Abstract
HSALR mice are the most broadly used animal model for studying myotonic dystrophy type I (DM1). However, so far, HSALR preclinical studies have often excluded female mice or failed to document the biological sex of the animals. This leaves an unwanted knowledge gap concerning the differential development of DM1 in males and females, particularly considering that the disease has a different clinical presentation in men and women. Here we compared typical functional measurements, histological features, molecular phenotypes and biochemical plasma profiles in the muscles of male and female HSALR mice in search of any significant between-sex differences that could justify this exclusion of female mice in HSALR studies and, critically, in candidate therapy assays performed with this model. We found no fundamental differences between HSALR males and females during disease development. Both sexes presented comparable functional and tissue phenotypes, with similar molecular muscle profiles. The only sex differences and significant interactions observed were in plasma biochemical parameters, which are also intrinsically variable in patients with DM1. In addition, we tested the influence of age on these measurements. We therefore suggest including female HSALR mice in regular DM1 studies, and recommend documenting the sex of animals, especially in studies focusing on metabolic alterations. This will allow researchers to detect and report any potential differences between male and female HSALR mice, especially regarding the efficacy of experimental treatments that could be relevant to patients with DM1.
Similar content being viewed by others
Main
Myotonic dystrophy type I (DM1) is a rare genetic disorder with autosomal dominant inheritance, which is estimated to affect 9–10 in 100,000 individuals1. It stems from the expansion of CTG repeats in the 3′ untranslated region (UTR) of the DM1 protein kinase (DMPK) gene2,3. A high number of repeats (starting at >50) produce long abnormal RNA, which becomes toxic for cell function4. Longer CUG repeats are associated with more severe symptoms in patients, particularly once these numbers reach the thousands5. Double-stranded RNA structures generated by abnormal DMPK transcripts with long CUG sequences result in nuclear sequestration of specific RNA-binding proteins, such as those of the muscleblind (MBNL) family, which mimic the loss of function of the sequestered proteins6,7,8,9. The expression of long CTG repeats in Drosophila10 and mice11, and MBNL1 loss of function in mice12, is sufficient to reproduce the DM1 condition. Typical symptoms of DM1 include progressive muscle weakness (myopathy), muscle stiffness and delayed muscle relaxation after contraction (myotonia), and progressive muscle wasting (atrophy), affecting mainly distal muscles and often appearing in adulthood6,13. These symptoms result in reduced strength, activity and survival, among other effects14,15. At a molecular level, MBNL proteins are required for proper alternative splicing of a subset of muscle, cardiac and brain transcripts16,17 and lead to substantial posttranscriptional modifications in many genes6,18. They participate in the switch from fetal and neonatal splicing forms to the adult ones necessary to develop mature muscles19,20,21. Spliceopathy is one of the main features of the DM1 muscle transcriptome. The complete miRNA content (miRNome) also exhibits changes in patients and animal models with DM1 (ref. 18). Indeed, miR-218 (ref. 22) and miR-7 (ref. 23) are strongly increased and decreased, respectively, in DM1 muscle biopsies and mice samples, with major consequences for muscle function. Several studies have indicated that the disease, which affects several systems, manifests differently in men and women. For example, muscle weakness in DM1 affects men more frequently, while obesity is more prevalent in women. However, these differences have not been investigated in animal models of DM1 (refs. 15,24,25).
So far, many different models have been developed to study DM1 (ref. 6). HSALR (from human skeletal actin – long repeat) mice were created in 2000 by Mankodi et al.11 to better understand the etiology of DM1. HSALR mice express an array of 250 CUG repeats in the 3′ UTR of the gene encoding human skeletal muscle actin. Analysis revealed that these animals develop DM1 symptoms such as myotonia and altered histology, with an increased number of central nuclei in affected myotubes, which is compatible with a pathogenic immature state26,27. Later studies showed that HSALR mice also replicate alternative splicing defects observed in the muscles of patients with DM128,29,30. A review of the literature identified 41 mouse studies on HSALR (according to a PubMed search that we performed on July 2023), of which 8 documented the use of only males, and 8 the use of both male and female mice. So far, to the best of our knowledge, no study has been performed only on females. Surprisingly, most studies (61%) did not report the sex of the animals used, probably assuming that standard biomedical research is performed by default in males31. Investigators tend to prefer males for research, as females are thought to introduce variability through factors such as hormones32,33. Consequently, females are underrepresented in biomedical research31,34. The exclusion of HSALR females has two main consequences: (1) there are no available data regarding female-specific characteristics related to DM1 development in assays conducted using these mice, which could be critical for translation of the findings to humans; (2) excluding females increases the overall number of animals needed for each experiment and disregards valuable animals that could otherwise be used for analysis35.
Compared with female patients, male patients with DM1 more frequently show severe muscular disability, with marked myotonia, muscle weakness and cardiac and respiratory involvement15,36. The decline of upper limb performance was also investigated in a 9-year follow-up study of the natural progression of the disease, which revealed that women lost less grip strength and gross dexterity than men37.
Therefore, in this study, we sought to investigate whether muscle-related phenotypes were similar between male and female HSALR mice. We examined multiple key phenotypes commonly investigated in the HSALR model of DM1 to identify any notable differences between males and females that could rationalize the exclusion of females from experiments. In addition, we investigated the possibility of interaction between the sources of variation analyzed: sex × genotype. The age of the mice was also taken into consideration for statistics.
Results and discussion
We selected a group of characteristics typically measured in DM1 studies and covering different aspects of disease alterations11. We analyzed muscle strength and myotonia, histological features and molecular genetics of muscle tissue, and plasma biochemical markers, with variable sample sizes (Table 1). To characterize the histological and molecular phenotypes of muscle tissue, we selected gastrocnemius and quadriceps muscle sections, because these muscles have been reported to exhibit the highest HSA transgene expression in the HSALR model38,39. As findings for both muscles were very similar, data for quadriceps and gastrocnemius were merged for graphs and statistical analyses (see Supplementary Dataset 1 for full details) for every individual (n representing the number of individuals; Table 1).
A total of 155 mice (100 male and 55 female; Fig. 1a,b) were included in this study. We first genotyped the animals to ensure that they carried the 250 CTG insertion (Supplementary Fig. 1a–d), and analyzed the HSA transgene expression (Fig. 1c), which was absent in wild type (WT) and slightly higher in female muscles (Supplementary Fig. 2a). To search for between-sex differences or possible sex × genotype interactions, we performed a two-way analysis of variance (ANOVA) for all phenotypes analyzed considering two sources of variation: genotype (WT versus HSALR) and sex (males versus females). Three-way ANOVA was performed to evaluate potential effects of mouse age.
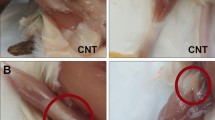
a,b, The size of male (a) and female (b) HSALR animals is increased compared with control WT. c, The human HSA transgene is expressed in the quadriceps and gastrocnemius of HSALR mice, but not in the muscles of WT mice. Slight but significant differences are detected in HSA transgene expression between female and male HSALR muscles (n = 10, n = 15, n = 10, n = 23, for WT males and females and HSALR males and females respectively). This observation explains the effect of sex and interaction genotype × sex observed with ANOVA (Supplementary Fig. 2). d, Strong myotonia is observed in HSALR animals (n = 27, n = 3, n = 42, n = 11). e,f, Weight measurements show that females are smaller than males (n = 37, n = 15, n = 42, n = 19) (e), and normalization to WT weight shows that weight increases consistently in both male and female HSALR animals (f). g, Forelimb strength, measured in seconds, is comparable between males and females and significantly reduced in HSALR animals (n = 14, n = 15, n = 17, n = 19). h, Forelimb strength normalized to weight shows significant differences between sexes and genotypes. The boxes indicate the two central quartiles, the midline represents median values and the whiskers indicate the minimum and maximum data value, excluding outliers. Statistical analysis was performed using two-way ANOVA (WT versus HSALR, males versus females). Differences between WT and HSALR are represented as black asterisks. Differences between sexes were confirmed using the Wilcoxon test and are represented as blue asterisks. ***P < 0.001; **P < 0.01; *P < 0.05.
