Abstract
Lysophosphatidic acid receptor 1 (LPA1) is one of the G protein-coupled receptors activated by the lipid mediator, lysophosphatidic acid (LPA). LPA1 is associated with a variety of diseases, and LPA1 agonists have potential therapeutic value for treating obesity and depression. Although potent nonlipid LPA1 agonists have recently been identified, the mechanisms of nonlipid molecule-mediated LPA1 activation remain unclear. Here, we report a cryo-electron microscopy structure of the human LPA1-Gi complex bound to a nonlipid basic agonist, CpY, which has 30-fold higher agonistic activity as compared with LPA. Structural comparisons of LPA1 with other lipid GPCRs revealed that the negative charge in the characteristic binding pocket of LPA1 allows the selective recognition of CpY, which lacks a polar head. In addition, our structure show that the ethyl group of CpY directly pushes W2716.48 to fix the active conformation. Endogenous LPA lacks these chemical features, which thus represent the crucial elements of nonlipid agonists that potently activate LPA1. This study provides detailed mechanistic insights into the ligand recognition and activation of LPA1 by nonlipid agonists, expanding the scope for drug development targeting the LPA receptors.
Similar content being viewed by others
Introduction
Lipid mediators function as intercellular signaling molecules and activate specific G-protein coupled receptors (GPCRs). Among lipid mediators, lysophospholipids are generated by removing one acyl chain from phospholipids. Lysophosphatidic acid (LPA), a typical lysophospholipid, activates LPA receptors (LPA1-6)1. Owing to their amino acid sequence identity, LPA1-3 belong to the endothelial differentiation gene (EDG) family along with sphingosine-1-phosphate receptors (S1P1-5), which recognize a different lysophospholipid, sphingosine-1-phosphate (S1P). In contrast, LPA4-6 belong to the non-EDG family and are more similar to the purinergic P2Y receptor family2. The dephosphorylated LPA derivative 2-arachidonoylglycerol (2-AG) activates cannabinoid receptors (CNRs), which are most closely related to the EDG family at the phylogenetic level3. In addition, unsaturated fatty acids such as oleic and linoleic acids, components of LPA, activate free fatty acid receptor 1 and 4 (FFAR1, 4)4,5. These diverse lipid-activated GPCRs precisely recognize the chemical structures of their lipid ligands.
Among the LPA receptors, LPA1 was the firstly identified GPCR, and LPA-LPA1 signaling has been extensively studied6,7. LPA1 is widely expressed in several organs and couples with multiple types of G-proteins, including Gαi, Gαq, and Gα12/13, to transduce various intracellular signals8. LPA1 is associated with a variety of diseases, including cancer, inflammation, and neuropathic pain, and thus is a pathologically important receptor and an essential drug target8,9. Several preclinical studies have indicated the potential therapeutic value of selective LPA1 agonists for obesity10 and urinary incontinence11. Furthermore, recent studies suggested that G-protein-biased LPA1 agonists are promising targets for the development of antidepressant drugs12. However, LPA is metabolically unstable and has a short half-life, thus complicating the functional characterization of LPA supplementation13. Therefore, more potent and stable agonists have been awaited to facilitate explorations of the potential therapeutic effects of LPA receptor agonists. Recently reported nonlipid LPA1-3 agonists are uniquely potent, selective, and especially stable compared to LPA, and are excellent tools for elucidating the LPA pathway14.
The crystal structure of inactive LPA115 and cryo-EM structures of the active LPA1-Gi complex bound to LPA or its analog16,17 were previously determined. However, as the structures of the endogenous agonist LPA and the nonlipid agonists might be quite different, their precise binding modes remain enigmatic. Therefore, the mechanisms by which the nonlipid agonists activate LPA1 are also unclear, limiting the design of useful nonlipid LPA receptor agonists. In this work, we report the cryo-EM structures of the LPA1-Gi signaling complex bound to a nonlipid agonist, revealing its unique binding and activation mechanisms that are not utilized by lipid agonists.
Results
Overall structure
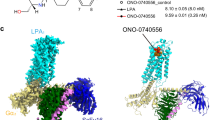
For the structural study, we used the previously reported nonlipid agonist, (2R)-2-(diethylamino)-2-(2-ethyl-3-methyl-benzofuran-7-yl)ethanol (CpY)14 (Fig. 1a). CpY is an alcohol conjugated with a diethylamino group and a modified benzofuran. Classic LPAR ligands contain a large nonpolar region and chiral hydroxyl, ester, and carboxylic acid groups. In contrast, CpY and derivatives are basic molecules at physiologic pH (pKa ≥8.8) and are less lipophilic (logP ≤3.9)14. In addition, the molecular weight of CpY is 275.4 Da, a diminutive size for a ligand activating lipid GPCRs. In a NanoBiT-G-protein dissociation assay18, CpY showed agonistic activity with an EC50 value of 0.95 nM for the human LPA1 receptor, surpassing LPA by 20-fold. Moreover, the Emax of CpY exceeds that of LPA by 1.5-fold (Fig. 1b and Supplementary Table 1). Notably, CpX, in which the ethyl group that modifies benzofuran of CpY is replaced by a methyl group, can activate LPA1, but its EC50 is double that of CpY (Fig. 1a, b and Supplementary Table 1).
