type (WT) and CGRPα knockout (KO) mice. (c) Volcano plot showing comparison of protein levels between WT and CGRPβ KO mice. (d) Volcano plot showing comparison of protein levels between CGRPα KO and CGRPβ KO mice. (e) Gene set enrichment analysis (GSEA) results showing comparison of protein expression between WT and CGRPKα KO mice, with positive normalized enrichment score indicating higher expression in the CGRPα KO mice and negative normalized enrichment score indicating higher expression in the WT mice. (f) GSEA of protein expression between WT and CGRPβ KO mice, with positive normalized enrichment score terms indicating higher expression in CGRPβ KO mice and negative terms indicating higher expression in WT mice. (g) GSEA of protein expression between CGRPα KO and CGRPβ KO mice, with positive normalized enrichment score terms indicating higher expression in CGRPβ KO mice and negative terms indicating higher expression in CGRPα KO mice.
Immune proteasome is induced in CGRPβ knockout mice
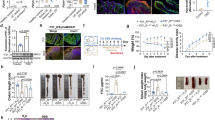
As mentioned in the previous sections, CGRPβ KO mice differed from WT and CGRPα KO mice in symptoms, such as faecal blood and diarrhoea. Although the symptoms of UC may be related to the immune system, based on our results at the endpoint of the experiment, we could not identify immune changes characteristic of CGRPβ KO mice and the mechanism by which CGRPβ suppresses UC. Therefore, we examined the possibility of identifying the factors that contribute to the aggravation of UC-like symptoms by analysing the colon proteome at 3 days after DSS consumption, when differences in symptoms started becoming obvious. First, a histological analysis of the colon was performed 3 days after DSS intake. No statistically significant differences were identified in CGRPβ KO mice at this point, but a trend towards an enhanced degree of damage was suggested (Fig. 3a, b). Interestingly, PCA results from proteomic analysis showed that the trend of protein expression in the colon tissue in CGRPβ KO mice differed significantly with regard the taxonomic characteristics from that in WT and CGRPα KO mice (Fig. 3c). Next, assessment of proteins with fluctuating expression on volcano plots indicated that the number of proteins fluctuating in CGRPβ KO mice was higher than compared with that in WT or CGRPα KO (Fig. 3d, Supplementary Fig. 2a, b). GSEA revealed proteins involved in antigen presentation, acquired immunity, and neutrophil activity, and those involved in the coagulation and fibrinolytic systems of the blood, which indicates that changes distinct from WT and CGRPα KO mice may have occurred in CGRPβ KO mice at day 3 of DSS consumption (Fig. 3e, Supplementary Fig. 2c, d). We attempted to identify factors contributing to the severity of UC-like symptoms, especially in CGRPβ KO mice. We hypothesised that, in addition to changes in the expression levels, changes in protein groups interacting with each other might be important factors. Therefore, we drew Venn diagrams of the proteins that were characteristically altered in CGRPβ KO mice compared with that in WT mice (Fig. 3f). Enrichment analysis of proteins that were characteristically up-regulated in CGRPβ KO mice confirmed that they were indeed enriched in immune system proteins (Supplementary Fig. 3a). Representation of interactions using STRING showed the presence of proteasome proteins (Fig. 3g). Interestingly, the proteome data revealed increased expression of many proteasome core subunits, especially PSMB8, PSMB9, and PSMB10, in CGRPβ KO mice, which highlighted the possibility that the expression of specialised proteasome core subunits called immunoproteasomes could be induced (Supplementary Fig. 3b). The immunoproteasome is formed by the replacement of the catalytically active subunits of the 20 S proteasome, PSMB6, PSMB7, and PSMB5, by PSMB9, PSMB10, and PSMB8, respectively, upon IFN-γ stimulation31. The activities of immunoproteasomes differ from those of the 20 S proteasome, and the antigen peptides presented by MHC class I are believed to be modified, which affects the activity of the immune system and the production of inflammatory cytokines after antigen presentation. Therefore, we focused on these immunoproteasomes and reconfirmed, using western blotting, that expression of these proteins was indeed increased in CGRPβ KO (Fig. 3h). These results indicate that the expression of proteins related to the immune system may already have been fluctuating in CGRPβ KO mice at 3 days after DSS consumption, and in particular, the increased expression of immunoproteasome components was confirmed. Immunoproteasomes contribute to the exacerbation of UC. Therefore, we expected that immunoproteasome expression might be a major factor in UC exacerbation in CGRPβ KO mice. On the contrary, induction of immunoproteasome expression in CGRPβ KO mice was observed 3 days after DSS consumption, and it