Phenotypes of HSALR mice are similar in males and females
The expression of the HSA transgene was linked to a notable delay in skeletal muscle relaxation, a phenomenon called myotonia (Fig. 1d), which is a prominent pathological feature that also occurs in muscles of patients with DM1 (refs. 4,40,41). We recorded body weight and found it to be influenced by both genotype and sex (Fig. 1e and Supplementary Fig 2a). On average, HSALR animals were around 11% heavier than WT ones, and WT and HSALR males were considerably larger than females (on average, 19–23%; Table 1). The differences in size between males and females were also confirmed using the Wilcoxon test. Interestingly, when weight was normalized to the WT average weight for each sex, the fold increase due to genotype remained constant for each sex (Fig. 1f). Despite the size differences between males and females, we noted no significant differences between sexes in forelimb strength determined by the grip resistance test (Fig. 1g and Methods); however, there was an apparent reduction of around 30% in grip strength in HSALR mice compared with WT mice (Supplementary Fig. 2a). Many strength measurements used to study muscle dystrophy in mice42 are normalized to body weight. In Fig. 1h, we show the ratio between grip strength and weight. As body weight is a sex-dependent characteristic, when we normalized the grip measures to the weight, the resulting data were also sex dependent. Importantly, we detected no interaction between sex and genotype in this set of in vivo measurements (Supplementary Fig. 2a). Further analysis considering the age of the animals showed that the weight, strength and myotonia of animals significantly increased in older animals (Supplementary Fig. 2b–h).
On average, we observed that males were larger than females, and that weight gain associated with the HSALR genotype affected both sexes proportionally. Previous studies have examined grip strength in male and female HSALR mice, although they did not provide detailed information on the calculation methods used or whether they observed any differences between sexes43,44,45,46. In our study, we found no significant differences in grip strength between males and females. Grip strength was reduced in HSALR animals, but there was no notable difference between sexes. We further confirmed this observation using a new different set of animals of intermediate ages, measuring both hindleg and foreleg strength (Supplementary Fig. 3). Many different devices are available for evaluating the strength of laboratory animals, some of which can be affected by the sex or weight of the animals. When measuring strength alterations, the devices require prior testing to determine whether males and females perform differently in different settings. In vivo phenotypes can therefore be studied in both male and female HSALR, always taking animal weight and weight-derived parameters into account.
Central nuclei and histology
We prepared quadriceps and gastrocnemius muscle sections (Supplementary Fig. 4a) from male and female mice of both genotypes (Table 1) and used hematoxylin and eosin staining to reveal muscle fibers and their nuclei (Fig. 2a and Supplementary Fig. 4b). Nuclei are known to migrate to the periphery of the myotubes during differentiation, and the presence of centralized myonuclei in adults is a sign of myofiber immaturity26. We obtained the percentage of muscle fibers with centrally located nuclei from at least 100 fibers per mouse in both quadriceps and gastrocnemius (before combining data from both muscles), which was dramatically increased in HSALR animals compared with WT (Fig. 2b and Supplementary Fig. 4c). The myofiber immaturity phenotype is clearly visible in our sample (Fig. 2a,b and Supplementary Fig. 4b), as previously described44,45,47 and in agreement with the original description of HSALR mice in the report of Mankodi et al.11. As expected, age had a significant effect on the percentage of fibers with central nuclei (Supplementary Fig. 4d,e). The shape of muscle fibers seemed altered in HSALR mice, with typical increased fat infiltration (Fig. 2a and Supplementary Fig. 4b), as observed elsewhere48,49,50. However, we could not confirm differences in the cross-sectional area of gastrocnemius fibers among groups, neither between sexes nor between genotypes (Supplementary Fig. 4f). The accumulation of transcripts with long CTG repeats and muscleblind 1 (MBNL1) proteins into the nucleus form so-called foci, which we detected in the muscles of HSALR mice using a (CUG)n probe (Fig. 2c). We also observed aggregates of the abnormal mRNA in the nucleus in HSALR muscles (but rarely in WT), which were slightly more abundant in females (Fig. 2d and Supplementary Fig. 4c), possibly corresponding to the slightly higher HSA transgene expression observed by quantitative reverse-transcription polymerase chain reaction (qRT–PCR) in HSALR females (Fig. 1c). As expected, MBNL1 protein seemed to accumulate in the nuclei in muscles of HSALR mice (Fig. 2e), as a result of being sequestered by the CUG repeats, while in WT muscles, MBNL1 seemed to be distributed mostly in the cytoplasm. Quantification of Mblnl1 fluorescence in the subset of samples analyzed revealed high variation from one sample to another (Fig. 2f), not allowing any conclusion on potential differences. We conclude that, despite the high level of variation found among muscles using this technique, we clearly observed a different localization of the MBNL1 protein in HSALR muscles compared with WT, presumably forming the foci.
a, Representative bright-field microscopy pictures (200× magnification; scale bar, 50 µm) of hematoxylin–eosin staining of quadriceps sections showing an increase in the amount of central nuclei (black arrows) in HSALR mice. b, The percentage of fibers with central nuclei is dramatically increased in HSALR muscles (quadriceps and gastrocnemius combined; WT males n = 21, WT females n = 19, HSALR males n = 37, HSALR females n = 24). c, Representative confocal images (gastrocnemius; 400× magnification) showing the detection of CUG repeats (green) and agglutinin (red) as a marker of cell membrane. d, The percentage of nuclei with foci in the muscles studied (quadriceps and gastrocnemius combined). The individual quantification of each slide is shown (seven slides per muscle per mouse; WT males n = 5, WT females n = 5, HSALR males n = 5, HSALR females n = 5). e, Representative confocal images (gastrocnemius; 400× magnification) where MBNL1 was stained (green). f, MBNL1 fluorescence was quantified (quadriceps and gastrocnemius combined). The mean quantification of the slides is shown (seven slides per muscle per mouse; WT males n = 5, WT females n = 5, HSALR males n = 5, HSALR females n = 5). The boxes indicate the two central quartiles, the midline represents median values and the whiskers indicate the minimum and maximum data value, excluding outliers. Statistical analysis was performed using two-way ANOVA (WT versus HSALR, males versus females). Differences between WT and HSALR are represented as black asterisks. Differences between sexes were confirmed using the Wilcoxon test and are represented as blue asterisks. ***P < 0.001; **P < 0.01; *P < 0.05.
Mbnl1 and Mbnl2 expression is similarly altered in HSALR males and females
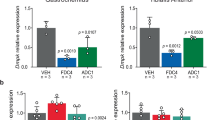
In patients with DM1, the high number of CUG repeats in the DMPK transcript produces aggregates with MBNL1 in the nucleus, called foci, which can be detected microscopically9,51. MBNL1 sequestration in these ribonuclear structures mimics protein loss of function, which is reported to have severe consequences for muscle development12,52. We measured the expression of Mbnl1 (Fig. 3a) and Mbnl2 (Fig. 3b)—two genes involved in DM1 phenotype53,54—by qRT–PCR in WT and HSALR muscles (quadriceps and gastrocnemius). Mbnl1 expression was upregulated in HSALR muscles, as previously shown22, probably as part of a response of the tissue to restore normal Mbnl1 levels. By contrast, we found significantly lower levels of Mbnl2 in HSALR tissue compared with WT, as previously reported55; however, the cause of this reduction is unclear. Previous studies have also reported increased or unchanged Mbnl2 expression in the muscles of these mice22,45. Despite the increase in Mbnl1 transcription, we observed no increase in MBNL1 protein (Fig. 3c,e,f), suggesting that its translation or protein stability could be somehow compromised. In fact, we observed slightly, but significantly, reduced MBNL1 levels in HSALR animals, which has been observed before in HSALR and DM1 cells50,56.
a, Mbnl1 gene expression, measured by qRT–PCR, is significantly increased in HSALR muscles compared with WT. b, By contrast, Mbnl2 expression is reduced in HSALR muscles. c, MBNL1 protein levels, as measured by quantitative dot blot, show a significant decrease in HSALR muscles compared with WT. d, miR-218 levels are increased in the muscles of HSALR animals compared with WT. The boxes indicate the two central quartiles, the midline represents median values and the whiskers indicate the minimum and maximum data value, excluding outliers. e,f, Representative western blot membranes depicting MBNL1 protein levels in quadriceps (e) and gastrocnemius (f). Statistical analysis was performed using two-way ANOVA (black asterisks) ***P < 0.001; *P < 0.05. Sample sizes for the whole figure: n = 10, n = 15, n = 17 and n = 24 for WT males and females and HSALR males and females, respectively. Quadriceps and gastrocnemius were analyzed for each individual.
