Abstract
The low immunogenicity of pancreatic cancer inhibits effective antitumor immune responses, primarily due to the immune evasion mediated by low expression of the major histocompatibility complex (MHC). Through comprehensive analysis, our study identifies UPK3B as a gene closely associated with low MHC expression and low immunogenicity in pancreatic cancer. UPK3B has been reported as a marker of primary mesothelial cells, mature epicardium and promotes extracellular matrix signaling. However, the role of UPK3B in pancreatic cancer remain unclear. We found that UPK3B is highly predictive of overall survival (OS) in patients with pancreatic ductal adenocarcinoma (PDAC) and is significantly related to clinical features, immune cell infiltration, and response to immune checkpoint inhibitor (ICI) therapy. Gene enrichment analysis revealed significant downregulation of immune regulatory and BCR signaling pathways in the UPK3B high-expression group. Additionally, UPK3B is positively correlated with immunosuppressive cells, suggesting that high UPK3B expression may inhibit antitumor immune responses by promoting low MHC expression. UPK3B is also positively correlated with immune checkpoints, indicating that tumors with high UPK3B expression may not benefit from ICI therapy. Therefore, UPK3B may serve as a novel biomarker and therapeutic target for pancreatic cancer.
Similar content being viewed by others
Introduction
Pancreatic cancer, a malignant tumor of the digestive tract with poor prognosis, has been exhibiting an increasing incidence rate annually, ranking as the seventh leading cause of cancer-related mortality worldwide. The five-year survival rate remains a mere 12%1. Pancreatic ductal adenocarcinoma (PDAC) is the most prevalent type of pancreatic cancer, accounting for approximately 90% of cases. Historical evidence suggests that surgical resection is the optimal method for curing pancreatic cancer. Unfortunately, early-stage pancreatic cancer is asymptomatic, and most patients present with advanced disease upon symptom onset. Additionally, without further treatment, over 90% of patients may succumb to recurrence post-surgery2. Gemcitabine is currently the frontline chemotherapeutic agent for pancreatic cancer, yet its adjuvant efficacy is limited3 and accompanied by severe side effects, including hepatic and renal toxicity, underscoring the urgent need for more effective treatments4.
The advent and development of immunotherapy and targeted therapy strategies have significantly altered the prognosis of many malignancies, improving survival rates to varying degrees. Despite the success of these therapies in various solid tumors over the past decades, they have not shown significant benefits for pancreatic cancer patients5. This discrepancy may be attributed to the unique tumor microenvironment of pancreatic cancer6, characterized by dense fibrotic stroma, accumulation of myeloid-derived suppressor cells (MDSCs)7, low immunogenicity8, a paucity of predicted neoantigens, and low immune infiltration9, contributing to its resistance to various treatments. High immunogenic neoantigens and substantial CD8 + T cell infiltration have been detected in long-term survivors of PDAC, suggesting that neoantigen-based cancer immunotherapy could potentially benefit the survival of pancreatic cancer patients10. The major histocompatibility complex (MHC), also known as the human leukocyte antigen (HLA) complex, comprises a set of genes whose corresponding molecules play critical roles in immune responses against various intracellular pathogens11. Numerous studies have demonstrated that MHC plays a pivotal role in tumor immune evasion in malignancies such as diffuse large B-cell lymphoma, cholangiocarcinoma, and pancreatic ductal carcinoma12,13,14. While MHC class I molecules are ubiquitously expressed, 40–90% of human tumor cells exhibit downregulation or loss of MHC class I expression, leading to impaired antigen presentation and subsequently disrupting CD8 + T cell recognition of tumor antigens, which diminishes their ability to attack tumor cells15. For instance, autophagy promotes immune evasion in pancreatic cancer by degrading MHC-I in PDAC, thereby weakening the response to immunotherapy16. Consequently, elucidating the mechanisms underlying low MHC-I expression in pancreatic cancer and identifying novel effective therapeutic strategies are of paramount importance.
In this study, we identified UPK3B as an MHC-I-related prognostic gene by comparing normal pancreatic tissue and PDAC tissue. We subsequently investigated the expression, prognostic significance, and immunological associations of UPK3B in pancreatic cancer. Additionally, we employed the TIDE algorithm to indicate UPK3B as a predictive marker for ICI therapy efficacy. This research facilitates the development of UPK3B as a prognostic biomarker and predictor of immunotherapy response in pancreatic cancer patients, potentially introducing new avenues for PDAC treatment.
