Abstract
Fortilin, a 172-amino acid polypeptide, is a multifunctional protein that interacts with various protein molecules to regulate their functions. Although fortilin has been shown to interact with cytoskeleton proteins such as tubulin and actin, its interactions with the components of adherens junctions remained unknown. Using co-immunoprecipitation western blot analyses, the proximity ligation assay, microscale thermophoresis, and biolayer interferometry, we here show that fortilin specifically interacts with CTNNA3 (α-T-catenin), but not with CTNNA1, CTNNA2, or CTNNB. The silencing of fortilin using small interfering RNA (siRNAfortilin) promotes the proteasome-mediated degradation of CTNNA3 in 293T cells. Using both fortilin-deficient THP1 cells and 293T cells that overexpress wild-type (WT), phospho-null (5A), and phospho-mimetic (5D) CTNNA3s, we also show that the absence of fortilin accelerates the phosphorylation of CTNNA3, leading to its ubiquitination and proteasome-mediated degradation. Further, the silencing of CTNNA3 using siRNACTNNA3 causes 293T cells to undergo apoptosis. These data suggest that fortilin guards the cells against apoptosis by positively regulating the pro-survival molecule CTNNA3 by protecting it against phosphorylation, ubiquitination, and proteasome-mediated degradation.
Similar content being viewed by others
Introduction
Fortilin (also known as translationally controlled tumor protein, histamine-releasing factor, and TPT1) is a ubiquitously expressed and highly conserved 172-amino acid, 20-kDa protein. Fortilin is present in intracellular1 and extracellular spaces2. Intracellularly, fortilin exists in both the nucleus and the cytosol1. Although it was originally cloned in 1988 by Gross et al. as a molecule abundantly expressed in tumor cells3, the function of fortilin remained unknown until 2001, when we and others reported that fortilin blocks apoptosis1,4,5. The mechanism by which fortilin does so is multifactorial; it positively regulates anti-apoptotic molecules such as peroxiredoxin-1 (PRX1)6 and myeloid cell leukemia 1 protein (MCL1)7,8 and negatively regulates pro-apoptotic molecules such as inositol-requiring transmembrane kinase/endoribonuclease 1α (IRE1α)9 and tumor suppressor protein p5310,11. Fortilin has also been recognized as a multi-functional protein with roles in various cellular processes, including cell-cycle progression12 and immunoglobulin (Ig) E-mediated histamine release and allergic reactions13,14.
Fortilin often exerts its biological activities through molecular interactions with “executioner” proteins. In the cytosol, fortilin binds PRX1, an enzyme that detoxifies a reactive oxygen species, and protects PRX1 from deactivating phosphorylation6. Fortilin also binds to MCL1, an anti-apoptotic Bcl-2 family member, and protects it from proteasome-mediated degradation7,8. Furthermore, fortilin interacts with and mitigates the pro-apoptotic activity of phosphorylated IRE1α, an endoplasmic reticulum stress sensor9. In the nucleus, fortilin binds to p53 and prevents it from binding the promoters of the BAX, PUMA, and NOXA genes and activating them transcriptionally10,11. Interestingly, fortilin can be secreted from the cell2 via the non-classical pathway15, and it circulates in the blood of humans and mice2. Fortilin binds TGF-β1 in the extracellular space and prevents it from activating the TGF-β1 receptor16.
In this study, we report that fortilin specifically interacts with CTNNA3 (also known as α-T-catenin). We show that fortilin-CTNNA3 binding prevents CTNNA3 from being phosphorylated, ubiquitinated, and proteasomally degraded. We also provide evidence that CTNNA3 is not only a cell adhesion protein but also a pro-survival molecule and that CTNNA3 deficiency leads to apoptosis. These findings highlight the diverse role of fortilin in protein homeostasis and apoptosis regulation.
