Abstract
The demand for demographically and geographically diverse, high-quality, fit-for-purpose real-world data has been increasing to support regulatory and other healthcare decision making. Accessing and sharing healthcare data across sites, regions, and countries while ensuring data privacy has been a long-standing challenge. We discuss synthetic data and federated data networks as examples of emerging privacy-preserving technologies and provide real-life use cases from government, industry, and academia with their opportunities and challenges.
Similar content being viewed by others
Background
With the growing adoption of real-world evidence (RWE) for regulatory and healthcare decision-making, data privacy in healthcare has become an even more critical consideration for healthcare providers, researchers, regulators, and patients. Researchers seek real-world data (RWD) to understand disease epidemiology, generate insights regarding treatments, and improve health outcomes. Healthcare providers and payers are responsible for protecting patient privacy and security while balancing the need for data use, access, and control. Many countries implemented privacy legislations to enable safe and secure secondary use of healthcare data. For example, in the United States (US), the Health Insurance Portability and Accountability Act (HIPAA) was passed in 1996 to create standards that protect the privacy of identifiable health information through privacy and security rules, setting an initial legal framework for safeguarding biomedical data in healthcare and research1. In Europe, General Data Protection Regulation (GDPR) was put into effect in 2018 to ensure the fundamental rights of data protection and strengthen oversight over the collection, sharing, and use of personal information. Additionally, with advances in artificial intelligence and growing need for data, other countries such as Canada, Singapore, India, and China have also recently published similar data privacy laws to protect personal health information2,3,4,5.
With increasing needs for data from diverse geographic locations and multiple formats (e.g., images, clinical notes), several privacy-preserving approaches, or privacy-enhancing technologies (PETs) have emerged within the context of healthcare data use. Synthetic data and federated networks provide promising venues to access real-world healthcare data in a privacy-preserving manner for evidence generation, and have also made large and diverse datasets available to train machine learning /artificial intelligence models6,7,8. Synthetic health data are artificial data that are intended to mimic the properties and relationships seen in real patient data. It is simulated or computationally derived data that preserves the statistical properties of the original data rather than acquired from a human subject by a physical system9. Additionally, federated data networks allow identifiable data to remain under the control of its original stewards, behind their firewall, but enable users to run analysis across multiple sites without centralizing or aggregating the data. In this paper, we focus the discussion on synthetic health data and federated data networks because they have the most applications and successful demonstrations in practical setting compared with other emerging privacy-preserving approaches. Specifically, for synthetic data, we describe the generation methods and use cases in public sector, life sciences industry, and academia. For federated data networks, we present well-known networks worldwide, describe the various statistical tasks implemented on federated data networks, and give examples on emerging applications.
Synthetic health data
Overview and methods to generate synthetic data
Various methods can be used to generate synthetic data. Two prominent ones are statistical-based methods and machine learning-based methods. The choice between the two often depends on the specific needs of the analysis, data complexity, and available resources. Statistical-based methods use statistical models such as Monte Carlo simulations, bootstrapping, and resampling techniques to generate data that mimic the characteristics of real datasets. These type of approaches do not require individual-level training data when the generation is based on distributions known a priori and informed by background knowledge, published summary statistics, and published risk calculators10,11,12,13. Thus, this approach offers ease of implementation (e.g., through standard distributions, parameter adjustments) and greater privacy protection as it relies on aggregate statistics. Since it relies on assumptions about the underlying distributions, which may not always hold, and could have limited application in the context of high-dimensional data or maintaining intricate relationships among data elements.
The other set of approaches to synthesize health data are machine learning-based, such as sequential decision trees, Bayesian network and generative adversarial networks (GAN)13,14,15,16. This type of technique can adequately maintain high-dimensional relationships and dependencies in the data and can be trained on existing datasets to produce data that are realistic and diverse. This machine learning-based method arguably has higher utility and can also have a low privacy risk when it is generated using additional privacy methods to prevent overfitting and duplication of the individual patient-level data in the training process, but it also requires more computational power and expertise than statistical simulation. Lastly, both approaches are limited in the generalizability of the synthetic data by the input parameters or training data used in the synthetic data generation model. Figure 1 illustrates a sample synthetic data generation process and its potential applications.
Use of synthetic data in public and government sector
The use of synthetic data in the public and government sector can promote innovation, collaboration, and responsible research practices. Government agencies can use synthetic data and even provide such data to citizen scientists, to model and simulate the potential impacts of policy changes on diverse populations, helping to inform decision-making without the risks associated with using real data. In this way, synthetic data can pave the way for different government agencies to share insights and collaborate on cross-agency initiatives without risking data breaches. These outcomes collectively enable better-informed decisions and policies while safeguarding individual privacy which can enhance public trust in data-driven decision-making processes.