Materials and methods
Data collection
We downloaded bulk RNA-seq and clinical data for pancreatic cancer patients from the TCGA and GTEx projects via the UCSC Xena database (http://xena.ucsc.edu/). Additionally, we included two GEO datasets, GSE132956 and GSE28735. Additional validation datasets were obtained from the ICGC (https://dcc.icgc.org/projects/PACA-CA) and EBI (E-MTAB- 6134). Only tumor samples were retained, and samples lacking survival information (including survival time and survival status) were excluded. scRNA-seq data were sourced from previously published study17. The protein expressions in PDAC tissues were analyzed using the HPA online database (https://www.proteinatlas.org/).
Differential gene expression analysis
We performed differential gene expression analysis using the limma18 package to identify significantly differentially expressed genes in pancreatic cancer tissues compared to normal tissues. Genes with a p-value < 0.05 and |LogFC| > 1 were considered differentially expressed. Additionally, we stratified the samples based on high and low MHC molecule expression levels to identify genes significantly negatively correlated with MHC expression. Among these, UPK3B emerged as a particularly interesting candidate for further investigation due to its distinct expression pattern.
Survival analysis and Cox regression analysis
Kaplan-Meier (K-M) survival analysis was performed using the survminer and survival packages to evaluate the differences between high and low expression groups based on optimal cut-off expression levels. Additionally, univariate and multivariate Cox regression analyses were conducted to investigate the relationship between UPK3B expression, clinical prognostic indicators, and survival time in TCGA-PAAD patients (Tables 1 and 2). Only significant variables from the univariate analysis were included in the multivariate analysis.
Functional enrichment analysis
We utilized the R package clusterProfiler19 to perform Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and Gene Set Enrichment Analysis (GSEA) to explore the gene functions and potential signaling pathways associated with UPK3B.
Immune infiltration analysis
We estimated the proportions of infiltrated immune cells using various algorithms20, such as XCELL, CIBERSORT, and MCP-counter. We analyzed the correlation between UPK3B expression levels and the abundance of various immune cell types to identify significant associations. Additionally, we evaluated the tumor microenvironment using the ESTIMATE21 package, which calculates the stromal, immune, and total ESTIMATE scores based on the gene expression profiles of each patient. Furthermore, we used the TIDE score (http://tide.dfci.harvard.edu/) to predict the efficacy of immunotherapy in high- and low-UPK3B expression groups.
scRNA-seq analysis
The Seurat22 package was used to perform the single-cell RNA sequencing (scRNA-seq) analysis. We specifically investigated the expression patterns of UPK3B across different cell types, aiming to elucidate its role and potential impact on the tumor microenvironment.
Cell lines and culture
AsPC- 1, BxPC- 3, Capan2, HPAC, PANC1, Miapaca2, KPC, and CFPAC1 cell lines were purchased from ATCC as described previous23. All cell lines were incubated in DMEM (Wisent, China) supplemented with 10% fetal bovine serum (Wisent, China), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen, USA) in an incubator at 37 °C and 5% CO2. The KPC cell line was used for in vivo experiment. Proteins from cell lines were used for the immunoblot analysis.
Small-interfering RNA (siRNA) transfection of UPK3B
All siRNAs were purchased from Origen (SR325143, USA). Transfections were carried out according to the manufacturer’s instructions. Briefly, siRNA was diluted in culture medium without serum. Lipo3000 (Thermo Fisher, L3000001, USA) transfection reagent was added to diluted siRNA and mixed by vortexing. Then, the complexes were added to the cells. The efficacy of siRNA was checked by immunoblot analysis after 72 h of transfection.
Stable cell line construction
Two shRNAs (short hairpin RNAs) targeting the mouse UPB3B (KD) locus were cloned into pLV3 ltr-ZsGreen-neo-U6-UPK3B (Corues, Nanjing, China). We used a lentivirus packaging system to construct stable transformants. The medium was changed to fresh serum-free and antibiotic-free DMEM before transfection. To generate lentiviral particles, transfection was performed with pLV3 ltr (7.5 µg), pSPAX2 (3.75 µg), and pMD2. G (3.75 µg) at 70% HEK293 T cell density (Lipo3000 Invitrogen, USA). Fresh medium containing 10% FBS was added after 8 h. The supernatant containing lentiviral particles was collected at 48 h, centrifuged at 1200 rpm, filtered, and stored at − 80 °C. Cells were transfected by mixing virus solution with 2–10 µg/ml polybrene. After 48 h, we screened with antibiotic at a concentration of 2–5 µg/ml. We placed single cells into 96-well plates to obtain UPK3B knockdown clones.