Results
Fortilin physically interacts with CTNNA3
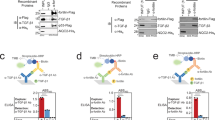
Although fortilin is known to interact with some of the cytoskeleton proteins, including α-tubulin17, β-tubulin17, and actin18, it remained unknown if fortilin interacted with catenins, which are a component of adherens junctions. To explore the interaction between fortilin and catenins19,20, we first optimized the binding and wash conditions of the experiment using both negative (glyceraldehyde 3-phosphate dehydrogenase, known not to interact with fortilin) and positive (p53, known to interact with fortilin10) controls (Supplementary Figs. S1a and S13a). We then subjected total cell lysates from 293T cells to standard co-immunoprecipitation-western blot (Co-IP-western) assays by first immunoprecipitating fortilin by anti-fortilin monoclonal antibody (α-fortilin mAb) and probing for co-immunoprecipitated catenins—CTNNA1 (α-E-catenin), CTNNA2 (α-N-catenin), CTNNA3 (α-T-catenin), and CTNNB (β-catenin)—using anti-CTNNA1 (α-CTNNA1), α-CTNNA2, and α-CTNNA3 mAbs and α-CTNNB polyclonal antibody (pAb), respectively (Fig. 1a–d). Although fortilin was consistently immunoprecipitated by α-fortilin mAb (Fig. 1a-B3, b-B3, c-B3, & d-B3), only CTNNA3 (Fig. 1c-A3)—not CTNNA1, CTNNA2, or CTNNB (Fig. 1a-A3, 1b-A3, & d-A3; Supplementary Figs. S6a–c, S7a)—was co-immunoprecipitated. To confirm the direct interaction between fortilin and CTNNA3, we subjected a buffer containing recombinant FLAG-tagged fortilin (fortilin-FLAG), CTNNA3-FLAG, p53-FLAG, and hexa-histidine-tagged NQO2 (His6-NQO2) to Co-IP-western using α-fortilin mAb. NQO2 is known not to interact with fortilin6,9. We found that fortilin, when immunoprecipitated by α-fortilin mAb (Fig. 1e-A3, Supplementary Fig. S7b) co-immunoprecipitated CTNNA3 (Fig. 1e-B3, Supplementary Fig. S7b) and p53 (Fig. 1e-C3, Supplementary Fig. S7b), but not NQO2 (Fig. 1e-D3, Supplementary Fig. S7b). We also subjected the same cell lysates from 293T cells to a reverse Co-IP-western assay in which we first immunoprecipitated CTNNA3 using α-CTNNA3 mAb and then probed for co-immunoprecipitated fortilin. We found that immunoprecipitated CTNNA3 (Fig. 1f-A3) co-precipitated with fortilin (Fig. 1f-B3; Supplementary Fig. S7c). These data suggest that fortilin specifically interacts with CTNNA3 but not with CTNNA1, CTNNA2, or CTNNB (Fig. 1a–f; Supplementary Figs. S1a, S6a–c, S7a–c and S13a) and that CTNNA3 is unique among the catenin family proteins in that it specifically interacts with fortilin.
IP immunoprecipitation, IB immunoblot, TCE 2,2,2-trichloroethanol, α-fortilin anti-fortilin antibody (Ab), α-CTNNA1 anti-CTNNA1 Ab, α-CTNNA2 anti-CTNNA2 Ab, α-CTNNA3 anti-CTNNA3 Ab, α-CTNNB anti-CTNNB Ab, α-FLAG anti-FLAG (DYKDDDDK) Ab, α-His6 anti-hexahistidine Ab, PLA proximity ligation assay, DAPI 4’,6-diamidino-2-phenylindole, MST microscale thermophoresis, Fnorml normalized fluorescence value, Kd dissociation constant. In vivo co-IP experiments in which fortilin was immunoprecipitated from 293T total cell lysates by α-fortilin Ab and the presence of co-immunoprecipitated catenin family proteins, namely (a) CTNNA1, (b) CTNNA2, (c) CTNNA3, and (d) CTNNB, was assessed by immunoblot analyses using α-CTNNA1, α-CTNNA2, α-CTNNA3, and α-CTNNB Abs. e Co-IP experiments using recombinant proteins where recombinant human FLAG-tagged fortilin protein was immunoprecipitated by α-fortilin Ab. Successful IP of fortilin as well as successful co-IP of FLAG-tagged CTNNA3 was confirmed by immunoblot analysis using α-FLAG Ab. f Reverse co-IP experiment in which CTNNA3 was immunoprecipitated from 293T total cell lysates by α-CTNNA3 Ab, and the presence of co-immunoprecipitated fortilin was assessed by immunoblot analyses using α-fortilin Ab. In a–d, f, total protein was visualized by incorporating TCE into the gel. g PLA to show the fortilin-CTNNA3 interaction. Top panels: PLA was performed using rabbit α-fortilin and mouse α-CTNNA3 Abs in U2OS cells, and the nuclei were counterstained with DAPI (blue). Red dots indicate that fortilin and CTNNA3 are located within 30 nm of each other. Bottom panels: PLA was performed using rabbit α-fortilin and mouse α-CTNNB mAbs in U2OS cells. No PLA (red) signals were observed. Each panel in rows 2 and 4 shows a magnified view of the white-boxed area in the panel above in rows 1 and 3, respectively, highlighting the specific interaction signals. Scale bar (long): 100 μm. Scale bar (short): 10 μm. h Miscroscale thermophoresis (MST) to characterize the fortilin–CTNNA3 interaction. Red-maleimide-labeled fortilin was mixed with varying concentrations (30 pM to 250 nM) of recombinant human CTNNA3 and loaded to the glass capillaries. Laser-induced, heat-mediated thermophoresis of fortilin, in the presence of varying concentrations of CTNNA3, was assessed using Monolith NT.115 Pico MST system (Nanotemper Technologies GmbH) and expressed as the relative Fnorml (%). The dissociation constants (Kd) were calculated using the Nanotemper analysis software based on the relative Fnorml values.