was unclear whether immunoproteasome function at this time would have a dominant effect on the subsequent UC exacerbation phase. Therefore, we surmised that it would be useful to examine the pathological significance of DSS consumption for 3 days in interpreting the phenotype of CGRPβ KO mice, considering the changes in protein expression occurring during the exacerbation phase of UC. Toward this end, we performed proteomic analysis of the colon tissue of WT mice from day 0 to day 5 of DSS consumption (Supplementary Fig. 4a). A heatmap of the overall proteome over time showed a significant change in the protein expression profile starting 3 days after DSS consumption (Supplementary Fig. 4b). In particular, cluster 1, comprising proteins with increased expression, was mainly related to immunity, whereas cluster 5, which showed a decreasing trend, represented a group of proteins related to DNA replication and cell cycle. In addition, cluster 2, comprising proteins with increased expression after 4 days of DSS consumption, cluster 3, comprising proteins whose expression was increased temporarily, and cluster 4, comprising proteins with decreased expression after approximately 4 days of DSS consumption, were composed of proteins not directly related to immunity (Supplementary Fig. 4c). Thus, DSS consumption induced stepwise changes in protein expression in the colon tissue, and in particular, the expression of many proteins related to the immune system was changed after 3 days. Therefore, the regulation of immunoproteasome expression by CGRPβ may be sufficiently noteworthy with regard to the mechanism of exacerbation of UC.
Immunoproteasome expression is induced in CGRPβ knockout (KO) mice 3 days after consumption of dextran sulphate sodium (DSS). (a) Images of haematoxylin and eosin (HE)-stained colon tissue sections from wild-type (WT), CGRPα KO, and CGRPβ KO mice with DSS treatment. The black scale bar represents 100 μm. (b) Pathology scores were calculated based on HE-stained images performed on colon tissue after 3 days of DSS treatment. For histological analysis, four WT mice, three CGRPα KO mice and three CGRPβ KO mice were used. (c) Results of the principal component analysis based on the colon proteome analysis of samples prepared after 3 days of DSS consumption. For proteomic analysis, four WT mice, three CGRPα KO mice and four CGRPβ KO mice were used. (d) Volcano plot showing comparison of protein levels between WT and CGRPβ KO mice. (e) GSEA of protein expression between WT and CGRPβ KO mice, with positive normalized enrichment score terms indicating higher expression in CGRPβ KO mice and negative terms indicating higher expression in WT mice. (f) Venn diagram of proteins whose expression was up-regulated in CGRPα KO and CGRPβ KO mice compared with that in WT mice in the proteome analysis. Proteins whose expression was specifically up-regulated in CGRPβ KO mice were analysed. (g) Results of STRING analysis: STRING focused on protein–protein interactions, and reactome enrichment analysis was applied to colour the protein nodes belonging to each term. (h) Western blot analysis of the expression of immunoproteasome subunits PSMB8, PSMB9, and PSMB10 in the colon samples after 3 days of DSS consumption.
Inhibition of immunoproteasomes reduces symptoms of DSS-induced colitis in CGRPβ KO mice
Because our data suggested that increased expression of immunoproteasomes during the early stages of UC formation may be involved in the exacerbation of colitis symptoms in CGRPβ KO mice, we investigated the possibility of ameliorating colitis symptoms by immunoproteasome inhibition. Mice subjected to DSS treatment were daily administered a subcutaneous injection of ONX-091418, which is a selective inhibitor of immunoproteasomes, and examined for UC-like symptoms using the DAI score (Fig. 4a). A significant decrease in DAI score and improvement in symptoms was observed in the CGRPβ KO-treated group (Fig. 4b). Although there was no significant improvement in weight loss (Fig. 4c), symptoms of bloody and soft stools were significantly improved only in the inhibitor-treated CGRPβ KO group (Fig. 4d, e). Furthermore, we observed the histology of the colon tissue using HE staining, and quantified the disappearance of the crypts and enlargement of the sub-mucosa using pathological scores. A trend towards improvement in the degree of tissue damage was evident (Fig. 4f, g). Finally, we performed proteomic analysis of the colon tissue on day 3 of DSS consumption. The results indicate that selective inhibition of the immunoproteasome may underlie the improvement of symptoms in CGRPβ KO mice, as well as functional changes in the machinery related to antigen presentation contributed by the immunoproteasome (Supplementary Fig. 5a, b). These studies indicate that CGRPβ contributes to the suppression of exacerbation in UC via suppression of immunoproteasome expression.