Immunoblot
Following a PBS wash, the cells were extracted using RIPA lysates that contained phosphatase and proteolytic enzyme inhibitors. The BCA Kit was used to measure the protein concentration (Vazyme: E112 - 01, China). To balance the buffer, SDS-PAGE loading buffer (CWBio: CW0027, China) was utilized. After loading the samples with 20 µg of protein onto 10% SDS-PAGE gels, they were electrophoresed at 80–120 V and then transferred at 250 mA to PVDF membranes. PVDF membranes were incubated with the primary antibody for one hour at 4 °C after being treated with 5% skim milk. After rinsing the membranes with TBST, they were exposed to a secondary antibody conjugated with horseradish peroxidase (HRP) for one hour at room temperature. Vazyme’s ECL Western Blotting Detection Reagent (E412 - 02-AB, China) was used to detect the signal after the membranes were cleaned with TBST. Protein bands were observed using the ChemiDoc MP Imaging System (Bio-Rad, USA). UPK3B (Protein-tech, 15709 - 1-AP, USA), Beta-actin (Protein-tech, 66009 - 1- 1 g, USA) were used in the study. Densitometric analysis of UPK3B and beta-actin bands was performed using Image J software and normalized to beta-actin expression.
Immunohistochemistry
The immunochemistry was performed as reported previously23. In summary, the tissue sections were rehydrated using xylene and graded alcohols after being deparaffinized in distilled water for five minutes. Citrate buffer (0.01 mol/L, pH: 6.0) was used for antigen exposure and repair, and the microwave oven was set to high for two minutes and low for ten minutes. An immunohistochemistry pen was used to circle the tissue sections once they had cooled to room temperature. For ten minutes, tissue slices were treated at room temperature with Triton X- 100. The tissue sections were incubated in deionized methanol containing 3% hydrogen peroxide at room temperature for a duration of 10 min, followed by a wash with TBST solution. Next, the sections were treated with 10% goat serum for 60 min at room temperature to inhibit nonspecific antibody binding. The sections were subsequently incubated overnight with the primary antibody at 4 °C. After a washing step, the sections were exposed to horseradish peroxidase-conjugated goat anti-rabbit or mouse secondary antibodies for 1 h at room temperature. The color reaction was developed using DAB (ZSGB-BIO; ZLI- 9019) combined with hematoxylin. Finally, following dehydration through a graded series of alcohols and xylene, the sections were mounted using neutral resin. UPK3B (Protein-tech, 15709 - 1-AP, USA) was used in the study. IHC semi quantification was performed according to previous study24.
Cell viability assay
CCK8 assay (Dojindo, Kumamoto, Japan) was performed according to the manufacturer’s instructions. Briefly, 5000 cells were seeded in 96 well plates. After 24 h, 48 h, and 72 h incubation. Then cells were incubated with 10 µL CCK8 for 2 h at 37 °C. The absorbance was recorded at 450 nm.
In vivo experiment
The portal vein injection models were performed according to a previous protocol25. Equal numbers of KPC/KPC UKD cells were injected in situ into C57BL/6 mice at 8 weeks of age. Mice were anesthetized using isoflurane at a constant flow rate, and after local shaving and disinfection, a small longitudinal incision was made in the left upper abdomen, and 50 µl of cell suspension (1 * 106 cells) was slowly injected into the tail of the pancreas. The success of injection is confirmed by forming liquid bubbles at the injection site. 200ug/mouse PD-L1 (BioXcell, BE0101, USA) was provided by intraperitoneal injection. Starting from the seventh day of orthotopic tumor implantation, the drug was given again at an interval of two days after each administration. Mice were observed regularly and analyzed 21 days later.
Trans-well/migration assay
Cells were cultured until they reach 70–80% confluence. Detach cells using Trypsin-EDTA, neutralize with complete media, and resuspend in serum-free medium at 1 × 10^5 cells/mL. Place the inserts in a 24-well plate. Put 100–200 µL of cell suspension in the top chamber and 600–800 µL of serum-free medium in the lower chamber. Incubate for 24–48 h at 37 °C with 5% CO2. Use crystal violet as a stain. The staining cell was counted and analyzed. The number of cells was counted under a light microscope. At least 5 pictures were captured each well with 20x magnificent, then the number of the cells were counted and analyzed.