The miRNome is also altered in muscles of HSALR mice and patients with DM1 (refs. 18,23). It has been reported that miR-218 is upregulated and could impinge on Mbln1 and Mbln2 expression22,56. We found that miR-218 was consistently upregulated in the muscles of HSALR male and female mice (Fig. 3d). miR-218 inhibition of Mbnl1 mRNA translation56 could explain why the higher Mbnl1 transcript levels in HSALR mice were not reflected in MBNL1 protein levels (Fig. 3a,c).
Statistical analysis indicated that Mbnl1 and Mbnl2 expression, MBNL1 expression and miR-218 expression were similar between males and females (Supplementary Fig. 5a), as there was no influence of sex or interaction between genotype and sex for any of these parameters.
There is a wide between-study variation in MBNL1 and MBNL2 levels. Although differences in the age of the animals included in the studies could explain this variation, we did not detect any influence of age on Mbnl1, MBNL1 and miR-218 levels (Supplementary Fig. 5b,d,e). In the case of Mbnl2, we detected increased expression due to age only in WT muscles, while Mbnl2 expression remained constant in HSALR mice (Supplementary Fig. 5c,f). Determining how these genes are regulated and how this variability is controlled could be an interesting direction for future research. Conceivably, increasing sample sizes and widening the age range analyzed could contribute to a better understanding of this variability.
Defects in alternative splicing are precisely replicated in HSALR females
Together with other RNA-binding proteins, Muscleblind proteins are involved in the alternative splicing and polyadenylation processing of numerous genes in muscle cells from patients with DM1 (refs. 17,21,57,58,59,60,61) and HSALR muscle tissue55,56,62. When analyzing the quadriceps and gastrocnemius of the mice, we investigated whether the splicing pattern of a few chosen genes with clearly documented alternative splicing defects in DM1 and HSALR males were also altered in female HSALR mouse muscles57,63,64 (Fig. 4). Three of these genes are characterized by increased exon inclusion (Nfix, Mbnl1 and Clcn1) and three others by increased exon exclusion (Atp2a1, Bin1 and Cacna1s).
Comparison of typical missplicing events in HSALR muscles in males (green) and females (orange). a–f, Percentages of abnormal inclusion of exon 7 in Nfix (a), exon 5 in Mbnl1 (b) and exon 7a in Clcn1 (c) transcripts and abnormal exclusion of exon 22 in Atp2A1 (d), exon 11 in Bin1 (e) and exon 29 in Cacna1s (f). The boxes indicate the two central quartiles, the midline represents median values and the whiskers indicate the minimum and maximum data value, excluding outliers. g, Representative images of agarose gel electrophoresis from semiquantitative PCR determinations of the indicated alternative exons (Ex). Statistical analysis was performed using two-way ANOVA (black asterisks). Minor effects are observed due to the sex of the animals, which are confirmed only in Atp2a1 using the Wilcoxon test (blue asterisks) ***P < 0.001; **P < 0.01; *P < 0.05. Sample sizes for the whole figure: n = 10, n = 15, n = 17 and n = 24 for WT males and females and HSALR males and females, respectively, except Clcn1 (n = 7, n = 14, n = 17 and n = 24 for WT males and females and HSALR males and females, respectively). Quadriceps and gastrocnemius were both analyzed for each individual.
Nfix, which encodes a transcription factor involved in extracellular matrix remodeling during myogenesis, presents an abnormal inclusion of exon 7 in 68% of DM1 muscle samples45,61,65, in accordance with previous observations in HSALR mice45,66. We found inclusion of exon 7 of Nfix in HSALR mice to be around 50%, compared with 13–15% in control samples (Fig. 4a,g), with no differences between males and females (Supplementary Fig. 6a). Nfix exon 7 inclusion is controlled by MBNL2 (ref. 60), suggesting a possible link between the observed reduction in Mbln2 expression (Fig. 3b and Supplementary Fig. 5a) and Nfix missplicing. Exon 5 inclusion in Mbnl1 transcripts, an event that rarely occurs in control samples, was dramatically increased (around 30%) in HSALR animals compared with control ones (Fig. 4b,g). With ANOVA, we detected a very weak influence of sex in this phenotype (Supplementary Fig. 6a), which was not confirmed with the Wilcoxon test. Clcn1 encodes a chloride channel regulating the electric excitability of the skeletal muscle membrane. The anomaly in the splicing pattern of Clcn1 mRNA underlies the prominent myotonia found in muscles of patients with DM1 and HSALR mice45,67,68,69. We observed an increase of exon 7a inclusion in HSALR samples, on average 20–25% more than controls, in both males and females (Fig. 4c,g).
Abnormal exclusion of Atp2a1 exon 22, Bin1 exon 11 and Cacna1s exon 29 are typical signatures in DM1 and HSALR muscle transcriptome. Atp2a1, also known as SERCA1, encodes a sarcoplasmic reticulum calcium-channel protein essential for proper muscle contraction64,65,66. In our study, exon 22 was always included in control animals, while in HSALR muscle samples only 20–30% of the transcripts retained this exon (Fig. 4d,g). Compared with the impact of the genotype, ANOVA (Supplementary Fig. 6a) revealed modest differences between males and females HSALR that we confirmed using the Wilcoxon test. Bin1 encodes a protein necessary for the biogenesis of muscle T tubules, which are specialized skeletal muscle structures essential for excitation–contraction coupling; exon 11 exclusion could contribute to the presence of central nuclei in HSALR mice. We observed a small but significant decrease of exon 11 inclusion in HSALR mice (Fig. 4e,g and Supplementary Fig. 6a), similar to what has been previously described in muscular biopsies from patients with DM1 (ref. 70). Finally, Cacna1s exon 29 exclusion enhances L-type Ca2+ channel conductance and voltage sensitivity in mouse muscle fibers. While exon 29 was always included in control mice, we observed an increase in Cacna1s missplicing in HSALR animals (Fig. 4f,g) consistent with published data71.
Interestingly, data analysis showed that the age of the animals was an important source of variation for splicing pattern (Supplementary Fig. 6b–h), with older HSALR animals displaying splicing levels closer to a WT situation, particularly older females, with Nfix being the only exception. In this study, males kept similar values of abnormal inclusion or exclusion of analyzed exons for all ages, but this finding could change for animals older than 6 months. Our observations fit with previously published data59,65 that show that the HSALR missplicing degree in the muscles of young animals is stronger than in old ones for Clcn1 and Atp2a1, among other genes. Some investigators have suggested that splice switching from fetal to postnatal patterns is temporarily delayed in HSALR mice, but not prevented72. In our hands, the missplicing events were comparable for males and females mostly in young animals (around 3 months) and differences appeared later with age. These differences could be considered negligible owing to the remarkable differences between genotypes, which are still excellent molecular markers of the disease.
Biochemical markers in blood plasma
Biochemical analysis revealed significantly increased plasma cholesterol in HSALR animals compared with WT (Fig. 5a and Supplementary Fig. 7a). Interestingly, cholesterol was significantly lower in females than in males, particularly in HSALR animals. Triglyceride levels were highly variable and did not present significant alterations among groups analyzed (Fig. 5b and Supplementary Fig. 7a). Creatine phosphokinase (CPK), lactate dehydrogenase (LDH) and aspartate transferase (AST) levels showed very similar patterns (Fig. 5c–e), with very high values in WT males. Statistically significant differences were observed between males and females in WT animals for LDH, with a similar pattern being observed for CPK, and AST levels. Notably, for all these parameters, differences between sexes were absent in HSALR mice (Fig. 5c–e and Supplementary Fig. 7a). A different pattern was observed for glucose levels (Fig. 5f and Supplementary Fig. 7a), showing especially low values in control males.
a–f, Plasma levels of cholesterol (a), triglycerides (b), CPK (c), LDH (d), AST (e) and glucose (f) were analyzed in WT and HSALR mice. The boxes indicate the two central quartiles, the midline represents median values and the whiskers indicate the minimum and maximum data value, excluding outliers. Statistical analysis was performed using two-way ANOVA (black asterisks). Detected differences between sexes were confirmed using the Wilcoxon test (blue asterisks) ***P < 0.001; **P < 0.01; *P < 0.05. Sample sizes for the whole figure: n = 12, n = 13, n = 19 and n = 13 for WT males and females and HSALR males and females, respectively.