Using phylogenetic and homology analyses, we consistently found that CTNNA1 and CTNNA2 are phylogenetically and structurally similar to each other but that CTNNA3 is less similar to either CTNNA1 or CTNNA2 (Supplementary Fig. S3a, b). We also found that CTNNB is not homologous to any of the CTNNA proteins (Supplementary Fig. S3a, b). Finally, we aligned the protein sequence of CTNNA3 to those of CTNNA1 and CTNNA2 and found that the region spanning between Q149 and I296 of CTNNA3 is the region least homologous to both CTNNA1 and CTNNA2, although there are several other smaller regions where the homology between CTNNA3 and CTNNA1/CTNNA2 is also low (Supplementary Fig. S3c). These results suggest that fortilin might interact with CTNNA3 through one of these regions.
To further characterize the interaction between fortilin and CTNNA3, we performed an in-situ proximity ligation assay (PLA) in U2OS cells using rabbit α-fortilin and mouse α-CTNNA3 and α-CTNNB mAbs. In this assay, each red dot indicated the presence of a fortilin molecule located within approximately 30 nm of an CTNNA3 molecule21. Confocal microscopic examination revealed numerous red dots in both the nucleus and cytosol when PLA was performed using α-fortilin and α-CTNNA3 mAbs (Fig. 1g, Fortilin + CTNNA3). Consistent with the Co-IP experiments that shows no interaction between fortilin and CTNNA1, CTNNA2, or CTNNB (Fig. 1a, b, d), we observed no red dots when PLA was performed using α-fortilin and their respective mAbs (Fig. 1g, Fortilin + CTNNB; Supplementary Fig. S1e, Fortilin + CTNNA1, Fortilin + CTNNA2, Fortilin + CTNNB). The α-CTNNA1, α-CTNNA2, and α-CTNNA3 mAbs used in the PLAs were specific for their respective proteins and did not cross react with other isoforms (Supplementary Figs. S1b–d and S13c). These data suggest that fortilin interacts with CTNNA3—but not CTNNA1, CTNNA2, or CTNNB—and does so in both the nucleus and cytosol.
To further characterize the interaction of fortilin with CTNNA3 and its isoforms—CTNNA1 and CTNNA2, we performed microscale thermophoresis (MST)22,23 using the NanoTemper Monolith NT.115 Pico system (NanoTemper Technologies GmbH, München, Germany) where the thermophoretic movement of recombinant human fortilin (rh-fortilin) labeled with red-maleimide fluorescent dye was measured in the presence of varying concentrations of CTNNA1, CTNNA2, or CTNNA3. We found that fortilin specifically binds CTNNA3 with an equilibrium dissociation constant (Kd) of 33.23 nM (Fig. 1h) but does not bind CTNNA1 or CTNNA2 (Supplementary Fig. S1f). We also performed a bio-layer interferometry (BLI) assay24,25 using rh-fortilin and rh-CTNNA3 in the BLItz® system (Sartorius, Göttingen, Germany). We found that rh-fortilin and rh-CTNNA3 bind to each other at a dissociation constant (Kd) of 36.1 nM (Supplementary Fig. S1g).
Fortilin protects CTNNA3 against proteasome-mediated degradation
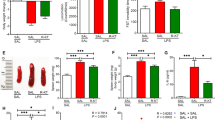
To investigate the impact of fortilin on CTNNA3 in the cell, we first used CRISPR-Cas9 technology26 to generate THP1 human monocyte-macrophage cells lacking the fortilin gene (THP1KO-fortilin) and characterized them using western blot analysis. We found that THP1KO-fortilin cells did not express fortilin (Fig. 2a-A2; Supplementary Fig. S8a), whereas the control cells THP1WT-fortilin did (Fig. 2a-A1; Supplementary Fig. S8a). Next, we seeded THP1WT-fortilin and THP1KO-fortilin cells in a chamber slide, immunostained these cells using anti-CTNNA3 and anti-fortilin mAbs, counterstained the nucleus with 4’, 6-diamidino-2-phenylindole (DAPI), and examined the distribution of CTNNA3 within the cells using confocal microscopy. Under these staining conditions, fortilin signals were lower in THP1KO-fortilin cells than in THP1WT-fortilin cells (Fig. 2b, Fortilin). Strikingly, however, we also found that CTNNA3 signals—present in both cytosol and nucleus—were also drastically lower in THP1KO-fortilin cells than in THP1WT-fortilin cells (Fig. 2b, CTNNA3).
