Abstract
Although aging clocks predicting the age of individual organisms have been extensively studied, the age of individual cells remained largely unexplored. Most recently single-cell omics clocks were developed for the mouse, however, extensive profiling the age of human cells is still lacking. To fill this gap, here we use available scRNA-seq data of 1,058,909 blood cells of 508 healthy, human donors (between 19 and 75 years), for developing single-cell transcriptomic clocks and predicting the age of human blood cells. By the application of the proposed cell-type-specific single-cell clocks, our main observations are that (i) transcriptomic age is associated with cellular senescence; (ii) the transcriptomic age of classical monocytes as well as naive B and T cells is decreased in moderate COVID-19 followed by an increase for some cell types in severe COVID-19; and (iii) the human embryo cells transcriptomically rejuvenated at the morulae and blastocyst stages. In summary, here we demonstrate that single-cell transcriptomic clocks are useful tools to investigate aging and rejuvenation at the single-cell level.

Similar content being viewed by others
Introduction
An aging society poses enormous challenges to both healthcare and the economy. To handle this growing problem we need to uncover the internal mechanisms of aging. Until now, it has not been possible to measure the aging process with a precision that can be used for practical applications. However, the recently developed epigenetic aging clocks, i.e., machine learning models that predict an individual’s age using epigenetic markers, have emerged as promising tools for measuring the aging process1,2,3,4,5,6,7,8. In addition to epigenetic clocks, transcriptomic clocks were also developed for the estimation of age9,10,11,12,13. Common in these “omics” aging clocks is that they have been trained on data originating from tissue-level (“bulk”) samples (e.g. blood, saliva, skin, and multi-tissue) that contain a mixture of multiple cells of various cell types14. Given that cell types exhibit distinct methylation and gene expression patterns, the predictions of the aging clocks might be skewed by age-related changes in cell compositions4,15,16.
Single-cell omics, especially single-cell transcriptomics is, revolutionizing cell biology by profiling individual cells of different cell types hence serves as a promising tool to measure the aging process at the single-cell level17,18,19. Importantly, using single-cell data instead of bulk samples excludes cell composition bias14. This has the promise to dissect the role of tissue heterogeneity in biological aging at the cellular level for multicellular organisms.
Recently, an algorithm (scAge) was developed that can determine the age of single cells based on methylation data and evaluated the age of different cells (such as hepatocytes, muscle stem cells, and various embryo cells) from mice20. As scAge was trained using bulk tissue data, the development of a cell-level aging clock directly trained on single-cell data remained a challenge.
Filling this gap a cell-type-specific single-cell transcriptomic aging clock was developed for neurogenic regions of the mouse and revealed that heterochronic parabiosis and exercise reverse transcriptomic aging in neurogenic regions21. Furthermore a very recent study developed single-cell transcriptomic aging clocks using human peripheral blood mononuclear cells (PBMC) and they revealed that supercentenarians were much younger than their actual age22. Importantly, the above-mentioned transcriptomic clocks, although using single-cell data, mainly focused on the age prediction of individuals (mice or humans) instead of cells.
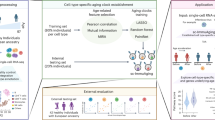
Here, we developed human cell-type-specific aging clocks directly trained on single-cell transcriptomic data and focused on the age prediction of single cells rather than the age of individuals. We also applied our clocks to different external datasets and, at the first time, evaluated the age of single cells originated from COVID-19 patients and human embryos (Fig. 1a).
a Schematics of the present study. Cell-type-specific single-cell transcriptomic clocks were built based on human blood samples. In addition, the clocks were applied to human blood samples originated from healthy and COVID-19 donors, and to human pre-implantation embryo cells and human embryonic stem cells. The figure was created with BioRender.com. b Number of genes whose expression profile shows significant negative/positive correlation with chronological age (light blue/red bars), and the ones remained significant after Bonferroni correction (dark blue/red bars). c Genes were showing the strongest correlations with age. The bars show the strength of the correlation (all of them are significant even after Bonferroni correction) and the cell types in which the specific correlations are detected are also shown (IL: innate lymphoid cell, naive B: naive B cell, CD8+ T: CD8-positive, alpha-beta T cell). d The performance of cell-type-specific single-cell clocks. The blue bars represent significant, while the gray bars the non-significant correlations between chronological and predicted age after Bonferroni correction. e Age predictions for the individual cells by the 9 best-performing cell-type-specific single-cell clocks. Each point corresponds to a cell of a given cell type. The number of samples (n), the regression lines as well as r and MAE calculated between chronological and predicted ages are provided for each panel.
Results
Profiling the age of human single-cells by cell-type-specific clocks
For the development of single-cell aging clocks, we used scRNA-seq data from the Asian Immune Diversity Atlas (AIDA) v123, which comprises 1,058,909 PBMCs in total from 508 healthy, human donors with age between 19 and 75 years (Supplementary Fig. 1a, b).
Considering the cells from 33 cell types and the 36,266 genes in the AIDA dataset, we calculated the Spearman correlation coefficient (ρ) between log-normalized gene expressions and ages for each cell type and gene separately (Fig. 1b). We observed more positively than negatively correlated genes in the majority of the cell types (p = 0.0005), suggesting that genes tend to be upregulated during aging. The strongest positive or negative correlation with age was observed for the expression of PPP1R15A, JUND, FOS, PCBP1 and DUSP1 in innate lymphoid cells (ρ = 0.42, ρ = −0.395, ρ = 0.391, ρ = −0.343, ρ = 0.347, respectively) as well as for KLF6, MT-ATP8 and GNAS in CD8-positive alpha-beta T cells (ρ = 0.33, ρ = −0.325 and ρ = −0.313, respectively), see Fig. 1c. The expression of FOS and DUSP1 positively correlated with age in naive B cells as well (ρ = 0.339, ρ = 0.309, respectively).
To contribute to the understanding of the process of single-cell transcriptomic aging, we created cell-type-specific aging clocks. We trained ElasticNet regression models with 5-fold cross-validation using the log-normalized gene expression counts of single cells to predict chronological age (for details see the Methods). 31 (94%) of the 33 cell-type-specific clocks showed a significant correlation between age and predicted age with Pearson correlation coefficients (r) between 0.22 and 0.5 (Fig. 1d). The MAE was between 8.64 and 10.48 years. Nine clocks showed an r above 0.4 and mean absolute error (MAE) below 10.1 years (Fig. 1e). The CD8-positive alpha-beta T cell clock was the best-performing model with r = 0.5036 and MAE = 8.6426 and interestingly eight of the top nine were based on some type of T cells. The average number of non-zero coefficients, i.e. the average number of genes affecting the predicted age is mostly less than 100.
We measured the variability of gene expressions, which is often referred to as transcriptional noise24,25, and their associations with the proposed single-cell clocks. We calculated the standard deviation (std) and interquartile range (IQR) of the normalized expressions of each gene for cells from a particular cell type and donor in the AIDA dataset. Then we tested whether the variability of the genes was significant (i.e., std/IQR significantly differed from zero, see the section “Methods”). We found that a large number of genes in each cell type was transcriptionally noisy, in other words, the expression values of cells from a given cell type varied considerably. We found that much fewer genes were transcriptionally noisy based on IQR than std (Supplementary Fig. 1j). We also measured the correlation of chronological age and transcriptional noise (based on IQR) for cell type and gene. We found at least one significantly correlated gene for 23 of the 33 cell types. We also found that the transcriptional noise rather decreased for 10 cell types and increased for 13 cell types with age (Supplementary Fig. 1k), meaning that in 10 cell types, gene expressions tended to stabilize showing less variability during aging. This means that the relation of aging and transcriptional noise varies among cell types to a great extent, and aging does not necessarily increase variability in every cell type. Finally, we checked if the genes with significant transcriptional noise overlapped with the important clock features (i.e. genes with non-zero regression coefficient). Among the nine best-performing single-cell clocks only two had features with zero transcriptional noise (double negative T regulatory clock: 3 of 637 important features; innate lymphoid clock: 236 of 677 important features) showing that genes that were found to be important in the age predictions were typically showed high variability in gene expression.