Immunoproteasome inhibitor treatment mitigates ulcerative colitis (UC) symptoms in CGRPβ knockout (KO) mice. (a) Schematic of the experimental model: Wild-type (WT), CGRPα KO, and CGRPβ KO mice were treated with 3% dextran sulphate sodium (DSS) for 5 days to generate the UC model, and the immunoproteasome inhibitor ONX-0914 was injected subcutaneously at 10 mg/kg daily. (b) Disease activity index (DAI) scores over 5 days. Percent body weight loss (c), blood stool score (d), and diarrhoea score (e) over 5 days. For these pathological assessment experiments in UC experiment, n = 8, for all the mice groups. (f) Images of haematoxylin and eosin (HE)-stained colon tissue sections from WT, CGRPα KO, and CGRPβ KO mice with or without ONX-0914 treatment. The black scale bar represents 100 μm. (g) Pathological scores were calculated based on the HE-stained images and were performed on four mice in the WT Inhibitor (-) group, three mice in the CGRPβ KO Inhibitor (-) group, three mice in the WT Inhibitor (+) group and five mice in the CGRPβ KO Inhibitor (+) group. Error bars represent SD. p-values were determined using one-way ANOVA with Tukey’s post hoc correction. *p < 0.05 for CGRPβ KO Inhibitor (-) vs. CGRPβ KO Inhibitor (+), **p < 0.01 for CGRPβ KO Inhibitor (-) vs. CGRPβ KO Inhibitor (+), ***p < 0.0001 for CGRPβ KO Inhibitor (-) vs. CGRPβ KO Inhibitor (+). #p < 0.05 for CGRPβ KO Inhibitor (-) vs. WT Inhibitor (-), ##p < 0.01 for CGRPβ KO Inhibitor (-) vs. WT Inhibitor (-).
Discussion
UC is one of the most intractable diseases. Repeated exacerbation and remission of this disease lead to a decline in the quality of life of patients. As such, elucidation of the pathomechanism of UC is warranted for its effective management. While various factors contribute to the pathogenesis of UC, the immune system is believed to play an important role, and elucidation of the mechanism of its dysregulation is expected to open avenues for an effective solution. In this study, we investigated the role of CGRP, a neuropeptide with a wide range of physiological effects, including modulation of the functions of the immune and nervous systems. Using CGRPα and CGRPβ KO mice in parallel experiments, we found that CGRPβ specifically suppresses UC exacerbation. Although the involvement of CGRP in the exacerbation phase of UC has been previously reported, based on our findings regarding the suppression of immunoproteasome expression, we propose a new core system in which regulation of the immune system is involved in the suppression of UC symptoms.
Bloody and soft stools were exacerbated in DSS-induced CGRPβ KO mice. The results of proteome analysis after 5 days of DSS consumption also indicated changes in the coagulation-fibrinolytic system relevant to support the UC symptoms. Functional analysis of CGRP in UC has been performed in several studies. Thompson et al. reported worsened UC-like symptoms in CGRPα and CGRPβ KO mice32. However, they used DSS with molecular weight of 36–50 kDa, whereas in the present study, we used DSS with a molecular weight of 5 kDa. The molecular weight of DSS is a factor that affects the severity of the symptoms of induced enteritis. Therefore, we also used high-molecular weight DSS and tested whether UC-like symptoms were exacerbated in CGRPα KO mice. Although the symptoms induced by high-molecular weight DSS were more severe, no significant difference was noted between the two groups (Supplementary Fig. 6). Therefore, the difference in molecular weight of DSS cannot be the cause of the phenotypic difference in CGRPα KO mice. On the contrary, no significant difference in exacerbation was observed in CGRPβ KO mice. This indicates that significant exacerbation of UC-like symptoms in CGRPβ KO mice can be achieved under relatively mild conditions. Wu et al. reported that CGRPα contributed to the recovery of UC-like symptoms in CGRPα KO mice28. Their data on the exacerbation phase showed no significant difference in the rate of weight loss, histological appearance, and levels of various cytokines between WT and CGRPα KO mice. The inconsistent results in the three above-mentioned UC studies using CGRPα KO mice, including ours, may be due to some subtle factor. This issue needs to be addressed to clarify the differences in physiological functions of the two isoforms.