a Chemical structures of LPA, CpX, and CpY. b Ligand-induced Gi activation by LPA1. Gi activation was measured by the NanoBiT-G-protein dissociation assay. Concentration–response curves are shown as means ± s.e.m. (standard error of the mean) from six independent experiments. Individual replicates are shown as small dots. Note that, for most data points, error bars are smaller than the size of the symbols and thus are not visualized in the panels. c Composite cryo-EM map and refined structures.
To efficiently purify the stable GPCR-G-protein complex, we used a ‘Fusion-G system’ by combining two complex stabilization techniques19. In the fusion-G system, LgBiT was fused to the C-terminus of the receptor, and HiBiT, a modified 13-amino acid peptide with potent affinity to LgBiT (Kd = 700 pM), was fused to the C-terminus of Gβ via the GS linker. The proteins were co-expressed in Sf9 cells, solubilized, and purified by GST affinity chromatography. After an incubation with scFv16, the complex was purified by size exclusion chromatography. The antibody fragment scFv16, which binds and stabilizes the GPCR-G protein complexes, is widely used for cryo-EM structural analysis of GPCR-G protein complexes20. The structure of the purified complex was determined by single-particle cryo-EM analysis with an overall resolution of 2.9 Å (Table 1, Supplementary Fig. 1, and “Methods”). As the extracellular portion of the receptor was poorly resolved, the receptor was masked and locally refined, yielding a structure with a nominal resolution of 3.1 Å, and then combined with the overall refined map. (Fig. 1c, and Supplementary Fig. 1). The structure of the receptor superimposes well with the two previously reported active LPA1 structures (LPA- and ONO-0740556-bound)16,17, with Cα atom root mean square deviations of 0.712 Å and 0.671 Å, respectively (Supplementary Fig. 2a, b). Furthermore, there are few differences in the degree of TM6 bending and the Gi binding mode. Superimposition of the receptors revealed slight differences in the position of the Gi trimer, but they are within the reported structural polymorphism and dynamics of the LPA1-Gi complex16,17 (Supplementary Fig. 2a, b), thereby ensuring that the most stable LPA1-Gi complex remains unchanged for different agonists.
CpY binding site
Previous studies have shown that the binding site of LPA and LPA analogs comprises the N-terminus, transmembrane helices (TMs) 2, 3, 5, 6, 7, and extracellular loops (ECLs) 1, 2. The binding site is broadly divided into a hydrophilic side that recognizes the phosphate group and a hydrophobic pocket that recognizes the acyl group16,17. We observed unambiguous density in the hydrophobic pocket and modeled CpY (Fig. 2a and Supplementary Fig. 2c). The hydrophobic pocket is negatively charged by D1293.33 (superscripts indicate Ballesteros–Weinstein numbers) residue deep inside the pocket, which forms a salt bridge with the positively-charged diethylamino group of CpY (Fig. 2b–d). Moreover, the hydroxy group forms hydrogen bonds with the carbonyl oxygen of the Q1253.29 main chain (Fig. 2c, d). The modified benzofuran of CpY is sandwiched between two leucine residues, L2786.55 and L2977.39, in a similar manner to the aromatic ring in the middle of the acyl chain of ONO-074055617 (Fig. 2c, d and Supplementary Fig. 2d). Among these leucine residues, L2977.39 forms a CH–π interaction with the furan ring of the benzofuran, which enables LPA1 to accept a ring structure that is not present in the endogenous agonist, LPA.
a Surface representation of the ligand binding pockets in LPA1. CpY and LPA (18:1) (PDB 7TD0) are represented by stick models. b Surface charge of the binding pocket. Structure diagram (c), and schematic representation (d) of the interactions between CpY and the receptor within 4.5 Å. The dashed lines indicate the salt bridge with D1293.33 and hydrogen bonds, and the line indicate the CH–π interaction. e NanoBiT-G-protein dissociation assays for LPA1 and its mutants. Concentration–response curves of CpY-dependent G-protein dissociation signals for LPA1 are shown as means ± s.e.m. Individual replicates are shown as small dots. f Conservation of the CpY binding site in LPA1-3.