Flow cytometry
For the staining procedure, cells were harvested, washed one time, and subsequently incubated with blocking agent Fc-block (CD16/32 Fc-block (eBiosciences, 14–0161- 85, USA)) in PBS/1% FCS for 20 min/4 °C. After centrifugation and an additional wash-step, cell suspensions were stained in PBS/1% FCS with the following antibodies for 30 min/dark/4 °C: MHC1 (Biolegend, 329708, USA). Single stains for fluorescence compensation corrections and unstained controls for exclusion of false positive measurements were incorporated. Flow cytometry analysis was performed using a Gallios flow cytometer (Beckman Coulter, USA). Flow cytometry analysis was performed. Data were analyzed using a software package (FlowJo, LLC, version 10.4). Experiments were repeated three times.
Statistical analysis
All data analysis and visualization were performed using R software. For data with non-normal distributions and uncertain variances, group differences were analyzed using the Wilcoxon rank-sum test. Univariate and multivariate analyses were conducted using the Cox regression model. Survival differences were assessed using the log-rank test. Spearman correlation analysis was employed to analyze the relationships between variables that did not follow a normal distribution. A p-value of less than 0.05 was considered statistically significant.
Ethic statement
The methodology of this investigation adhered to the requirements established in the Helsinki Declaration. The ethics committee and institutional review board for clinical research at Zhongda Hospital (ZDKYSB077) accepted the study protocol. All human tumor tissue samples were obtained in conformity with national and institutional ethical standards. All participants signed informed consent prior to their inclusion in the study.
Results
Identification of differentially expressed genes negatively correlated with MHC
The TCGA-PAAD and GSE132956 datasets were divided into tumor and normal groups, identifying 586 and 1333 upregulated genes in the tumor groups, respectively. Furthermore, the TCGA data were categorized into high and low expression groups based on the median expression of several MHC molecules, and upregulated genes in the low MHC expression group were identified. Subsequently, using paired clinical survival data from TCGA, univariate Cox regression analysis was conducted on all genes to identify potential prognostic factors. The intersection of these gene sets resulted in a single gene, UPK3B (Fig. 1).
The clinical significance of UPK3B in pancreatic Cancer
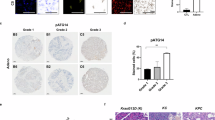
Immunohistochemical staining from the HPA database confirmed the high expression of the UPK3B protein in tumor tissues (Fig. 2). By integrating clinical information from the TCGA database, we observed that UPK3B is highly expressed in patients with pancreatitis (Fig. 3A), in high-grade pathological specimens, and in advanced pancreatic cancer (Fig. 3B). Kaplan-Meier survival analysis indicated that the high UPK3B expression group had significantly worse prognosis (Fig. 3C, D) (Tables 1 and 2).
Immunohistochemistry images of UPK3B in pancreatic cancer. (A) The immunohistochemistry picture shows the UPK3B expression in pancreatic cancer. (B) The immunohistochemistry picture shows the UPK3B expression in normal pancreas tissue. (C) The bar chart shows the semi-quantification of UPK3B expression in normal pancreas tissue and pancreatic cancer.
Clinical significance of UPK3B in pancreatic cancer patients. (A) UPK3B is highly expressed in pancreatitis and tumor samples. (B) UPK3B is highly expressed in high-grade pathological and advanced samples. (C, D) Patients with high UPK3B expression have worse overall survival (OS) and progression-free survival (PFS).
Biological pathways and immune infiltration associated with UPK3B
TCGA samples were stratified into high and low expression groups based on the median expression of UPK3B for GSEA analysis. The results demonstrated that several immune regulatory pathways were downregulated in the high UPK3B expression group (Fig. 4A, B). Immune infiltration analysis indicated that both immune and stromal scores were significantly lower in the high UPK3B expression group (Fig. 4C). Furthermore, the abundance of cytotoxic cells, macrophages, mast cells, and T cells was markedly reduced (Fig. 4D). Correlation analysis revealed that UPK3B expression was significantly positively correlated with immunosuppressive cells, such as MDSCs and CAFs, while it was significantly negatively correlated with monocytes, macrophages, and B cells (Fig. 4E).