Overall, we found statistically significant differences between WT and HSALR for cholesterol, CPK, LDH and AST values. Abnormally low LDH circulating levels are rare but considered not harmful73. Taken together, we observed a slight increase in cholesterol levels, and a reduction in CPK, LDH and AST levels, which affected mostly HSALR males. Further work is needed to gain a greater insight into the role of these enzymes in DM1. Plasma biochemical features are intrinsically variable in patients with DM1. Nevertheless, it is well documented that these patients show increased body weight, characterized by adipose tissue expansion, despite muscle fiber atrophy4,74,75. A sedentary lifestyle and related muscle weakness contribute to the onset of metabolic syndrome in these patients. Patients with DM1 often exhibit increased plasma cholesterol and triglyceride levels, insulin resistance and an increased risk of developing metabolic syndrome76,77,78,79,80,81. Patients with DM1 often develop hyperglycemia76,79. Sex differences have been reported for several metabolic features in patients with DM1 (refs. 79,81). So far, the biochemical profile of HSALR mice has not been described in detail, but previous studies have shown that they develop insulin resistance to some degree and often present altered plasma lipid profile42. We showed above that HSALR animals are generally heavier than control (Fig. 1a,b,e,f), with an increase in adipocytes and infiltrated fat in muscle composition (Fig. 2a and Supplementary Fig. 4b), similar to the muscles of patients with DM1 (refs. 76,82). HSALR mice also exhibit features compatible with insulin resistance, such as increased cholesterol in both sexes and higher glucose levels in circulating plasma of males. LDH variations have not been previously characterized in HSALR mice, but it would be interesting to further determine to what extent they are linked to impaired glucose metabolism in muscle. CPK catalyzes the conversion of creatine to create phosphocreatine and has been reported to be slightly elevated in the blood of patients with DM1 (ref. 41). Unlike the findings from patients with DM1, we found that CPK average levels in HSALR mice were consistently reduced in a genotype-dependent manner (Fig. 5c and Supplementary Fig. 7a). Both CPK and LDH leak from damaged muscle and can be used as an indicator of muscular metabolic state83. Abnormal liver tests in patients with DM1 (ref. 84) show high AST levels, while we observed reduced AST levels for HSALR mice compared with WT mice (Fig. 5e). The implications of CPK correlation with AST and LDH levels in HSALR mice and the role of muscle metabolism in plasma glucose levels are unclear and warrant further investigation.
When taking the age of the animals into consideration for the analysis (Supplementary Fig. 7b–h), we also observed that triglycerides and LDH tend to increase and glucose tends to decrease with age (Supplementary Fig. 7c,e,g,h), with the trend being stronger in males. Cholesterol, CPK and AST did not show a clear correlation with age (Supplementary Fig. 7b,d,f,h).
To summarize, in HSALR mice, we identified changes in the levels of several markers related to the development of metabolic syndrome and insulin resistance observed in patients with DM1, including cholesterol, CPK, AST, LDH and blood glucose79. Interestingly, in HSALR mice expressing the CUG repeats only in muscle tissue11, we observed an impact on tissues beyond muscle, including plasma and liver markers. These observations indicate that HSALR mice are an excellent model for studying how altered muscle function originating from RNA toxicity can have a nonautonomous effect on other organs. Muscle tissue has a major role in whole-body metabolism85,86, and its dysfunction in DM1 leads progressively to a metabolic disease that could be studied in HSALR mice. While some effects of sex on disease symptoms, such as obesity and metabolic syndrome, seem partially mimicked in the HSALR mice, others are not. These discrepancies could result from the intrinsic limitation of HSALR mice that express the expanded CUG RNA only in skeletal muscles, and several aspects such as pain, gastrointestinal alterations or cognitive symptoms are not expected to be replicated in the model. The transgenic nature of the model probably impacts the extent to which sex-influenced parameters are recapitulated. Patients with DM1 carry the pathogenic CUG repeats in the DMPK transcript, while the HSALR mice model expresses the repeats in the 3′ UTR of the HSA transgene. The different genetic context, the absence of the DMPK background influencing repeat transcription and the fact that the expression of the repeats in this mouse model is limited to just skeletal muscle most likely represent intrinsic limitations of the HSALR model when it comes to biochemical parameters in blood plasma and related parameters, such as obesity.
Conclusion
In summary, muscle disease in HSALR mice is comparable between males and females, which show similar increases in body weight, strength reduction and myotonia. Histologically, both sexes presented a dramatic increase in the percentage of fibers containing central nuclei, and this phenotype seemed to worsen with age. Analysis of the MBNL family of proteins showed similar changes in males and females (with the highest variability detected in the amount of MBNL1 protein). Likewise, analysis of missplicing events revealed comparable findings between sexes. Slight differences in misspicing events were found between male and female HSALR mice, which are negligible when compared with WT versus HSALR differences. Nevertheless, further research is warranted to determine whether other RNA-binding proteins reported to be altered in the HSALR model, such as CELF1 and Staufen1 (refs. 43,87), are similarly altered in males and females. In addition, it might be useful for further studies to characterize a broader, more in-depth set of missplicing events that could account for or relate to other disease phenotypes and abnormalities.
An overview of how all phenotypes examined in this study respond to the expression levels of the HSA transgene is shown in Supplementary Fig. 8, for all samples for which every parameter was measured. These correlations reveal how measured phenotypes behave differently in response to varying amounts of transgene expression. Whereas some parameters like myotonia and Atp2a1 exon 22 inclusion displayed a sort of all-or-nothing effect (Supplementary Fig. 8b,l), others exhibited a more graded correlation with transgene expression, such as miR-218 relative expression and Bin1 exon 11 inclusion (Supplementary Fig. 8h,m). Such differences indicate that for some parameters even small amounts of transgene expression are enough to induce DM1-like phenotypes in the HSALR model, while for other phenotypes, their severity depends on the levels of HSA expression. It must be noted that, for some phenotypes, reduced sample sizes made it difficult to recapitulate all findings, as was the case for most blood plasma markers (Supplementary Fig. 8o–t), for which most samples were not available for analysis of transgene expression.
Biochemical parameters are intrinsically variable according to sex, genotype and other factors such as age. This high variability made it challenging to reach consistent conclusions, and further studies should be performed to clarify the metabolic features of HSALR mice. Attention to sex and gender aspects in biomedical research has been a major initiative of the gender equality policy for research88,89. Females should, therefore, be included in biomedical research experimental designs, and their inclusion should be recommended in general guidelines for the use of laboratory animals90. Using HSALR females in DM1 studies has multiple advantages. Including females in preclinical and clinical assays can identify possible off-targets of the assayed therapies that could affect men and women differently38,91. For example, previous studies have reported differences in the degree of phenotype recovery between males and females following treatment with a combination of AMPK activators in HSALR mice92. Taken together with our observations that no major differences can be seen in the HSALR model between sexes in basal conditions, these previous findings suggest that differences between males and females might arise in the physiological response to future therapeutic candidates, further warranting the inclusion of females in future studies. Finally, experiments with HSALR using both sexes could help reduce the number of animals required in DM1 studies, which is in line with the 3R (reduce, replace and refine) principles. Therefore, we encourage researchers working with HSALR animals to include females in their studies and register and report their observations, which will contribute toward building a comprehensive understanding of DM1 disease in both sexes.
Methods
Animals
All parameters were studied in homozygous transgenic HSALR mice (line 20b), provided by Prof. C. Thornton (University of Rochester Medical Center, Rochester, NY, USA), and friend virus B (FVB) control mice, which provide the corresponding genetic background (named WT within this paper). Animals were housed under a 12-h light/12-h dark cycle with unrestricted access to food and water. Temperature and humidity were kept within the recommended range (20–24 °C and 40–60%, respectively).