The importance of these residues was validated by evaluating the functionality of alanine mutants (Fig. 2e, Supplementary Fig. 3a). The carboxy group of CpY forms hydrogen bonds only with the oxygen atoms of the main chain of Q1253.29. However, the Q1253.29A mutant have a reduced response, suggesting that the side chain of Q1253.29 also plays some role in CpY binding. The D1293.33A mutant completely lost the response to CpY, although its expression level was similar to that of the wild type, indicating the functional significance of the electrostatic interaction for CpY binding. In addition, both the L2786.55A and L2977.39A mutations reduced the activity, indicating that these leucines contribute to the establishing a suitable environment for the ring motif binding. In conclusion, CpY is well adapted to the residues in the LPA1 pocket, particularly D1293.33, which plays a more significant role than other residues due to its strong electrostatic interactions. The residues involved in the CpY, including D1293.33, binding are highly conserved among the EDG family LPA receptors (LPA1–3) (Fig. 2f), suggesting a similar mechanism for CpY recognition and explaining the strong activating capacity of CpY for LPA1-314.
Structural insight into CpY selectivity
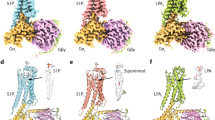
Various lipid GPCRs recognize lipids including hydrocarbon tails. CNRs (CB1, 2) are activated by a dephosphorylated LPA derivative, 2-arachidonoylglycerol (2-AG)3. FFAR4 is activated by oleic acid (OA)21, which is a type of acyl chain found in LPA. Similar to 2-AG and OA, CpY lacks a phosphate group but exhibits no activity against CNRs or FFARs (Supplementary Fig. 4). To elucidate the mechanism by which CpY selectively activates LPA1-3, we compared the CpY-bound LPA1 structure with the previously determined agonist-bound lipid GPCR structures including S1PRs, CNRs and FFARs (Fig. 3a–d and Supplementary Fig. 5)2,21,22,23,24,25,26,27,28,29,30,31,32,33,34. Locations of the binding pockets in FFAR1 (GPR40), LPA6, LPS1, 3 (GPR34 and GPR174, respectively), and prostaglandin receptors (EP2, 3, and FP) significantly differ from that in LPA1(Supplementary Fig. 5). Especially, the FFAR1 pocket is shallower than other receptors and has an unorthodox activation mechanism. S1PRs, which belong to the same EDG family as LPA1, has an allosteric site in almost the same position as LPA1, but its hydrophobic pocket extends deeper into TM5 (Fig. 3b and Supplementary Fig. 5). The acyl chain of the ligand enters deep into this pocket and interacts directly with W6.48, a conserved residue crucial for GPCR activation35. These elongated pockets are suitable for accommodating linear acyl chains, but not rounded molecules such as CpY.
CB1, which is phylogenetically close to the EDG family3, has a similar location and shape of the ligand-binding pocket to those of LPA1. The TM5 side of the pocket is occluded by a tryptophan residue at position 5.43, which characterizes LPA1-3 and CNRs as class A GPCRs15 (Fig. 3a, c). F268ECL2 in CB1 replaces A199ECL2 in LPA1, resulting in a pocket curvature to constrain the shape and orientation of the ligand (Fig. 3a, c and Supplementary Fig. 5). Like LPA1, FFAR4 (GPR120) also has a bulging pocket and appears capable of accepting CpY (Fig. 3a, d). Nonetheless, FFAR4 activation requires a direct interaction with the activation switch W2776.48 at the end of TM521, and the shape of CpY would hinder its interactions with W2776.48. Furthermore, closer inspections of the residues forming the pockets show that CNRs and FFAR4 lack the acidic residues in the pockets like D1293.33 of LPA1 (Supplementary Fig. 6a, b). Consequently, the pocket of LPA1 is negatively charged, compared to the pockets of these receptors (Supplementary Fig. 6c).
In summary, these structural comparisons of the ligand-binding pockets indicate that the hydrophobic pocket of LPA1 is characterized by a particularly strong negative charge and a small, compact spherical shape, among lipid GPCRs. The small, non-chain molecule CpY fits capably into this characteristic pocket of LPA1. In addition, the positively charged diethylamino group of CpY is recognized by the characteristic negatively charged pocket of LPA1 and forms a salt bridge with D1293.33. These factors collectively enable CpY to selectively activate LPA1.
