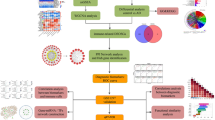
Abstract
Alzheimer’s disease (AD) is a common neurodegenerative disorder; however, its molecular complexity remains poorly understood. Single-cell analysis can reveal the molecular changes in AD in different types of brain cells. In this study, we integrated single-cell sequencing and transcriptome data to explore the molecular mechanism of integrated stress response (ISR) in AD. Analysis of the GSE264648 (49 cases) and GSE48350 (253 cases) datasets showed that the integrated stress response (ISR) activity of endothelial cells in patients with AD was significantly increased compared with normal control group. Six key genes (BTG1, EPB41L4A, HERPUD1, SLC3A2, SLC7A11, and SLC7A5) were screened by combining the Least Absolute Shrinkage and Selection Operator (LASSO) regression and the random forest algorithm. Urine test for β-amyloid protein, Clinical Dementia Rating, modified Hachinski Ischemia Scale, Hamilton Depression Scale, Hamilton Anxiety Scale and head magnetic resonance imaging were used to screen cilinical subjects, and then verified the six key genes in their blood samples. These key genes are enriched in inflammatory pathways such as NF-κB and TNF, and are closely related to immune cell infiltration (e.g., M2 macrophages and neutrophils). This research also revealed the association between key and core genes of AD (e.g., APOE) and their clinical predictive value, providing new clues for mechanistic research and targeted therapy of AD.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a global health concern that causes loss of neuropsychological function and cognitive ability. The prevalence of AD has been increasing annually with the advent of an aging society. Among the ten most common diseases that cause death worldwide, it is a disease that lacks effective prevention and treatment programs. It is of great scientific value and practical significance to study the pathogenesis of AD and explore effective prevention and treatment methods. The pathology of AD mainly involves the formation of amyloid plaques and neurofibrillary tangles, leading to neuronal damage and death1. Abnormal metabolism of amyloid precursors produces beta-amyloid proteins, which accumulate in the brain, while abnormal phosphorylation of tau protein results in the formation of tangles, affecting the function of nerve cells. In terms of epidemiology, AD is the most common type of dementia, with approximately 50 million patients worldwide, and it is expected to increase to 152 million by 2050. The incidence of AD increases with age, and approximately 10% of people over 65 years of age suffer from the disease. The incidence rate in women is higher than that in men, and both genetic and environmental factors are thought to affect the risk of the disease2.
Treatments for AD mainly includes drug therapy and non-drug interventions. So far, the drugs approved by the U.S. Food and Drug Administration for the treatment of AD mainly include acetylcholinesterase inhibitors, n-methyl-D-aspartate receptor antagonists, and anti-beta-amyloid (β-amyloid, Aβ) monoclonal antibodies3.
However, these drugs can only improve some of the clinical symptoms of AD, and it is crucial to mine more potential key genes to explore the mechanism of AD and develop AD management drugs. Non-drug interventions involve cognitive training and social activities to improve quality of life. In prognostic management, regular monitoring of cognitive and daily living abilities is essential, and family support and education are also important to help provide better care for patients. The overall goal is to delay the course of the disease and improve functional status and quality of life4.
Bioinformatics is an emerging discipline widely used in various biomedical research fields5. Bioinformatics involves the integrated analysis of multiple databases using a computer6, which provides a relatively quick and convenient research method for exploring the pathogenesis and potential targets of diseases, with particular significance for the diagnosis and treatment of patients. With continuous progress in science and technology, single-cell RNA transcriptome (scRNA-seq) technology has become increasingly mature and has been applied to embryo, tumor, stem cell, nervous system, and other fields7,8. scRNA-seq can detect differences in cellular gene transcription levels at the single-cell level, which is conducive to the discovery of new cells. Differences between tissues and cells can be determined by accurately measuring the levels of intracellular gene expression, providing a basis for the precise treatment of AD. In early embryonic research, this technology was used to track the developmental processes of embryonic cells and reveal their developmental trajectory. This technique can also be used to study cell-cell interactions and identify co-regulatory gene modules9. In terms of data processing, how to more accurately describe the biological function of cells at the molecular level and how to accurately determine cell types and define cell subsets through transcriptome data requires greater room for progress10. The application of scRNA-seq technology provides a deeper understanding of the pathophysiological mechanism of diseases and provides new ideas and targets for the diagnosis and treatment of diseases. Microarray gene chip technology has been used to hybridize DNA samples with gene chips covered with probes to detect chromosomal abnormalities at the whole-genome level. Gene chips can not only detect chromosome aneuploidy abnormalities, but can also detect chromosome copy number variations and heterozygosity loss over 50–100 kb11,12, which is of great significance for the early diagnosis of neurodegenerative diseases.
Materials and methods
Bioinformatics analysis
Data acquisition
Data from 49 GSE264648 samples (25 AD samples and 24 control samples) were downloaded from the NCBI GEO public database (https://www.ncbi.nlm.nih.gov/geo/info/datasets.html) for single-cell analyses13. The Series Matrix file of GSE48350 was downloaded from the NCBI GEO public database, and the annotation file GPL570 included the expression profile data of 253 patients, with 173 in the control group and 80 in the AD group.
Single-cell analysis
The expression profile was read in through the Seurat package, in which we filtered the cells according to the total number of unique molecular identifiers (UMIs) in each cell, the number of expressed genes, and the mitochondrial and ribosome expression ratios of each cell14. The median absolute deviation (MAD) was used for quality control15. Data with more than three times the MAD were generally considered outliers. DoubletFinder (V2.0.4) was used to filter the double cells of each sample separately, thereby completing cell quality control.
Data standardization
We used the global normalization LogNormalize method to adjust the total expression of each cell to 10,000 by multiplying a coefficient s0, and then took the logarithm for standardization16. CellCycleScoring was used to calculate the cell cycle score, and FindVariableFeatures was used to find highly variable genes17. The ScaleData function was used to remove gene expression fluctuations caused by the expression ratio of mitochondrial genes, the expression ratio of ribosomal genes, and the different cycles of cells18. RunPCA was employed to perform linear dimensionality reduction on the expression matrix and select the principal components for subsequent analysis19. Harmony was used to remove batch effects, and RunUMAP unified manifold approximation and projection (UMAP) was used for nonlinear dimensionality reduction20. The clusters were annotated using the CellMarker database, and cells that were important to the disease were annotated21. SingleR software automated annotation was used to identify the cell types and corresponding marker genes in the corresponding tissues for cell annotation22.
Ligand-receptor interaction analysis (CellChat)
In this analysis, we used the standardized single-cell expression profile as input data and the cell subtypes obtained from the single-cell analysis as cell information23. We analyzed cell-related interactions and quantified the closeness of the interaction relationship using the interaction strength (weights) and count between cells to observe the activity and influence of each cell type on the disease14.
Least absolute shrinkage and selection operator (LASSO) regression and random forest feature selection
LASSO logistic regression and random forest algorithms were used to select features for the diagnostic markers of the diseases. The LASSO algorithm uses the “glmnet” package14. We evaluated the importance of features using the random forest algorithm and %IncMSE, and selected the top ten features and LASSO feature genes for subsequent analysis.
Gene set enrichment analysis (GSEA)
Patients were divided into the high-expression group and the low-expression group based on the median expression values of key genes, and the differences in signaling pathways between the two groups were further analyzed using GSEA14. The background gene set was the annotation gene set downloaded from the MsigDB database as the annotation gene set of the subtype pathway, and differential expression analysis of the pathways between subtypes was performed. The significantly enriched gene set (adjusted p < 0.05) was ranked according to the consistency score.
Gene set variation analysis (GSVA)
In this study, we downloaded gene sets from the Molecular Signatures database and used the GSVA algorithm to comprehensively score each gene set to evaluate the differences24.