All mouse handling and procedures followed the European law regarding laboratory animal care and experimentation (2003/65/CE) and were approved beforehand by Conselleria de Agricultura, Generalitat Valenciana. CEEA reference numbers for mice are as follows: 2016/VSC/PEA/0015, 2020/VSC/PEA/0164, 2020/VSC/PEA0166, 2020/VSC/PEA/0203, A1529567788818, A1458832800370, A20200902200603, A20200717201633, A20210218542822 and A20220322163151.
Electromyography studies
Electromyography studies were performed blindly under general inhalation anesthesia (5% isoflurane for induction, 2% for maintenance) before euthanasia, as previously described28. Five needle insertions were made in each quadriceps muscle of both forelimbs, grading myotonia on a 0–10 scale: 0, no myotonia; 1–5, occasional myotonic discharge in <50% of total needle insertions; 6–7, myotonic discharge in >50% of total needle insertions; 8–9, myotonic discharge observed in nearly all of the insertions (80%); and 10, when the myotonic discharge occurred in all insertions.
Grip strength test
The foreleg and hindleg grip strength test was performed blindly, using a Grip Strength Meter (BIO-GS3; Bioseb). The maximum pull force in grams was recorded on a digital force transducer when the mice grasped the bar, and the gauge of the force transducer was reset to 0 g after every measurement. The tension was recorded by the gauge at the time the mouse released its forepaws from the bar. Three consecutive measurements were taken for each mouse, letting the mouse rest for 30 s between each measurement. The body weight for each mouse was recorded in parallel and blindly, and used to normalize average grip strength. The data analyzed correspond to the average of the three measures. Both grip strength and grip strength normalized by weight were analyzed. We tested foreleg strength in the first set of animals, the same ones used to perform the whole phenotypical study (Fig. 1g). We measured both foreleg and hindleg strength in a second set of age-matched noneuthanized mice to confirm our observations in hindleg (Supplementary Fig. 3 and Supplementary Dataset 2).
Tissue and sample collection
Tissue collection was performed after euthanasia. Each tissue of interest (that is, quadriceps and gastrocnemius) was appropriately collected and divided into two pieces. One of these pieces was immediately frozen in liquid nitrogen and later used for molecular studies, while the other was snap-frozen in liquid-nitrogen-chilled isopentane and then stored at −80 °C until tissue mounting for histology. Blood was collected by cardiac puncture exsanguination and collected into two different vials: a K3-EDTA vial (SARSTEDT, MICROVETTE 500, cat. no. 20.1341.100) or a vial treated with coagulation activators (KIMA MICRO TEST, cat. no. 811020). K3-EDTA vials were subjected to thorough shaking and −80 °C storage, while serum collection in vials containing coagulation activators needed 10 min centrifugation at 5,000 rpm, upon which supernatant (serum) was stored at −80 °C and the pellet discarded. Liver samples were used for animal genotyping.
DNA extraction and genotyping
The genotype of every HSALR and FVB mouse used was confirmed by PCR analysis of genomic DNA from liver samples following the protocol described by Difranco et al.91, consisting of two PCRs with specific conditions and primers. Genomic DNA extraction was performed with the DNeasy Blood & Tissue kit (Qiagen, cat. no. 69504), following the manufacturer instructions but implementing modifications in the user-developed protocol ‘Purification of total DNA from soft tissues using the TissueLyser II (Qiagen) and the DNeasy® Blood &Tissue Kit’.
The first PCR reaction detects the presence of murine skeletal actin (MSA, Acta1) and the human skeletal actin transgene (HSA, ACTA1) using specific primers located in the 3′ noncoding region of both genes (MSA1, MSA2, HSA23 and HSA24 primers). A volume of 2 μL of genomic DNA (about 50 ng/μL) was analyzed in a 50 μL PCR containing 10 μL of 5× Colorless GoTaq Flexi Buffer (Promega), 5 μL of MgCl2 (25 mM), 1 μL of dNTPs (10 mM), 0,25 μL of GoTaq G2 DNA polymerase (Promega) and 1 μL each of the four primers described below. The GoTaq G2 DNA polymerase was activated at 95 °C for 2 min, followed by 28 cycles of denaturation at 95 °C for 30 s, annealing at 54.5 °C for 31 s and extension at 72 °C for 30 s. After the 28 cycles, DNA synthesis was completed by incubating the reaction at 72 °C for 5 min.
The second PCR reaction confirms the length of the CTG repeats (approximately 250) using primers that flanked the 5′ and 3′ ends of the repeat (HSA10 and HSA18 primers). In this case, 2 μL of genomic DNA (about 50 ng/μL) was analyzed in a 50 μL PCR containing 10 μL of 5× Green GoTaq Flexi Buffer (Promega), 5 μL of MgCl2 (25 mM), 1 μL of dNTPs (10 mM), 0,25 μL of GoTaq Flexi DNA Polymerase (Promega) and 1 μL each of the two primers described below. The GoTaq Flexi DNA polymerase was activated at 95 °C for 2 min and then submitted to 21 cycles of denaturation at 95 °C for 30 s, annealing at 64 °C for 1 min and extending at 72 °C for 2 min. After the 21 cycles, DNA synthesis was completed by incubating the reaction at 72 °C for 5 min.
All reactions were done on a DNA thermal cycler (either GeneAmp PCR System 9700, Applied Biosystems, or Gene Max Tc-s-B, Bioer). All reaction products were analyzed by electrophoresis through 1.7% agarose gels. A volume of 10 μl of 5× Green GoTaq Buffer (Promega) was added to the first PCR reaction products. The MSA1/MSA2 product is a 310 bp fragment, and the HSA23/HSA24 product, if positive, is a 249 bp fragment. If positive, the HSA10/HSA18 product is a fragment of around 1,200 bp (approximately 250 repeats).
Primer sequences used can be found in the Supplementary Information.
Histological preparations and nucleus analysis
For quadriceps and gastrocnemius histology, muscles were embedded in OCT mounting medium (Leica) and 10-µm sections were obtained using a Leica CM1950 cryostat. Frozen sections were stained with the conventional hematoxylin and eosin protocol and mounted with DPX Mountant for histology (Sigma). Images were taken at 200× magnification with an Automated Upright LEICA DM4000 LED Microscope. The total number of fibers and the percentage of fibers containing central nuclei were quantified in 100 fibers per muscle. The number of mice analyzed per group (genotype × sex) can be found in Table 1. The total number of fibers and the fibers with central nuclei were counted using the ImageJ cell counter tool.
Fluorescence in situ hybridization and immunofluorescence methods
Gastrocnemius and quadriceps muscles from five mice per group (genotype × sex) aged 4.5 months were used to analyze the foci presence and MBNL1 localization and intensity. Around seven sections were analyzed for each individual.
Localization of CUGexp RNA (foci) by fluorescence in situ hybridization and localization of MBNL1 by immunofluorescence were carried out according to Bisset et al.38. In brief, 10-µm sections were obtained using a Leica CM1950 cryostat from both muscles embedded in OCT mounting medium. Sections were fixed in 4% paraformaldehyde for 15 min, washed with sterile phosphate-buffered saline (PBS) 1× and permeabilized with 0.5% Triton X-100/PBS for 5 min, all at room temperature.
For MBNL1 immunofluorescence, sections were blocked in 5% normal goat serum/PBS for 30 min at room temperature and incubated overnight at 4 °C with the primary antibody anti-MBNL1 (ab45899, 1/2,500 in 1% BSA/PBS). Then, sections were washed with PBS and incubated for 45 min in the dark at room temperature with the secondary antibody goat anti-rabbit Alexa Fluor 488 (A11008, Thermofisher, 1/400 in 1× PBS), washed with PBS and mounted with DAPI-containing VECTASHIELD mounting medium (Vector). MBNL1 signal was quantified using the ImageJ software, dividing green channel intensity by the muscle area. ImageJ software measures intensity and pixel size area from the confocal images.