Activation mechanism by CpY
To examine how CpY activates LPA1 and stabilizes the active conformation, we compared the LPA1 structures in the CpY-bound active and the antagonist-bound inactive states15. The antagonist ONO9780307 binds to the same position as CpY and competitively inhibits LPA1 (Fig. 4a, b). The indane moiety of the antagonist prevents the inward movement of TM7. In contrast, the ethyl group of CpY permits its inward shift (Fig. 4b). As TM7 shifts into the core of LPA1, E3017.43 and Y1022.57 swap side chain positions to stabilize the structure, and A3007.42 and N3037.45 push W2716.48 inward (Fig. 4b, c). Furthermore, the ethyl group of CpY penetrates deeper than the indane of ONO9780307 and interacts directly with W2716.48. As a result, the ethyl group secures W2716.48 and stabilizes the active conformation. The lower activity of CpX than CpY supports the importance of the ethyl group (Fig. 1b). The movement of W2716.48 upon agonist binding causes the structural reconfiguration of F2676.44 in the P5.50I3.40F6.44 motif, which is also essential for receptor activation35(Supplementary Fig. 2e). The movement of the F2185.51 is responsible for the large outward movement of the intracellular portion of TM6, which is essential for Gi binding35 (Fig. 4a, c, d). In addition, the binding of the diethylamino group of CpY induces a conformational change in the rotamer of L1323.36 and W2105.43 (Fig. 4b, c). L1323.36 works with W2716.48 as a twin-toggle switch, and W2105.43 pushes out L1323.36 and N2145.47. N2145.47 changes the position of F2185.51, thereby facilitating the movement of F2766.44 within the P5.50I3.40F6.44 motif (Fig. 4b).
a–d Superimposition of the CpY- and antagonist-bound LPA1 structures, colored aquamarine and gray (PDB 4Z34), respectively, a viewed from the membrane plane. b Structural changes of the intramolecular interactions induced by agonist binding. c Rearrangements of the central hydrophobic core and d the P5.50I3.40F6.44 motifs and movement of TM6. e Superimposition of the CpY-, LPA-, and ONO-0740556-bound LPA1 structures, colored aquamarine and blue (PDB 7TD0), and orange (PDB 7YU3), respectively. f Changes in EC50 and Emax of NanoBiT-G-protein dissociation assays due to the W2105.43A mutation are shown as means ± s.e.m. from three independent experiments. The W2105.43A mutant completely lost the response for ONO-0740556. g Interaction of CpY with W(F)2716.48 in WT and W271F mutant. W2716.48F is a predicted structure with a residue substitution. CpY and F2676.44 and W(F)2716.48 residues are represented by stick models and cpk models. h Changes in EC50 of NanoBiT-G-protein dissociation assays due to the W2716.48F mutation are shown as means ± s.e.m. from three independent experiments.
To gain insights into the structural rearrangement of the hydrophobic core by CpY, we compared the structure of active LPA1 bound to each agonist. The LPA and ONO-0740556 agonists have limited interactions with W2716.48 (Fig. 4e). Previous studies revealed that those lipid agonists binding causes the inward movement of TM7 and rotamer changes in W2105.43 and L1323.36, which cooperatively induce the movement of W2716.4816,17,. The W2105.43A mutant completely lost the response to ONO-0740556 and reduced the Emax to LPA, indicating that W2105.43 is required for strong activation by ONO-0740556 (Fig. 4f and Supplementary Fig. 3b). In contrast, the W2105.43A mutant is responsive to CpX and CpY, and the activation is independent on W2105.43. To explore the importance of the direct interaction of CpY with W2716.48, we measured the response of the W2716.48 mutants. The W2716.48A mutant lost its role as an activation motif and significantly decreased in response to all agonists (Supplementary Fig. 3b). Therefore, we designed W2716.48F mutant, which has little interaction with CpY (Fig. 4g). The W2716.48F mutant exhibited a reduction in response to CpX and CpY, with minimal alteration in response to LPA and ONO-0740556 (Fig. 4g, h and Supplementary Fig. 3c). These results indicate that the direct interaction of CpY and CpX with W2716.48 is crucial for activation, in contrast to LPA and ONO-0740556, which exhibit weak interaction with W2716.48.
Discussion
Previous studies of LPA receptors suggested that the positively-charged residues on the hydrophilic side recognize the phosphate head group, thereby contributing to the LPA selectivity16,17. The recognition of a polar head group is responsible for accommodating large lipid agonists within the binding pocket, as mutants of the positively-charged residues showed reduced activity with lipid agonists LPA and LPA analogs16,17. Despite its small size and lack of a polar group recognized by the positively-charged residues, CpY forms an electrostatic interaction with D1293.33 in the hydrophobic pocket, ensuring its high affinity and selectivity for LPA1. In drug development, compounds have frequently been designed with an acidic head group analogous to that of LPA to ensure affinity for LPA1. The D1293.33 residue may represent a novel point of interest when developing drugs targeting LPA1 to increase binding capacity. Moreover, D1293.33 is located within the hydrophobic pocket, close to the hydrophobic core, which is present in the lower part of the pocket and where the rearrangement of residues takes place during activation. In LPA, a long, flexible carbon chain connects the phosphate group to the interaction site in the hydrophobic core, whereas in CpY, a rigid benzofuran connects the diethylamino group to the ethyl group. Therefore, CpY enters the hydrophobic core by using its diethylamino group as a fulcrum, and its ethyl group acts effectively on W2716.48 to stabilize the active conformation. This mechanism allows CpY to function as a potent LPA1 agonist with high Emax and EC50 values (Fig. 5a–d).
a–d Schematic representations of the recognition and activation of each LPA1 agonist, highlighting the crucial residues and interactions involved. The thickness of the dashed lines and arrows represents the strength and importance of the interactions. Residues marked with squares are responsible for holding the agonist, while those marked with ovals transmit the binding signal of the compound into the cell.