GSEA enrichment analysis and immune infiltration analysis based on UPK3B. (A, B) Immune activation-related pathways are downregulated in the UPK3B high expression group. (C) The stromal and immune scores are lower in the UPK3B high expression group. (D, E) UPK3B expression is negatively correlated with immune-activated cells and positively correlated with immunosuppressive cells.
Prediction and validation of immunotherapy response
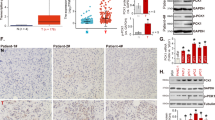
Further investigation into the correlation between UPK3B and immune checkpoints revealed that UPK3B expression was significantly positively correlated with CD274, PDCD1, and CTLA4 (Fig. 5A). TIDE analysis indicated that the high UPK3B expression group exhibited significantly elevated immune exclusion scores and MDSC infiltration, while the IFNG signaling pathway score was significantly reduced (Fig. 5B). Subsequently, the prognostic significance of UPK3B was validated in three external datasets, confirming that, consistent with the TCGA data, the high UPK3B expression group had a significantly poorer prognosis (Fig. 5C).
The characteristics of UPK3B in scRNA-seq
To further investigate the expression pattern of UPK3B in PDAC, we analyzed published single-cell datasets, revealing that UPK3B is specifically expressed in malignant cells (Fig. 6A, B). Malignant cells were then divided into UPK3B-positive and UPK3B-negative groups based on UPK3B expression. Genes highly expressed in the UPK3B-positive group were identified and subjected to enrichment analysis, which demonstrated that these genes were significantly enriched in glycolysis-related pathways (Fig. 6C, D). GSEA results indicated that immune-related pathways were significantly downregulated in the UPK3B-positive group, such as Interferon gamma response, Interferon alpha response (Fig. 6E).
Validation of UPK3B in scRNA-seq. (A) UMAP dimensionality reduction plots for various cell types. (B) UPK3B expression in malignant cells. (C, D) KEGG and GO enrichment analysis results for marker genes in UPK3B-positive malignant cells. (E) GSEA results show that immune activation-related pathways are downregulated in UPK3B-positive malignant cells.
Silencing of UPK3B inhibits cell proliferation, invasion, and migration in human pancreatic cancer cells
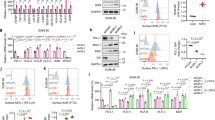
To elucidate the functional role of UPK3B in pancreatic cancer cells, we conducted comparative experiments after silencing UPK3B expression using siRNA. Considering that UPK3B is expressed at relatively high levels in Aspc- 1 and PANC1 cells (Fig. 7A), we selected these two cell lines for transfection and subsequent experiments, comparing them with a negative control group. Following siRNA transfection, UPK3B protein expression was significantly reduced in the Aspc- 1 and PANC1 cells (Fig. 7B). CCK8 assay showed that Silencing UPK3B markedly reduced the proliferation ability of Aspc- 1 and PANC1 cells compared to the control group (Fig. 7C, D). Following this, the transwell and invasion assay was applied to test the migration and invasion ability of selected cells, and the transwell assay proved that the migration ability of tumor cells was significantly reduced when UPK3B was down regulated in PANC1, which was also evident in Aspc- 1 (Fig. 7E-H). Specifically, silencing UPK3B leads to a significant increase in the expression of MHC on the surface of tumor cells (Fig. 7I, J). Furthermore, to test the impact of UPK3B in vivo, mouse KPC cell were orthotopically transplanted into C57BL/6 mice. The UPK3B knockdown KPC cell was established (Fig. 7K). Orthotopic transplantation experiment identified UPK3B knockdown significantly inhibited tumor growth than control cells, and PD-L1 treatment significantly decreased tumor growth (Fig. 7L-N). Moreover, UPK3B knockdown significantly sensitized tumors to PD-L1 treatment (Fig. 7L-N).
Influence of UPK3B on the phenotypic characteristics of pancreatic cancer cells. (A) Protein expression of UPK3B in pancreatic cancer cells. (B) The protein expression of UPK3B decreases in Ascp- 1 and PANC1 cell lines following the silencing of UPK3B. (C, D) Cell proliferation decreases after transfection of si-UPK3B. (E-H) Cell invasion and migration are inhibited after transfection of si-UPK3B. (I, J) Inhibition of UPK3B increases the expression of MHC on the cell surface. (K) The expression of UPK3B decreases in KPC cell line following knockdown of UPK3B. (L) Gross appearance of the tumor in the orthotopic injection tumor model in the con, shUPK3B, ICB, and ICB + shUPK3B groups. (M-N) Quantitative analysis showed the volume and weight of the tumor. *p < 0.05;**p < 0.01;***p < 0.001.