We also examined the relation of predicted age of single cells to the process of cellular senescence. We performed gene set enrichment analysis (GSEA) using the predictions given by the top 5 best-performing clocks as phenotypes and the REACTOME_CELLULAR_SENESCENCE gene set from the Molecular Signatures Database (MSignDB). In all 5 cases, the cellular senescence-related genes were enriched among the genes positively correlating with the predicted ages (Normalized Enrichment Score = 1.09, 1.01, 2.1, 1.62, 1.55 for the CD8-positive, alpha-beta T cells, innate lymphoid cells, CD8-positive, alpha-beta memory T cells, effector memory CD4-positive, alpha-beta T cells and gamma-delta T cells, respectively). Consequently, predicted age of cells given by the proposed single-cell clocks may be able to measure the process of cellular senescence.
We investigated the overlap between cell-type-specific marker genes and the important clock features. We found a small overlap between the whole set of marker genes and important clock features. Only 20 of the 79 marker genes were features in at least one clock, and these 20 genes were less than 8% of the whole set of important clock features (Supplementary Fig. 2a). In the case of 20 of the 32 cell types (62.5%) there was no overlap between the markers and the clock features of the given cell type (Supplementary Fig. 2b, c). The largest proportion of marker genes among clock features was 11.11% while the largest proportion of clock features among marker genes was 28.57%. Consequently, the marker genes, which characterize the different cell types were mostly distinct from the clock feature genes, which characterize aging.
Estimating the age of donors
In addition to cell-level age prediction, we also used multiple different methods to give estimate on donor age.
Cell-type-level averaging of single-cell age predictions for each donor resulted in a slight increase in correlation between chronological and predicted age and also a slight decrease in MAE (19 out of the 33 cell types) compared to the single-cell results (Supplementary Fig. 3a). Similarly, calculating donor-level predicted ages by averaging the predictions over all cells for each donor resulted in similar performance (Supplementary Fig. 3b).
To estimate donor age, we also constructed pseudo-bulk samples22 and trained regression models (see section “Methods”) on these donor-level averaged gene expression levels. Our cross-validation-based results (Supplementary Fig. 3c) indicate strong predictive performance with MAE = 5.97. Furthermore, we constructed cell-type-specific pseudo-bulk samples and trained cell-type-specific pseudo-bulk models (Supplementary Fig. 3d). Here particular cell types, notably CD4-positive, alpha-beta T cells derived pseudo-bulk samples provide the best donor age estimates.
Single-cell transcriptomics data allow the examination of the cell-type composition of samples that could also reveal important characteristics of aging. We calculated cell type proportions for each donor in the AIDA dataset (see Methods, Supplementary Fig. 4a), and examined how they change during aging. Proportions of 14 cell types out of the 33 correlated significantly with chronological age (based on Spearman’s rank correlation with Bonferroni correction to the total number of cell types). We observed that among these 14 cell types, 7 showed significant increases and 7 showed significant decreases in their proportion with aging, which suggests balanced changes in cell type proportions (Supplementary Fig. 5a). The proportion of the naive thymus-derived CD8-positive alpha-beta T cells showed the strongest correlation with age (ρ = −0.62). These results recapitulate the findings of Zhu et al.22 and are in alignment with the observation about the reduction in thymic output with age26,27. The proportions of naive CD8-positive T cells in samples of 20-year-old young individuals was around 12%, while for 70-year-old people their proportions dropped down to around 2% (Supplementary Fig. 4b). We also found that the proportions of naive CD8- and naive CD4-positive alpha-beta T cells decreased with age, and the proportions of the memory T cell types increased with age, which supports previous findings about T cell productions in the elderly27,28.
To explore the association of cell type proportions with chronological age, we trained ElasticNet models with 5-fold cross-validation to predict the age of donors using their cell type proportions (see section “Methods”). The age prediction performance were MAE = 8.75 and r = 0.49 (Supplementary Fig. 5b), thus cell type proportions had similar predictive power as single-cell gene expressions. Regarding the number of cell types, that contributed to the predictions (i.e. having non-zero coefficients in the model), there were usually 11–13 such cell types in each cross-validation clock, and there were 6 types, which contributed to all clocks (Supplementary Fig. 5c). The aggregation of cell types into larger categories, i.e. CD8-positive, alpha-beta T cells, CD4-positive, alpha-beta T cells, dendritic cells, natural killer cells, and monocytes, and training the model using the proportion of these larger types did not produce valuable predictions (r = −0.1092, MAE = 11).
We combined predictions of single-cell gene expression and cell type proportion clocks to check if such an ensemble approach could approximate the chronological age of donors more accurately than the individual models (see section “Methods”). We observed that in each case, the performance improved compared to the given single-cell expression clock, and many expression-based models combined with a proportion clock provided better results than simply considering the proportion-based model. The Pearson r of age and predicted age increased by around 0.1 in general (Supplementary Fig. 5d). Altogether, we observed that the combination of different types of aging clocks produced more accurate predictions of chronological age.
External validation of aging clocks on healthy samples
To assess the generalization power of the proposed aging clocks, we applied them to four independent PBMC datasets, the eQTL dataset29 and the healthy samples from the Yoshida et al.30, Liu et al.31, and the Stephenson et al.32 datasets (Supplementary Fig. 1c, d, f, h).
In the case of the healthy samples of the Yoshida et al. dataset, 18 (78.3%) of the 23 cell-type-specific single-cell clocks showed a significant positive correlation between age groups and predicted age with a Spearman ρ between 0.17 and 0.48 (Fig. 2a, and Supplementary Fig. 6a). We concluded that the correlation coefficients were comparable to that of the AIDA dataset with a slight decrease. MAE was not calculated as exact ages were not available for this dataset only age groups. Cells from aged adult donors were predicted to be significantly older than cells from adults for 13 cell types, while significant differences between adolescent and adult groups were observed for 19 cell types.
a The performance of cell-type-specific single-cell clocks in the healthy subset of the Yoshida et al. dataset. The blue bars represent significant, while the gray bars the non-significant correlations between chronological and predicted age after Bonferroni correction. b The performance of aging clocks in the healthy subset of the Liu et al. dataset. c The performance of aging clocks in the eQTL dataset. d The performance of aging clocks in the healthy subset of the Stephenson et al. dataset. Dark blue/gray bars correspond to the performance achieved on all available samples, light blue/gray bars and the shown correlation coefficients show the performance of the clocks when applied to samples in and above the 4th decade.
For the healthy samples of the Liu et al. dataset, 12 (63.2%) of the 19 cell-type-specific single-cell clocks showed significant positive correlation between chronological and predicted age with r between 0.07 and 0.4 (Fig. 2b). The MAE was between 10.61 and 23.48 years. The naive thymus-derived CD4-positive alpha-beta T cells showed the strongest association with r = 0.4 and MAE = 15 years. The best-performing clocks on AIDA, showed low performance on the Liu et al. dataset (Supplementary Fig. 6b). The predicted age of cells from adults above 65 years was significantly higher compared to the adults below 65 in 10 out of the 19 cases.