We have determined the cryo-EM structure of the LPA1-Gi complex bound to the nonlipid, potent agonist CpY. We elucidated the recognition and activation mechanisms of the nonlipid agonists and clarified their distinct features from the lipid agonists. This study will facilitate the design of small molecule agonists for LPA1 and drug development for other lipid GPCRs.
Methods
NanoBiT-G-protein dissociation assay
LPA1-induced Gi activation was measured by a NanoBiT-G-protein dissociation assay18, in which the LPA1-induced dissociation of a Gα subunit from a Gβγ subunit was monitored by a NanoBiT system (Promega). Specifically, a NanoBiT-Gi1 protein consisting of a large fragment (LgBiT)-containing Gαi1 subunit and a small fragment (SmBiT)-fused Gγ2 subunit with the C68S mutation, along with the untagged Gβ1 subunit, was expressed with a test LPA1 construct, and the ligand-induced change in the luminescent signal was measured. We used the N-terminal FLAG (DYKDDDDK)-tagged constructs of human LPA1. Of the test compounds, LPA (18:1 Lyso PA) was purchased from Sigma-Aldrich, while CpX, CpY and ONO-074055617 were synthesized by Ono Pharmaceutical Co., Ltd. HEK293T cells were seeded in a 6-well culture plate at a concentration of 2 × 105 cells ml−1 (2 ml per well in DMEM supplemented with 10% fetal bovine serum), 1 d before transfection. The transfection solution was prepared by combining 2.5 µl (per well hereafter) of Lipofectamine 2000 (ThermoFisher Scientific) and a plasmid mixture consisting of 100 ng LgBiT-containing Gαi subunit, 500 ng Gβ1, 500 ng SmBiT-fused Gγ2 with the C68S mutation, and 200 ng LPA1 in 500 µL of Opti-MEM (ThermoFisher Scientific). After an incubation for 1 d, the transfected cells were harvested with 0.5 mM EDTA-containing PBS, centrifuged, and suspended in 2 ml of HBSS containing 0.01% bovine serum albumin (BSA fatty acid–free grade, SERVA) (assay buffer). The cell suspension was dispensed into a white 96-well plate at a volume of 80 µL per well and loaded with 20 μL of 50 μM coelenterazine diluted in the assay buffer. After 2 h of incubation, the plate was measured for baseline luminescence and then 20 µL of 6× test compound, diluted in the assay buffer, was manually added. After an incubation for 8–10 min at room temperature, the plate was read for the second measurement. The second luminescence counts were normalized to the initial counts, and the fold changes in the signals were plotted for the G-protein dissociation response. Using the Prism 9 software (GraphPad Prism), the G-protein dissociation signals were fitted to a four-parameter sigmoidal concentration–response curve, from which the pEC50 values (negative logarithmic values of half-maximum effective concentration (EC50) values) and Emax were used to calculate the mean and s.e.m.
S1PRs-, CNRs- and FFAR4-induced Gi activation was measured by the same method. We used the N-terminal FLAG (DYKDDDDK)-tagged constructs of human. As control compounds, S1P (Sphingosine 1-phosphate; Sigma) was used for S1PRs, AEA (Arachidonoylethanolamide; Sigma) for CNR and TUG-891 (Sigma) for FFAR4.
Expression and purification of scFv16
The gene encoding scFv16 was synthesized (GeneArt) and subcloned into a modified pFastBac vector36, with the resulting construct encoding the GP67 secretion signal sequence at the N terminus, and a His8 tag followed by a TEV cleavage site at the C terminus35. The His8-tagged scFv16 was expressed and secreted by Sf9 insect cells, as previously reported37. The Sf9 cells were removed by centrifugation at 5000 × g for 10 min, and the secreta-containing supernatant was combined with 5 mM CaCl2, 1 mM NiCl2, 20 mM HEPES (pH 8.0), and 150 mM NaCl. The supernatant was mixed with Ni Superflow resin (GE Healthcare Life Sciences) and stirred for 1 h at 4 °C. The collected resin was washed with buffer containing 20 mM HEPES (pH 8.0), 500 mM NaCl and 20 mM imidazole, and further washed with 10 column volumes of buffer containing 20 mM HEPES (pH 8.0), 500 mM NaCl and 20 mM imidazole. Next, the protein was eluted with 20 mM Tris (pH 8.0), 500 mM NaCl and 400 mM imidazole. The eluted fraction was concentrated and loaded onto a Superdex200 10/300 Increase size-exclusion column, equilibrated in buffer containing 20 mM Tris (pH 8.0) and 150 mM NaCl. Peak fractions were pooled, concentrated to 5 mg ml−1 using a centrifugal filter device (Millipore 10 kDa MW cutoff), and frozen in liquid nitrogen.