THP1WT-fortilin THP1 cells expressing wild-type fortilin, THP1KO-fortilin cells in which the fortilin genes have been deleted using Crispr-Cas9 technology, GFPstrep-tag strep-tagged recombinant green fluorescence protein, FTstrep-tag strep-tagged recombinant human fortilin protein, * degradation products, IB immunoblot, TCE 2,2,2-trichloroethanol, DAPI 4’,6-diamidino-2-phenylindole, siRNAfortilin small interfering RNA against fortilin, siRNAControl the non-targeting pool of siRNAs, A.U. arbitrary unit, IB immunoblot, α-CTNNA3 anti-CTNNA3 Ab, MG132 carbobenzoxy-L-leucyl-L-leucyl-L-leucine, a proteasome inhibitor. a Characterization by western blot analysis of THP1WT-fortilin and THP1KO-fortilin cells. Total proteins were visualized in the gel using TCE. b THP1WT-fortilin and THP1KO-foritlin cells were seeded on a chamber slide and subjected to immunocytochemistry using α-CTNNA3 and α-fortilin Abs. The nucleus was counter-stained by DAPI. The intensities of CTNNA3 and fortilin signals were evaluated by confocal microscopy using the same imaging conditions. Long scale bar = 100 µm. Short scale bar = 10 μm. c Silencing of fortilin by siRNAfortilin. Fortilin mRNA expression levels was evaluated by RT-qPCR normalized to that of 18S rRNA after 293T cells were transfected with the siRNAs. Data are expressed as means ± s.d. (n = 3 biological replicates), and they were analyzed by two-tailed, unpaired Student’s t test. d Western blotting analysis of fortilin protein expression levels after 293T cells were transfected with the siRNAs. Total proteins were visualized by TCE staining. e Densitometric quantification of fortilin protein signals using ImageJ software. The fortilin protein expression indices (in A.U.) were determined by dividing the signal intensity of the immunoblotted fortilin band by that of the total protein bands identified by TCE staining. f Time-course analysis using western blot analysis of CTNNA3 expression levels in 293T cells. g Densitometric quantification of CTNNA3 signals using ImageJ software. The CTNNA3 expression indices were calculated by dividing the signal intensities of the immunoblotted CTNNA3 bands by those of the total protein bands identified by TCE staining. They were expressed in A.U. after normalizing the indices to time 0. The percentage reduction of CTNNA3 proteins at 36 h is shown in the graph. Data are expressed as means ±s.d. (n = 3 biological replicates), and they were analyzed by two-tailed, unpaired Student’s t tests.
Fortilin binds MCL127 and PRX16 and protects these proteins against proteasome-mediated degradation6,27. CTNNB, a member of the catenin family to which CTNNA3 belongs, is degraded by the proteasome pathway28. To test the hypothesis that the binding of fortilin to CTNNA3 protects CTNNA3 against proteasome-mediated degradation, we silenced fortilin in 239T cells using small interfering RNA against fortilin (siRNAfortilin) and subjected the cells to a protein degradation assay. In this assay, we kept the cells exposed to the protein synthesis inhibitor cycloheximide (CHX), incubated them in the presence or absence of the proteasome inhibitor MG132, harvested them at various time points (0, 0.5, 1, 2, 4, 8, 12, 24, and 36 h), and subjected their lysates to both standard and JESS™ western blot analyses to quantify the expression levels of CTNNA3. JESS™ is an automated, capillary-based, highly quantitative western blot system (ProteinSimple®, San Jose, CA, USA)29,30. We found that the treatment of 293T cells with siRNAfortilin decreased both fortilin mRNA and proteins at time 0 (Fig. 2c–e, Supplementary Fig. S8b). Also at time 0, the CTNNA3 protein levels were significantly less in siRNAfortilin-treated, fortilin-deficient