We found that the expression of immunoproteasome was up-regulated in CGRPβ KO mice upon DSS consumption, and administration of ONX-0914, an immunoproteasome inhibitor, improved the symptoms. We also found that CGRPβ may contribute to the suppression of UC by avoiding excessive and inappropriate immune application through the suppression of immunoproteasome expression. Immunoproteasomes play an important role in antigen presentation in the periphery by promoting the generation of peptides suitable for binding to MHC class I through their unique enzymatic activity33 High expression of the immunoproteasome subunit PSMB9 has been reported in the mucosal layer of patients with active UC34, and its potential as a therapeutic target is also being investigated, as KO mice and inhibitors improve DSS-induced UC symptoms35,36. Immunoproteasomes are induced in response to several stimuli, and the contribution of INF-γ and TNF-α has attracted particular attention37,38. Nevertheless, the top factors contributing to immunoproteasome expression in UC have not been clarified as yet, and we believe that the proposed CGRPβ-mediated regulation of immunoproteasome expression by the nervous system could be a new immunomodulatory mechanism. It needs to be investigated as to how CGRPβ contributes to the suppression of immunoproteasome expression. This might involve the role of CGRP in suppressing the expression of TNF-α and INF-γ. It would be necessary to examine the effects of CGRPβ on the secretion of TNF-α and INF-γ by effector cells, such as macrophages and Th1 cells, during colon inflammation.
In this study, we found that CGRPβ may contribute to the alleviation of symptoms during the exacerbation phase of UC by suppressing the induction of immunoproteasome expression. However, several issues remain to be addressed. Among them, the degree of symptom exacerbation in UC differs among CGRP isoform knockout mice. We expect that the tissue expression patterns of CGRPα and CGRPβ will provide important clues in this regard. Exogenous sensory neurons from the dorsal root ganglion mainly express CGRPα and innervate all layers of the intestinal wall, including the muscular and sub-mucosal plexuses, vessels of the sub-mucosal plexus, annular and longitudinal smooth muscle layers, microfold cells of Payer’s patches, and mucus-producing goblet cells in the colon. Additionally, intrinsic enteric neurons mainly express CGRPβ and are present in both the enteric and sub-mucosal plexuses39. It is speculated that the distribution of these CGRP-containing nerves may modulate the activity of the immune system associated with UC-like symptoms. Moreover, the cells that receive CGRPβ and are involved in immunoproteasome expression have not been identified. Immunohistochemical staining of immunoproteasome-expressing cells suggests that immunoproteasome may be widely distributed in mucosal surface of the colon tissue (Supplementary Fig. 7). This suggests that the induction of immunoproteasome expression in CGRPβ KO mice may have an effect that spreads throughout the intestinal epithelium. scRNA-seq of the colon tissue revealed specific expression of RAMP1, a receptor for CGRP, in goblet cells, highlighting the possibility of mucosal protection through this receptor29. Based on these results, we infer that the induction of immunoproteasome expression in CGRPβ KO mice might be due to a decrease in the intestinal barrier function. RAMP1 is also expressed in various immune cells, such as T cells, and stimulation with CGRP suppresses the production of IFN-γ, which induces immunoproteasome expression40. Therefore, CGRPβ may suppress the induction of immunoproteasome expression through its effects on the immune system. In the future, we expect that spatiotemporal analysis of neurons and immune cells in the colon tissue will lead to the resolution of the aforementioned challenges.
Conclusion
In this study, we focused on deciphering the role of CGRPβ and its implications in the pathogenesis and exacerbation of UC. The deficiency of CGRPβ was found to aggravate UC symptoms, such as bloody stools and diarrhoea. Our findings suggest that CGRPβ plays a crucial protective role in modulating UC conditions. In CGRPβ KO mice, the expression of proteins related to immune responses, particularly the immunoproteasome and antigen presentation components, was markedly increased. This increase suggests a hyperactive immune response, contributing to the severity of the UC symptoms. Administration of ONX-0914, an immunoproteasome inhibitor, significantly decreased the expression of proteins related to immune responses. This intervention markedly improved UC-like symptoms. The outcomes of this study not only advance our understanding of the protective role of CGRPβ against UC but also pave the way for devising novel therapeutic strategies focusing on the modulation of the immunoproteasome. Our findings highlight the need for a broader approach to UC treatment, combining CGRPβ enhancement with immunoproteasome inhibition to potentially synergize the suppression of inflammatory responses in UC.