Discussion
Pancreatic cancer treatment faces significant challenges due to the highly fibrotic and immunosuppressive tumor microenvironment26, which impedes both chemotherapy and immunotherapy, complicating treatment. Although progress has been made with immunotherapy, the low immunogenicity and presence of immunosuppressive cells within the tumor largely result in a lack of response to treatment27. These factors contribute to poor prognosis and low survival rates, highlighting the urgent need for new, more effective therapeutic strategies.
Current research into immunosuppressive cells within the tumor microenvironment is burgeoning. Studies7,28 have shown that the abundance of Th2, M2 macrophages, Tregs, and myeloid-derived suppressor cells (MDSCs) in pancreatic cancer can block Th1 and CD8 + T lymphocyte antitumor responses, thereby promoting tumor growth. This study focuses on the low immunogenicity of pancreatic cancer, which depends on several indicators, including the major histocompatibility complex (MHC), predominantly composed of human leukocyte antigen (HLA) molecules. The interaction between MHC molecules and T cell receptors significantly influences the effectiveness of the immune response. For a robust immune response, antigens must be properly processed and presented by MHC molecules to T cells. Tumors can evade immune detection through various mechanisms, including downregulation of MHC molecules. Reduced expression of MHC Class I molecules on cancer cells impairs the presentation of tumor antigens to CD8 + T cells, leading to immune evasion29,30,31.
Recent therapeutic strategies have been developed to counteract MHC-I loss, including the use of immune checkpoint inhibitors (ICIs), autophagy inhibitors, and combinatory approaches23,32,33. Autophagy-mediated degradation of MHC-I molecules leads to reduced antigen presentation. Inhibiting autophagy has been proposed as a therapeutic strategy to restore MHC-I levels on tumor cells. For instance, chloroquine, an autophagy inhibitor, has been shown to reverse immune evasion by preventing MHC-I degradation33. Combining ICIs with autophagy inhibitors represents a promising strategy to enhance anti-tumor immunity, especially in tumors with low MHC-I expression34. Preclinical studies have demonstrated that autophagy inhibitors can improve the efficacy of ICIs by increasing MHC-I expression and promoting dendritic cell recruitment34. For example, combining autophagy inhibition with dendritic cell recruitment has shown potential in enhancing anti-tumor immunity. Additionally, combining autophagy modulators with ICIs has shown encouraging preclinical results. GNS561, a palmitoyl-protein thioesterase 1 (PPT1) inhibitor, is a promising autophagy modulator that has entered a phase 2 clinical trial in liver cancer, combined with atezolizumab and bevacizumab. This approach paves a new road, leading to the resurgence of anticancer autophagy inhibitors as an attractive therapeutic target in cancer35. Thus, targeting MHC- 1 is a promising method for the treatment of cancer.
Through comprehensive bioinformatics analysis, this study identified UPK3B as a potential key gene associated with low MHC expression. Survival analysis further validated the prognostic value of UPK3B. Gene enrichment analysis revealed significant downregulation of immune regulatory and BCR signaling pathways in the UPK3B high-expression group. Additionally, UPK3B was positively correlated with immunosuppressive cells, suggesting that high UPK3B expression may inhibit the antitumor immune response in pancreatic cancer by promoting low MHC expression. Furthermore, UPK3B was positively correlated with immune checkpoints, implying that tumors with high UPK3B expression may not benefit from immune checkpoint inhibitor therapy using checkpoint molecule-specific monoclonal antibodies. Therefore, enhancing MHC-I expression and function, and developing related targeted drugs, may improve the efficacy of immune checkpoint inhibitors in pancreatic cancer patients. scRNA-seq showed that the expression pattern of UPK3B is consistent with bulk RNA-seq. We demonstrated the role of UPK3B in promoting adverse phenotypes in pancreatic cancer cells through in vitro cellular experiments.
UPK3B has been reported as a marker of primary mesothelial cells36,37. Previous studies have mostly associated UPK3B with urothelial cells38, while a recent study indicated that UPK3B is a marker of mature epicardium and promotes extracellular matrix signaling39. Based on our findings, UPK3B may serve as a potential therapeutic target for pancreatic cancer. Its association with poor prognosis suggests that inhibiting or interfering with UPK3B function could help suppress pancreatic cancer growth and metastasis.