Globally, national statistical agencies have used synthetic data for sharing their data products17, and some large data custodians have shared synthetic data publicly such as the CMS Data Entrepreneur’s Synthetic Public Use files18, cancer data from Public Health England19, primary care data from the Clinical Practice Research Datalink20, synthetic variants of the French public health system claims and hospital dataset (SNDS)21, Norwegian hospitalization and prescription data22, and synthetic microdata from Israel’s National Registry of Live Births23. To the extent that this becomes the norm, it can significantly enable broader data access. Below are two examples of government use cases in US and UK.
NIH N3C experience in the US
In times of crisis and public health emergencies, synthetic data can facilitate rapid analysis and response planning, enabling agencies to simulate scenarios and evaluate potential interventions quickly. The National COVID Cohort Collaborative (N3C) provided valuable insights into the application of synthetic data in research, particularly in the context of health data during the pandemic24. The N3C synthetic data was generated using machine learning-based approach and emphasized the need for robust methods to ensure patient privacy. This included techniques like differential privacy to mask sensitive information while retaining its utility. Evaluations ensured that synthetic data accurately reflected real-world conditions and demonstrated the reliability of synthetic data through rigorous validation against actual datasets. Researchers showed that synthetic data allows for the exploration of various research questions that may not be feasible with real data or even HIPAA-deidentified data due to restrictions or availability. The N3C experience demonstrated that while synthetic data holds great promise for advancing research, its effectiveness relies on careful consideration of privacy, data quality, collaboration, and methodological rigor. These lessons can guide future initiatives in synthetic data applications across various fields.
MHRA experience in the UK: generation, applications and governance
The Medicines and Healthcare products Regulatory Agency (MHRA) is the UK’s regulator of medical products, including the regulation of Software as a Medical Device (SaMD) and AI as a Medical Device (AIaMD). It was in this context that the MHRA’s interest in synthetic data originally emerged in 2017, as a potential solution for external validation of machine learning algorithms, in the absence of an alternative real world data source. Given the intended purpose of synthetic data in this scenario, the requirement was high-fidelity synthetic data that was able to capture both the complex inter-relationships between various data fields and the statistical properties of real data. In the context of patient data, high-fidelity synthetic data would capture complex clinical relationships and be clinically indistinguishable from ‘real’ patient data. For the initial proof-of-concept project an extract of anonymized, coded, tabular primary care data from the MHRA’s real world data research service, the Clinical Practice Research Datalink (CPRD), was used as the ground truth data used for generating the synthetic data. An evaluation framework was developed to assess the fidelity, utility and privacy of the generated synthetic data25. The initial pilot demonstrated that it was possible to generate clinically validated, high-fidelity synthetic patient data26. Further testing of the synthetic data generation method was undertaken using other non-CPRD healthcare datasets including liver disease patient data and blood glucose monitoring data27. Two of the synthetic CPRD datasets generated as part of this work are available on license from CPRD20.
The MHRA team preferentially adopted Bayesian network (BN) approaches to synthetic data generation over GAN-based approaches as the former were more explainable to clinical experts undertaking the clinical validation of the synthetic datasets. Subsequent unpublished work has shown that in the context of tabular data, BN approaches have an advantage over GANs when dealing with small ground truth data samples and categorical data. The synthetic data generation approach was further refined to deal with the temporal nature of health datasets and missing fields in the ground truth data.
The MHRA has also developed a novel approach to detecting biases due to underrepresentation in real data using uncertainty analysis, and then correcting these via conditional boosting of underrepresented groups, using synthetic data28. More recently, the MHRA has been researching the application of high-fidelity synthetic data in the context of clinical trials, including data augmentation to boost small sample sizes and synthetic control arms. A validation study of this application is currently underway using data from a previously conducted clinical trial dataset29.
At present there is not a formal MHRA policy regarding synthetic data use for the validation of machine learning applications or for data augmentation and synthetic control arms in the context of clinical trials. However, the MHRA has published an Expert Group report on Regulatory Considerations when using Synthetic data for development of AI medical devices30. This report is intended to provide scenarios in which synthetic data use will be considered acceptable as well as quality considerations. The MHRA has also initiated a regulatory sandbox with selected industry partners to consider the use of synthetic data in the context of clinical trials. In the meantime, manufacturers of medical products wishing to explore these methods are invited to discuss their specific proposals with the MHRA’s scientific advice service.