Immune infiltration analysis
The CIBERSORT algorithm was used to analyze patient data to infer the relative proportions of 22 immune-infiltrating cells and to perform correlation analysis of gene expression and immune cell content25.
Relevant clinical importance score
AlzData (http://www.alzdata.org/) is a resource website focusing on AD and other cognitive disorders, which covers the following: (1) high-throughput omics data, such as genomics (GWAS and whole exome sequencing), transcriptomics, proteomics, and functional genomics; and (2) high-confidence functional data, such as neuroimaging screening, population-based longitudinal studies, and transgenic mouse phenotyping26.
Nomogram model construction
By constructing a multivariate regression model, each value level of each influencing factor was scored according to the contribution of each influencing factor in the model to the outcome variable (size of the regression coefficient)27. The scores were then added together to obtain the total score, thereby calculating the predicted value.
Developmental trajectory of key cell subtypes
Monocle introduces a strategy for sorting single cells in pseudotime (pseudo-sequential) using the asynchronous process of single cells to place them on trajectories corresponding to biological processes such as cell differentiation27.
Clinical validation
Diagnostic criteria
The diagnosis of AD was based on the AD Core Criteria (National Institute on Aging-Alzheimer’s Association [NIA-AA], 2011)28: (1) latent onset with a clear history of progressive deterioration of cognitive function; (2) history and examination confirming one of the earliest and most significant categories of cognitive impairment (amnestic and non-amnestic symptoms); and (3) ruling out other causes. Additionally, it was necessary for the patient’s urine test to be positive for Aβ amyloid protein.
The urine β-amyloid protein detection kit (colloidal Gold lateral immunochromatography method) is produced by Hunan Gankang Technology Co., Ltd. (batch number: A20241013, license number: Xiang Food and Drug Administration Medical Device Production License No. 20160047, Registration Certificate Number). The commercial test kits of Xiangjiang Medical Device Registration (20202401261) strictly follow the procedures specified in the kit manual during the testing process. This kit achieves qualitative detection by specifically binding colloidal gold-labeled anti-Aβ monoclonal antibody to β-amyloid protein in urine samples, forming a visible color band on the nitrocellulose membrane29.
Inclusion criteria
The inclusion criteria were as follows: (1) age ≥ 50 and ≤ 80 years; (2) met the NIA-AA 2011 diagnostic criteria for AD and the urine test for β-amyloid protein was positive; (3) The Clinical Dementia Rating (CDR) score is greater than 0 and less than 2; (4) modified Hachinski Ischemia Scale (m-HIS) total score < 4; (5) Hamilton Depression Scale (HAMD) (24 items) < 8, Hamilton Anxiety Scale (HAMA) < 7 ; (6) head magnetic resonance imaging (MRI) showing hippocampal atrophy consistent with AD; (7) no obvious positive signs on nervous system examination; (8) availability of a stable and reliable caregiver to accompany the patient and assist the researchers with completing the HAMD and HAMA; and (9) provision of written informed consent by the patients and their families.
Exclusion criteria
The exclusion criteria were as follows: (1) presence of other neurological diseases that can cause brain dysfunction; (2) presence of systemic diseases that can cause cognitive impairment; (3) presence of a condition that makes the patient unable to cooperate with the completion of cognitive tests; (4) presence of contraindications to MRI; (5) presence of mental retardation or neurological impairment; and (6) refusal to provide a blood sample.
Quality control in clinical research
Inclusion and exclusion criteria were established to minimize bias in the clinical studies. The healthy control group comprised patients screened at the Health Examination Center of the Zaozhuang Hospital of Traditional Chinese Medicine. Patients in the control group were required to have a CDR of 0, and were similar to patients in the AD group in terms of age, sex, HAMA, and HAMD scores. Brain information was collected using 1.5T/3.0T MRI, and MR reports were issued by two professional doctors in the radiology department of Zaozhuang Hospital of Traditional Chinese Medicine. The participants and their families were assessed by a qualified neuropsychologist in a quiet setting, followed by screening by two neurologists (including at least one lead physician) according to the inclusion and exclusion criteria. After participants were enrolled, venous blood was collected by a nurse, and peripheral blood mononuclear cells (PBMCs) were extracted.
The study was conducted in accordance with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the Zaozhuang Hospital of Traditional Chinese Medicine (2025-syyws-001).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 20 PBMC samples, from ten patients with AD and ten healthy controls using TRIzol reagent according to the manufacturer’s instructions. RNA (1 µL) was detected with a nanometer photometer N50. Reverse transcription of mRNA was performed using a SureScript First-Strand cDNA Synthesis Kit (GeneCopoeia Inc., Rockville, MD, USA). Amplification and dissolution curves were drawn, and cycle threshold (Ct) values were determined. The primer sequences are listed in Table 1. Finally, the relative expression was calculated using the 2-∆∆Ct method. Finally, the 2-∆∆Ct value was calculated and the p-value was calculated using GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA).The method refers to previous studies30.
Statistical analysis
All statistical analyses were performed using R language (version 4.3.2). SPSS (version 19.0; IBM Corp., Armonk, NY, USA) and GraphPad Prism (version 9.4.1; GraphPad Software, San Diego, CA, USA) were used to perform statistical analyses and data mapping. The statistical significance of the differences in sex between groups was assessed using Fisher’s exact test. Age, CDR score, HAM-A score, HAM-D score, and relative mRNA expression were normally distributed, and an independent sample t-test was used. P-values (*) < 0.05 were considered statistically significant, with p-values (**) < 0.01 flagged as highly significant.
Results
Preliminary processing of single-cell expression profile data
The single-cell expression profiles used in this analysis included 25 AD samples and 24 control samples. The DoubletFinder package was used to filter doublet cells, and the feature expression levels of 178,303 cells were included for subsequent analysis (Supplementary Fig. 1A,B). The ten genes with the highest standard deviations are displayed (Supplementary Fig. 1C).
Single-cell data cell subpopulation annotation
The optimal number of principal components (PCs) was observed through ElbowPlot: 20 (Supplementary Fig. 1); principal component analysis (PCA) dimensionality reduction analysis revealed a batch effect between samples (Supplementary Fig. 1E). Harmony analysis was further used to reduce the dimensionality and remove the batch (Supplementary Fig. 1F), and 17 subtypes were obtained through UMAP (Fig. 1A). We further annotated each subtype, and 17 clusters were annotated to nine cell categories, including astrocytes, endothelial cells, smooth muscle cells, microglia, inhibitory neurons, excitatory neurons, oligodendrocytes, oligodendrocyte precursor cells, and unknowns (Fig. 1B), bubble diagrams of the classic markers of the 9 cell types (Fig. 1C), and grouping proportions (Fig. 1D).
Annotation of cells. (A) Based on the important components available in principal component analysis (PCA), we divided the cells into 17 clusters through the UMAP algorithm. (B) Cell annotation situations of 17 clusters. The 17 clusters were annotated into nine cell types, namely astrocytes, endothelial cells, smooth muscle cells, microglias, inhibitory neurons and excitatory neurons, oligodendrocytes, oligodendrocyte precursor cells, and unknowns. (C) Bubble plots of dotplots for nine types of cells and cell markers. (D) The differences in the proportion of nine types of cells in the two groups of samples.
Quantitative integrated stress response scores and differential analysis of high and low scores and cellchat
We then obtained 129 genes related to the integrated stress response from the literature31 and used AUCell to quantify the integrated stress response genes in single cells. The integrated stress response score of endothelial cells was significantly higher in the disease group than in the control group, which was used as the key cell subtype (Fig. 2A), and endothelial cells were extracted for differential analysis (Fig. 2B). The screening conditions were p_val_adj < 0.05, |logfc| > 0.25, and 202 differentially expressed genes were identified (Table PbmcMarkers-all.txt). We used the software package CellChat to analyze the ligand-receptor relationship of the features in the single-cell expression profiles of these two groups. We observed complex interactions between these cell subtypes (Fig. 2C). The ligands of High_Endothelial cells and LowEndothelial cells are shown (Fig. 2D).