For the eQTL dataset, 16 of the 24 cell-type-specific single-cell clocks showed a significant positive correlation between chronological age and predicted age with r between 0.03 and 0.17 (Fig. 2c, and Supplementary Fig. 6c). The strongest correlation was Pearson r = 0.17 for the gamma-delta T cells. MAEs between chronological and predicted age were above 20 years. However, the predicted age of cells from aged adults was significantly higher compared to the adults in 14 out of the 24 cases. Altogether, the model performances highly decreased for the eQTL dataset compared to the AIDA dataset both in terms of MAE and correlation, but tendencies, i.e. significant (positive) correlation between chronological and predicted ages were still clearly shown.
In the case of the healthy samples of the Stephenson et al. dataset, surprisingly, the predicted age of cells from donors in the 3rd decade were higher than from samples of the other age groups, for the majority of the cell types, which often resulted in negative overall Spearman correlation between age groups and predicted age. However, considering samples from donors in and above the 4th decade, 15 of 26 cell types showed a significant positive Spearman correlation (ρ was between 0.08 and 0.4) between age groups and predicted age (Fig. 2d and Supplementary Fig. 6d). The best performance was shown by the effector memory CD4-positive alpha-beta T cell clock with ρ = 0.4.
The cell type proportion clock predicted ages on the Yoshida et al. dataset with Spearman ρ = 0.4268 with the age categories, that is performed slightly worse than the best single-cell clocks, but approached the performance on the training dataset (AIDA). Moreover, adults were predicted significantly older than adolescents, however, there was no significant difference between the predicted age of adult and aged adult groups, possibly because of the small sample size. The clock did not show a significant correlation between chronological and predicted age in the case of the Liu et al. dataset, but it is possibly because of the large number of missing cell types (compared to AIDA) in this dataset. The performance of this clock on the eQTL dataset was Pearson r = 0.6146 and MAE = 16.8782 along with a significant difference between adult and aged adult groups. In the case of the Stephenson et al. dataset, the Spearman correlation between age groups and predicted age was ρ = 0.6297, meaning that the clock outperformed its training performance on this dataset, although direct comparison was not possible due to the lack of information about exact ages. Consequently, the proposed cell-type proportion clocks generalized well to unseen data in terms of correlation between predicted and chronological age, however, the MAE highly increased in the case of the eQTL dataset (Supplementary Fig. 7a).
The combination of single-cell and cell-type proportion clocks resulted in improved age predictions on the Yoshida et al. dataset compared to the training dataset (AIDA). The Spearman correlation coefficient of predicted age and age groups was usually around 0.5, and in some cases the performance approached the training results (Supplementary Fig. 7b). We note that similarly to the case of the individual cell type proportion clock, there was no significant difference between the adult and aged adult groups. Due to the poor performance of the cell type proportion clock on the Liu et al. dataset, the combination of its predictions with single-cell predicted ages also did not produce satisfactory results (Supplementary Fig. 7c). The combined clock approach improved the performance of age prediction on the eQTL dataset. This way, the MAE also increased, but the difference between the adult and aged adult groups remained significant (Supplementary Fig. 7d). Finally, the combination of single-cell and cell-type proportion-based predictions resulted in greatly improved performance on the Stephenson et al. dataset (Supplementary Fig. 7e). In some cases the Spearman correlation between age groups and predicted age even exceeded 0.8.
To further evaluate the generalization power of the proposed single-cell clocks, we also applied them to cells derived from lung samples of healthy donors (Lung dataset of Sikkema et al.33). In the case of 10 of the 11 cell types (91%), the predicted ages correlated significantly and positively with the chronological age (Supplementary Fig. 8a). The Pearson r was between 0.12 and 0.51, and the MAE between 9.85 and 16.28 years. The best performance was achieved by the T-cell clock with r = 0.51 and MAE = 9.85. Among the 9 best-performing clocks on AIDA we could apply two and they showed a decreased performance on this external dataset (Supplementary Fig. 8b, c).
In summary, we observed that the cell-type-specific single-cell clocks generalized well to external data in the case of the healthy subset of the Yoshida et al. dataset and for the healthy samples above 3rd decade of the Stephenson et al. dataset. The clocks showed a moderate generalization ability in the case of the Liu et al. dataset, and the Lung dataset while they did not generalize well for the eQTL dataset, however, significant differences between age groups and significant correlations between chronological and predicted ages were shown in many cases. Additionally, we observed that the generalization power of the cell-type proportion and combined clocks were better than the cell-type-specific single-cell clocks. Altogether, the clocks possibly give insight into the process of aging when applied to unseen data, however, this may depend on the application dataset.
Decreased transcriptomic age of classical monocytes, naive B and T cells in moderate COVID-19 followed by an increase for some types in severe COVID-19
To examine the effect of SARS-CoV-2 infection on the transcriptomic age of single cells, we applied the proposed cell-type-specific single-cell aging clocks to COVID-19 and healthy samples of adults donors of the Yoshida et al., Liu et al. and Stephenson et al. datasets (Supplementary Fig. 1e, g, i). We compared the different severity groups and the healthy group based on the predicted age of cells for each cell type separately. In order to account the age distribution differences of the different groups we included chronological age as a confounder variable in the comparison of the groups (see section “Methods”). We only considered the findings consistent over all three examined datasets, to avoid drawing conclusions based on noise or the uncertainty of the clocks.
We observed that the predicted age of classical monocyte cells from COVID-19-afflicted donors was significantly lower than those originating from healthy donors, across all severity conditions (Fig. 3a). We also observed a decreased predicted age of naive thymus-derived CD4 and CD8-positive, alpha-beta T cells and naive B cells from donors in moderate severity compared to healthy controls (Fig. 3b–d). Moreover, the predicted age of samples from the severe group was significantly higher than those from the moderate group for the following four cell types: naive thymus-derived CD4-positive, alpha-beta T cell; CD8-positive, alpha-beta memory T cell; naive B cell; gamma-delta T cell (Supplementary Fig. 9). These findings were consistent for all of the three examined datasets.
Predicted age of a classical monocyte cells, b naive thymus-derived CD4-positive alpha-beta T cells, c naive thymus-derived CD8-positive alpha-beta T cells, and d naive B cells from healthy and COVID-19-afflicted donors in the Yoshida et al., Liu et al., and Stephenson et al. datasets, respectively, applying the correspondent cell-type-specific single-cell aging clocks. We compared the different severity stages to the healthy group. The p-value between the healthy and moderate groups was calculated by a generalized linear model fitted to the predicted age with severity as the independent variable and age as a confounder variable. The sample size (n) of each group is also shown for each panel.
We further examined the expression of clock feature genes for the cell types we observed decrease/increase in predicted ages of COVID-19 patients. We say that a gene “supports the rejuvenation” (i.e. the transcriptomic age decrease) if its expression changes in the opposite direction during rejuvenation compared to aging. In the case of the classical monocytes, we found 7 genes with decreased expression in cells derived from COVID-19 patients (Fig. 4a–c), while they were upregulated during aging according to the classical monocyte clock (Fig. 4d). Interestingly, 4 of 7 genes are involved in the inflammatory response: ILB1 and CXL8 are coding pro-inflammatory cytokines (IL-1β, and IL-8, respectively) while NFKBIA and NFKBIZ regulates pro-inflammatory cytokines through the NF-κB pathway34. The reduced expression of these genes over all severity conditions suggests suppressed inflammation. In the naive B cells, from donors in moderate condition, we observed 4 genes (DUSP1, FOS, FOSB, and JUN) with decreased expression compared to healthy samples (Fig. 4e–g), which genes were upregulated during aging (Fig. 4h). The decreased expression of the MT-ATP6 and PPP1R15A genes, and the increased expression of the GYPC gene in the naive thymus-derived CD4-positive alpha-beta T cells of moderate condition donors were found to be supporting the rejuvenation of these cells, while in the case of the naive thymus-derived CD8-positive alpha-beta T cells, the decreased expression levels of the DUSP1 gene supported the rejuvenation (Supplementary Fig. 10).