Finally, as the MHRA’s CPRD RWD service also has a data custodian role, there has been separate work on synthetic data as a PET. This has entailed commissioning a legal review of whether synthetic data could be considered personal data and developing an approach to privacy assessments of synthetic data to determine whether they are suitable for release31. CPRD’s approach to privacy preservation and risk assessment to enable release of synthetic data has been outlined in Myles et al.32.
Use of synthetic data in life sciences industry
Synthetic data is a promising privacy-preserving technology in the life sciences industry to facilitate responsible data sharing, improve representation in existing data sources, and for augmenting clinical trial datasets. Multiple validation studies have been conducted to demonstrate the validity and utility of synthetic data in generating RWE in different disease areas and for different research problems8,33,34,35.
Getting access to individual-level patient data is an ongoing challenge in the life sciences industry. Access to RWD that is generated through the delivery, administration, and reimbursement of healthcare can be prohibitively expensive and time consuming. Annual access to large, closed network, third-party, private payer claims data in the United States can cost between 100K and 800K US dollars depending on the breadth of data required, whereas specialty structured EHR data can cost between 3 and 5 million US dollars36. However, not all RWD is readily available due to privacy constraints, especially in Europe where GDPR restricts the use of sensitive information37. Synthetic data is being used to facilitate access to real world data sources in regions with restrictive privacy regulations as described in the government sector above.
Synthetic versions of RWD allow researchers to conduct data feasibility assessments, develop study protocols and write analytic code on readily accessible datasets38. This allows researchers to get access to individual patient data more quickly and accelerate timeline to insights. However, the fidelity of synthetic data can vary and it is often recommended that once protocols and analytic code have been developed, the analysis is re-run on real data to finalize results when attempting to draw conclusions about patterns in health data19,21. While adoption of synthetic data to facilitate data sharing is growing, there is limited guidance from privacy regulators. The Singaporean Personal Data Protection Commission has issued a draft guidance document regarding the use of synthetic data as a PET5. Additionally, the European Innovative Health Initiative partnered with the European Federation of Pharmaceutical Industries and Associations to publish a data sharing playbook to provide actionable guidance on data sharing in the health industry39. This playbook lists synthetic data as a notable PET. This shows the growing traction for synthetic data not just from the health data community but also with privacy regulators and industry groups.
Beyond its applications as a PET, synthetic data is also being applied in the life sciences industry as an innovative tool to enhance existing data sources40, including amplification, and de-biasing. These involve using the synthetic data generation models to produce datasets that are larger or intentionally have different compositions of patient characteristics than the data the models were trained on.
Using synthetic data to amplify existing datasets can be particularly impactful when attempting to use machine learning or AI models on small healthcare datasets6,41,42,43. Having a larger training dataset can allow more generalizable models to be fit. For example, when working with colorectal cancer registry data, using a Bayesian network to synthesize a dataset 4x the original dataset size resulted in an improvement in model predictive ability44. This has also been applied to improve the performance of machine learning models to predict nocturnal hypoglycemic events in type 1 diabetic patients45. This allows innovative machine learning and artificial intelligence models to be trained on small datasets and could be used to promote innovation in rare diseases.
Synthetic data can also be used to address representation bias in datasets, such as racial or gender bias28,46. In June 2024, the FDA released a draft guidance document: Diversity Action Plans to Improve Enrollment of Participants from Underrepresented Populations in Clinical Studies47. This guidance document solidifies the need to ensure that health research is representative of diverse populations. Synthetic data could be used as a complementary approach to improve representation of underrepresented groups during the analysis of clinical trials. Synthetic data can be used to de-bias data sources by synthesizing additional records from underrepresented groups through Synthetic Minority Augmentation (SMA). SMA has shown in low to moderate bias settings, higher precision for parameter estimates, higher overall model AUC, and improved fairness relative to a ground truth46. This approach could be used to perform analytic adjustments to improve treatment effect estimation when clinical trials had poor participation from under-represented populations.
Use of synthetic data in academia
The use of synthetic data offers several advantages for academia and offers opportunity for innovation and training. Synthetic data are increasingly used for machine learning model training and application1,48. Synthetic data can allow for quick testing of hypotheses and machine learning models which facilitate rapid iteration and experimentation in research. In the context of rare diseases, simulated synthetic data can help researchers perform analyses that would otherwise be limited. In addition, synthetic data mimic real data’s statistical properties without containing identifiable personal information or protected health information, allowing researchers to share and analyze data across centers without compromising privacy. Synthetic datasets can also be used in educational settings to teach data analytics in courses, datathons or workshops without the ethical concerns associated with using real data.