Constructs for expression of LPA1 and Gi
The human LPA1 gene (UniProtKB, Q92633) was subcloned into a modified pFastBac vector36, with an N-terminal haemagglutinin signal peptide followed by the Flag-tag epitope (DYKDDDD) and the LgBiT fused to its C-terminus followed by a 3 C protease site and EGFP-His8 tag. A 15 amino sequence of GGSGGGGSGGSSSGG was inserted into both the N-terminal and C-terminal sides of LgBiT. Rat Gβ1 and bovine Gγ2 were subcloned into the pFastBac Dual vector. In detail, rat Gβ1 was cloned with a C-terminal HiBiT connected with a 15 amino sequence of GGSGGGGSGGSSSGG. Moreover, human Gαi1 was subcloned into the C-terminus of the bovine Gγ2 with a nine amino sequence GSAGSAGSA linker. The resulting pFastBac dual vector can express the Gi trimer.
Expression and purification of the human LPA1- Gi complex
The recombinant baculovirus was prepared using the Bac-to-Bac baculovirus expression system (Thermo Fischer Scientific). Spodoptera frugiperda Sf9 insect cells (Thermo Fischer Scientific) at a cell density of 3.0 × 106 cells per milliliter in Sf900 II medium (Gibco) were co-infected with the virus encoding the LPA1-LgBiT-EGFP and Gi trimer at the ratio of 1:1. After infection, the cells were incubated at 27 °C for 48 hours and then harvested. The harvested cell pellets were resuspended in 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM CaCl2, 5 mM MgCl2, 10% Glycerol, 5.2 μg/ml aprotinin, 2.0 μg/ml leupeptin, 1.4 μg/ml pepstatin A, and 100 μM PMSF and homogenized by douncing ~30 times. Apyrase was added to the lysis at a final concentration of 25 mU/ml. CpY was added at a final concentration of 2 µM. The lysate was incubated at room temperature for 1 h with flipping. The crude membrane fraction was collected by ultracentrifugation at 180,000 g for 1 h. The membrane fraction was solubilized in buffer, containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% n-dodecyl-beta-D-maltopyranoside (DDM) (Calbiochem), 0.2% CHS, 10% glycerol, 5.2 μg/ml aprotinin, 2.0 μg/ml leupeptin, 1.4 μg/ml pepstatin A, and 100 μM PMSF, 25 mU/ml Apyrase, 2 μM CpY for 1 h at 4 °C. The supernatant was separated from the insoluble material by ultracentrifugation at 180,000 × g for 20 min. GST fusion GFP enhancer38 was added and incubated with Glutathione Sepharose resin (Cytiva) for 1 h. The resin was washed with ten column volumes of buffer, containing 50 mM Tris-HCl, pH 8.0 150 mM NaCl, 0.05% glyco-diosgenin (GDN) (Anatrace), and 0.2 μM CpY. The receptor was eluted in buffer, containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.01% GDN, and 1 μM CpY, and 40 mM Glutathione (Reduced Form). The eluate was treated with 0.5 mg of HRV-3C protease (homemade). Then, cleavaged GFP-His8, GST fusion GFP enhancer, and HRV-3C protease were removed with Ni-NTA resin. The flow-through was incubated with the scFv16. The complex was purified by size exclusion chromatography on a Superose 6 increase (GE) column in 20 mM Tris-HCl, pH8.0, 150 mM NaCl, 0.01% GDN, and 0.1 μM CpY. CpY was added to the peak fraction at a final concentration of 10 µM and concentrated to 11 mg/ml.
Sample vitrification and cryo-EM data acquisition
The purified complex was applied onto a freshly glow-discharged Quantifoil UltraAu grid (R1.2/1.3, 300 mesh), and plunge-frozen in liquid ethane by using a Vitrobot Mark IV. Data collections were performed on a 300 kV Titan Krios G3i microscope (Thermo Fisher Scientific) and equipped with a BioQuantum K3 imaging filter and a K3 direct electron detector (Gatan).
First, 5013 movies were acquired with a calibrated pixel size of 0.83 Å pix−1 and with a defocus range of −0.8 to −1.6 μm, using the SerialEM software39. Each movie was acquired for 2.30 s and split into 48 frames, resulting in an accumulated exposure of about 50.362 e− Å−2 at the grid. In addition, 3,312 movies were acquired with a calibrated pixel size of 0.83 Å pix−1 and with a defocus range of −0.6 to −1.6 μm, using the EPU software (Thermo Fisher’s single-particle data collection software). Each movie was acquired for 2.50 s and split into 48 frames, resulting in an accumulated exposure of about 48.405 e− Å−2 at the grid.