Differences in ISR scores and cell communication. (A) The differences in ISR scores and the nine types of cells between the control and disease. Red represents the disease group score, and blue represents the control group score (ISR: Integrated stress score). (B) Volcano plots of differentially expressed genes, with blue indicating differentially downregulated genes and red indicating differentially up-regulated genes. (C) The cell interaction networks among nine types of cells, with the edge width representing the communication probability and intensity between cells. (D) Bubble diagrams of receptors and ligands between cells. In the figure, the communication intensity is represented by the color gradient (red to blue), and the bubble size reflects the significance. Among them, the ligand-receptor pairs marked by the red bubble (such as NAMPT-INBR) suggest the key intercellular regulatory relationship. The arrow direction indicates the signal transmission path, while the marked p-value and threshold line (such as x + 0.5) are used to determine the statistical significance of the interaction.
LASSO regression and random forest feature selection
To further identify the key genes that affect AD among the integrated stress-related genes, the 202 differentially expressed genes obtained in the previous step and the integrated stress-related genes were intersected to obtain 11 genes (Fig. 3A). Characteristic AD genes were screened using LASSO regression and random forest. LASSO regression identified seven genes as characteristic genes of AD (Fig. 3B,C); however, we screened the characteristic genes in AD through random forest, and took the top ten genes as the characteristic genes of AD (Fig. 3D). We then took the intersection with the characteristic genes screened by the LASSO regression algorithm, and screened out six intersection genes in total (Fig. 3E), which were used as key genes for subsequent research: BTG1, EPB41L4A, HERPUD1, SLC3A2, SLC7A11, and SLC7A5.
Construction of LASSO and random forest models. (A) Venn diagram of the intersection of ISR genes and differentially expressed genes. (B) Take the intersection of the LASSO regression algorithm and the characteristic genes screened out by the random forest. (C) The distribution of LASSO coefficients and the gene combination at the minimum lambda value. (D) Ten cross-validations for the selection of tuning parameters in the LASSO model to determine the minimum lambda value. (E) Characteristic genes of the random forests.
Signaling pathways involved in key genes
Next, we studied the specific signaling pathways involved in key genes and explored the potential molecular mechanisms through which these genes affect disease progression. The GSEA results showed that BTG1 was enriched in signaling pathways such as the NF-kappa B, TNF, and IL-17 signaling pathways (Fig. 4A). EPB41L4A was enriched in signaling pathways such as the PPAR, NF-kappa B, and Notch signaling pathways (Fig. 4B). HERPUD1 was enriched in signaling pathways such as the NF-kappa B, IL-17, and p53 signaling pathways (Fig. 4C). SLC3A2 was enriched in signaling pathways such as the NF-kappa B, TNF, and PPAR signaling pathways (Fig. 4D). SLC7A11 was enriched in signaling pathways such as the PPAR, Notch, and NF-kappa B signaling pathways (Fig. 4E). SLC7A5 was enriched in signaling pathways such as the PI3K-Akt, NF-kappa B, and TGF-beta signaling pathways (Fig. 4F). The GSVA results were shown in Fig. 5. BTG1 was enriched in signaling pathways such as IL6_JAK_STAT3_signaling and P53_pathway. EPB41L4A was enriched in signaling pathways such as INTERFERON_ALPHA_RESPONSE and EPITHELIAL_MESENCHYMAL _TRANSITION. HERPUD1 is enriched in signaling pathways such as ANDROGEN_RESPONSE and WNT_BETA_CATENIN_signaling. SLC3A2 is enriched in signaling pathways such as ESTROGEN_RESPONSE_LATE and INTERFERON_GAMMA_RESPONSE. SLC7A11 is enriched in signal pathways such as COAGULATION and NOTCH_signaling. SLC7A5 is enriched in signal pathways such as APICAL_JUNCTION and HYPOXIA.
GSEA analysis of key genes. (A–F) The KEGG signaling pathways involved in the key genes, as well as the pathway regulation and the genes involved. KEGG is developed by Kanehisa Laboratories. Reference: Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. and Ishiguro-Watanabe, M.; KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672-D677 (2025). https://doi.org/10.1093/nar/gkae909.
Immune infiltration analysis
The microenvironment is mainly composed of fibroblasts, immune cells, the extracellular matrix, multiple growth factors, inflammatory factors, and unique physical and chemical characteristics. The microenvironment significantly affects the diagnosis, survival outcomes, and clinical treatment sensitivity of the disease. We demonstrated the distribution of immune cell infiltration levels and immune cell correlations in different forms (Fig. 6A, Supplementary Fig. 2). The levels of macrophages M2, neutrophils, and CD4 resting memory T cells in the disease group were significantly higher than those in the control group, while the levels of activated Natural killer (NK) cells and follicular helper T cells were significantly lower (Fig. 6B). We further explored the relationship between key genes and immune cells and found that BTG1 was significantly positively correlated with monocytes and neutrophils, and significantly negatively correlated with memory B cells and activated NK cells (Fig. 6C). EPB41L4A was significantly positively correlated with macrophages M2 and eosinophils, and significantly negatively correlated with follicular helper T cells and macrophages M0 (Fig. 6D). HERPUD1 was significantly positively correlated with CD4 resting memory T cells and neutrophils, and significantly negatively correlated with naive CD4 T cells and activated NK cells (Fig. 6E). SLC3A2 was significantly positively correlated with CD4 resting memory T cells and macrophages M1, and significantly negatively correlated with CD8 T cells and follicular helper T cells (Fig. 6F). SLC7A11 was significantly positively correlated with macrophages M2 and activated dendritic cells, and significantly negatively correlated with follicular helper T cells and macrophages M0 (Fig. 6G). SLC7A5 was significantly positively correlated with CD4 resting memory T cells and neutrophils, and significantly negatively correlated with CD8 T cells and dendritic cells (Fig. 6H). We also conducted co-expression analyses of key genes and different immune factors, including immunosuppressive factors, immunostimulatory factors, chemokines, and receptors. These analyses suggested that the key genes are closely related to the level of immune cell infiltration (Fig. 7A–E).
Analysis of immune infiltration. (A) The vertical axis represents the relative percentage of immune cell subsets, and the horizontal axis represents different samples. (B) The difference in immune cell content between the control and disease samples. (C–H) The correlation between the key genes and immune cells. The larger the point is, the greater the correlation is. The greener the point is, the smaller and more significant the p-value is.
Relationship between key genes and disease-related genes
Disease regulation-related genes were identified using the GeneCards database (https://www.genecards.org/). The expression levels of 20 genes ranked high in Relevance*score and expressed in the transcriptome were analyzed, and it was found that the expression of A2M, APP, BACE1, GRN, MAPT, PLAU, PRNP, PSEN2, SORL1, TARDBP, TOMM40, TREM2, VCP was significantly different between the two groups of patients. We then performed a correlation analysis of the key genes and disease-regulated genes. The expression levels of key genes were significantly correlated with those of the disease-regulated genes. Among them, SLC7A11 and APOE were significantly positively correlated (cor = 0.732), whereas SLC3A2 and PRNP were significantly negatively correlated (cor = − 0.494) (Fig. 8).
Key gene-related clinical importance scores and nomogram model
We analyzed the relevant clinical importance scores of key genes through the official AlzData website, obtained supplementary CFG scores and rankings, and displayed single-cell RNA sequencing of key genes in different cell types of the human brain (Fig. 9A–F). We present the results of the regression analysis in the form of a nomogram based on the expression of key genes. The results of the regression analysis showed that, in all of our samples, the values of different clinical indicators of AD and the distribution of key gene expression showed varying degrees of contribution in the entire scoring process (Fig. 10A). The nomogram model had good predictive performance, and the ROC curve was drawn (Fig. 10B).