For foci detection, sections were prehybridized in 30% formamide for 10 min at room temperature, hybridized with Cy3-(CAG)7-Cy3-labeled probe for 2 h in the dark at 37 °C (1:500, in hybridization buffer), posthybridized with formamide 30% for 30 min at 42 °C and then incubated with 1× SSC in 1× PBS for 30 min at room temperature and washed with PBS. Finally, sections were incubated with wheat germ agglutinin-FITC (1:400 in 1× PBS) for 45 min at room temperature to stain cell membranes, washed with PBS and mounted with DAPI-containing VECTASHIELD mounting medium (Vector). The percentage of ribonuclear foci was quantified with
Gastrocnemius foci images were also used to quantify the cross-sectional area of the fibers by the semi-automatic analysis tool of the ZEN software (Zeiss), which calculates the area of each fiber from the green channel. In both cases, images were acquired using the LSM800 confocal microscope (Zeiss) at 400× magnification.
RNA extraction, RT-PCR, semiquantitative PCR and qRT–PCR
Total RNA from quadriceps and gastrocnemius muscles was extracted using the miRNeasy Mini Kit (Qiagen, cat. no. 217084) following the manufacturer’s instructions. For qRT–PCR detection of Mbnl1 and Mbnl2 transcripts, and alternative splicing measures by PCR, 1 µg of total RNA was digested with DNase I (Invitrogen, cat. no. 4716728001) and reverse transcribed using SuperScript II reverse transcriptase (Invitrogen, cat. no. 18064-014) and random hexanucleotides (Roche). Expression levels of Mbnl1 and Mbnl2, as well as reference gene Gapdh, were determined by multiplex qPCR using the HOT FIREPol Multiplex qPCR Mix (Solisbiodyne #08-01-00001) and PrimeTime qPCR probe assays (IDT). Relative expression of Mbnl1 and Mbnl2 were measured and normalized to an endogenous control, Gapdh, using a 2−ΔΔCt methodology. Primer and probe sequences used can be found in the Supplementary Information.
Alternative splicing was assayed by standard PCR, using 20 ng of cDNA as template, GoTaq polymerase (Promega, cat. no. M3008) and specific primers for Atp2a1, Nfix, Clcn1, Mbnl1, Bin1 and Cacna1s alternative splicing events, as already described by Cerro-Herreros et al.56, Overby et al.50, Fugier et al.70 and Tang et al.71. The percentage of abnormal inclusion of the exons of each missplicing event was quantified using ImageJ. Levels of HSA transgene expression were determined by qRT–PCR, according to Wheeler et al.40.
For microRNA miR-218 detection by qPCR, 10 ng of total RNA was used for total microRNA retrotranscription using miRCURY LNA RT Kit (Qiagen, cat. no. 339340). A volume of 3 μL of diluted cDNA was used in the reaction with the miRCURY LNA SYBR Green PCR kit (Qiagen, cat. no. 339346) and Qiagen primers for qRT–PCR on the Applied Biosystems QuantStudio 5 Real-Time PCR System per the manufacturer’s protocol. Relative expression of hsa-miR-218-5p (Qiagen, cat. no. YP00206034) was measured and normalized to endogenous controls, RNU1A1 (Qiagen, cat. no. YP00203909) and U6 snRNA (Qiagen, cat. no. YP00203907) using the 2−∆∆Ct method.
Total protein extraction, quantitative dot blot and western blot
Mouse muscle samples were mechanically disaggregated with a TissueLyser II (Qiagen) and homogenized in RIPA Buffer (Thermo Scientific, cat. no. 89900) supplemented with protease and phosphatase inhibitors (Roche, cat. nos. 11873580001 and 4906845001). Total protein was quantified with Pierce BCA Protein Assay Kit (cat. no. 23225) using bovine serum albumin as the standard.
Immunodetection assay of MBNL1 and endogenous control tubulin was performed as described in Moreno et al.93. In brief, 2 µg/well of total protein, previously denatured at 100 °C for 5 min, was loaded in quantitative dot blot plates (Quanticision Diagnostics). Each sample was loaded in quadruplicate on three different dishes, of which one was incubated with mouse anti-MBNL1 antibody (1:200, MB1a(4A8), Developmental Studies Hybridoma Bank (DSHB)) and another with anti-α-tubulin antibody (1:1,000, 12G10, DSHB), at 4 °C overnight in both cases. Primary antibodies were detected using goat anti-mouse IgG (H + L) secondary antibody HRP-conjugated (1:3,500; Invitrogen). Immunoreaction was detected using Pierce ECL Western reagent (Thermo Scientific), and luminescence was acquired using an Infinite M200 PRO plate reader (Tecan).
Representative western blot was performed by denaturing 15 µg of samples for 5 min at 100 °C, protein separation by electrophoresis on 12% SDS–PAGE gels, transfer onto 0.45 µm nitrocellulose membranes (GE Healthcare) and blocking with 5% nonfat powdered milk in PBS-T (8 mM Na2HPO4, 150 mM NaCl, 2 mM KH2PO4, 3 mM KCl and 0.05% Tween 20, pH 7.4). Immunodetection of MBNL1 was achieved by incubating membranes at 4 °C with primary mouse anti-MBNL1 (1:200, MB1a(4A8), DSHB) and anti-Gapdh (1 h, 1:5,000, clone G-9, Santa Cruz) antibodies. Primary antibodies were detected with HRP-conjugated anti-mouse-IgG secondary antibody (1 h, 1:3,500, Sigma-Aldrich) and ECL Western Blotting Substrate (Pierce). Image acquisition was performed with an AMERSHAM ImageQuant 800 (GE Healthcare).
Determination of biochemical parameters in blood plasma
Biochemical blood parameters were analyzed by Montoro Botella Laboratories. Using a Cobas 600 CCE Modular Analyzer (Roche), serum biochemistry profiles were obtained to evaluate total cholesterol, triglyceride levels, CPK, LDH, glucose and AST.
Statistics
Phenotypes were measured in different sets of animals, euthanized on different dates, aged approximately 3–6 months; mice were individually identified. They were classified according to two factors: genotype (WT versus HSALR) and sex (males versus females), and the age in months registered. Two different muscles were analyzed: quadriceps and gastrocnemius, and data available were merged for both muscles considered as duplicates; so, the n indicated in Table 1 and figure legends corresponds to the number of individuals, not the number of muscles. All data are summarized in Supplementary Dataset 1. Two-way ANOVA tests were performed for all phenotypes to detect to what extent they were defined by genotype, sex or interaction between them (genotype × sex). The data from the two muscles were included together in the model, but considering the animal as a source of error. Two-way ANOVA results are depicted in Supplementary Figs. 2 and 4–7. Boxplots (green for males and orange for females) represent the distribution of the phenotypes. Each colored dot corresponds to single measures performed in different muscles or in the whole organism (green for males and orange for females). Median values and sample sizes are described in Table 1. All statistic estimators, including the interquartile range, maximum and minimum, are detailed in Supplementary Dataset 3. The Wilcoxon nonparametric test was used to analyze further differences between groups (blue asterisk inside the plots). The measures of all phenotypes are also represented by the age of the animals (in months) in supplementary figures, classified by sex and genotype, and a tendency line is added to the dot graphs using a linear model. Three-way ANOVA was used to analyze the effect of age on the phenotypes studied. The single effect of sex, genotype and age was considered, as well as the interaction between these sources of variation; the results are presented in supplementary data. All statistics and graphics were performed using R-4.2.1 for Windows and Desktop RStudio-2022.02.3-492.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Liao, Q., Zhang, Y., He, J. & Huang, K. Global prevalence of myotonic dystrophy: an updated systematic review and meta-analysis. Neuroepidemiology 56, 163–173 (2022).
Brook, J. D. et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68, 799–808 (1992).
Mahadevan, M. et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 255, 1253–1255 (1992).
Thornton, C. A. Myotonic dystrophy. Neurol. Clin. 32, 705–719 (2014).
LoRusso, S., Weiner, B. & Arnold, W. D. Myotonic dystrophies: targeting therapies for multisystem disease. Neurotherapeutics 15, 872–884 (2018).
Thomas, J. D., Oliveira, R., Sznajder, Ł. J. & Swanson, M. S. Myotonic dystrophy and developmental regulation of RNA processing. Compr. Physiol. https://doi.org/10.1002/cphy.c170002 (2018).
Miller, J. W. et al. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448 (2000).