Image processing
All acquired movies were dose-fractionated and subjected to beam-induced motion correction implemented in RELION 3.140,41. The contrast transfer function (CTF) parameters were estimated using cryoSPARC v3.342. A total of 6,061,154 particles were extracted. The particles were subjected to 2D classifications, Ab-initio reconstruction and several rounds of hetero refinement in cryoSPARC. Next, the particles were subjected to Bayesian polishing in RELION 3.143. The polished particles were subjected to 3D classifications with a mask on the receptor. The best class of particles was subjected to non-uniform refinement, yielding a map with a nominal overall resolution of 2.86 Å, with the gold standard Fourier Shell Correlation (FSC = 0.143) in cryoSPARC. Moreover, the 3D model was refined with a mask on the receptor. As a result, the receptor has a higher resolution with a nominal resolution of 3.12 Å. The overall and receptor focused maps were combined by phenix44. The processing strategy is described in Supplementary Fig. 1.
Model building and refinement
The density map was generated by servalcat45 and the quality of the density map was sufficient to build a model manually in COOT46,47. The model building was facilitated by the cryo-EM structure of the LPA1-Gi (PDB 7YU3)17. We manually modeled LPA1, the Gi heterotrimer and scFv16 into the map. We then manually readjusted the model into the density map using COOT and refined it using phenix.real_space_refine (v.1.19) with the secondary-structure restraints using phenix secondary_structure_restraints48.
Measurement of receptor cell-surface expression by ELISA
To measure the cell surface expression level of wild-type LPA1 and its mutants, HEK293T cells were transiently transfected in 12-well plates and incubated for 1d. Transfection was performed by following the same procedure as described in the NanoBiT-G-protein dissociation assay section, with a downscaled volume (250 µL transfection solution). The transfected cells were harvested with 0.5 mM EDTA-containing PBS, centrifuged, and blocked with 5% (w/v) BSA at room temperature for 1 h. Anti-Flag HRP conjugate (Sigma) was then added to a dilution of 1:20,000 and incubated for 2 h at room temperature. After washing with PBS, the cells were suspended in 100 µL of PBS and 10 µL portions were dispensed into the tubes. An 80 µL aliquot of TMB, HRP Microwell Substrate (SurModics, Inc.) was added. The reactions were quenched by adding an equal volume of 450 nm Liquid Stop Solution for TMB Microwell Substrate (SurModics, Inc.) and the optical density at 450 nm was measured.
Statistics and reproducibility
the Prism 9 software (GraphPad Prism) was used for data analysis. Data sets were of at least n = 3 and the experimental n number is stated for each data set in the corresponding figure legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM density map and atomic coordinates for the LPA1-Gi complex have been deposited in the Electron Microscopy Data Bank and the PDB, under accession codes: EMD-61154, EMD-61912, EMD-61913, and PDB 9J5V. Source data have been deposited with Communications Biology in Supplementary Data 1. All other data are available from the corresponding authors upon reasonable request.
References
Kihara, Y., Maceyka, M., Spiegel, S. & Chun, J. Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br. J. Pharmacol. 171, 3575–3594 (2014).
Taniguchi, R. et al. Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6. Nature 548, 356–360 (2017).
Hurst, D. P., Schmeisser, M. & Reggio, P. H. Endogenous lipid activated G protein-coupled receptors: emerging structural features from crystallography and molecular dynamics simulations. Chem. Phys. Lipids 169, 46–56 (2013).
Briscoe, C. P. et al. The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J. Biol. Chem. 278, 11303–11311 (2003).
Hirasawa, A. et al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 11, 90–94 (2005).
Hecht, J. H., Weiner, J. A., Post, S. R. & Chun, J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J. Cell Biol. 135, 1071–1083 (1996).
Choi, J. W. et al. LPA receptors: subtypes and biological actions. Annu. Rev. Pharmacol. Toxicol. 50, 157–186 (2010).
Yung, Y. C., Stoddard, N. C. & Chun, J. LPA receptor signaling: pharmacology, physiology, and pathophysiology. J. Lipid Res. 55, 1192–1214 (2014).
Houben, A. J. S. & Moolenaar, W. H. Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev. 30, 557–565 (2011).
Rancoule, C. et al. Involvement of autotaxin/lysophosphatidic acid signaling in obesity and impaired glucose homeostasis. Biochimie 96, 140–143 (2014).
Terakado, M. et al. Discovery of ONO-7300243 from a Novel Class of Lysophosphatidic Acid Receptor 1 Antagonists: From Hit to Lead. ACS Med. Chem. Lett. 7, 913–918 (2016).
Kajitani, N. et al. G protein-biased LPAR1 agonism of prototypic antidepressants: Implication in the identification of novel therapeutic target for depression. Neuropsychopharmacology 1–12 (2023) .