Federated data networks
Overview
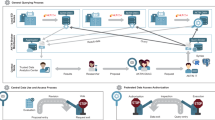
In recent years, federated data networks have emerged as a new privacy-protecting paradigm to query or analyze data from multiple sites without centralized pooling of individual-level data. Specifically for healthcare, federated data networks can facilitate access to sensitive health data across healthcare institutions, regional, and national borders, and have the potential to enable large cohort analysis on diverse populations. Compared to centralized data centers that are costly to maintain and subject to many constraints, federated networks can be nimbler and more scalable49. An illustration of federated data network and its differences to traditional centralized approach are outlined in Fig. 2.
Notable federated data networks include the Sentinel System funded by the US Food and Drug Administration50, the National Patient-Centered Clinical Research Network (PCORnet) funded by the Patient-Centered Outcomes Research Institute in the US51, the Canadian Network for Observational Drug Effect Studies (CNODES)52, the Observational Health Data Sciences and Informatics (OHDSI) in the US53, and the Data Analysis and Real World Interrogation Network (DARWIN-EU) funded by the European Medicines Agency54. See Table 1 for a selected list of federated data networks globally, many of which were funded by the government and run by academic institutions.
Tasks implemented on federated data networks
Federated data networks are increasingly used in machine learning tasks and certain types of statistical analyses55,56. For descriptive studies that aim to examine the utilization patterns of medical products or the natural history of diseases, the analysis can generally be performed using only summary-level information. For example, participating organizations will only need to share the number of incident outcome events and the number of at-risk individuals or at-risk person-times to estimate the incidence or incidence rate of the study outcome following initiation of a drug. This approach has been used to understand disease history and medication utilization in Sentinel, including studies that investigated the risk of arterial and venous thrombotic events associated with COVID-1957, the use of systemic corticosteroids for COVID-19 in the outpatient setting58, and the use of valsartan products containing nitrosamine impurities59.
For inferential studies that aim to assess the causal effects of medical treatments, meta-analysis of database-specific results had been the primary data-sharing and analytic option in the past. More recently, several federated data networks have exploited the properties of summary scores, such as propensity scores or disease risk scores, to perform sophisticated analysis without sharing individual-level data. For example, researchers can now perform propensity score matching, stratification, weighting, and outcome modeling using only summary-level information in federated data networks56. This approach has been used in Sentinel to examine the comparative safety of medical products, including studies that investigated the risk of neuropsychiatric events with montelukast60, the risk of hospitalized heart failure associated with anti-hyperglycemic agents61, and the risk of venous thromboembolism associated with oral contraceptives62.
Distributed regression has also been shown to be a viable privacy-protecting analytic method in these networks63,64. In distributed regression, participating organizations share summary-level intermediate statistics, often iteratively, with an analytic center (which could also be a data-contributing site) to produce the overall effect estimates. The approach has been piloted in PCORnet and in Sentinel65,66. In recent years, researchers have developed new distributed algorithms that do not require multiple exchanges of information across sites but still produce results highly comparable to those from pooled individual-level data analysis67,68.
Privacy safeguards and emerging applications in federated data networks
For certain complex analyses, such as analyses that require sophisticated adjustment for time-varying covariates, it may still be necessary to transfer de-identified individual-level data. To mitigate data privacy risks, most federated networks follow the “minimum necessary” standard, which only shares information that is needed for the study. Even for studies that only share summary-level information, there are additional safeguards that are put in place, such as suppressing small cell counts and additional masking of certain numbers to avoid back calculations. Some federated networks also mask the identities of the data-providing organizations to protect patient and institutional privacy. Although additional layers of privacy protection, such as differential privacy or homomorphic encryption, can be applied, these approaches are not widely used in existing federated networks.
Some networks have employed privacy-preserving record linkage to improve data completeness while preserving patient privacy. For example, PCORnet developed a privacy-preserving record linkage solution to identify overlap between EHR systems (i.e., individuals who appear in more than one EHR systems) and link EHR with medical claims69,70.
Federated networks and infrastructure can also be used in combination with other privacy-preserving technologies. For example, one study compared federated analysis with partial synthesis48,71. Partial synthesis is when real data is pooled with synthetic data to create a final dataset that is a combination of both. When there are multiple nodes, one would be designated as the analysis node and others as the contributing nodes. The contributing nodes would create synthetic variants of the datasets and send these to the analysis node, which would combine the synthetic datasets with their real data. Then an analysis is performed on this pooled and partially synthetic dataset. The results of that study, which only had two nodes, showed that the analysis using the partially synthetic dataset produced the same findings as the results using the federated analysis system. The main difference was that the portion of the project which used partial synthesis was completed in a fraction of the time due to the complexity of setting up a federated analysis system across jurisdictions and institutions.