cells than in siRNAcontrol-treated, fortilin-present cells (Supplementary Figs. S2a–d, S13b). In that system and in the absence of MG132, CTNNA3 protein was found to be stable with only 4–22% loss after 36 h in siRNAcontrol-treated, fortilin-present cells (Fig. 2f, g; MG132(–), siRNAcontrol; Supplementary Fig. S8c; Supplementary Fig. S2a, e; MG132(–), siRNAcontrol; Supplementary Fig. S13b). Strikingly, CTNNA3 disappeared very quickly in the siRNAfortilin-treated, fortilin-deficient cells, in which the majority (75 – 90%) of CTNNA3 degraded within 36 h in the absence of MG132 (Fig. 2f, g, MG132(–), siRNAfortilin; Supplementary Fig. S8c; Supplementary Figs. S2a, e; MG132(–), siRNAfortilin; Supplementary Fig. S13b). CTNNA3 degradations were significantly greater in siRNAfortilin-treated cells than in siRNAcontrol-treated cells at and after 8 h (P = 0.002 at 8 h, P = 0.001 at 12 h, P = 0.000 at 24 h, P = 0.000 at 36 h) (Fig. 2f, g, MG132(–); Supplementary Fig. S8c) by the standard western blots and at and after 2 h (P = 0.006 at 2 h, P = 0.023 at 4 h, P = 0.003 at 8 h, P = 0.006 at 8 h, P = 0.000 at both 24 and 36 h; Supplementary Fig. S2a, e, MG132 (–); Supplementary Fig. S13b) by JESS™. In contrast, no significant differences were observed between siRNAfortilin-treated and siRNAcontrol-treated cells in the presence of MG132 (Fig. 2f, g, MG132(+); Supplementary Fig. S8c; Supplementary Fig. S2c, e, MG132(+); Supplementary Fig. S13b). These data suggest that fortilin prevents CTNNA3 from being degraded by the proteasome pathway.
Phosphorylation of CTNNA3 triggers its ubiquitination and proteasome-mediated degradation
Although the above data suggested that the lack of fortilin causes CTNNA3 to be degraded by the proteasome pathway (Fig. 2), we still did not know what triggers its ubiquitination in the absence of fortilin. Because the ubiquitination of CTNNB is triggered by its phosphorylation31, we hypothesized that the lack of fortilin promotes the phosphorylation of CTNNA3 and its subsequent ubiquitination. To test this premise, we transiently transfected THP1WT-fortilin and THP1KO-fortilin cells (Fig. 2a, b) with the pEZ-M39-CTNNA3WT-FLAG mammalian-expression plasmid vector, incubated the cells for 48 h, lysed the cells, and subjected the cleared total cell lysates to immunoprecipitation using anti-FLAG M2 magnetic beads. Subsequently, we subjected the eluates to SDS-PAGE. We stained the gel first with Pro-Q Diamond32 and then with SYPRO Ruby33 staining solutions to visualize phosphorylated CTNNA3 and total CTNNA3, respectively. CTNNA3WT-FLAG was successfully immunoprecipitated as visualized by SYPRO-Ruby (Fig. 3a-B3, B4; Supplementary Fig. S9a). Strikingly, however, CTNNA3WT-FLAG was found to be phosphorylated only in the lysates from THP1KO-fortilin cells (Fig. 3a-A4; Supplementary Fig. S9a) but not in those from THP1WT-fortilin cells (Fig. 3a-A3; Supplementary Fig. S9a). These data suggest that CTNNA3 is more heavily phosphorylated in cells lacking fortilin.
FLAG-CTNNA3WT (also WT) a mammalian plasmid containing the construct for FLAG-tagged wild-type human CTNNA3 (pEZ-CTNNA3WT-FLAG), Pro-Q Diamond phosphoprotein gel staining, SYPRO Ruby total protein gel staining, CTNNA35D (also 5D) CTNNA3 protein with the following phosphomimetic mutations—S637D, S647D, T649D, S650D, and T653D; CTNNA35A (also 5A), CTNNA3 protein with the following phospho-null mutations—S637A, S647A, T649A, S650A, and T653A, THP1WT-fortilin THP1 cells expressing wild-type fortilin, THP1KO-fortilin THP1 cells in which the fortilin gene was deleted using Crispr-Cas9 technology, IP immunoprecipitation, α-FLAG anti-FLAG antibody (Ab), α-His6, anti-His6 Ab, JESS™ an automated