Salous, A. K. et al. Mechanism of rapid elimination of lysophosphatidic acid and related lipids from the circulation of mice. J. Lipid Res. 54, 2775–2784 (2013).
Guillot, E. et al. Lysophosphatidic acid receptor agonism: discovery of potent non-lipid benzofuran ethanolamine structures. J. Pharmacol. Exp. Ther. 374, 283–294 (2020).
Chrencik, J. E. et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell 161, 1633–1643 (2015).
Liu, S. et al. Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate. Nat. Commun. 13, 731 (2022).
Akasaka, H. et al. Structure of the active Gi-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist. Nat. Commun. 13, 5417 (2022).
Inoue, A. et al. Illuminating G-Protein-Coupling Selectivity of GPCRs. Cell 177, 1933–1947.e25 (2019).
Sano, F. K., Akasaka, H., Shihoya, W. & Nureki, O. Cryo-EM structure of the endothelin-1-ETB-Gi complex. eLife 12, e85821 (2023).
Maeda, S. et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 9, 3712 (2018).
Mao, C. et al. Unsaturated bond recognition leads to biased signal in a fatty acid receptor. Science 380, eadd6220 (2023).
Krishna Kumar, K. et al. Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell 176, 448–458.e12 (2019).
Li, X. et al. Structural basis of selective cannabinoid CB2 receptor activation. Nat. Commun. 14, 1447 (2023).
Kumari, P., Inoue, A., Chapman, K., Lian, P. & Rosenbaum, D. M. Molecular mechanism of fatty acid activation of FFAR1. Proc. Natl Acad. Sci. USA 120, e2219569120 (2023).
Xu, Z. et al. Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism. Nat. Chem. Biol. 18, 281–288 (2022).
Chen, H. et al. Structure of S1PR2-heterotrimeric G13 signaling complex. Sci. Adv. 8, eabn0067 (2022).
Maeda, S. et al. Endogenous agonist-bound S1PR3 structure reveals determinants of G protein-subtype bias. Sci. Adv. 7, eabf5325 (2021).
Yuan, Y. et al. Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition. Cell Res. 31, 1263–1274 (2021).
Wang, N. et al. Structural basis of leukotriene B4 receptor 1 activation. Nat. Commun. 13, 1156 (2022).
Liang, J. et al. Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and activation. Nat. Commun. 14, 1012 (2023).
Xia, A. et al. Cryo-EM structures of human GPR34 enable the identification of selective antagonists. Proc. Natl Acad. Sci. USA 120, e2308435120 (2023).
Qu, C. et al. Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E2 receptor EP2 subtype. Sci. Adv. 7, eabf1268 (2021).
Morimoto, K. et al. Crystal structure of the endogenous agonist-bound prostanoid receptor EP3. Nat. Chem. Biol. 15, 8–10 (2019).
Wu, C. et al. Ligand-induced activation and G protein coupling of prostaglandin F2α receptor. Nat. Commun. 14, 2668 (2023).
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013).
Fukuhara, S. et al. Structure of the human secretin receptor coupled to an engineered heterotrimeric G protein. Biochem. Biophys. Res. Commun. 533, 861–866 (2020).
Okamoto, H. H. et al. Cryo-EM structure of the human MT1-Gi signaling complex. Nat. Struct. Mol. Biol. 28, 694–701 (2021).
Katoh, Y., Nozaki, S., Hartanto, D., Miyano, R. & Nakayama, K. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351–2362 (2015).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Crystallogr. Sect. Struct. Biol. 77, 1282–1291 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. Struct. Biol. 74, 531–544 (2018).
Acknowledgements
We thank K. Ogomori and C. Harada for technical assistance, Inoue. A. for the advice on the assay experiment and provision of the plasmids. This work was supported by grants from the JSPS KAKENHI, grant numbers 21H05037 (O.N.), 22K19371 and 22H02751 (W.S.), 23KJ0491 (F.K.S.), and 24KJ0906 (H.A.); the Takeda Science Foundation (W.S.); the Lotte Foundation (W.S.); the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from the Japan Agency for Medical Research and Development (AMED), grant numbers JP22ama121002 (support number 3272, O.N.) and JP22ama121012 (O.N.).
Author information
Authors and Affiliations
Contributions
H.A. performed all the experiments. F.K.S. assisted with the cryo-EM data collection and the single particle analysis. The manuscript was mainly prepared by H.A. with assistance from W.S. and O.N.
Corresponding authors
Ethics declarations
Competing interests
O.N. is a co-founder and scientific advisor for Curreio. All other authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Huan Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Janesh Kumar and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Akasaka, H., Sano, F.K., Shihoya, W. et al. Structural mechanisms of potent lysophosphatidic acid receptor 1 activation by nonlipid basic agonists. Commun Biol 7, 1444 (2024). https://doi.org/10.1038/s42003-024-07152-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07152-y