Davis, B. M., McCurrach, M. E., Taneja, K. L., Singer, R. H. & Housman, D. E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA 94, 7388–7393 (1997).
Taneja, K. L., McCurrach, M., Schalling, M., Housman, D. & Singer, R. H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128, 995–1002 (1995).
Llamusi, B. et al. Muscleblind, BSF and TBPH are mislocalized in the muscle sarcomere of a Drosophila myotonic dystrophy model. Dis. Model Mech. 6, 184–196 (2013).
Mankodi, A. et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289, 1769–1772 (2000).
Kanadia, R. N. et al. A muscleblind knockout model for myotonic dystrophy. Science 302, 1978–1980 (2003).
Smith, C. A. & Gutmann, L. Myotonic dystrophy type 1 management and therapeutics. Curr. Treat. Options Neurol. 18, 52 (2016).
Mateos-Aierdi, A. J. et al. Muscle wasting in myotonic dystrophies: a model of premature aging. Front. Aging Neurosci. 7, 125 (2015).
Dogan, C. et al. Gender as a modifying factor influencing myotonic dystrophy type 1 phenotype severity and mortality: a nationwide multiple databases cross-sectional observational study. PLoS ONE 11, e0148264 (2016).
Ho, T. H. et al. Muscleblind proteins regulate alternative splicing. EMBO J. 23, 3103–3112 (2004).
Pascual, M., Vicente, M., Monferrer, L. & Artero, R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74, 65–80 (2006).
Fernandez-Costa, J. M. et al. Expanded CTG repeats trigger miRNA alterations in Drosophila that are conserved in myotonic dystrophy type 1 patients. Hum. Mol. Genet. 22, 704–716 (2013).
Day, J. W. & Ranum, L. P. W. RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord. 15, 5–16 (2005).
Todd, P. K. & Paulson, H. L. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 67, 291–300 (2010).
Lin, X. et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 15, 2087–2097 (2006).
Cerro-Herreros, E. et al. Preclinical characterization of antagomiR-218 as a potential treatment for myotonic dystrophy. Mol Ther. Nucleic Acids 26, 174–191 (2021).
Sabater-Arcis, M., Bargiela, A., Furling, D. & Artero, R. miR-7 restores phenotypes in myotonic dystrophy muscle cells by repressing hyperactivated autophagy. Mol. Ther. Nucleic Acids 19, 278–292 (2020).
Perna, A. et al. High prevalence and gender-related differences of gastrointestinal manifestations in a cohort of DM1 patients: a perspective, cross-sectional study. Front. Neurol. 11, 394 (2020).
Solbakken, G. et al. Pain in adult myotonic dystrophy type 1: relation to function and gender. BMC Neurol. 21, 101 (2021).
Folker, E. S. & Baylies, M. K. Nuclear positioning in muscle development and disease. Front. Physiol. 4, 363 (2013).
Amack, J. D. & Mahadevan, M. S. Myogenic defects in myotonic dystrophy. Dev. Biol. 265, 294–301 (2004).
Kanadia, R. N. et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl Acad. Sci. USA 103, 11748–11753 (2006).
Nakamori, M., Kimura, T., Fujimura, H., Takahashi, M. P. & Sakoda, S. Altered mRNA splicing of dystrophin in type 1 myotonic dystrophy. Muscle Nerve 36, 251–257 (2007).
Kimura, T. et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 14, 2189–2200 (2005).
Wald, C. & Wu, C. Of mice and women: the bias in animal models. Science 327, 1571–1572 (2010).
Check Hayden, E. Sex bias blights drug studies. Nature 464, 332–333 (2010).
Shansky, R. M. Are hormones a ‘female problem’ for animal research? Science 364, 825–826 (2019).
Nowogrodzki, A. Inequality in medicine. Nature 550, S18–S19 (2017).
Franco, N. H. & Olsson, Ia. S. Scientists and the 3Rs: attitudes to animal use in biomedical research and the effect of mandatory training in laboratory animal science. Lab Anim. 48, 50–60 (2014).
Garibaldi, M. et al. Gender effect on cardiac involvement in myotonic dystrophy type 1. Eur. J. Neurol. 28, 1366–1374 (2021).
Raymond, K., Levasseur, M., Mathieu, J., Desrosiers, J. & Gagnon, C. A 9-year follow-up study of the natural progression of upper limb performance in myotonic dystrophy type 1: a similar decline for phenotypes but not for gender. Neuromuscul. Disord. 27, 673–682 (2017).
Bisset, D. R. et al. Therapeutic impact of systemic AAV-mediated RNA interference in a mouse model of myotonic dystrophy. Hum. Mol. Genet. 24, 4971–4983 (2015).
Hicks, S. M. et al. Alternative splicing dysregulation across tissue and therapeutic approaches in a mouse model of myotonic dystrophy type 1. Mol. Ther. Nucleic Acids 35, 102338 (2024).
Wheeler, T. M. et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325, 336–339 (2009).
Turner, C. & Hilton-Jones, D. Myotonic dystrophy: diagnosis, management and new therapies. Curr. Opin. Neurol. 27, 599–606 (2014).
Sharp, L., Cox, D. C. & Cooper, T. A. Endurance exercise leads to beneficial molecular and physiological effects in a mouse model of myotonic dystrophy type 1. Muscle Nerve 60, 779–789 (2019).
Jones, K. et al. GSK3β mediates muscle pathology in myotonic dystrophy. J. Clin. Invest. 122, 4461–4472 (2012).
Wei, C. et al. Correction of GSK3β at young age prevents muscle pathology in mice with myotonic dystrophy type 1. FASEB J. 32, 2073–2085 (2018).
Chen, G. et al. Phenylbutazone induces expression of MBNL1 and suppresses formation of MBNL1-CUG RNA foci in a mouse model of myotonic dystrophy. Sci Rep. 6, 25317 (2016).
Wang, M. et al. Correction of glycogen synthase kinase 3β in myotonic dystrophy 1 reduces the mutant RNA and improves postnatal survival of DMSXL mice. Mol. Cell. Biol. 39, e00155-19 (2019).
Wei, C., Jones, K., Timchenko, N. A. & Timchenko, L. GSK3β is a new therapeutic target for myotonic dystrophy type 1. Rare Dis. 1, e26555 (2013).
Banks, G. B., Combs, A. C., Chamberlain, J. R. & Chamberlain, J. S. Molecular and cellular adaptations to chronic myotendinous strain injury in mdx mice expressing a truncated dystrophin. Hum. Mol. Genet. 17, 3975–3986 (2008).
Bargiela, A. et al. Quantitative magnetic resonance imaging assessment of muscle composition in myotonic dystrophy mice. Sci. Rep. 13, 503 (2023).
Overby, S. J. et al. Proof of concept of peptide-linked blockmiR-induced MBNL functional rescue in myotonic dystrophy type 1 mouse model. Mol. Ther. Nucleic Acids 27, 1146–1155 (2022).
Fardaei, M., Larkin, K., Brook, J. D. & Hamshere, M. G. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 29, 2766–2771 (2001).
Jiang, H., Mankodi, A., Swanson, M. S., Moxley, R. T. & Thornton, C. A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 13, 3079–3088 (2004).
Holt, I. et al. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am. J. Pathol. 174, 216–227 (2009).
Lee, K.-Y. et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 5, 1887–1900 (2013).
Bargiela, A. et al. Increased Muscleblind levels by chloroquine treatment improve myotonic dystrophy type 1 phenotypes in in vitro and in vivo models. Proc. Natl Acad. Sci. USA 116, 25203–25213 (2019).
Cerro-Herreros, E. et al. miR-23b and miR-218 silencing increase Muscleblind-like expression and alleviate myotonic dystrophy phenotypes in mammalian models. Nat. Commun. 9, 2482 (2018).
López-Martínez, A., Soblechero-Martín, P., de-la-Puente-Ovejero, L., Nogales-Gadea, G. & Arechavala-Gomeza, V. An overview of alternative splicing defects implicated in myotonic dystrophy type I. Genes 11, 1109 (2020).
Suenaga, K. et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PLoS ONE 7, e33218 (2012).
Du, H. et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17, 187–193 (2010).