capillary-based quantitative western blot system, Ni-NTA pulldown using Ni-NTA Superflow beads, A.U. arbitrary unit, Ubn-CTNNA3 polyubiquitinated CTNNA3, α-fortilin anti-fortilin Ab, TCE 2,2,2-trichloroethanol. a Top panel: Pro-Q Diamond staining of phosphorylated CTNNA3WT in the presence (THP1WT-fortilin) or absence (THP1KO-fortilin) of fortilin in the cell. THP1 cells were transfected with the pEZ-CTNNA3WT-FLAG plasmid. CTNNA3WT-FLAG in the total cell lysates was immunoprecipitated by anti-FLAG Ab and subjected to SDS-PAGE. Phosphorylated CTNNA3WT-FLAG was visualized by Pro-Q Diamond staining. Bottom panel: SYPRO Ruby staining of total CTNNA3WT in the presence (THP1WT-fortilin) or absence (THP1KO-fortilin) of fortilin in the cell. The same eluates from the above experiment were subjected to SDS-PAGE. Total CTNNA3WT-FLAG was visualized by SYPRO Ruby staining. b Wild-type, phosphomimetic (5D), and phospho-null (5A) CTNNA3 mutant constructs. c Top panel: Pro-Q Diamond staining of phosphorylated CTNNA3WT and CTNNA35A proteins in the presence (THP1WT-fortilin) or absence (THP1KO-fortilin) of fortilin in the cell. THP1 cells were transfected by the pEZ-CTNNA3WT-FLAG plasmid. Bottom panel: SYPRO Ruby staining of total CTNNA3WT and CTNNA35A proteins in the presence (THP1WT-fortilin) or absence (THP1KO-fortilin) of fortilin in the cell. The same eluates from the above experiment were subjected to SDS-PAGE. Total CTNNA3WT-FLAG and CTNNA35A proteins were visualized by SYPRO Ruby staining. d Impact of CTNNA3 phosphorylation on the degree of ubiquitination in 293T cells. 293T cells were transfected by the pEZ-CTNNA3WT-FLAG, pEZ-CTNNA35D-FLAG, or pEZ-CTNNA35A-FLAG plasmid. Ubiquitinated wild-type and mutant CTNNA3s were visualized by first pulling down the ubiquitinated proteins using Ni-NTA beads and then immunoblotting CTNNA3 within the pulled-down proteins using α-FLAG Ab in the JESS™ system. e The CTNNA3 ubiquitination indices of wild-type and mutant CTNNA3s. The indices (in A.U.) were calculated by dividing the area under the curve of the ubiquitinated CTNNA3-FLAG by that of the input CTNNA3-FLAG. Data are expressed as means ± s.d. (n = 3 biological replicates), and they were analyzed by one-way ANOVA and Tukey–Kramer multiple-comparisons. f, g Impact of CTNNA3 phosphorylation on its proteasome-mediated degradation. 293T cells were transfected by the pEZ-CTNNA3WT-FLAG, pEZ-CTNNA35D-FLAG, or pEZ-CTNNA35A-FLAG plasmid and incubated in the presence of CHX and with or without MG132 for various durations. CTNNA3-FLAGs were detected by anti-FLAG Ab, and total proteins were visualized using the JESS™ total protein detection module. CTNNA3 expression indices (in A.U.) were calculated using Compass software by dividing the area under the curve of a CTNNA3 peak by the total proteins loaded in the same capillary (“in-capillary normalization”). Data are expressed as means ± s.d. (n = 3 biological replicates). Statistical analyses were performed using one-way ANOVA with Fisher’s pairwise comparisons. h, i Co-IP analysis of the strength of the interaction between fortilin and wild-type and mutant CTNNA3s. The total cell lysates from 293T cells transfected by the pEZ-CTNNA3WT-FLAG, pEZ-CTNNA35D-FLAG, or pEZ-CTNNA35A-FLAG plasmid were subjected to IP by α-FLAG Ab. Co-immunoprecipitated fortilin was then detected by immunoblotting using α-fortilin Ab. The fortilin-CTNNA3 interaction indices (in A.U.) were calculated using ImageJ software by dividing the signal intensities of co-immunoprecipitated fortilin bands by the immunoprecipitated CTNNA3-FLAG bands. Data are expressed as means ± s.d., (n = 3 biological replicates), and they were analyzed by one-way ANOVA and Tukey–Kramer tests.