Materials & methods
Mice
C57BL/6 N mice (WT) were purchased from Charles River Laboratories Japan, Inc. and bred for use in experiments. The CGRPα KO mice were previously created by Dr. Hiroki Kurihara and were acquired [41]. To create CGRPβ KO mice, we decided to delete exon 2, which contains the start codon of the CGRPβ gene, and exon 3, which has the potential to frameshift the start codon sequence. The construct shown in Supplementary Fig. 1 was created and incorporated into the pNT1.1 vector. The targeting vector was transfected into ES cells from 129/SvJ mice by electroporation, and neomycin and ganciclovir were added to the ES cell culture medium. Positive and negative selection was performed by neomycin resistance gene and thymidine kinase gene, respectively. Selected ES cells were injected using manipulators into the cavity where they could be fertilised in the placental follicle stage of C57BL/6 and placed in provisional parents to create chimeric mice. mRNA was recovered from the brains of CGRPβ-deficient and wild-type mice and genotyping was performed by RT-PCR. For genotyping, 5’-GCAGAAGTGTAAGTAGCTCC-3’ was used as forward primer and 5’-CCCGTGATATTGCTGAAGAG-3’ as reverse primer. CGRPα KO mice were backcrossed to C57BL/6 N mice for more than 10 generations. CGRPβ KO mice were backcrossed to C57BL/6 N mice for more than 8 generations. All mice were used at 8–10 weeks of age. Mice and specimens used for pathological experiments, histological validation, and proteomics may have been procured from individual mice. The number of mice used in each experiment is given in the figure legend. Mice were euthanased by inhalation of isoflurane. Animal experiments were conducted in accordance with the Osaka University Animal Experiment Regulations after application to the Animal Experiment Committee of the Graduate School of Pharmaceutical Sciences, Osaka University, and approval by review in accordance with the ARRIVE guidelines (https://arriveguidelines.org).
Dextran sodium sulphate-induced colitis
Colitis was induced in 8–10-weeks-old mice by adding 3% DSS (Fujifilm Wako Pure Chemicals) in drinking water for 5 days. Body weight, haematochezia, and faecal characteristics were measured, and DAI scores were assigned according to the score table (Table 1). These measurements were made on days 0–5 after DSS consumption. Mice were sacrificed on day 3 or day 5, and their colons were analyzed.
HE staining
Glass slides with tissue sections were dried at 37 °C for 1 h, fixed in 10% neutral buffered formalin solution (Fujifilm Wako Pure Chemicals) for 30 min at room temperature, and washed by immersing in PBS for 1 min three times. The slides were then immersed in Meyer’s haematoxylin solution (Fujifilm Wako Pure Chemicals) for 5 min and rinsed under running tap water for 5 min. They were subsequently immersed in 0.5% eosin solution for 3 min, rinsed under running water for 30 s, immersed for 1 min each in 70%, 90%, and 100% four times, and then twice in Lemosol (Fujifilm Wako Pure Chemicals Corporation) for 5 and 3 min, in this order. A cover glass (Matsunami Glass Industry) was placed on the sections and sealed using MountQuick (Daido Sangyo). The slides were dried at room temperature for 1 day and observed with a Research Slide Scanner VS200 (EVIDENT). Pathological evaluation of UC was based on HE-stained images and was quantified by scoring the status of inflammatory cell infiltration, absence of goblet cells, crypt density, crypt hyperplasia, thickening of the muscularis layer, inflammation of the sub-mucosal layer, crypt abscess, and ulcer formation.
Extraction and quantitation of proteins from the colon tissue
The colon tissue (approximately 10 mg) was lysed in 100 µL of RIPA lysis buffer containing 1 µL of protease inhibitor cocktail (Nacalai Tesque) by sonicating for 20–60 s using Ultrasonic Disrupter UD-100 (TOMY). The lysate was incubated on ice for 30 min, and centrifuged at 10,000 × g for 15 min at 4 °C. The supernatant was transferred to a 1.5 mL tube and centrifuged again under the same conditions. The supernatant, thus obtained, was transferred to another 1.5 mL tube and used as the protein extract.
Western blot analysis
The tissue lysate, adjusted to 10 µg protein content, was mixed with 6× SDS sample buffer, and heat-denatured at 95 °C for 3 min. The sample was subjected to SDS polyacrylamide gel electrophoresis (PAGE) on a 12% gel at a constant current of 10 mA. The electrophoresed proteins were then transferred from the gel onto a polyvinylidene di-fluoride (PVDF) membrane (Millipore) treated with MeOH (Fuji Film Wako Pure Chemicals) using a semi-dry system (BIORAD) at a constant voltage of 12 V for 1 h. The PVDF membrane was then blocked with a 5% skim milk in TBS-T solution for 1 h, and then incubated overnight with anti-PSMB9 (Proteintech, #14544-1-AP, 1:1000), anti-PSMB10 (Proteintech, #15976-1-AP, 1:1000), or anti-PSMB8 (Proteintech, #14859-1-AP, 1:1000) antibody at 4 °C. The next day, the membrane was washed three times with TBS-T, and incubated with HRP-linked anti-rabbit or anti-mouse IgG (Cell Signaling Technology) for 1 h at room temperature. The membrane was washed again with TBS-T, the bands were visualised using ECL Prime Western Blotting Detection Reagents (GE Healthcare), and photographed using Amersham Imager 680 (GE Healthcare).