Normalized expression of genes in classical monocyte cells from the a Yoshida et al., b Liu et al., and the c Stephenson et al. dataset supporting the decreased predicted age of classical monocytes in the moderate stage of COVID-19 compared to healthy individuals. d The regression coefficient of the supporting genes in the five classical monocyte cross-validation clocks. The bars illustrate the distribution of the coefficients. Normalized expression of genes in naive B cells from the e Yoshida et al. f Liu et al., and g Stephenson et al. dataset supporting the decreased predicted age of naive B cells of donors in the moderate stage of COVID-19 compared to healthy individuals. h The regression coefficient of the supporting genes in the naive B cell cross-validation clocks. The bars illustrate the distribution of the coefficients. The p-value between the two groups was calculated by a generalized linear model fitted to the gene expression with severity as the independent variable and age as a confounder variable. The sample size (n) of each group is also shown for the relevant panels.
For the cell types, we observed consistently increased transcriptomic age in severe COVID-19, none of the related clock feature genes showed the same behavior as during aging of healthy individuals.
Overall, our data surprisingly show that the transcriptomic age of classical monocyte cells decreased in COVID-19-afflicted subjects, and the suppressed expression of multiple inflammatory response-related genes may explain this phenomena. Furthermore, the transcriptomic age of naive B cells and naive T cells also decreased in moderate COVID-19.
Transcriptomic rejuvenation of human embryo cells at the morulae and blastocyst stages
To reveal the transcriptomic age dynamics of human embryogenesis at the single-cell level, here, we applied the proposed single-cell transcriptomic aging clocks to three datasets of early development (Yan et al.35, Petropoulos et al.36, and Meistermann et al.37). We examined the predictions of the top 9 best-performing cell-type-specific single-cell clocks based on the AIDA dataset (presented in Fig. 1d). The majority of clocks showed a significant transcriptomic age decrease of the morulae stage compared to the 8-cell stage (6 of 9 cases in the Yan et al. dataset and 7 of 9 cases in Petropoulos et al. datasets), followed by a further significant decrease at the blastocyst stage (8 of 9 cases in the Yan et al. dataset, 6 of 9 cases in Petropoulos et al. datasets, and 5 of 9 in the Meistermann et al. dataset; see Fig. 5a, b, and Supplementary Fig. 11a). We observed similar dynamics when we aggregated the predictions of all clocks, except for the dendritic cell and erythrocyte clocks due to their non-significant performance in the AIDA dataset (Fig. 5c, d, Supplementary Fig. 11b). In addition, in the case of the Yan et al. dataset, all of the clocks showed an increased transcriptomic age of human embryonic stem cells (hESCs), representing post-implantation epiblast cells, compared to late blastocyst cells (Fig. 5a). Altogether, the data show the transcriptomic rejuvenation of the human pre-implantation embryo cells at the morulae and blastocyst stages.
a Predicted ages of cells from different embryonic development stages in the Yan et al. dataset, given by the 9 best-performing cell-type-specific clocks based on the AIDA dataset. b Predicted ages of cells from different embryonic development stages in the Petropoulos et al. dataset. c The average predicted age of cells in the Yan et al. dataset, derived from the predictions of all the cell-type-specific clocks (except for the dendritic cell and erythrocyte clocks). Embryonic days 3, 4, and 5–7 correspond to the 8-cell, morulae, and the (pre-implantation) blastocyst stage, respectively. d The average predicted age of cells in the Petropoulos et al. dataset.
Additionally, we observed that the standard deviation (std) of the transcriptomic age of hESCs was remarkably higher in passage 10 compared to passage 0 (primary outgrowth), while passaging did not alter transcriptomic age. The average std of the 9 clocks for passage 10 (std = 3.6) was significantly higher than that of passage 0 (std = 1.2) based on a one-sided, two-sample t-test; there was no significant difference between the predicted age of passage 0 and passage 10 cells in 6 cases out of 9.
To explain the observed rejuvenation event during early development, we further examined the expression of clock feature genes in the embryo datasets. We found four genes (FOS, LGALS1, IER2, and PCBP1) supporting the rejuvenation of the morulae in all dataset (Fig. 6, Supplementary Fig. 11c). That is IER2 and PCBP1 expression decreased with age (based on the AIDA dataset) but increased during the observed transcriptomic rejuvenation (based on the Yan et al. and Petropoulos et al. datasets). In contrast, FOS and LGALS1 expression increased with age (based on the AIDA dataset) but decreased during the observed transcriptomic rejuvenation (based on the Yan et al. and Petropoulos et al. datasets).
Discussion
Here, we provided a comprehensive analysis of the transcriptomic age of single cells by using cell-type-specific scRNAseq clocks. While the age prediction performance for single cells was considerably lower than that of bulk or pseudo-bulk samples22, profiling the age of individual single cells provides a unique insight into the aging process at the single-cell level. For instance, we found that the predicted age of individual cells may be able to measure the process of cellular senescence. We also found that T cells, especially CD8-positive T cells have a strong relation to aging (both in terms of gene expression and cell type proportion), which cell type plays a major role in adaptive immunity. The large difference in age prediction performance between single cells and bulk or pseudo-bulk samples suggests an important role of cell-type proportion in the prediction of aging clocks over molecular changes (including molecular damage).
Accelerated epigenetic age based on bulk samples was associated with the incidence of COVID-1938,39. It was also shown that severe COVID-19 caused a reversible increase in DNA methylation age40. In contrast, another study concluded that epigenetic clocks were not accelerated in COVID-19 patients41. As cell fractions (especially leukocyte fraction) could be changed due to a SARS-CoV-2 infection it could affect previous results based on bulk samples. Single-cell clocks are free of cell fraction bias, and also allow cell-type-specific examinations. Interestingly, the transcriptomic age of monocytes, as well as, naive B and T cells decreased in moderate COVID-19 compared to healthy individuals. If we consider transcriptomic age as a proxy of biological age the results of the present study show that these cells rejuvenated due to the infection. An explanation of this interesting possibility could be that the infection increased the proportion of newly born cells compared to the older ones. Another possible explanation could be as follows. The study of Liu et al. revealed a negative association of IFN-I expression signatures with COVID-19 severity, but elevated expression compared to healthy controls in multiple cell types, including classical monocytes31. Here, we found a set of genes explaining the observed rejuvenation, which are also involved in the inflammatory response. However, their expression was decreased in moderate COVID-19 cases compared to the healthy condition. This result suggests that some genes regulating inflammation may be underactive due to the infection, already at the early stages, which could negatively impact adaptive responses as raised by Liu et al.31.