Expression profile of key genes in single cell data and immunometabolic pathways
In this study, we analyzed the expression of key genes in single cells and demonstrated the expression of BTG1, EPB41L4A, HERPUD1, SLC3A2, SLC7A11, and SLC7A5 in astrocytes, endothelial cells, smooth muscle cells, microglia, inhibitory neurons, excitatory neurons, oligodendrocytes, oligodendrocyte precursors. cells, and unknowns (Fig. 11A, B). We used AUCell to quantitatively score immune metabolism-related pathway genes in single cells and employed bubble charts to display the expression differences of key genes in immune metabolism-related pathways32. The results showed that HERPUD1 was enriched in pathways such as hedgehog and hedgehog signaling; SLC7A5 was enriched in hedgehog signaling; EPB41L4A was enriched in pathways such as hedgehog_signaling; BTG1 was enriched in pathways such as hedgehog_signaling; SLC7A11 was enriched in pathways such as notch_signaling; and SLC3A2 was enriched in pathways such as hedgehog_signaling (Fig. 11C).
Differences in the expression of key genes in single cells and the activity of immune metabolic pathways. (A) Scatter plot of the expression profile of key genes in a single cell. Orange-red indicates that the gene is expressed in this cell. (B) A violin plot of the expression profile of key genes in a single cell. One dot represents one cell. (C) Differences in the activity of key genes and immune metabolic pathways, with blue indicating low expression and red indicating high expression. The larger the dot, the smaller the p-value.
Pseudo-chronological analysis of key cells
Endothelial cells were extracted in the next step of analysis. First, we calculated the similarity between the cells and constructed a cell differentiation trajectory. Subsequently, by visualizing the trajectory, a picture of the cell differentiation trajectory constructed over time can be generated to show the developmental process of the cells, which can be used to study the process of cell differentiation and gene expression patterns at different time points. The pseudotime values are output. Pseudotime is the probability calculated by monocle2 based on cell gene expression information, indicating the order of time. Images of cells colored according to cell type (Fig. 12A,B). These genes were differentially expressed in different branches. However, this cannot be displayed on the overall heat map. We selected all branch points, identified genes with large differences in cell expression before and after the branch point, and visualized them in a branch heat map. The trajectory starts from the middle and differentiates to both sides, representing the differences at both ends of the branch points respectively. The pre-branch of the Cell Type in the legend represents the data before the branch point. Cell fate1 is the side with the smallest state after the branch point. Cell fate2 is the side with the largest state after the branch point. The default classification was based on gene changes. Six clusters (Fig. 12C) showed gene expression changes in key genes from the beginning to the end of the pseudo-time course (Fig. 12D).
Validation of key gene expression in patients with AD
To further verify the changes in the expression levels of key genes in the AD population, we screened and tested patients who visited the hospital and healthy controls who underwent physical examinations at the physical examination center. Ten patients with AD and ten healthy controls were screened. The baseline information is shown in Table 2. There were no significant differences between the two groups in terms of age, gender, HAMA score and HAMD score. The CDR score of AD patients was higher than that of the control group (p** < 0.01). The RT-qPCR results (Fig. 13) showed that the relative expression level of HERPUD1 mRNA in AD patients was higher than that in the control group (p **< 0.01), and the relative expression level of BTG1, EPB41L4A, SLC3A2, SLC7A11, and SLC7A5 mRNA in AD patients was lower than that in the control group (p*<0.05), which was consistent with the results of bioinformatics analysis.
Discussion
Neurodegenerative diseases involve a series of complex changes that occur at the molecular level during the evolution, development, maturation, and aging of neural tissue. Researchers do not fully understand this series of dynamic changes and their mechanisms, and effective treatment methods remain limited33. AD progresses from undetectable brain changes in patients to severe memory and thinking disorders, ultimately leading to a decline in living ability, known as the Alzheimer’s continuum34. An increasing number of studies have shown that the occurrence, development, and phenotypes of AD are associated with genetics35. Indeed, genetic susceptibility plays an important role in the occurrence and development of AD under the same environmental conditions36. There are also studies that have found Aβ-protein-precursor (AβPP)-derived intraneuronal Aβ (iAβ) accumulated to neuronal integrated stress response (ISR)-eliciting levels trigger AD, and iAβ produced independently of AβPP drives the disease37. Therefore, extensive research needs to be conducted to discover and explore key hub genes related to AD. With advancements in science and technology, bioinformatics, and the human genome, many AD-related genes have been discovered. However, due to differences in research methods, populations, disease classifications, and phenotypes, more rigorous studies are needed to verify the relevant conclusions. Currently, life science and computer analytical methods are widely applied in genetic analysis and screening.
This study was based on the GEO database. Single-cell data (GSE264648) were downloaded. The integrated stress response genes were quantitatively scored in single cells using AUCell, and endothelial cells were determined as the key cell subtype. By taking the intersection of 202 differentially expressed genes and integrated stress-related genes, six AD-characteristic genes were screened through LASSO regression and random forest: BTG1, EPB41L4A, HERPUD1, SLC3A2, SLC7A11, and SLC7A5. Further verification was performed by collecting blood samples from clinical patients. The PCR results showed a gene expression trend consistent with the results of the bioinformatics analysis.
BTG1 is a negative regulator of the cell cycle that inhibits cyclin D1 and controls the transition from G1 to S38. BTG1 is essential for maintaining the dentate gyrus in the hippocampus. Knockout (KO) of the BTG1 gene can present characteristics of early neurogenic aging. Adult BTG1 KO mice showed impaired associative memory in the hippocampus. The increase in progenitor cell proliferation leads to the generation of new neurons, which integrate into existing memory circuits and enhance the performance of the hippocampus in memory-dependent and learning tasks, including spatial memory and spatial pattern separation, novel object recognition, and situational fear-conditioned reflexes39,40,41.
EPB41L4A is a long non-coding RNA (lncRNA). LncRNAs play an important role in the physiological and pathological processes of AD by interacting with transcription factors, epigenetic alterations, and the separation of RNA or proteins39. Defective autophagy is a pathogenic factor of neurodegenerative diseases. In patients with AD, autophagy plays a role in clearing intracellular Aβ and MAPT/TAU tangles. Downregulation of EPB41L4A in patients with AD is associated with lower expression of multiple autophagy-related genes42,43,44. This is achieved by reducing the expression level of histone acetyltransferase GCN5L2, thereby inhibiting the enrichment of acetylation, barbuylation, and lactation of these autophagy-related genomic proteins, and thus achieving the inhibition of autophagy-mediated Aβ clearance45. The clearance and accumulation of Aβ-mediated by autophagy may be due to the dysfunction of the lysosomal pathway. When EPB41L4A is downregulated, the expression of the lysosomal markers LAMP1, CTSB, and CTSD is inhibited, suggesting that EPB41L4A-AS1 plays a role in maintaining lysosomal function46.
Homocysteine induces the conversion of endoplasmic reticulum protein (HERPUD1) into an ER-associated degradation complex (ERAD) with ubiquitin-like domain 1, which participates in the ubiquitination and relocalization of ERAD substrates47,48,49,50. HERPUD1 is involved in the regulation of inflammation-related diseases via ERS, including neurodegenerative changes caused by unknown neuroinflammation-related proteins51. In terms of regulating endothelial cell apoptosis, upregulation of HERPUD1 promotes the induction of this injury event by angiotensin II52. In neurons and glioma cells, HERPUD1 promotes cell survival under endoplasmic reticulum stress by inhibiting apoptosis53,54. KO of HERPUD1 can increase the expression of caspase-3, the most important terminal-cleaving enzyme during apoptosis, reduce the activity of caspase-12, and alleviate caspase-12-mediated apoptosis55.