Next, we tested the hypothesis that phosphorylation of CTNNA3 leads to its ubiquitination. Reitz et al. reported that CTNNA3 is highly phosphorylated in the hearts of patients with dilated cardiomyopathy—especially in three serine (S637, S647, S650) and two threonine (T649, T653) residues clustered within a span of 14 amino acids34. First, we generated pEZ-M39-CTNNA35A-FLAG and pEZ-M39-CTNNA35D-FLAG plasmid vectors in which all five sites were mutated to alanine (5A; phospho-null mutations; S637A, S647A, T649A, S650A, and T653A) or to aspartic acid (5D; phospho-mimetic mutations; S637D, S647D, T649D, S650D, and T653D), respectively (Fig. 3b). We then transiently transfected THP1WT-fortilin or THP1KO-fortilin cells with the pEZ-M39-CTNNA3WT-FLAG or pEZ-M39-CTNNA35A-FLAG plasmid vectors, immunoprecipitated CTNNA3WT-FLAG and CTNNA35A-FLAG using anti-FLAG M2 magnetic beads, subjected the eluates to SDS-PAGE, and stained the gel first with Pro-Q Diamond32 and then with SYPRO Ruby33 staining solutions to visualize phosphorylated CTNNA3 and total CTNNA3, respectively. Both CTNNA3WT-FLAG and CTNNA35A-FLAG were successfully immunoprecipitated (Fig. 3c-B1–4; Supplementary Fig. S9b). In that system, Pro-Q Diamond staining showed a CTNNA3WT-FLAG band (Fig. 3c-A3; Supplementary Fig. S9b) but not a CTNNA35A-FLAG band (Fig. 3c, A4, Supplementary Fig. S9b) in the THP1KO-fortilin lysates, suggesting that the majority of phosphorylation of CTNNA3 occurs at the five phosphosites (S637, S647, T649, S650, and T653). To further verify the negative regulatory role of fortilin on CTNNA3 phosphorylation, we immunoprecipitated CTNNA3 from THP1KO-foritlin and THP1WT-fortilin cells using anti-CTNNA3 mAb and visualized co-immunoprecipitated total CTNNA3 and phosphorylated CTNNA3 by immunoblotting the eluates with anti-CTNNA3 mAb and anti-phosphoserine/threonine pAb, respectively. We found that CTNNA3 was significantly more phosphorylated in the absence of fortilin than in its presence (Supplementary Fig. S4a, b; Supplementary Fig. S14a). Finally, we tested if fortilin impacts the status of the acetylation of CTNNA3 using THP1KO-fortilin and THP1WT-fortilin cells and by immunoprecipitating CTNNA3 from the total cell lysates and immunoblotting the eluates using anti-CTNNA3 mAb and anti-acetylated lysine pAb. We found no obvious difference in the status of CTNNA3 in the presence or absence of fortilin (Fig. S4c, d; Supplementary Fig. S14b).
siRNAcontrol non-targeting pool of small interfering RNAs, siRNACTNNA3 siRNA against CTNNA3, siRNAfortilin siRNA against fortilin, IB immunoblot, α-CTNNA3 anti-CTNNA3 antibody (Ab), α-fortilin anti-fortilin Ab, NA no knockdown target, A.U. arbitrary unit. a–c JESS ™ Western blot analysis of CTNNA3 and fortilin protein levels following siRNA treatments. a The total cell lysates from 293T cells transfected by siRNAcontrol, siRNACTNNA3, siRNACTNNA3/fortilin, or siRNAfortilin were subjected to JESS™ western blot analysis using α-CTNNA3 (upper panel) and α-fortilin (middle panel) Abs while total proteins were visualized and used as the loading control. b The fortilin expression level (in A.U.) was calculated by dividing the area under the curve (AUC) of the fortilin signals by that of corresponding total protein signals. c The CTNNA3 expression level (in A.U.) was calculated by dividing the AUC of the CTNNA3 signals by that of corresponding total protein signals. d Trypan blue positivity (in A.U.) of 293T cells transfected by siRNAcontrol, siRNACTNNA3, siRNACTNNA3/fortilin, or siRNAfortilin. e Activated caspase 3 levels (in A.U.) of 293T cells transfected by siRNAcontrol, siRNACTNNA3, siRNACTNNA3/fortilin, or siRNAfortilin. f DNA fragmentation levels (in A.U.) of 293T cells transfected by siRNAcontrol, siRNACTNNA3, siRNACTNNA3/fortilin, or siRNAfortilin. d–f The values indicate fold-change of the respective levels of the siRNACTNNA3, siRNACTNNA3/fortilin, and siRNAfortilin-treated cells relative to that of siRNAcontrol-treated cells. Data are expressed as means ± s.d. (n = 3 biological replicates), and they were analyzed by one-way ANOVA and Tukey–Kramer tests.