Immunoproteasome Inhibition in mice
ONX-0914 (Cayman Chemical Company), dissolved in a mixture of 10% (w/v) Captisol (SBE-β-CD; Selleck Biotech Corporation) and 10 mM sodium citrate (pH 6), was administered subcutaneously to mice daily for 5 days at 10 mg/kg body weight.
Proteome sample Preparation
Protein (100 µg) extracted from the rectal tissue was re-dissolved in 100 µL of 100 mM Tris pH 8.5 solution and reduced with 5 µL of 200 mM tris(2-carboxyethyl)phosphine (ThermoFisher Scientific) for 1 h at 55 °C. Thereafter, 5 µL of 375 mM iodoacetamide (ThermoFisher Scientific) was added and the mixture was allowed to react for 30 min at room temperature. After adding 400 µL of methanol (Fujifilm Wako Pure Chemicals), 100 µL of chloroform (Wako Pure Chemicals), and 300 µL of water, the mixture was shaken and centrifuged at 14,000 × g for 2 min. The upper layer (80%) was removed, mixed with 300 µL of methanol, and the mixture was centrifuged at 14,000 × g for 3 min. The supernatant was removed and allowed to air dry. It was re-dissolved in 50 mM ammonium bicarbonate, and the proteins were digested by overnight reaction with 2.5 µg of trypsin (ThermoFisher Scientific) at 37 °C. An equivalent to 500 ng of peptides was subjected to nLC-MS/MS.
nLC-MS/MS analysis of the proteome
Online nLC-MS/MS was performed using an Easy-nLC 1200 system coupled with an Orbitrap Eclipse Tribrid mass spectrometer (ThermoFisher Scientific). Samples were trapped on a C18 guard-desalting column (ThermoFisher Scientific; Acclaim PepMap 100, 75 μm × 2 cm, nanoViper, C18, 5 μm, 100 Å), and separated on a 12 cm long C18 column (Nikkyo Technos, C18, 3 μm, 75 μm × 12 cm). The nano capillary solvent A consisted of 100% water and 0.1% formic acid; solvent B consisted of 20% water, 80% acetonitrile, and 0.1% formic acid. At a constant flow rate of 0.3 µL/min, the curved gradient increased from 6% B to 31% B, followed by a steep increase to 90% B over 10 min. The eluted peptide samples were analysed using a data-independent acquisition (DIA) method. For MS1, the m/z range was set from 500 to 1100, mass resolution was 24,000, AGC target was 500%, maximum injection time was 50 ms, and the data type was centroid. MS2 was acquired in the quadrupole isolation mode with the isolation window set to 10 m/z, 25% HCD collision energy, 120,000 mass resolution with Orbitrap detection, 2000% AGC target, maximum injection time, and centroid data type. Raw data files were processed in the library-free mode in DIA-NN (version 1.8.1). Trypsin/P allowed a maximum of one missed cleavage, N-terminal methionine removal for variable modifications, and fixed carbamidomethylation on cysteine. Peptide lengths ranged from 7 to 30 amino acids. The precursors ranged from 300 to 1800, and the fragment ions ranged from 200 to 1800. Both MS1 and MS2 mass accuracies were set to automatic determination. Protein inference was set to ‘Protein names (from FASTA)’ and the ‘Heuristic protein inference’ option and MBR (between-run) were employed. The RT-dependent and Robust LC (high precision) options were selected for quantification. The MS data have been deposited in the jPOST repository (jPOST ID: JPST003301).