SLC3A2 is a type 2 transmembrane glycoprotein that forms disulfide-linked heterodimers with amino acid exchangers, including SLC7A5 and SLC7A1156, bringing N-glycans into the heterodimers. SLC3A2 is involved in mediating cystine uptake in exchange for glutamic acid output, providing cysteine for the biosynthesis of glutathione (GSH), and alleviating oxidation-induced ferroptosis- or pyroptosis-induced cell death57,58,59. SLC7A5 balances essential intracellular amino acids and metabolic requirements. A recent study has shown that the absence of SLC3A2 can lead to a serious imbalance in amino acid levels within cells; more specifically, it can cause a decrease in leucine concentration. Changes in SLC3A2 levels may affect cell growth and survival by regulating leucine-mediated mTOR activation. The degradation of SLC3A2 further activates the iron deposition pathway, promoting endothelial cells (EC) damage, and ultimately accelerating atherosclerosis. The regulation of system XC−, involving SLC3A2 and SLC7A11, plays an important role in regulating the intracellular transport of cysteine. Cysteine, as a substrate, supports GSH synthesis in cells. Activation of SLC3A2 and SLC7A11 enables cells to restore REDOX homeostasis and maintain survival under stress conditions60. Conversely, disrupting the XC-system can lead to glutathione depletion, endoplasmic reticulum stress, and ultimately, cell death61.
Through GSEA, it was found that the NF-kappa B signaling pathway is a common molecular mechanism of these six characteristic genes. The NF-κB signaling pathway is related to multiple mechanisms for the formation of AD, including the beta-amyloid protein cascade reaction, the neuroinflammatory hypothesis, and the gut microbiota theory. Studies have shown that peroxisome proliferator-activated receptor (PPARγ) is a ligand-activated transcription factor, and NF-κB is the key factor exerting the anti-inflammatory activity of PPARγ62. Experiments on Aβ overactivated microglia models and APPswe/PS1E9 transgenic mice revealed that PPARγ reduced the number of activated microglia in mice by inhibiting the AMPK/NF-κB pathway, suggesting that the NF-κB signaling pathway is related to the deposition of Aβ63. NF-κB proteins usually form dimer complexes, including an inhibitor of NF-κB (IκB)α and other related anchor protein repeat sequences. When cells are stimulated accordingly, IκB kinase is phosphorylated, leading to the degradation of IκB and the release of NF-κB dimers, which then translocate into the cell nucleus. After the activated NF-κB dimers enter the cell nucleus, they induce the transcription of related inflammatory mediators and immune genes, thereby promoting the expression of inflammatory factors and chemokines in cells, ultimately activating the inflammatory response64,65. Inhibiting the NF-κB-related signaling pathway can reduce the production of inflammatory factors and improve cognitive impairment in patients with AD66. Some bacteria in the intestinal flora secrete large amounts of lipopolysaccharide (LPS). LPS is a potent inducer of the pro-inflammatory transcription factor NF-κB(p50/p65) complex, which can bind to the TLR4 receptor and activate the downstream NF-κB pathway, thereby releasing inflammatory cytokines, such as IL-1 and IL-6, ultimately leading to the occurrence and development of AD67.
This study focused on immune infiltration in AD and presented the distribution of immune infiltration levels and the correlation between immune cells in different forms. These analyses suggest that the key genes are closely related to the infiltration level of immune cells and play an important role in the immune microenvironment. An increasing number of studies have found that AD is closely related to immune inflammation, providing a new direction and potential therapeutic target for the treatment of AD68. The inflammatory response is a defense response of the body to abnormal substances, with the aim of eliminating harmful substances and promoting tissue repair. Multiple studies have suggested an important role of T cell immune infiltration in the occurrence and development of AD; that is, T cells enter the central nervous system through processes such as the blood-brain barrier from the peripheral circulatory system, triggering abnormal neuroimmune responses69. Research on neurological diseases has shown that microglial activation in patients with AD damages blood vessels and parenchymal tissue. The use of immunosuppressive drugs may suppress brain inflammation, Aβ deposition, and tau hyperphosphorylation in AD. Further clarification of the immune mechanisms of AD may contribute to its treatment70. Currently, many studies have been conducted on the mechanism, regulation, and potential therapeutic targets of immunity against AD. However, further studies are needed to clarify the detailed mechanisms and provide more accurate treatment ideas for clinical practice. Through immune infiltration analysis, we found that the infiltration degrees of macrophages M2, neutrophils, and CD4 resting memory T cells in AD were significantly higher than those in the control group, with key genes playing a role. BTG1, EPB41L4A, HERPUD1, SLC3A2, SLC7A11, and SLC7A5 may be involved in the occurrence and development of AD through immune inflammation. However, there are currently no studies on key genes related to AD in the immune aspect. Therefore, it is worth exploring the related mechanisms of key genes in AD.
We analyzed the expression of key genes in single cells and quantitatively scored the genes related to immune metabolic pathways in single cells using AUCell. The results showed that the key genes were generally highly active in hedgehog-signaling and notch-signaling. Typical Hh signals are spontaneously initiated by active Hh ligands. After the mature Hh ligand binds to Ptch1/271, it inhibits key protein kinases promoted by growth inhibitory specificity 1 (GAS1), oncogene (CDO), CDO sibling genes (BOC), and low-density lipoprotein receptor-related protein 2 (LRP2)., including protein kinase A (PKA), casein kinase 1 (CK1), and synthase kinase 3β (GSK3β)72,73, and deprives them of the ability to mediate the phosphorylation of the SUFU-Gli complex. Hh signals include PTCH-HH-mediated (Type I), SMO-dependent (Type II), and GLI-activated (Type III) signals74. Ptch1 can regulate apoptosis by recruiting pro-apoptotic elements, such as caspase-9, dependent-related receptor transmembrane (DRAL), and tumor-upregulated caspase-9 antagonists (TUCAN) into its C-terminal domain. Most atypical Hh signaling axes do not rely on Ptch binding. By coupling SMO, a G protein-coupled receptor, to heterotrimeric G proteins of the G inhibition (Gi) family75, the activities of several key protein kinases, including PKA, Ras homologous A (RhoA), RAS-related C3 botulinum toxin substrate 1 (Rac1), Neurofibromatosis type 1 (NF-1), and Src are altered. PI3K/PLCγ and calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2)76,77,78,79 promote changes in actin rearrangement, immune synaptic formation, and glycolytic capacity80,81.
Notch and Hh signals play a synergistic role in the cellular development of multiple systems such as the digestive system, nervous system, and immune system82,83,84. Notch signaling regulates Hh85, and Hh signaling can directly or indirectly control Notch signaling through downstream activity86. Regulation of the Notch pathway by Hh includes the following aspects: downstream effectors of Hh control the transcription of ligands and regulators of the Notch pathway, such as marginal proteins. The Gli protein exerts immediate and Notch-independent transcriptional regulation on certain Notch target genes87,88. Hh is controlled by Notch signaling in the following ways: The downstream effectors of Notch control the transport of Hh components (e.g., Ptch and Smo) to PC89. The downstream effector of Notch controls Gli levels independent of transcription. Notch intracellular domain (NICD) plays a direct role in the transcriptional regulation of Gli genes90. Furthermore, both Notch and Hh signals are inhibited by NUMB87, which helps Notch1 to interact directly with the WW domain of itchy E3 ubiquitin ligase (ITCH) through ubiquitination and degradation of its polypyrimidine beam-binding protein (PTB) domain91. Similarly, NUMB directly inhibits the hedgehog pathway, while ITCH promotes the ubiquitination of Glis192.
Conclusion
In this study, the genetic information of AD was analyzed using life science and computer analysis methods. The six key screened genes were verified in clinical samples. However, there was a limitation to the clinical sample size of this study. Moreover, the regulatory roles of the NF-kappa B signaling pathway, hedgehog-signaling, and notch-signaling in the AD continuum were not further elucidated. We plan to improve the research design in future studies and explore the disease mechanisms and potential intervention targets in greater depth.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available because they protect patient privacy, but are available from the corresponding author upon reasonable request.