Discussion
Both synthetic data and federated networks have several limitations and challenges. Operationally, a federated network is subject to the data infrastructure readiness, the extent of data harmonization, and coordination complexity. Scientifically, heterogeneity across data silos may complicate integration of site-specific information and generalizability of results. Similarly, synthetic data approaches also have challenges: the trade-off between privacy and utility from different synthetic data generation methods is a concern for users48,72; a particular synthetic data generation method could have limited generalizability across datasets and populations; and the unpredictability of what information is preserved in synthetic dataset could also complicate privacy assessment and data quality73.
Besides synthetic data and federated data networks, there are other types of PETs. For example, homomorphic encryption and differential privacy are also being tested in biomedical research, either independently or combined with other PETs. Differential privacy is an approach in which “random noise” is added until it becomes technically impossible to identify any individual in a dataset. Homomorphic encryption is a mathematical operation done on top of encrypted data1. It generates an encrypted result which, when decrypted, matches the result of the operations as if they had been performed on unencrypted data. For example, differential privacy and homomorphic encryption can add more rigorous privacy guarantee within a federated data network (e.g., enhancing institutions’ privacy), although doing so while maintaining the accuracy of the model can be challenging due to additional noise1,74. Synthetic data can also be enhanced with differential privacy guarantees, such as the N3C example discussed in the earlier section. Additionally, homomorphic encryption can be used to ensure privacy for computational tasks involving highly sensitive data, such as sensitive medical and genomic data75.
While many of these algorithms are proprietary, fostering cross-discipline collaborations can enhance the validation and demonstration of these methodologies, leading to more effective harmonization projects. Moreover, transparency in the generation processes of high-fidelity synthetic data is essential to assure stakeholders of patient privacy as well as validity and quality of data generated. From a legal standpoint, synthetic data may still be regarded as personal data, depending on various factors such as the source of the training data, the generation method (whether through perturbation or entirely synthetic means), the identifiability of the output, and the surrounding data environment, including technical and organizational measures against reverse engineering threats. Different legislative frameworks and the varying level of legal maturity in different countries could also impact the application of PETs. Therefore, consensus standards and country-specific guidance are needed to evaluate the privacy vulnerability, utility, and validity for synthetic data use. On the other hand, in federated network models, the main privacy-preserving principle is that data stays local. Nonetheless, robust data anonymization and security measures are needed to protect patient confidentiality as sharing certain types of patient data across different institutions and borders can raise privacy concerns.
Conclusions
As RWE becomes increasingly utilized in regulatory and healthcare decision-making, the protection of patient privacy and security while navigating the complexities of data use, access, and control has emerged as a crucial consideration for healthcare providers, researchers, regulators, and patients alike. Synthetic data presents a promising avenue for advancing research while addressing privacy concerns associated with real patient data; however, it should not be viewed as a simple and easy alternative to robust privacy management strategies. The balance and trade-off between privacy and utility need to be carefully considered for each use case76. Federated data networks also demonstrated opportunities to collaborate and access data cross different institutions in a privacy-preserving manner, but the upfront investments in proper infrastructure and coordination can be significant. Regardless of privacy-preserving technology type, a cross-sector collaborative approach, coupled with rigorous validation and transparency, should be the essential path forward in leveraging data responsibly while safeguarding patient privacy.
Data availability
No datasets were generated or analysed during the current study.
References
Cho, H. et al. Privacy-enhancing technologies in biomedical data science. Annu Rev. Biomed. Data Sci. 7, 317–343 (2024).
Government of Canada. Modernizing Canada’s Privacy Act, https://www.justice.gc.ca/eng/csj-sjc/pa-lprp/modern.html (2024).
Bloomberg Law. Consumer Data Privacy: EU’s GDPR vs. China’s PIPL. https://pro.bloomberglaw.com/insights/privacy/consumer-data-privacy-eus-gdpr-vs-chinas-pipl/ (2023).
DLA PIPER. Data protection laws in India. https://www.dlapiperdataprotection.com/?t=law&c=IN (2025).
Personal Data Protection Commission Singapore. Privacy Enhancing Technology (PET) Proposed Guide on Synthetic Data Generation (2024).
van der Ploeg, T., Austin, P. C. & Steyerberg, E. W. Modern modelling techniques are data hungry: A simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. 14, 137 (2014).
Geraci, J. et al. Current opportunities for the integration and use of artificial intelligence and machine learning in clinical trials: Good clinical practice perspectives. J. Soc. Clin. Data Manag. 5 (2025).