This study mainly relies on the analysis of gene expression and its correlation with clinical data. Although functional validation experiments have confirmed the detrimental effects of UPK3B, further animal studies are still lacking, underscoring the necessity for further experimental research to validate our conclusions. Future research directions may focus primarily on the mechanisms by which UPK3B regulates MHC-I expression.
Conclusion
We identified UPK3B as a gene strongly associated with low immunogenicity in pancreatic cancer. UPK3B exhibits a strong predictive ability for overall survival (OS) in PDAC patients and is closely related to clinical characteristics, immune cell infiltration, and responses to immune checkpoint inhibitor (ICI) therapy. Overall, our study reveals the significant role of UPK3B in pancreatic cancer and provides insights into its potential mechanisms. These findings contribute to a better understanding of pancreatic cancer progression and prognosis evaluation. UPK3B may serve as a novel biomarker and therapeutic target for PDAC.
Data availability
The datasets generated and/or analysed during the current study are available in the online repository, We downloaded bulk RNA-seq and clinical data for pancreatic cancer patients from the TCGA and GTEx projects via the UCSC Xena database (http://xena.ucsc.edu/). Additionally, we included two GEO datasets, GSE132956 and GSE28735. Additional validation datasets were obtained from the ICGC (https://dcc.icgc.org/projects/PACA-CA) and EBI (E-MTAB- 6134). Only tumor samples were retained, and samples lacking survival information (including survival time and survival status) were excluded. scRNA-seq data were sourced from previously published study[15]. The protein expressions in PDAC tissues were analyzed using the HPA online database (https://www.proteinatlas.org/).
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. Cancer J. Clin. 73 (1), 17–48 (2023).
Neoptolemos, J. P. et al. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 15 (6), 333–348 (2018).
McGuigan, A. et al. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 24 (43), 4846–4861 (2018).
Gu, Z., Du, Y., Zhao, X. & Wang, C. Tumor microenvironment and metabolic remodeling in gemcitabine-based chemoresistance of pancreatic cancer. Cancer Lett. 521, 98–108 (2021).
Henriksen, A., Dyhl-Polk, A., Chen, I. & Nielsen, D. Checkpoint inhibitors in pancreatic cancer. Cancer Treat. Rev. 78, 17–30 (2019).
Di Federico, A. et al. Immunotherapy in pancreatic cancer: why do we keep failing?? A focus on tumor immune microenvironment, predictive biomarkers and treatment outcomes. Cancers (Basel) 14(10), 24-29 (2022).
Sharma, V. et al. IL-6 is associated with expansion of myeloid-derived suppressor cells and enhanced immunosuppression in pancreatic adenocarcinoma patients. Scand. J. Immunol. 94 (6), e13107 (2021).
Schizas, D. et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat. Rev. 86, 102016 (2020).
Carstens, J. L. et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun. 8, 15095 (2017).
Balachandran, V. P. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551 (7681), 512–516 (2017).
Algarra, I., Garrido, F. & Garcia-Lora, A. M. MHC heterogeneity and response of metastases to immunotherapy. Cancer Metastasis Rev. 40 (2), 501–517 (2021).
Bourne, C. M. et al. Unmasking the suppressed immunopeptidome of EZH2-mutated diffuse large B-cell lymphomas through combination drug treatment. Blood Adv. 6 (14), 4107–4121 (2022).
Sabbatino, F. et al. PD-L1 and HLA class I antigen expression and clinical course of the disease in intrahepatic cholangiocarcinoma. Clin. Cancer Res. 22 (2), 470–478 (2016).
Imai, D. et al. The prognostic impact of programmed cell death ligand 1 and human leukocyte antigen class I in pancreatic cancer. Cancer Med. 6 (7), 1614–1626 (2017).
Yarchoan, M., Johnson, B. A. 3, Lutz, E. R., Laheru, D. A., Jaffee, E. M. & rd,, Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer. 17 (4), 209–222 (2017).
Yamamoto, K. et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581 (7806), 100–105 (2020).
Peng, J. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell. Res. 29 (9), 725–738 (2019).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16 (5), 284–287 (2012).