References
Rajendran, K. & Krishnan, U. M. Biomarkers in alzheimer’s disease. Clin. Chim. Acta. 562, 119857. https://doi.org/10.1016/j.cca.2024.119857 (2024).
Preethy, H. A., Rajendran, K., Sukumar, A. J. & Krishnan, U. M. Emerging paradigms in alzheimer’s therapy. Eur. J. Pharmacol. 981, 176872. https://doi.org/10.1016/j.ejphar.2024.176872 (2024).
Cummings, J., Osse, A. M. L., Cammann, D., Powell, J. & Chen, J. Anti-amyloid monoclonal antibodies for the treatment of alzheimer’s disease. BioDrugs 38 (1), 5–22. https://doi.org/10.1007/s40259-023-00633-2 (2024).
Newby, D. et al. Artificial intelligence for dementia prevention. Alzheimers Dement. 19 (12), 5952–5969. https://doi.org/10.1002/alz.13463 (2023).
Suvà, M. L. & Tirosh, I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell. 75 (1), 7–12. https://doi.org/10.1016/j.molcel.2019.05.003 (2019).
Giladi, A. & Amit, I. Single-cell genomics: A stepping stone for future immunology discoveries. Cell 172 (1–2), 14–21. https://doi.org/10.1016/j.cell.2017.11.011 (2018).
Zhang, S., Li, X., Lin, J., Lin, Q. & Wong, K. C. Review of single-cell RNA-seq data clustering for cell-type identification and characterization. RNA 29 (5), 517–530. https://doi.org/10.1261/rna.078965.121 (2023).
Su, M. et al. Data analysis guidelines for single-cell RNA-seq in biomedical studies and clinical applications. Mil Med. Res. 9 (1), 68. https://doi.org/10.1186/s40779-022-00434-8 (2022). Published online.
Lieberman, B. et al. Toward uncharted territory of cellular heterogeneity: advances and applications of single-cell RNA-seq. J. Transl Genet. Genom. 5, 1–21. https://doi.org/10.20517/jtgg.2020.51 (2021).
Birnbaum, K. D. Power in numbers: single-cell RNA-seq strategies to dissect complex tissues. Annu. Rev. Genet. 52, 203–221. https://doi.org/10.1146/annurev-genet-120417-031247 (2018).
Singh, A. & Hermann, B. P. Bulk and single-cell RNA-Seq analyses for studies of spermatogonia. Methods Mol. Biol. 2656, 37–70. https://doi.org/10.1007/978-1-0716-3139-3_4 (2023).
Fishman, L. et al. Cell-type-specific mRNA transcription and degradation kinetics in zebrafish embryogenesis from metabolically labeled single-cell RNA-seq. Nat. Commun. 15 (1), 3104. https://doi.org/10.1038/s41467-024-47290-9 (2024).
Fancy, N. N. et al. Characterisation of premature cell senescence in alzheimer’s disease using single nuclear transcriptomics. Acta Neuropathol. 147 (1), 78. https://doi.org/10.1007/s00401-024-02727-9 (2024).
Bohai Feng, D. et al. Ligand-receptor interactions combined with histopathology for improved prognostic modeling in HPV-negative head and neck squamous cell carcinoma. NPJ Precis Oncol. 9, 57. https://doi.org/10.1038/s41698-025-00844-6 (2025).
Voloh, B., Watson, M. R., König, S. & Thilo Womelsdorf. MAD saccade: statistically robust saccade threshold estimation via the median absolute deviation. J. Eye Mov. Res. 12 (8). https://doi.org/10.16910/jemr.12.8.3 (2019).
Soumita Seth, S., Mallik, T., Bhadra, Z. & Zhao Dimensionality reduction and Louvain agglomerative hierarchical clustering for Cluster-Specified frequent biomarker discovery in Single-Cell sequencing data. Front. Genet. 13, 828479. https://doi.org/10.3389/fgene.2022.828479 (2022).
Kotb Abdelmohsen, K. et al. Identification of senescent cell subpopulations by CITE‐seq analysis. Aging Cell. 23 (11), e14297. https://doi.org/10.1111/acel.14297 (2024).
Zhang, T. Q., Chen, Y., Liu, Y., Lin, W. H. & Wang, J. W. Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat Commun. 12, 2053. https://doi.org/10.1038/s41467-021-22352-4 (2021).
Pei, Y. et al. Aging and head and neck cancer insights from single cell and spatial transcriptomic analyses. Discov. Oncol. 15, 801. https://doi.org/10.1007/s12672-024-01672-z (2024).
Yuhan Hao, S. et al. Integrated analysis of multimodal single-cell data. Cell 184 (13), 3573–3587e29. https://doi.org/10.1016/j.cell.2021.04.048 (2021).
Hu, C. et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51 (D1), D870–D876. https://doi.org/10.1093/nar/gkac947 (2023).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172. https://doi.org/10.1038/s41590-018-0276-y (2019).
Rachael, G. et al. Clustering-independent Estimation of cell abundances in bulk tissues using single-cell RNA-seq data. Cell. Rep. Methods. 18 (11), 100905. https://doi.org/10.1016/j.crmeth.2024.100905 (2024).
Zun Mao, Z. et al. Mitotic catastrophe heterogeneity: implications for prognosis and immunotherapy in hepatocellular carcinoma. Front. Immunol. 15, 1409448. https://doi.org/10.3389/fimmu.2024.1409448 (2024).
Craven, K. E., Gökmen-Polar, Y. & Badve, S. S. CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer. Sci. Rep. 11 (1), 4691. https://doi.org/10.1038/s41598-021-83913-7 (2021).
Yingying Shi, Z., Chen, L., Huang, Y., Gong & Lu Shi. A network pharmacology approach to reveal the key ingredients in Scrophulariae Radix (SR) and their effects against Alzheimer’s disease. Heliyon 10 (3), e24785. https://doi.org/10.1016/j.heliyon.2024.e24785 (2024).
Jing Du, Y. et al. Single-cell transcriptomics reveal the prognostic roles of epithelial and T cells and DNA methylation-based prognostic models in pancreatic cancer. Clin. Epigenetics. 16, 188. https://doi.org/10.1186/s13148-024-01800-0 (2024).
Jack, C. R. Jr. et al. NIA-AA research framework: toward a biological definition of Alzheimer‘s disease. Alzheimers Dement. 14 (4), 535–562. https://doi.org/10.1016/j.jalz.2018.02.018 (2018).
Gu, B. J. et al. Fragments of amyloid precursor protein in urine are novel and practical biomarkers for early screening of dementia. Alzheimers Dem. 19 (S14), e075205. https://doi.org/10.1002/alz.075205 (2023).
Zhang, P. L. D. K. X. et al. Temporal Effects of disease signature genes and core mechanisms in the hyperacute phase of acute ischemic stroke: a bioinformatics analysis and experimental validation. Mediators Inflamm. 2025 6808184. https://doi.org/10.1155/mi/6808184 (2025).
Han, S. et al. Mitochondrial integrated stress response controls lung epithelial cell fate. Nature 620 (7975), 890–897. https://doi.org/10.1038/s41586-023-06423-8 (2023).
Xu, Y., Lin, P., Zhu, Y., Zhang, Q. & Zhou, J. Applying integrated transcriptome and single-cell sequencing analysis to develop a prognostic signature based on M2-like tumor-associated macrophages for breast cancer. Discov Oncol. 16 (1), 389. https://doi.org/10.1007/s12672-025-02161-7 (2025).
Scheltens, P. et al. Alzheimer‘s disease. Lancet 397 (10284), 1577–1590. https://doi.org/10.1016/S0140-6736(20)32205-4 (2021).