Wang, E. et al. Validation assessment of privacy-preserving synthetic electronic health record data: Comparison of original versus synthetic data on real-world COVID-19 vaccine effectiveness. Pharmacoepidemiol Drug Saf. 33, e70019 (2024).
U.S. Food and Drug Administration (FDA), Digital Health and Artificial Intelligence Glossary – Educational Resource, https://www.fda.gov/science-research/artificial-intelligence-and-medical-products/fda-digital-health-and-artificial-intelligence-glossary-educational-resource (2024).
Walonoski, J. et al. Synthea: An approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J. Am. Med Inf. Assoc. 25, 230–238 (2018).
Jeanson, F. et al. Medical calculators derived synthetic cohorts: a novel method for generating synthetic patient data. Sci. Rep. 14, 11437 (2024).
Al-Dhamari, I., Abu Attieh, H. & Prasser, F. Synthetic datasets for open software development in rare disease research. Orphanet J. Rare Dis. 19, 265 (2024).
Goncalves, A. et al. Generation and evaluation of synthetic patient data. BMC Med Res Methodol. 20, 108 (2020).
Park, N. et al. Data synthesis based on generative adversarial networks. Proc. VLDB Endow. 11, 1071–1083 (2018).
Ghosheh, G. O., Li, J. & Zhu, T. A survey of generative adversarial networks for synthesizing structured electronic health records. ACM Comput. Surv. 56, 147 (2024).
Choi, E. et al. in Proceedings of the 2nd Machine Learning for Healthcare Conference 68 (eds Doshi-Velez F. et al.) 286-305 (PMLR, Proceedings of Machine Learning Research, 2017).
United Nations Economic Commission for Europe (UNECE). Synthetic Data for Official Statistics - A Starter Guide. https://unece.org/statistics/publications/synthetic-data-official-statistics-starter-guide (2023).
Centers for Medicare & Medicaid Services (CMS). CMS 2008-2010 Data Entrepreneurs’ Synthetic Public Use File (DE-SynPUF). https://www.cms.gov/data-research/statistics-trends-and-reports/medicare-claims-synthetic-public-use-files/cms-2008-2010-data-entrepreneurs-synthetic-public-use-file-de-synpuf (2024).
The Simulacrum. The Simulacrum https://simulacrum.healthdatainsight.org.uk/available-data/ (2025).
Clinical Practice Research Datalink (CPRD). CPRD Synthetic Data, https://www.cprd.com/synthetic-data (2024).
Synthétiques, S. Systeme national des donnees de sante., https://documentation-snds.health-data-hub.fr/formation_snds/donnees_synthetiques/ (2022).
Chauhan, P. Synthetic version of anonymized Norway Registry data containing prescriptions and hospitalization of the patients. https://doi.org/10.18710/YABAGM, DataverseNO, V1 (2024).
Hod, S. & Canetti, R. In 2025 IEEE Symposium on Security and Privacy (SP) 100-100. (IEEE Computer Society, 2025).
Foraker, R. et al. The National COVID Cohort Collaborative: Analyses of original and computationally derived electronic health record data. J. Med Internet Res 23, e30697 (2021).
Wang, Z. C., Myles, P. & Tucker, A. Generating and evaluating cross-sectional synthetic electronic healthcare data: Preserving data utility and patient privacy. Comput Intell. -Us 37, 819–851 (2021).
Tucker, A., Wang, Z. C., Rotalinti, Y. & Myles, P. Generating high-fidelity synthetic patient data for assessing machine learning healthcare software. Npj Digital Med. 3 (2020).
Wang Z. et al. Evaluating a Longitudinal Synthetic Data Generator using Real World Data. IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS). 259–264 (2021).
Draghi, B., Wang, Z. C., Myles, P. & Tucker, A. Identifying and handling data bias within primary healthcare data using synthetic data generators. Heliyon 10, e24164 (2024).
UK Department for Business, E. I. S. Projects selected for the Regulators’ Pioneer Fund, https://www.gov.uk/government/publications/projects-selected-for-the-regulators-pioneer-fund/projects-selected-for-the-regulators-pioneer-fund-2022#month-projects-1 (2022).
Valena R. et al. Synthetic data for development of AI as a medical device. Regulatory considerations. MHRA & PHG Foundation (2025).
Mitchell, C., Hill, E. R., Are synthetic health data ‘personal data’? PHG Foundation, Cambridge UK. https://www.phgfoundation.org/media/886/download/Are%20synthetic%20health%20data%20%E2%80%98personal%20data%E2%80%99.pdf?v=1&inline=1 (2023).