It was proposed that germline cells may be rejuvenated in the offspring after conception42,43. Recently, we tested this idea and revealed an epigenetic rejuvenation during embryogenesis in mice, and humans20,44,45. We found that the epigenetic age of early post-implantation embryos was lower compared to the pre-implantation stages. We also found that this rejuvenation event was followed by aging during embryogenesis44. In the present study, we examined the transcriptomic age dynamics of early human embryogenesis at the single-cell level. We observed a transcriptomic rejuvenation at the morulae and blastocyst cells compared to earlier stages, and also an increased transcriptomic age of hESCs compared to pre-implantation embryos. A gene expression analysis of Yan et al. study suggested that hESCs are more similar to mouse epiblast stem cells (mEpiSCs), which derive from post-implantation epiblast cells, than to mouse embryonic stem cells (mESCs), which derive from blastocyst-stage pre-implantation epiblast cells35. As hESC represents human post-implantation epiblast cells, our data show a decrease of transcriptomic age of pre-implantation cells followed by an increase in post-implantation cells. These results compared to our epigenetic studies suggest that the rejuvenation of the embryo start earlier in the transcriptomic level compared to the epigenetics level.
Although the applied single-cell clocks were trained on PBMCs, which cells cannot be found at the early stages of embryo development, multiple studies showed the relevance of cross-tissue application of aging clocks21,46, and that blood clocks can show similar dynamics in embryogenesis as multi-tissue clocks20,44,45. In the present study, different cell-type-specific clocks showed the same dynamics consistently in multiple embryogenesis datasets suggesting that the observed rejuvenation event is not a cell-type-specific but a more general process.
In summary, our large-scale study shows that single-cell transcriptomic clocks can contribute to the understanding of human aging and draws attention to the importance of single-cell aging clocks. These clocks can help in the development of targeted rejuvenation strategies by serving as a tool for evaluation, and allow deeper, cell-level investigation of the aging process.
Limitations
A limitation of the proposed cell-type-specific single-cell clocks is that while they generalized well in one external dataset (the Yoshida et al. dataset) they did not generalize well in other datasets. The present clocks may be largely impacted by the method of collection and pre-processing of the cell data used for training. The prediction performance was especially low on the eQTL dataset possibly caused by the smaller variability of expressions of clock feature genes. The clocks may also be impacted by the training population, which, in our case, is largely different from that of the external validation datasets (Asian vs mostly Caucasian).
Another limitation of the clocks may be that they are based on a linear model. Although ElasticNet is a preferred model to develop aging clocks, as it can provide accurate age predictions on the test sets even with very small sample size and very high number of features, it may not be the most suitable model for the data it is applied to, since it cannot capture non-linear feature-target interactions47.
A limitation in assessing the performance of the proposed clocks is the lack of information about the exact age of donors in two of the four external datasets. Finally, while the sample sizes of some application datasets used in this study were relatively small we made conclusions based on consistent (and statistically significant) results on multiple independent datasets.
Methods
Description of the input datasets
This study used publicly available datasets, all of which were produced by single-cell RNA sequencing of human cells (Supplementary Table 1). The collection and processing of samples were done by the original authors of the studies, this study relies on the available gene expression datasets. The main dataset we used was The Asian Immune Diversity Atlas (AIDA, https://explore.data.humancellatlas.org/projects/f0f89c14-7460-4bab-9d42-22228a91f185)23, which comprises 1,058,909 peripheral blood mononuclear cells (PMBCs) in total from 508 healthy, human donors. The age of the donors ranges from 19 to 75 years, with a mean of 42 years and a median of 41 years. Cell type annotation was done by the AIDA team in multiple steps in a hierarchical manner and was available for the dataset. Clustering and annotation of cells were performed on four levels, the marker genes identifying each cell type are available in the Supplementary Table S2 of the preprint of the AIDA dataset23.
The study of Yazar et al. presents a dataset of 1,248,980 PBMCs from 981 healthy, human donors with ages ranging from 19 to 97 years29 (referred to as “eQTL dataset”).
Yoshida et al. analyzed single-cell samples from healthy, COVID-19 infected and post-COVID-19 patients30. The COVID-19 set includes donors with RT-qPCR test positive for SARS-CoV-2 and in asymptomatic, mild, moderate, or severe conditions. The post-COVID-19 set consists of donors recovered from COVID-19, including symptomatic patients as well. Although Yoshida et al. carried out multi-omic analyses, here we only used the provided PBMC dataset, which consists of 422,220 cells from 75 donors (referred to as “Yoshida et al. dataset”), and we focus on the healthy and COVID-19 samples. The dataset does not contain information about the exact chronological age of donors, but they were assigned to age groups. It contains both pediatric and adult samples, the developmental stage of the donors ranges from the newborn to the elderly (aged adult) stage. Liu et al. collected and analyzed PBMC samples from healthy and COVID-19 infected patients31. The COVID-19 set includes donors with positive nasopharyngeal swab and/or positive serology for SARS-CoV-2 infection31 and in moderate, severe or critical conditions. The dataset consists of 372,081 cells from 46 donors with age ranging from 24 to 84 years (referred to as “Liu et al. dataset”). The study of Stephenson et al. presents a dataset of 647,366 PBMCs from healthy and COVID-19 infected, non-COVID-19 and IV-LPS patients32 (referred to as “Stephenson et al. dataset”). The COVID-19 group includes patients in asymptomatic, mild, moderate, severe and critical conditions. In this study, we focus on the healthy and COVID-19 cases. The dataset does not contain information about the exact chronological age of every donor, but they were assigned to age groups from the 3rd to the 10th decade.
Sikkema et al. presented a large-scale, integrated single-cell reference atlas of the human lung33. Here, we only used data of cells derived from the lung parenchyma of healthy donors, which dataset consists of 333,468 cells from 66 donors with age ranging from 15 to 76 years (referred to as “Lung dataset”).
Single-cell RNA sequencing of 124 human pre-implantation embryos and embryonic stem cells was done by Yan et al.35 (referred to as “Yan et al. dataset”). The following development stages were present in the dataset (number of cells in parentheses): oocyte (3), zygote (3), 2-cell embryo (6), 4-cell embryo (12), 8-cell embryo (20), morulae (16), late blastocyst (30), hESC passage 0 (8), hESC passage 10 (26). The study of Petropoulos et al. presents a dataset consisting of 1529 human preimplantation embryo cells36 (referred to as “Petropoulos et al. dataset”). The following development stages (embryonic days) were present in the dataset (number of cells in parentheses): embryonic day 3 (81), embryonic day 4 (190), embryonic day 5 (377), embryonic day 6 (415), embryonic day 7 (466). Finally, the dataset of Meistermann et al. contains 150 human preimplantation embryo cells37 (referred to as “Meistermann et al. dataset”). The following development stages (branches) were present in the dataset (number of cells in parentheses): pre-morula (1), morula (30), early blastocyst (5), inner cell mass (9), early trophectoderm (62), epiblast (36), primitive endoderm (1), TE.NR2F2- (5), TE.NR2F2+ (1).
The AIDA, eQTL, Yoshida et al., Liu et al., Stephenson et al. and Lung datasets were downloaded from https://cellxgene.cziscience.com/. The Yan et al. data was retrieved from the Gene Expression Omnibus48, the Petropoulos et al. dataset from BioStudies49 and the Meistermann et al. dataset from Mendeley Data (https://data.mendeley.com).
Data pre-processing
Each single-cell PBMC and lung dataset (AIDA23, eQTL dataset29, Yoshida et al. dataset30, Liu et al. dataset31, Stephenson et al. dataset32 and Lung33) were downloaded as h5ad files containing AnnData objects. The raw gene expression counts were accessed either through the main layer of the object or through the .raw layer when transformed counts were assigned to the main. In the case of each dataset, the raw, gene-by-cell count matrix was considered as the base data for our analyses. Then, raw counts were transformed by a log-normalization step, i.e. for each cell, each gene expression count value was divided by the total expression count of the given cell, multiplied by 10,000, and transformed with log1p transformation. Formally: \({\hat{c}}_{ij}=\log (1+[\frac{{c}_{ij}}{{c}_{i}}\cdot 10,000])\), where cij is the raw expression count of gene j in cell i, ci is the total expression count in cell i, \(\log\) indicates the natural logarithm and \({\hat{c}}_{ij}\) denotes the resulting log-normalized count.