Statistical analysis
For data related to UC standard deviations are indicated using error bars. Statistical analysis was performed using ANOVA in Python version 3.11.8, followed by Tukey’s post-hoc test for comparisons, and graphs were prepared using Python or Excel (Microsoft). Proteome analysis data were processed using LFQ values obtained from DIANN. These LFQ values were log-transformed, and missing values were imputed before PCA. Proteins with an FDR < 0.05 were identified using the ANOVA function in Perseus42. Proteins were clustered and visualized using a heatmap. Comparisons between two groups were conducted using the R package MS-DAP43. Significant differences were estimated using MS-EmpiRe, a function within MS-DAP. For enrichment analysis, fold change values were used to conduct GSEA with WebGestalt, employing the Reactome or the KEGG pathway database or R package genekitr44,45.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. The MS data have been deposited in the jPOST repository (jPOST ID: JPST003301).
References
Le Berre, C. & Honap, S. Peyrin-Biroulet, L. Ulcerative colitis. Lancet 402, 571–584 (2023).
Ordás, I., Eckmann, L., Talamini, M., Baumgart, D. C. & Sandborn, W. J. Ulcerative colitis. Lancet 380, 1606–1619 (2012).
Neurath, M. Current and emerging therapeutic targets for IBD. Nat. Rev. Gastroenterol. Hepatol. 14, 269–278 (2017).
Lawrance, I. C., Fiocchi, C. & Chakravarti, S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum. Mol. Genet. 10, 445–456 (2001).
Linggi, B. et al. Meta-analysis of gene expression disease signatures in colonic biopsy tissue from patients with ulcerative colitis. Sci. Rep. 11, 18243 (2021).
Beaudoin, M. et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723 (2013).
Ge, L. et al. Psychological stress in inflammatory bowel disease: psychoneuroimmunological insights into bidirectional gut-brain communications. Front. Immunol. 13, 1016578 (2022).
Sun, Y. et al. Stress triggers flare of inflammatory bowel disease in children and adults. Front. Pediatr. 7, 432 (2019).
Zhu, S. et al. Composition and diverse differences of intestinal microbiota in ulcerative colitis patients. Front. Cell. Infect. Microbiol. 12, 953962 (2022).
Nishihara, Y. et al. Mucosa-associated gut microbiota reflects clinical course of ulcerative colitis. Sci. Rep. 11, 13743 (2021).
Luu, L. D. W. et al. Profiling the colonic mucosal response to fecal microbiota transplantation identifies a role for GBP5 in colitis in humans and mice. Nat. Commun. 15, 2645 (2024).
Brazil, J. C., Louis, N. A. & Parkos, C. A. The role of polymorphonuclear leukocyte trafficking in the perpetuation of inflammation during inflammatory bowel disease. Inflamm. Bowel Dis. 19, 1556–1565 (2013).
Hart, A. L. et al. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology 129, 50–65 (2005).
Matsuno, H. et al. CD103 + dendritic cell function is altered in the colons of patients with ulcerative colitis. Inflamm. Bowel Dis. 23, 1524–1534 (2017).
Heller, F., Fuss, I. J., Nieuwenhuis, E. E., Blumberg, R. S. & Strober, W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 17, 629–638 (2002).
Heller, F., Fromm, A., Gitter, A. H., Mankertz, J. & Schulzke, J. D. Epithelial apoptosis is a prominent feature of the epithelial barrier disturbance in intestinal inflammation: effect of pro-inflammatory interleukin-13 on epithelial cell function. Mucosal Immunol. 1 (Suppl 1), S58–S61 (2008).
Chen, L. et al. The role of Th17 cells in inflammatory bowel disease and the research progress. Front. Immunol. 13, 1055914 (2023).
Labastida-Ramírez, A. et al. Mode and site of action of therapies targeting CGRP signaling. J. Headache Pain. 24, 125 (2023).
Edvinsson, L., Haanes, K. A., Warfvinge, K. & Krause, D. N. CGRP as the target of new migraine therapies - successful translation from bench to clinic. Nat. Rev. Neurol. 14, 338–350 (2018).
Balcziak, L. K. & Russo, A. F. Dural immune cells, CGRP, and migraine. Front. Neurol. 13, 874193 (2022).
Assas, B. M., Pennock, J. I. & Miyan, J. A. Calcitonin gene-related peptide is a key neurotransmitter in the neuro-immune axis. Front. Neurosci. 8, 23 (2014).
Rochlitzer, S. et al. The neuropeptide calcitonin gene-related peptide affects allergic airway inflammation by modulating dendritic cell function. Clin. Exp. Allergy. 41, 1609–1621 (2011).
Levite, M. Nerve-driven immunity. The direct effects of neurotransmitters on T-cell function. Ann. N Y Acad. Sci. 917, 307–321 (2000).
Mikami, N. et al. Calcitonin gene-related peptide is an important regulator of cutaneous immunity: effect on dendritic cell and T cell functions. J. Immunol. 186, 6886–6893 (2011).