Myles, P., Mitchell, C., Redrup Hill, E., Foschini, L. & Wang, Z. High-fidelity synthetic patient data applications and privacy considerations. J. Data Prot. Priv. 6, 344–354 (2024).
Lun, R., Siegal, D., Ramsay, T., Stotts, G. & Dowlatshahi, D. Synthetic data in cancer and cerebrovascular disease research: A novel approach to big data. PLoS One 19, e0295921 (2024).
Reiner Benaim, A. et al. Analyzing medical research results based on synthetic data and their relation to real data results: Systematic comparison from five observational studies. JMIR Med. Inf. 8, e16492 (2020).
Foraker, R. E. et al. Spot the difference: Comparing results of analyses from real patient data and synthetic derivatives. JAMIA Open 3, 557–566 (2020).
Dagenais, S., Russo, L., Madsen, A., Webster, J. & Becnel, L. Use of real-world evidence to drive drug development strategy and inform clinical trial design. Clin. Pharm. Ther. 111, 77–89 (2022).
Kalkman, S., Mostert, M., Gerlinger, C., van Delden, J. J. M. & van Thiel, G. Responsible data sharing in international health research: A systematic review of principles and norms. BMC Med Ethics 20, 21 (2019).
Gatto, N. M., Vititoe, S. E., Rubinstein, E., Reynolds, R. F. & Campbell, U. B. A structured process to identify fit-for-purpose study design and data to generate valid and transparent real-world evidence for regulatory uses. Clin. Pharm. Ther. 113, 1235–1239 (2023).
Chlebus, M. & Laverty, H. Data Sharing Playbook. European Federation of Pharmaceutical Industries and Associations, (2024).
Myles, P. et al., High-Fidelity Synthetic Data Applications for Data Augmentation. in Deep Learning - Recent Findings and Research (eds Manuel Jesus Domínguez-Morales, Javier Civit-Masot, Luis Muñoz-Saavedra, & Robertas Damaševičius) (IntechOpen, 2024).
Sabay, A., Harris, L., Bejugama, V. & Jaceldo-Siegl, K. Overcoming small data limitations in heart disease prediction by using surrogate data. SMU Data Sci. Rev. 1, 12 (2018).
Ahmadian, M. et al. Overcoming data scarcity in radiomics/radiogenomics using synthetic radiomic features. Comput Biol. Med. 174, 108389 (2024).
Zhao, Y. & Duangsoithong, R. in 2019 16th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology (ECTI-CON). 814-817.
Kim, H. et al. Synthetic data improve survival status prediction models in early-onset colorectal cancer. JCO Clin. Cancer Inf. 8, e2300201 (2024).
Noguer, J., Contreras, I., Mujahid, O., Beneyto, A. & Vehi, J. Generation of individualized synthetic data for augmentation of the type 1 diabetes data sets using deep learning models. Sensors (Basel) 22 (2022).
Juwara, L., El-Hussuna, A. & El Emam, K. An evaluation of synthetic data augmentation for mitigating covariate bias in health data. Patterns (N. Y) 5, 100946 (2024).
Administration., U. S. F. A. D. Diversity Action Plans to Improve Enrollment of Participants from Underrepresented Populations in Clinical Studies. Draft Guidance for Industry. (2024).
Rajotte, J. F. et al. Synthetic data as an enabler for machine learning applications in medicine. Iscience 25 (2022).
Mahajan, P., Macias, C., Barda, A. & Fung, C. M. Federated data health networks hold potential for accelerating emergency research. J. Am. Coll. Emerg. Physicians Open 4, e12968 (2023).
Brown, J. S. et al. The US Food and Drug Administration Sentinel System: a national resource for a learning health system. J. Am. Med Inf. Assoc. 29, 2191–2200 (2022).
Fleurence, R. L. et al. Launching PCORnet, a national patient-centered clinical research network. J. Am. Med Inf. Assoc. 21, 578–582 (2014).
CNODES. Canadian Network for Observational Drug Effect Studies (CNODES) https://www.cnodes.ca/ (2025).
Hripcsak, G. et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud. Health Technol. Inf. 216, 574–578 (2015).
The European Medicines Agency (EMA), DARWIN EU, https://www.darwin-eu.org/ (2024).
Wong, J. et al. Applying machine learning in distributed data networks for pharmacoepidemiologic and pharmacovigilance studies: Opportunities, challenges, and considerations. Drug Saf. 45, 493–510 (2022).
Toh, S. Analytic and data sharing options in real-world multidatabase studies of comparative effectiveness and safety of medical products. Clin. Pharm. Ther. 107, 834–842 (2020).