The embryogenesis data of Yan et al.35 were downloaded as individual txt files for each of the 124 cells, containing the raw expression count of the detected genes. These files were processed to obtain a cell-by-gene matrix containing the corresponding gene expression values. Missing values in this matrix, i.e. expression counts of genes that were not detected in a given cell were treated as 0 values. Then the resulting raw count matrix was log-normalized in the same way as the other datasets described above. The Petropoulos et al. dataset36 were accessed as a single txt file containing the gene by cell count matrix of raw expression values. After transposing, it was log-normalized in the same way as described above. The dataset of Meistermann et al.37 was accessed through the Supplementary data repository of Radley et al.50. The tsv file containing the raw gene expression values was filtered to obtain the data of the cells introduced in the study of Meistermann et al. The resulting count matrix was then log-normalized. The development stages (branches) present in this dataset were adjusted to have similar stages as the other embryogenesis datasets namely, a blastocyst stage has been created and all cells of the following branches were assigned to it: inner cell mass, early trophectoderm, epiblast, primitive endoderm, TE.NR2F2-, TE.NR2F2+. Furthermore, the pre-morula stage has been removed from the dataset due to the insufficient number of samples (1 cell).
Calculation of transcriptional noise
The variability of cells, i.e. transcriptional noise in the AIDA dataset was calculated for each (donor, cell type, gene) triplet. Standard deviation (std) and interquartile range (IQR) of the log-normalized gene expressions of a given gene were calculated (for each donor and cell type). One-sample, two-sided t-tests were calculated with each metric separately to test if the transcriptional noise of a given gene in a given cell type significantly differs from 0. In addition, Spearman's correlation between chronological age and transcriptional noise was measured for each cell type and gene.
Gene set enrichment analysis
Gene set enrichment analysis was performed with the GSEA software51,52, for each of the top 5 best-performing single-cell clocks, separately. The data of normalized expression values of cells of the given cell type was used as the expression dataset, the REACTOME_CELLULAR_SENESCENCE gene set from the Molecular Signatures Database (MSignDB) as the gene set and the predicted age of the cells as phenotype labels. The genes in the expression data were mapped to the cellular senescence gene set prior to the analysis, consequently the analysis was run with the “No_Collapse” setting. The number of permutations was set to 100 and the metric for ranking genes to Spearman correlation, otherwise the default parameter setting was used.
Age prediction models
Chronological age prediction models (i.e., aging clocks) were developed using the AIDA dataset. Single-cell models were developed for each cell type, separately, resulting in cell-type-specific aging clocks. Pseudo-bulk data-based models were trained on cells averaged by donor, cell-type-specific pseudo-bulk models were trained on cells averaged by donor and cell type. We used the categorization of cell types created and assigned to the cells by the original authors of the dataset, and available in the retrieved AnnData object. For each cell type, ElasticNet models were trained on the log-normalized gene expressions of single cells to predict chronological age. 5-fold cross-validation (CV) on the donor level was used to prevent overfitting, i.e. a model was trained on cells from \(\frac{4}{5}\) of the donors, and tested on the remaining cells (coming from the remaining \(\frac{1}{5}\) of the donors), thus there was no overlapping of the train and test set of a given model, even on the donor level. The cross-validation resulted in five models (for each cell type and model type) and the predicted age of a given cell was considered to be the prediction of the model that used it as a test sample. The ElacticNet model was trained by using the default parameter settings of the scikit-learn implementation of the model (l1_ratio = 0.5, alpha = 1). We have also experimented with the glmnet implementation of the ElasticNet model, which fine-tunes the alpha parameter (lambda with the glmnet terminology), however, as we observed a similar performance along with a much higher computational demand, we relied on the default scikit-learn model in this study.
Using the single-cell predictions, we additionally calculated donor-level age predictions by averaging the predicted age of cells of a given donor. The averaging was done for each cell type, separately, resulting in cell-type-specific age predictions for each donor. The average of the age predictions of all cells of a given donor resulted in the general (non-cell-type-specific) age prediction of the donor.
Besides the single-cell aging clocks, cell-type proportion-based models were also developed motivated by the work of Zhu et al.22. For this purpose, the proportion of each cell type compared to the total number of cells of the donor was calculated and used to predict chronological age. The glmnet implementation of the ElasticNet algorithm was trained on the cell type proportion values using 5-fold cross-validation. We have also experimented with Partial Least Square Regression and XGBoost models, but due to their similar or weaker performance compared to ElasticNet, we excluded these results from the study.
Combined chronological age predictions were calculated based on the single-cell gene expression clocks and cell-type proportion clocks. To predict the donor age in this way, the average of the donor-level prediction given by a cell-type-specific single-cell clock and the predicted age given by a cell type proportion-based clock was calculated. Formally:
where \({\,{\mbox{PredAge}}\,}_{{{\rm{scE}}}}^{d,i}\) is the predicted age of cell i of donor d given by a cell-type-specific single-cell clock, \({\,{\mbox{PredAge}}\,}_{{{\rm{P}}}}^{d}\) is the predicted age of donor d given by a cell type proportion clock and \({\,{\mbox{PredAge}}\,}_{{{\rm{combined}}}}^{d}\) denotes the resulting combined predicted age of donor d.
For the evaluation of the described aging clocks, we measured the Mean Absolute Error (MAE) and the Pearson correlation of the chronological and predicted age.
Model validation on external datasets
For the external validation of the developed aging clocks, we used the eQTL dataset (Yazar et al.29), the healthy samples of the Yoshida et al. dataset30, the Liu et al. dataset31, and the Stephenson et al. dataset32. In the case of all datasets cell-type-specific single-cell clocks were applied to the common cell types, based on the categorization of cell types available for each dataset. Additionally, CD8-positive alpha-beta memory T cell clock was applied to the narrower types, i.e. to the central and effector memory CD8-positive alpha-beta T cell cells in the case of the eQTL dataset, the central, effector, and terminally differentiated effector memory CD8-positive alpha-beta T cells in the case of the Yoshida et al. dataset, and the effector memory CD8-positive, alpha-beta T cell in the case of the Stephenson et al. dataset. In the case of the Stephenson et al. dataset the innate lymphoid cell clock was applied to the group 2 innate lymphoid cell and ILC1 cell types, the plasma cell clock to the IgM, IgA and IgG plasma cells, and the memory B cell clock to the class switched and unswitched memory B cells. Moreover, the CD8-positive, alpha-beta cytotoxic T cell clock was applied to the effector CD8-positive, alpha-beta T cell-typed cells in this dataset. The CD14-positive monocyte clock was applied to the classical, the CD14-low, CD16-positive monocyte clock to the non-classical monocyte cells and the monocyte clock to the intermediate monocytes (Yoshida et al. and Liu et al. datasets). In the case of the Liu et al. dataset both the effector and central memory CD4-positive, alpha-beta T cell clocks were applied to the CD4-positive, alpha-beta memory T cells. Consequently, 23 cell-type-specific clocks were applied to the eQTL dataset, 21 to the Yoshida et al. dataset, 20 to the Liu et al. dataset and 26 to the Stephenson et al. dataset. In all datasets the primary ID of genes was their Ensembl ID, thus it was possible to directly map the genes to clock features in the external validation datasets. During the application of clocks, missing values were handled with average imputation, that is the expression value of a gene that was not presented in the validation set but presented among the clock features was imputed with the average (log-normalized) expression value of the gene in the training (AIDA) dataset, where the average was taken over all cells of the given cell type. In all cases, all of the five clocks of the cross-validation of the training set (AIDA) were applied to the external datasets, and for each cell, the average of the five predictions was considered as the final predicted age of the cell.