Alzheimer’s Association report. 2024 alzheimer’s disease facts and figures. Alzheimers Dement. 20 (5), 3708–3821. https://doi.org/10.1002/alz.13809 (2024).
Sargurupremraj, M. et al. Genetic complexities of cerebral small vessel disease, blood pressure, and dementia. JAMA Netw. Open. 7 (5), e2412824. https://doi.org/10.1001/jamanetworkopen.2024.12824 (2024). Published online.
Livingston, G. et al. Dementia prevention, intervention, and care: 2020 report of the lancet commission. Lancet 396 (10248), 413–446. https://doi.org/10.1016/S0140-6736(20)30367-6 (2020).
Volloch, V. & Rits-Volloch, S. Alzheimer’s is a multiform disease of sustained neuronal integrated stress response driven by the C99 fragment generated independently of AbetaPP; proteolytic production of Abeta is suppressed in AD-Affected neurons: evolution of a theory. Int. J. Mol. Sci. 26 (9), 4252. https://doi.org/10.3390/ijms26094252 (2025).
Ceccarelli, M. et al. Altered cerebellum development and impaired motor coordination in mice lacking the Btg1 gene: involvement of Cyclin D1. Dev. Biol. 408 (1), 109–125. https://doi.org/10.1016/j.ydbio.2015.10.007 (2015).
Cooper, C., Moon, H. Y. & van Praag, H. On the run for hippocampal plasticity. Cold Spring Harb Perspect. Med. 8 (4), a029736. https://doi.org/10.1101/cshperspect.a029736 (2018).
Kempermann, G. Adult neurogenesis: an evolutionary perspective. Cold Spring Harb Perspect. Biol. 8 (2), a018986. https://doi.org/10.1101/cshperspect.a018986 (2015).
Jung, H. Y. et al. Age-associated alterations in constitutively expressed cyclooxygenase-2 immunoreactivity and protein levels in the hippocampus. Mol. Med. Rep. 15 (6), 4333–4337. https://doi.org/10.3892/mmr.2017.6512 (2017).
Liao, W. et al. Persistent high glucose induced EPB41L4A-AS1 inhibits glucose uptake via GCN5 mediating crotonylation and acetylation of histones and non-histones. Clin. Transl Med. 12 (2), e699. https://doi.org/10.1002/ctm2.699 (2022).
Yang, T. et al. Down-regulation of EPB41L4A-AS1 mediated the brain aging and neurodegenerative diseases via damaging synthesis of NAD+ and ATP. Cell. Biosci. 11 (1), 192. https://doi.org/10.1186/s13578-021-00705-2 (2021).
Xu, Y., Zhang, S. & Zheng, H. The cargo receptor SQSTM1 ameliorates neurofibrillary tangle pathology and spreading through selective targeting of pathological MAPT (microtubule associated protein tau). Autophagy 15 (4), 583–598. https://doi.org/10.1080/15548627.2018.1532258 (2019).
Chai, W. et al. Elevated exosome-transferrable LncRNA EPB41L4A-AS1 in CD56bright NK cells is responsible for the impaired NK function in neuroblastoma patients by suppressing cell Glycolysis. Clin. Immunol. 250, 109322. https://doi.org/10.1016/j.clim.2023.109322 (2023).
Liao, M. et al. LncRNA EPB41L4A-AS1 regulates Glycolysis and Glutaminolysis by mediating nucleolar translocation of HDAC2. EBiomedicine 41, 200–213. https://doi.org/10.1016/j.ebiom.2019.01.035 (2019).
Schulz, J. et al. Conserved cytoplasmic domains promote Hrd1 ubiquitin ligase complex formation for ER-associated degradation (ERAD). J. Cell. Sci. 130 (19), 3322–3335. https://doi.org/10.1242/jcs.206847 (2017).
Américo-Da-Silva, L. et al. A new role for HERPUD1 and ERAD activation in osteoblast differentiation and mineralization. FASEB J. 32 (9), 4681–4695. https://doi.org/10.1096/fj.201701229RR (2018).
Leitman, J. et al. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol. Biol. Cell. 25 (7), 1050–1060. https://doi.org/10.1091/mbc.E13-06-0350 (2014).
Joly, P. et al. ERAD defects and the HFE-H63D variant are associated with increased risk of liver damages in alpha 1-antitrypsin deficiency. PLOS One. 12 (6), e0179369. https://doi.org/10.1371/journal.pone.0179369 (2017).
, L. T. M. et al. Deletion of Herpud1 enhances Heme oxygenase-1 expression in a mouse model of Parkinson‘s disease. Parkinsons Dis. 2016 (6163934). https://doi.org/10.1155/2016/6163934 (2016).
Lin, H. et al. MicroRNA-384-mediated Herpud1 upregulation promotes angiotensin II-induced endothelial cell apoptosis. Biochem. Biophys. Res. Commun. 488 (3), 453–460. https://doi.org/10.1016/j.bbrc.2017.05.035 (2017).
Chan, S. L. et al. Herp stabilizes neuronal Ca2 + homeostasis and mitochondrial function during Endoplasmic reticulum stress. J. Biol. Chem. 279 (27), 28733–28743. https://doi.org/10.1074/jbc.M404272200 (2004).
Kwon, K. et al. Adaptive cellular response of the substantia Nigra dopaminergic neurons upon age-dependent iron accumulation. Aging Cell. 21 (9), e13694. https://doi.org/10.1111/acel.13694 (2022).
Hetz, C., Zhang, K. & Kaufman, R. J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell. Biol. 21 (8), 421–438. https://doi.org/10.1038/s41580-020-0250-z (2020).
Bröer, S. Amino acid transporters as targets for cancer therapy: why, where, when, and how. Int. J. Mol. Sci. 21 (17), 6156. https://doi.org/10.3390/ijms21176156 (2020).
Jiang, L. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520 (7545), 57–62. https://doi.org/10.1038/nature14344 (2015).
Bergsbaken, T., Fink, S. L. & Cookson, B. T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7 (2), 99–109. https://doi.org/10.1038/nrmicro2070 (2009).
Leu, J. I. J., Murphy, M. E. & George, D. L. Mechanistic basis for impaired ferroptosis in cells expressing the African-centric S47 variant of p53. Proc. Natl. Acad. Sci. U S A. 116 (17), 8390–8396. https://doi.org/10.1073/pnas.1821277116 (2019).
Koppula, P., Zhang, Y., Zhuang, L. & Gan, B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. (Lond). 38 (1), 12. https://doi.org/10.1186/s40880-018-0288-x (2018).
Liu, H. et al. Identification of SLC3A2 as a potential therapeutic target of osteoarthritis involved in ferroptosis by integrating bioinformatics, clinical factors and experiments. Cells 11 (21), 3430. https://doi.org/10.3390/cells11213430 (2022).
Colle, R. et al. PPAR-γ agonists for the treatment of major depression: a review. Pharmacopsychiatry 50 (2), 49–55. https://doi.org/10.1055/s-0042-120120 (2017).
Geng, L., Zhang, T., Liu, W. & Chen, Y. Inhibition of miR-128 abates Aβ-Mediated cytotoxicity by targeting PPAR-γ via NF-κB inactivation in primary mouse cortical neurons and Neuro2a cells. Yonsei Med. J. 59 (9), 1096–1106. https://doi.org/10.3349/ymj.2018.59.9.1096 (2018).
Varrone, A. & Nordberg, A. Molecular imaging of neuroinflammation in Alzheimer‘s disease. Clin. Transl Imaging. 3 (6), 437–447. https://doi.org/10.1007/s40336-015-0137-8 (2015).
Guo, Q. et al. Immune activation of murine RAW264.7 macrophages by sonicated and alkalized paramylon from Euglena gracilis. BMC Microbiol. 20 (1), 171. https://doi.org/10.1186/s12866-020-01782-y (2020).