Lo Re, V. et al. Association of COVID-19 vs influenza with risk of arterial and venous thrombotic events among hospitalized patients. JAMA 328, 637–651 (2022).
Bradley, M.C. et al. Systemic corticosteroid use for COVID-19 in US outpatient settings from April 2020 to August 2021. JAMA 327, 2015–2018 (2022).
Eworuke, E. et al. Exposure to valsartan products containing nitrosamine impurities in the United States, Canada, and Denmark. Pharmacoepidemiol Drug Saf. 33, e5849 (2024).
Sansing-Foster, V., et al. Risk of psychiatric adverse events among Montelukast users. J. Allergy Clin. Immunol. Pract. 9, 385–393.e312 (2021).
Toh, S. et al. Risk for hospitalized heart failure among new users of saxagliptin, sitagliptin, and other antihyperglycemic drugs: A retrospective cohort study. Ann. Intern Med. 164, 705–714 (2016).
Li, J. et al. Association of risk for venous thromboembolism with use of low-dose extended- and continuous-cycle combined oral contraceptives: A safety study using the sentinel distributed database. JAMA Intern Med. 178, 1482–1488 (2018).
Karr, A. F., Xiaodong, L., Sanil, A. P. & Reiter, J. P. Secure regression on distributed databases. J. Comput. Graph. Stat. 14, 263–279 (2005).
Fienberg, S. E., Fulp, W. J., Slavkovic, A. B. & Wrobel, T. A. in Privacy in Statistical Databases. PSD 2006. 4302 (Springer, Berlin, Heidelberg, 2006).
Her, Q. et al. Distributed regression analysis application in large distributed data networks: Analysis of precision and operational performance. JMIR Med. Inf. 8, e15073 (2020).
Toh, S. et al. Privacy-protecting multivariable-adjusted distributed regression analysis for multi-center pediatric study. Pediatr. Res. 87, 1086–1092 (2020).
Duan, R., Boland, M. R., Moore, J. H. & Chen, Y. ODAL: A one-shot distributed algorithm to perform logistic regressions on electronic health records data from multiple clinical sites. Pac. Symp. Biocomput 24, 30–41 (2019).
Luo, C. et al. ODACH: A one-shot distributed algorithm for Cox model with heterogeneous multi-center data. Sci. Rep. 12, 6627 (2022).
Marsolo, K. et al. Assessing the impact of privacy-preserving record linkage on record overlap and patient demographic and clinical characteristics in PCORnet(R), the National Patient-Centered Clinical Research Network. J. Am. Med Inf. Assoc. 30, 447–455 (2023).
Kiernan, D. et al. Establishing a framework for privacy-preserving record linkage among electronic health record and administrative claims databases within PCORnet((R)), the National Patient-Centered Clinical Research Network. BMC Res Notes 15, 337 (2022).
Azizi, Z. et al. A comparison of synthetic data generation and federated analysis for enabling international evaluations of cardiovascular health. Sci. Rep. 13, 11540 (2023).
Kaabachi, B. et al. A scoping review of privacy and utility metrics in medical synthetic data. NPJ Digit Med. 8, 60 (2025).
Stadler,T., Bristena, O. & Troncoso, C. Synthetic Data–Anonymisation Groundhog Day in 31st USENIX Security Symposium (2022).
Cho, H. et al. Secure and federated genome-wide association studies for biobank-scale datasets. Nat. Genet 57, 809–814 (2025).
Bos, J. W., Lauter, K. & Naehrig, M. Private predictive analysis on encrypted medical data. J. Biomed. Inf. 50, 234–243 (2014).
Adams, T. et al. On the fidelity versus privacy and utility trade-off of synthetic patient data. iScience 28, 112382 (2025).
Acknowledgements
This work did not receive any financial support from any funding agency in the public, commercial, or not-for-profit sectors. The views and opinions expressed in this paper are those of the authors and may not necessarily reflect the views or opinions of any other public or private entity.
Author information
Authors and Affiliations
Contributions
E.H.W. and M.B. conceived the paper. E.H.W., P.M., R.F., S.T., L.M., K.E.E., and M.B. contributed to the content and writing of the paper and have reviewed and approved the content.
Corresponding author
Ethics declarations
Competing interests
E.H.W. and M.B. are current employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA, and may hold equity interest in Merck & Co., Inc., Rahway, NJ, USA. L.M. is a current employee of Aetion, a Datavant Company.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, E.H., Myles, P., Foraker, R. et al. Crossing borders securely: synthetic data and federated networks for privacy-preserving access to real-world data and emerging use cases. npj Digit. Med. 8, 758 (2025). https://doi.org/10.1038/s41746-025-02126-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02126-8