The application of the cell-type proportion clocks was done similarly to the single-cell clocks, namely, all five fitted models of the cross-validation were applied to the external validation data, and the average of their prediction was considered to be the final predicted age of a donor. Missing value imputation was done with the average proportion of a given cell type in the training set (AIDA), where the average was taken over all donors in AIDA. Proportions of narrower cell types mentioned above were summarized thus average imputation was not performed in these cases. Combined chronological age predictions were calculated on the external validation sets, according to Equation (1).
For the further validation of the proposed single-cell aging clocks, we used the Lung dataset (Sikkema et al.33). Similarly to the application of the clocks to PBMC data, the cell-type-specific clocks were applied to the common cell types. In the case of higher-level categorization of cell types in the Lung dataset, all the corresponding narrower cell-type-specific clocks were applied to the cells and their predictions were averaged for each cell (e.g. B cell, memory B cell, naive B cell and mature B cell clocks were all applied to the B cells in the Lung dataset). In total, predicted age for cells of 11 cell types were calculated.
The evaluation of the clocks on the external datasets was done based on MAE and Pearson correlation of the chronological and predicted ages when exact ages were assigned to donors (in the case of the eQTL, Liu et al. and Lung datasets). When only age groups were available (in the case of the Yoshida et al., and the Stephenson et al. datasets), we calculated Spearman correlation coefficients between the age group and the predicted age variables, as well as t-tests for the comparison of two groups. In addition to the exact ages in the eQTL and Liu et al. datasets, we assigned the donors to two age categories, motivated by the groups of the Yoshida et al. dataset, to adult (18 ≤ age < 65) and aged adult (65 ≤ age) groups, and the clock predictions were compared between these age groups.
Application of clocks to additional datasets
The created single-cell clocks were applied to the COVID-19-infected samples of the Yoshida et al., Liu et al., and Stephenson et al. datasets as well. The ElasticNet-based clocks were applied to the samples the same way as in the case of the healthy samples, described above in the Model validation on external datasets section. Predicted age of samples in the different severity groups and from healthy individuals were compared to assess the effect of SARS-CoV-2 infection on biological age captured by the proposed aging clocks. In order to account the age distribution differences of the different groups we included chronological age as a confounder variable in the comparison of the groups. In the Yoshida et al. dataset more precise chronological age was available to a few donors, but to properly compare the different groups, every donor was assigned either to the adult (18 ≤ age < 65) or aged adult (65 ≤ age) groups. Similarly, in the case of the Stephenson et al. dataset, every donor was assigned to an age group from 3rd to 10th decade.
In the case of the three embryogenesis datasets, we could apply only the single-cell clocks. Multiple ElasticNet-based cell-type-specific clocks were applied to the samples in a similar way as described above, namely, missing values were imputed with the corresponding average expression value in the training (AIDA) dataset, and the average prediction of the five CV clocks was considered as the final predicted age of a sample (see the Model validation on external datasets section for more details). In contrast to the other datasets used in this study, where genes were identified with their Ensembl ID, here they were represented by their name (in all three datasets). To map the genes to clock features two approaches were applied. Direct mapping was used if a gene could be mapped directly by its name to the gene set of the training dataset (AIDA) where gene names were also available. Otherwise, using the mapping function of UniProt (https://www.uniprot.org), gene names were mapped to Ensembl IDs and matched with the gene set of AIDA through these IDs. Duplicates, i.e. when more than one gene name were mapped to an Ensembl ID, were checked manually to make sure that the correct name was assigned to the ID.
Explanation of changes in predicted ages
In the case of the changes in predicted ages shown by the single-cell clocks consistent over all three COVID-19 datasets (Yoshida et al., Liu et al., Stephenson et al.), we searched for the genes supporting these changes. For a given cell type, the genes having non-zero regression coefficient in all five CV clocks were selected, and their normalized expression in each dataset were further examined. The different severity groups were compared based on the expression of a given gene, similarly as described above, accounting for the age distribution differences by including age as a confounder variable. A gene was selected to support the decrease in predicted ages between two groups, if its expression decreases too, while the gene is upregulated with aging according to the clock (i.e. shows opposing pattern to aging), and similarly if its expression increases. Based on the same considerations, a gene was selected to support the increase in predicted ages between two groups, if its expression increases too and the gene is also upregulated with aging according to the clock (i.e. shows the same pattern as during aging), and similarly if its expression decreases.
In the case of the embryogenesis analyses, we searched for clock feature genes supporting the decrease of predicted ages at the morulae stage. Since all the clocks show a similar aging (and rejuvenation) process during early development, some common genes are assumed to drive this process. Genes having the 10 highest (in absolute value) regression coefficient in all five CV clocks for more than one cell-type-specific single-cell clocks have been selected for further examination. A gene is assumed to be supporting the rejuvenation process, if its expression decreases at morulae, but increases during aging, and similarly, if its expression increases at morulae, but decreases with aging.
Statistics
The significance of the differences between the two different groups in the external validation analyses was calculated by two-sample, two-sided Student’s t-tests. In the case of the COVID-19-related analyses, the significance of the difference between two groups was calculated by a generalized linear model fitted to the predicted age/gene expression with severity as the independent variable and age as a confounder variable. The significance of the differences between the two different groups in the embryogenesis analyses was calculated by two-sample, two-sided Welch’s t-tests. ****: p ≤ 0.0001, ***: 0.0001 < p ≤ 0.001, **: 0.001 < p ≤ 0.01, *: 0.01 < p ≤ 0.05, ns:0.05 < p.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The AIDA Data Freeze v1 dataset23, containing the gene expression counts is available at https://cellxgene.cziscience.com/collections/ced320a1-29f3-47c1-a735-513c7084d508. The processed PBMC data of Yazar et al.29 are available at https://cellxgene.cziscience.com/collections/dde06e0f-ab3b-46be-96a2-a8082383c4a1. All processed data of Yoshida et al.30 are available at https://cellxgene.cziscience.com/collections/03f821b4-87be-4ff4-b65a-b5fc00061da7 (in the present study the PBMC dataset was used). The data of Liu et al.31 is available at https://cellxgene.cziscience.com/collections/ed9185e3-5b82-40c7-9824-b2141590c7f0. The dataset of Stephenson et al.32 is available at https://cellxgene.cziscience.com/collections/ddfad306-714d-4cc0-9985-d9072820c530. The Lung dataset of Sikkema et al.33 is available at https://cellxgene.cziscience.com/collections/6f6d381a-7701-4781-935c-db10d30de293. Raw gene expression counts of embryo cells and embryonic stem cells studied by Yan et al.35 are available at Gene Expression Omnibus under accession number GSE36552. The preimplantation embryo dataset of Petropoulos et al.36 is available at BioStudies under accession number E-MTAB-3929. The dataset of Meistermann et al.37 was made publicly available by Radley et al.50 at https://data.mendeley.com/datasets/689pm8s7jc/1. The cellular senescence gene set used for the gene set enrichment analysis can be accessed at https://www.gsea-msigdb.org/gsea/msigdb/human/geneset/REACTOME_CELLULAR_SENESCENCE.html. The set of cell type marker genes for the AIDA dataset is available in the supplementary material of the work of Kock et al.23. Data generated in this study are available at Zenodo under https://doi.org/10.5281/zenodo.10405106.