Batra, R. et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell 56, 311–322 (2014).
Nakamori, M. et al. Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 74, 862–872 (2013).
Li, M. et al. HNRNPA1-induced spliceopathy in a transgenic mouse model of myotonic dystrophy. Proc. Natl Acad. Sci. USA 117, 5472–5477 (2020).
Hu, N., Kim, E., Antoury, L. & Wheeler, T. M. Correction of Clcn1 alternative splicing reverses muscle fiber type transition in mice with myotonic dystrophy. Nat. Commun. 14, 1956 (2023).
Wagner, S. D. et al. Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 12, e1006316 (2016).
Yamashita, Y. et al. Four parameters increase the sensitivity and specificity of the exon array analysis and disclose 25 novel aberrantly spliced exons in myotonic dystrophy. J. Hum. Genet. 57, 368–374 (2012).
Mondragon-Gonzalez, R., Azzag, K., Selvaraj, S., Yamamoto, A. & Perlingeiro, R. C. R. Transplantation studies reveal internuclear transfer of toxic RNA in engrafted muscles of myotonic dystrophy 1 mice. eBioMedicine 47, 553–562 (2019).
Lueck, J. D., Mankodi, A., Swanson, M. S., Thornton, C. A. & Dirksen, R. T. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 129, 79–94 (2007).
Charlet-B, N. et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 10, 45–53 (2002).
Mankodi, A. et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 10, 35–44 (2002).
Fugier, C. et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 17, 720–725 (2011).
Tang, Z. Z. et al. Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of CaV1.1 calcium channel. Hum. Mol. Genet. 21, 1312–1324 (2012).
Hu, N. et al. Antisense oligonucleotide and adjuvant exercise therapy reverse fatigue in old mice with myotonic dystrophy. Mol. Ther. Nucleic Acids 23, 393–405 (2021).
Farhana, A. & Lappin, S. L. Biochemistry, Lactate Dehydrogenase (StatPearls, 2023).
Perseghin, G. et al. Postabsorptive and insulin-stimulated energy and protein metabolism in patients with myotonic dystrophy type 1. Am. J. Clin. Nutr. 80, 357–364 (2004).
Aitkens, S., Kilmer, D. D., Wright, N. C. & McCrory, M. A. Metabolic syndrome in neuromuscular disease. Arch. Phys. Med. Rehabil. 86, 1030–1036 (2005).
Vujnic, M. et al. Metabolic syndrome in patients with myotonic dystrophy type 1. Muscle Nerve 52, 273–277 (2015).
Rakocevic Stojanovic, V. et al. Leptin and the metabolic syndrome in patients with myotonic dystrophy type 1. Acta Neurol. Scand. 121, 94–98 (2010).
Shieh, K., Gilchrist, J. M. & Promrat, K. Frequency and predictors of nonalcoholic fatty liver disease in myotonic dystrophy. Muscle Nerve 41, 197–201 (2010).
Spaziani, M. et al. Hormonal and metabolic gender differences in a cohort of myotonic dystrophy type 1 subjects: a retrospective, case-control study. J. Endocrinol. Invest. 43, 663–675 (2020).
Moxley, R. T. et al. Decreased insulin sensitivity of forearm muscle in myotonic dystrophy. J. Clin. Invest. 62, 857–867 (1978).
Heatwole, C. R., Miller, J., Martens, B. & Moxley, R. T. Laboratory abnormalities in ambulatory patients with myotonic dystrophy type 1. Arch. Neurol. 63, 1149–1153 (2006).
Heskamp, L. et al. Lower extremity muscle pathology in myotonic dystrophy type 1 assessed by quantitative MRI. Neurology 92, e2803–e2814 (2019).
Gagliano, M. et al. Low-intensity body building exercise induced rhabdomyolysis: a case report. Cases J. 2, 7 (2009).
Achiron, A. et al. Abnormal liver test results in myotonic dystrophy. J. Clin. Gastroenterol. 26, 292–295 (1998).
Baskin, K. K., Winders, B. R. & Olson, E. N. Muscle as a ‘mediator’ of systemic metabolism. Cell Metab 21, 237–248 (2015).
Argilés, J. M., Campos, N., Lopez-Pedrosa, J. M., Rueda, R. & Rodriguez-Mañas, L. Skeletal muscle regulates metabolism via interorgan crosstalk: roles in health and disease. J. Am. Med. Dir. Assoc. 17, 789–796 (2016).
Ravel-Chapuis, A. et al. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 196, 699–712 (2012).
Klinge, I. Gender perspectives in European research. Pharmacol. Res. 58, 183–189 (2008).
Yakerson, A. Women in clinical trials: a review of policy development and health equity in the Canadian context. Int. J. Equity Health 18, 56 (2019).
Jones-Bolin, S. Guidelines for the care and use of laboratory animals in biomedical research. Curr. Protoc. Pharmacol. https://doi.org/10.1002/0471141755.pha04bs59 (2012).
DiFranco, M., Yu, C., Quiñonez, M. & Vergara, J. L. Age-dependent chloride channel expression in skeletal muscle fibres of normal and HSA(LR) myotonic mice. J. Physiol. 591, 1347–1371 (2013).
Misquitta, N. S., Ravel-Chapuis, A. & Jasmin, B. J. Combinatorial treatment with exercise and AICAR potentiates the rescue of myotonic dystrophy type 1 mouse muscles in a sex-specific manner. Hum. Mol. Genet. 32, 551–566 (2023).
Moreno, N., González-Martínez, I., Artero, R. & Cerro-Herreros, E. Rapid determination of MBNL1 protein levels by quantitative dot blot for the evaluation of antisense oligonucleotides in myotonic dystrophy myoblasts. Methods Mol. Biol. 2434, 207–215 (2022).
Acknowledgements
This work was funded by research grant HR17-00268 (TATAMI project) from the ‘la Caixa’ Bank Foundation awarded to R.A. Additional funding was from PROMETEO/2020/081 and CIPROM/2023/22 from the Conselleria de Educación, Universidades y Empleo of the Generalitat Valenciana, and DTS19/00128 from the Instituto de Salud Carlos III (ISCIII). ARTHEx has the financial support of the CREATEC-CV (Creation of Technology-Based Companies) program from GVA-IVACE for the execution of the project ‘First preclinical phases for the development of an innovative drug against myotonic dystrophy type 1’ (MCBTA/ 2019/47) with co-financing from the European Union through the European Regional Development Fund. Part of the equipment used in this work has been acquired by Generalitat Valenciana and co-financed with ERDF funds (OP ERDF of Comunitat Valenciana 2014-2020). I.G.-M. was supported by predoctoral fellowship FDEGENT/2020/001 from Generalitat Valenciana. MDA Monoclonal Antibody Resources provided antibody MB1a (4A8).
Author information
Authors and Affiliations
Contributions
The authors confirm the following contributions to the paper. Conceptualization: B.L.; formal analysis: R.P.-G. and E.C.-H.; data curation: M.C.-S., E.C.-H., R.P.-G. and I.G.-M.; investigation: E.C.-H., M.C.-S., A.C.-R., I.G.-M., A.G.-R. and D.P.-L.; supervision: B.L., E.C.-H. and R.A.; visualization: R.P.-G., E.C.-H., A.C.-R. and A.G.-R.; writing—original draft: R.P.-G., with contributions by M.C.-S., A.G.-R. and A.C.-R.; writing—review and editing: R.A., R.P.-G., B.L., E.C.-H., I.G.-M., A.C.-R. and M.C.-S.; funding acquisition, project administration and resources: R.A. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Lab Animal thanks Kun Huang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary material of Methods section: primer list used for HSA transgene quantification and for qRT–PCR and Supplementary Figs. 1–8.
Supplementary Dataset 1
Raw data of all experiments in a matrix used for statistical analysis.
Supplementary Dataset 2
Foreleg and hindleg strength measures in a new set of mice.
Supplementary Dataset 3
Descriptive statistics of statistical analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carrascosa-Sàez, M., Colom-Rodrigo, A., González-Martínez, I. et al. Use of HSALR female mice as a model for the study of myotonic dystrophy type I. Lab Anim 54, 92–102 (2025). https://doi.org/10.1038/s41684-025-01506-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41684-025-01506-7