Sturm, G. et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 35 (14), i436–i445 (2019).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 (13), 3573–3587e3529 (2021).
Zhang, Z. et al. NDRG1 overcomes resistance to immunotherapy of pancreatic ductal adenocarcinoma through inhibiting ATG9A-dependent degradation of MHC-1. Drug Resist. Updates: Reviews Commentaries Antimicrob. Anticancer Chemother. 73, 101040 (2024).
Crowe, A. R. & Yue, W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio-protocol 9(24), e3465 (2019).
Zhang, Z. et al. AGR2-Dependent nuclear import of RNA polymerase II constitutes a specific target of pancreatic ductal adenocarcinoma in the context of Wild-Type p53. Gastroenterology 161 (5), 1601–1614e1623 (2021).
Guo, S., Contratto, M., Miller, G., Leichman, L. & Wu, J. Immunotherapy in pancreatic cancer: unleash its potential through novel combinations. World J. Clin. Oncol. 8 (3), 230–240 (2017).
Halbrook, C. J., Lyssiotis, C. A., Pasca di Magliano, M. & Maitra, A. Pancreatic cancer: advances and challenges. Cell 186 (8), 1729–1754 (2023).
Lafaro, K. J. & Melstrom, L. G. The Paradoxical web of pancreatic cancer tumor microenvironment. Am. J. Pathol. 189 (1), 44–57 (2019).
Carretero, F. J. et al. Frequent HLA class I alterations in human prostate cancer: molecular mechanisms and clinical relevance. Cancer Immunol. Immunother. 65 (1), 47–59 (2016).
Pulido, M. et al. Restoration of MHC-I on tumor cells by Fhit transfection promotes immune rejection and acts as an individualized immunotherapeutic vaccine. Cancers (Basel) 12(6), 1563 (2020).
Lim, S. Y. et al. The molecular and functional landscape of resistance to immune checkpoint Blockade in melanoma. Nat. Commun. 14 (1), 1516 (2023).
Hazini, A., Fisher, K. & Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer 9(8), e002899 (2021).
Wu, X. et al. Targeting MHC-I molecules for cancer: function, mechanism, and therapeutic prospects. Mol. Cancer. 22 (1), 194 (2023).
Oyama, K. et al. Combined autophagy Inhibition and dendritic cell recruitment induces antitumor immunity and enhances immune checkpoint Blockade sensitivity in pancreatic cancer. Cancer Res. 84 (24), 4214–4232 (2024).
Bestion, E., Raymond, E., Mezouar, S. & Halfon, P. Update on autophagy inhibitors in cancer: opening up to a therapeutic combination with immune checkpoint inhibitors. Cells 12(13), 1702 (2023).
Kanamori-Katayama, M. et al. LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS One. 6 (10), e25391 (2011).
Lennartz, M. et al. Analysis of more than 16,000 human tumor and normal tissues identifies uroplakin 3B as a useful diagnostic marker for mesothelioma and normal mesothelial cells. Diagnostics (Basel) 12(10). 2516 (2022).
Babu Munipalli, S. & Yenugu, S. Uroplakin expression in the male reproductive tract of rat. Gen. Comp. Endocrinol. 281, 153–163 (2019).
Du, J. et al. Single-cell and Spatial heterogeneity landscapes of mature epicardial cells. J. Pharm. Anal. 13 (8), 894–907 (2023).
Funding
This research was supported by grants from the National Natural Science Youth Foundation of China (82203330 to ZH.Z., 82002590 to T.C.), China Postdoctoral Science Foundation (2022M711589 to ZH.Z.). Distinguished Youth Project of Nanjing Drum Tower Hospital. Nanjing Health Science and Technology Development Project (YKK22065, YKK22267).
Author information
Authors and Affiliations
Contributions
ZH.Z. Conception and design of the research. ZH.Z., T.C., ZY.J., T.P., RJ.L., and HZ.Z. were acquisition of data. JL.S. WY. Z. was analysis and interpretation of data. NP.S. performed the statistical analysis. ZH.Z. and T.C. obtained the funding. ZH.Z. was a major contributor to writing the manuscript. ZY.J. and T.P. were revision of manuscript for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jian, Z., Pan, T., Li, R. et al. Comprehensive analysis of UPK3B as a marker for prognosis and immunity in pancreatic adenocarcinoma. Sci Rep 15, 12716 (2025). https://doi.org/10.1038/s41598-025-97213-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97213-x