Code availability
Data and codes generated in this study are available at Zenodo under https://doi.org/10.5281/zenodo.1040510653. The pipeline for the application of single-cell clocks to new data can also be found at https://github.com/polyake/scaging. All analyses were performed by using Python 3.8.17. Here, we listed the most important packages with the version numbers in parentheses: • Basic packages for data manipulation: numpy (1.24.4), pandas (1.5.3) • Packages to handle AnnData: scanpy (1.9.3) • Statistics and machine learning packages: scipy (1.10.1), scikit-learn (1.3.0), glmnet (2.2.1), xgboost (1.7.6), statsmodels (0.14.0) • Packages and software for figure generation: matplotlib (3.3.4), seaborn (0.11.2), statannotations (0.6.0), Tableau (2022.4).
References
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, 1–20 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573 (2018).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018).
Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303 (2019).
Bell, C. G. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20, 1–24 (2019).
Duan, R., Fu, Q., Sun, Y. & Li, Q. Epigenetic clock: A promising biomarker and practical tool in aging. Ageing Res. Rev. 81, 101743 (2022).
Moqri, M. et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758–3775 (2023).
Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 1–14 (2015).
Fleischer, J. G. et al. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 19, 1–8 (2018).
Mamoshina, P. et al. Machine learning on human muscle transcriptomic data for biomarker discovery and tissue-specific drug target identification. Front. Genet. 9, 242 (2018).
Shokhirev, M. N. & Johnson, A. A. Modeling the human aging transcriptome across tissues, health status, and sex. Aging Cell 20, e13280 (2021).
Meyer, D. H. & Schumacher, B. BiT age: A transcriptome-based aging clock near the theoretical limit of accuracy. Aging Cell 20, e13320 (2021).
Li, X. & Wang, C.-Y. From bulk, single-cell to spatial RNA sequencing. Int. J. Oral. Sci. 13, 36 (2021).
Tomusiak, A. et al. Development of an epigenetic clock resistant to changes in immune cell composition. Commun. Biol. 7, 934 (2024).
Zhang, Z. et al. Deciphering the role of immune cell composition in epigenetic age acceleration: Insights from cell-type deconvolution applied to human blood epigenetic clocks. Aging Cell 23, e14071 (2024).
Regev, A. et al. The human cell atlas. elife 6, e27041 (2017).
Aldridge, S. & Teichmann, S. A. Single cell transcriptomics comes of age. Nat. Commun. 11, 4307 (2020).
Jovic, D. et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl. Med. 12, e694 (2022).
Trapp, A., Kerepesi, C. & Gladyshev, V. N. Profiling epigenetic age in single cells. Nat. Aging 1, 1189–1201 (2021).
Buckley, M. T. et al. Cell-type-specific aging clocks to quantify aging and rejuvenation in neurogenic regions of the brain. Nat. Aging 3, 121–137 (2023).
Zhu, H. et al. Human PBMC scRNA-seq–based aging clocks reveal ribosome to inflammation balance as a single-cell aging hallmark and super longevity. Sci. Adv. 9, eabq7599 (2023).
Kock, K. H. et al. Single-cell analysis of human diversity in circulating immune cells. Preprint at bioRxiv (2024). https://www.biorxiv.org/content/early/2024/07/01/2024.06.30.601119 (2024).
de Jong, T. V., Moshkin, Y. M. & Guryev, V. Gene expression variability: the other dimension in transcriptome analysis. Physiol. Genomics 51, 145–158 (2019).
Ibañez-Solé, O., Ascensión, A. M., Araúzo-Bravo, M. J. & Izeta, A. Lack of evidence for increased transcriptional noise in aged tissues. Elife 11, e80380 (2022).
Yayon, N. et al. A spatial human thymus cell atlas mapped to a continuous tissue axis. Preprint at bioRxiv https://doi.org/10.1101/2023.10.25.562925 (2023).
Gui, J., Mustachio, L. M., Su, D.-M. & Craig, R. W. Thymus size and age-related thymic involution: early programming, sexual dimorphism, progenitors and stroma. Aging Dis. 3, 280 (2012).
Posnett, D. N. et al. Oligoclonal expansions of antigen-specific CD8+ T cells in aged mice. Ann. N Y Acad. Sci. 987, 274–279 (2003).
Yazar, S. et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376, eabf3041 (2022).
Yoshida, M. et al. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature 602, 321–327 (2022).
Liu, C. et al. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell 184, 1836–1857 (2021).
Stephenson, E. et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 27, 904–916 (2021).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Chen, L. et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 9, 7204 (2018).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013).
Petropoulos, S. et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016).
Meistermann, D. et al. Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell 28, 1625–1640 (2021).
Corley, M. J. et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J. Leukoc. Biol. 110, 21–26 (2021).
Cao, X. et al. Accelerated biological aging in COVID-19 patients. Nat. Commun. 13, 2135 (2022).
Poganik, J. R. et al. Biological age is increased by stress and restored upon recovery. Cell Metab. 35, 807–820 (2023).
Franzen, J. et al. Epigenetic clocks are not accelerated in COVID-19 patients. Int. J. Mol. Sci. 22, 9306 (2021).
Ashapkin, V. V., Kutueva, L. I. & Vanyushin, B. F. Aging as an epigenetic phenomenon. Curr. Genomics 18, 385–407 (2017).
Gladyshev, V. N. The ground zero of organismal life and aging. Trends Mol. Med. 27, 11–19 (2021).
Kerepesi, C., Zhang, B., Lee, S.-G., Trapp, A. & Gladyshev, V. N. Epigenetic clocks reveal a rejuvenation event during embryogenesis followed by aging. Sci. Adv. 7, eabg6082 (2021).
Kerepesi, C. & Gladyshev, V. N. Intersection clock reveals a rejuvenation event during human embryogenesis. Aging Cell 22, e13922 (2023).
Petkovich, D. A. et al. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 25, 954–960 (2017).
de Lima Camillo, L. P., Lapierre, L. R. & Singh, R. A pan-tissue DNA-methylation epigenetic clock based on deep learning. npj Aging 8, 4 (2022).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Sarkans, U. et al. The BioStudies database—one stop shop for all data supporting a life sciences study. Nucleic Acids Res. 46, D1266–D1270 (2018).
Radley, A., Corujo-Simon, E., Nichols, J., Smith, A. & Dunn, S.-J. Entropy sorting of single-cell RNA sequencing data reveals the inner cell mass in the human pre-implantation embryo. Stem Cell Rep. 18, 47–63 (2023).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. Pgc-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Zakar-Polyák, E. & Kerepesi, C. Data for “Profiling the transcriptomic age of single-cells in humans”. https://doi.org/10.5281/zenodo.10405106 (2024).
Acknowledgements
E.Z.-P. and C.K. were supported by the European Union project RRF-2.3.1-21-2022-00004 within the framework of the Artificial Intelligence National Laboratory, Hungary, and the “OTKA” Young Researcher Excellence Program (FK-146113), Hungary. Figure 1a was created with BioRender.com.
Author information
Authors and Affiliations
Contributions
C.K. conceived the study and supervised the work. E.Z.-P. retrieved data, developed the single-cell clocks and performed the data analysis. R.P. developed the pseudo-bulk clocks. All authors interpreted the results. E.Z.-P. and C.K. wrote the manuscript with the contribution of A.C and R.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: George Inglis, Aylin Bircan and Tobias Goris.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zakar-Polyák, E., Csordas, A., Pálovics, R. et al. Profiling the transcriptomic age of single-cells in humans. Commun Biol 7, 1397 (2024). https://doi.org/10.1038/s42003-024-07094-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07094-5