Jusek, G., Reim, D., Tsujikawa, K. & Holzmann, B. Deficiency of the CGRP receptor component RAMP1 attenuates immunosuppression during the early phase of septic peritonitis. Immunobiology 217, 761–767 (2012).
Ma, W., Dumont, Y., Vercauteren, F. & Quirion, R. Lipopolysaccharide induces calcitonin gene-related peptide in the RAW264.7 macrophage cell line. Immunology 130, 399–409 (2010).
Li, F. J. et al. Calcitonin gene-related peptide is a promising marker in ulcerative colitis. Dig. Dis. Sci. 58, 686–693 (2013).
Wu, W. et al. The CGRP/macrophage axis signal facilitates inflammation recovery in the intestine. Clin. Immunol. 245, 109154 (2022).
Yang, D. et al. Nociceptor neurons direct goblet cells via a CGRP-RAMP1 axis to drive mucus production and gut barrier protection. Cell 185, 4190–4205e25 (2022).
de Jong, P. R. et al. TRPM8 on mucosal sensory nerves regulates colitogenic responses by innate immune cells via CGRP. Mucosal Immunol. 8, 491–504 (2015).
Ferrington, D. A. & Gregerson, D. S. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol. Biol. Translat Sci. 109, 75–112 (2012).
Thompson, B. J. et al. Protective roles of alpha-calcitonin and beta-calcitonin gene-related peptide in spontaneous and experimentally induced colitis. Dig. Dis. Sci. 53, 229–241 (2008).
Murata, S., Takahama, Y., Kasahara, M. & Tanaka, K. The Immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat. Immunol. 19, 923–931 (2018).
Scalavino, V. et al. A novel mechanism of Immunoproteasome regulation via miR-369-3p in intestinal inflammatory response. Int. J. Mol. Sci. 23, 13771 (2022).
Basler, M., Dajee, M., Moll, C., Groettrup, M. & Kirk, C. J. Prevention of experimental colitis by a selective inhibitor of the Immunoproteasome. J. Immunol. 185, 634–641 (2010).
Schmidt, N. et al. Targeting the proteasome: partial Inhibition of the proteasome by bortezomib or deletion of the immunosubunit LMP7 attenuates experimental colitis. Gut 59, 896–906 (2010).
Morozov, A. V. & Karpov, V. L. Proteasomes and several aspects of their heterogeneity relevant to cancer. Front. Oncol. 9, 761 (2019).
McCarthy, M. K. & Weinberg, J. B. The immunoproteasome and viral infection: a complex regulator of inflammation. Front. Microbiol. 6, 21 (2015). (2015).
Russo, A. F. & Hay, D. L. CGRP physiology, pharmacology, and therapeutic targets: migraine and beyond. Physiol. Rev. 103, 1565–1644 (2023).
Tsujikawa, K. et al. Hypertension and dysregulated Proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc. Natl. Acad. Sci. U S A. 104, 16702–16707 (2007).
Oh-hashi, Y. et al. Elevated sympathetic nervous activity in mice deficient in AlphaCGRP. Circulation Res. 89, 983–990 (2001).
Tyanova, S. et al. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13, 731–740 (2016).
Koopmans, F., Li, K. W., Klaassen, R. V. & Smit, A. B. MS-DAP platform for downstream data analysis of label-free proteomics uncovers optimal workflows in benchmark data sets and increased sensitivity in analysis of Alzheimer’s biomarker data. J. Proteome Res. 22, 374–386 (2023).
Elizarraras, J. M. et al. WebGestalt 2024: faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 52, W415–W421 (2024).
Liu, Y. & Li, G. Empowering biologists to Decode omics data: the Genekitr R package and web server. BMC Bioinform. 24, 214 (2023).
Acknowledgements
We would like to give our thanks to Dr. Kurihara for providing us with the CGRPα knock out mice.
Author information
Authors and Affiliations
Contributions
Conceptualization: TS, HH, and KT. Investigation; TS, HH, KM, and AU. Resources: KK and YU. Writing—Original Draft: TS and HH. Writing—Review & Editing: KT. Supervision: KJ. Project administration: KT. Funding acquisition: KT.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shibao, T., Hase, H., Mizokami, K. et al. CGRPβ suppresses the pathogenesis of ulcerative colitis via the immunoproteasome. Sci Rep 15, 7224 (2025). https://doi.org/10.1038/s41598-025-91933-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91933-w