Heneka, M. T. et al. Neuroinflammation in Alzheimer‘s disease. Lancet Neurol. 14 (4), 388–405. https://doi.org/10.1016/S1474-4422(15)70016-5 (2015).
Zhao, W. J., Dong, F., Wang, J., Guo, X. & P Research progress on the relationship between intestinal flora and alzheimer’s disease. Adv. Clin. Med. 9 (5), 703–710. https://doi.org/10.12677/ACM.2019.95107 (2019).
Afsar, A., Chen, M., Xuan, Z. & Zhang, L. A glance through the effects of CD4 + T cells, CD8 + T cells, and cytokines on Alzheimer‘s disease. Comput. Struct. Biotechnol. J. 21, 5662–5675. https://doi.org/10.1016/j.csbj.2023.10.058 (2023).
Dai, L. & Shen, Y. Insights into T-cell dysfunction in Alzheimer‘s disease. Aging Cell. 20 (12), e13511. https://doi.org/10.1111/acel.13511 (2021).
Wang, Y. & He, L. Research progress on microglia regulating microtubule-associated proteins in Alzheimer‘s disease. Cent. S Pharm. 22 (04), (2024).
Holtz, A. M. et al. Secreted HHIP1 interacts with Heparan sulfate and regulates Hedgehog ligand localization and function. J. Cell. Biol. 209 (5), 739–757. https://doi.org/10.1083/jcb.201411024 (2015).
Wen, J. & Hadden, M. K. Affinity-based protein profiling identifies vitamin D3 as a heat shock protein 70 antagonist that regulates Hedgehog transduction in murine basal cell carcinoma. Eur. J. Med. Chem. 228, 114005. https://doi.org/10.1016/j.ejmech.2021.114005 (2022).
Blotta, S. et al. Canonical and noncanonical Hedgehog pathway in the pathogenesis of multiple myeloma. Blood 120 (25), 5002–5013. https://doi.org/10.1182/blood-2011-07-368142 (2012).
Hewavitharana, T. & Wedegaertner, P. B. Non-canonical signaling and localizations of heterotrimeric G proteins. Cell. Signal. 24 (1), 25–34. https://doi.org/10.1016/j.cellsig.2011.08.014 (2012).
Akhshi, T., Shannon, R. & Trimble, W. S. The complex web of canonical and non-canonical Hedgehog signaling. BioEssays 44 (3), e2100183. https://doi.org/10.1002/bies.202100183 (2022).
Jäger, T., Ocker, M., Kiesslich, T., Neureiter, E. & Neureiter, D. Thoughts on investigational Hedgehog pathway inhibitors for the treatment of cancer. Expert Opin. Investig. Drugs. 26 (2), 133–136. https://doi.org/10.1080/13543784.2017.1274392 (2017).
Praktiknjo, S. D., Saad, F., Maier, D., Ip, P. & Hipfner, D. R. Activation of smoothened in the Hedgehog pathway unexpectedly increases Gαs-dependent cAMP levels in drosophila. J. Biol. Chem. 293 (35), 13496–13508. https://doi.org/10.1074/jbc.RA118.001953 (2018).
Polizio, A. H. et al. Heterotrimeric Gi proteins link Hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J. Biol. Chem. 286 (22), 19589–19596. https://doi.org/10.1074/jbc.M110.197111 (2011).
Qu, C. et al. Trimeric G protein-CARMA1 axis links smoothened, the Hedgehog receptor transducer, to NF-κB activation in diffuse large B-cell lymphoma. Blood 121 (23), 4718–4728. https://doi.org/10.1182/blood-2012-12-470153 (2013).
Yam, P. T., Langlois, S. D., Morin, S. & Charron, F. Sonic Hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron 62 (3), 349–362. https://doi.org/10.1016/j.neuron.2009.03.022 (2009).
Tukachinsky, H., Lopez, L. V. & Salic, A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell. Biol. 191 (2), 415–428. https://doi.org/10.1083/jcb.201004108 (2010).
Sphyris, N., Hodder, M. C. & Sansom, O. J. Subversion of niche-signalling pathways in colorectal cancer: what makes and breaks the intestinal stem cell. Cancers (Basel). 13 (5), 1000. https://doi.org/10.3390/cancers13051000 (2021). Published online.
Jacobs, C. T. & Huang, P. Complex crosstalk of Notch and Hedgehog signalling during the development of the central nervous system. Cell. Mol. Life Sci. 78 (2), 635–644. https://doi.org/10.1007/s00018-020-03627-3 (2021).
Martelli, A. M. et al. Understanding the roles of the Hedgehog signaling pathway during T-cell lymphopoiesis and in T-cell acute lymphoblastic leukemia (T-ALL). Int. J. Mol. Sci. 24 (3), 2962. https://doi.org/10.3390/ijms24032962 (2023).
Pelullo, M. et al. Wnt, Notch, and TGF-β pathways impinge on Hedgehog signaling complexity: an open window on cancer. Front. Genet. 10, 711. https://doi.org/10.3389/fgene.2019.00711 (2019).
Chatterjee, S. & Sil, P. C. Targeting the crosstalks of Wnt pathway with Hedgehog and Notch for cancer therapy. Pharmacol. Res. 142, 251–261. https://doi.org/10.1016/j.phrs.2019.02.027 (2019).
Jacobs, C. T. & Huang, P. Notch signalling maintains Hedgehog responsiveness via a Gli-dependent mechanism during spinal cord patterning in zebrafish. eLife 8, e49252. https://doi.org/10.7554/eLife.49252 (2019).
Patni, A. P. et al. Comprehending the crosstalk between Notch, Wnt and Hedgehog signaling pathways in oral squamous cell carcinoma - clinical implications. Cell. Oncol. (Dordr). 44 (3), 473–494. https://doi.org/10.1007/s13402-021-00591-3 (2021).
Wall, D. S. et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell. Biol. 184 (1), 101–112. https://doi.org/10.1083/jcb.200805155 (2009).
Marczenke, M. et al. GAS1 is required for NOTCH-dependent facilitation of SHH signaling in the ventral forebrain neuroepithelium. Development 148 (21), dev200080. https://doi.org/10.1242/dev.200080 (2021).
Luo, Z. et al. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J. Mol. Cell. Biol. 12 (5), 345–358. https://doi.org/10.1093/jmcb/mjz088 (2020).
Ma, L. et al. WNT/NOTCH pathway is essential for the maintenance and expansion of human MGE progenitors. Stem Cell. Rep. 12 (5), 934–949. https://doi.org/10.1016/j.stemcr.2019.04.007 (2019).
Acknowledgements
This work was supported by the [Natural Science Foundation of China] under Grant [Number 82205064] and [Hunan Provincial Science and Technology Innovation Program] under Grant [2023RC3215], the [Hunan Provincial Natural Science Foundation] under Grant [2024JJ5236], and the [Furong Laboratory Science and Technology Research Project] under Grant [2023SK2113-2].
Funding
This work was supported by the [Natural Science Foundation of China] under Grant [Number 82205064] and [Natural Science Foundation of Shandong Province] under Grant [Number ZR2021QH110].
Author information
Authors and Affiliations
Contributions
NS: Original draft. HYW: Project administration, Visualization. KS: Project administration, Formal analysis. YZ: Visualization. ZYZ: Formal analysis. JWG: Methodology. DHW: Methodology, Supervision. YHW: Writing—review & editing, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the Zaozhuang Hospital of Traditional Chinese Medicine (2025-syyws-001). All enrolled subjects signed an informed consent form (Date:2025/02/27/ version.2.0).
Consent for publication
Written informed consent was obtained from the clinical participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sheng, N., Wang, HY., Song, K. et al. Uncovering the role of integrated stress in Alzheimer’s disease through single-cell and transcriptomic analysis. Sci Rep 16, 4779 (2026). https://doi.org/10.1038/s41598-026-34997-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-026-34997-6