Next, we transiently transfected 293T cells with (a) pReceiver-His6-Ub3 and (b) pEZ-M39-CTNNA3WT-FLAG, pEZ-M39-CTNNA35A-FLAG, or pEZ-M39-CTNNA35D-FLAG. We then pulled down His6-tagged-ubiquitins (His6-Ub) using Ni-NTA Superflow® beads in the presence of MG132 and subjected the eluates to the ProteinSimple® JESS™ system, an automated capillary-based protein separation and detection system. Immunodetection using anti-His6 mAb showed that His6-Ub were expressed equally in all four groups of 293T cells transfected with the pReceiver-His6-Ub3 plasmid (Fig. 3d-C6–9; Supplementary Fig. S10). Expression levels of CTNNA3WT-FLAG, CTNNA35A-FLAG, and CTNNA35D-FLAG—either un-ubiquitinated or mono/oligo-ubiquitinated—were similar to each other (Fig. 3d-B7, B8, &B9; Supplementary Fig. S10). When pulling down His6-Ub using Ni-NTA Superflow® beads and subjecting them to JESS™ to quantify the amount of ubiquitinated CTNNA3, we found that CTNNA5D-FLAG was significantly more ubiquitinated than CTNNA3WT-FLAG or CTNNA35A-FLAG (Fig. 3d, e; Supplementary Fig. S10; CTNNAWT-FLAG, CTNNA5A-FLAG, & CTNNA5D-FLAG = panels A7, A8, and A9; 1.0 ± 0.00, 0.85 ± 0.04, and 1.28 ± 0.51, respectively; P < 0.0001 for CTNNA5D-FLAG vs. CTNNA35A-FLAG; P = 0.0003 for CTNNA5D-FLAG vs. CTNNA3WT-FLAG, N = 3, ANOVA with Tukey–Kramer’s pairwise comparison). These data suggest that the phosphorylation of CTNNA3—especially in S637, S647, T649, S650, and T653—accelerates the ubiquitination of CTNNA3.
To test if phosphorylation of the five phosphosites of CTNNA3 in fact causes the protein to be proteasomally degraded, we transiently transfected 293T cells with pEZ-M39-CTNNA3WT-FLAG, pEZ-M39-CTNNA35A-FLAG, or pEZ-M39-CTNNA35D-FLAG, harvested the cells at 0, 4, 8, 12, and 24 h after exposure to CHX and in the presence or absence of MG132, subjected the total cell lysates to JESS™, and quantified the amounts of CTNNA3WT-FLAG, CTNNA35A-FLAG, and CTNNA35D-FLAG in the cells using anti-FLAG pAb. The expression levels of FLAG-tagged CTNNA3s were calculated first by dividing the signal intensity of each FLAG-tagged CTNNA3 band at a given time point by the total protein signal intensity of the same time point, and then normalizing these values to their respective time 0 values, which were set to 1. In the absence of MG132, the levels of CTNNA35D-FLAG were significantly lower than those of CTNNA3WT-FLAG and CTNNA35A-FLAG at 4, 8, and 24 h (Fig. 3f, g, MG132(–); Supplementary Fig. S11a). In the presence of MG132, however, there was no significant degradation in any CTNNA3-FLAGs (Fig. 3f, g, MG132(+); Supplementary Fig. S11a). When taken together, these data suggest that the lack of fortilin in the cells promotes the phosphorylation of CTNNA3 mainly at its major phosphosites (S637, S647, T649, S650, and T653), which in turn triggers its ubiquitination and proteasome-mediated degradation.
Pinkaew et al. showed that fortilin binds with a higher affinity to phosphorylated IRE1α than to un-phosphorylated IRE1α9. To evaluate the impact of the phosphorylation of CTNNA3 on its interaction with fortilin, we transiently transfected 293T cells with pEZ-M39-CTNNA3WT-FLAG, pEZ-M39-CTNNA35A-FLAG, or pEZ-M39-CTNNA35D-FLAG, immunoprecipitated FLAG-tagged CTNNA3s from total cell lysates using anti-FLAG pAb and evaluated the presence of co-immunoprecipitated fortilin in the eluates by western blot analyses. All three transfection groups contained similar amounts of FLAG-tagged CTNNA3 and fortilin in their total cell lysates (Fig. 3h; INPUT; A1–3, B1–B3, and C1–3; Supplementary Fig. S11b). Anti-FLAG pAb immunoprecipitated similar amounts of CTNNA3WT-FLAG, CTNNA35A-FLAG, and CTNNA35D-FLAG (Fig. 3h, IP: α-FLAG; A4–6; Supplementary Fig. S11b). Strikingly, CTNNA35D-FLAG co-immunoprecipitated fortilin significantly less than did CTNNA3WT-FLAG and CTNNA35A-FLAG (Fig. 3h–B6 vs. B4 & B5; Fig. 3i; Supplementary Fig. S11b). These data suggest that fortilin preferentially binds unphosphorylated CTNNA3 over phosphorylated CTNNA3.
Depletion of CTNNA3 induces apoptotic cell death
Goossens et al. reported that constitutional CTNNA3 knockout mice exhibit decreased left ventricular (LV) function at 3 months of age35. Cardiac-specific CTNNA1 knockout causes cardiac cells to apoptose, leading to LV dysfunction at 15 months36. Based on these observations, we focused on apoptosis as a pivotal consequence of CTNNA3 deficiency in the cell. To test if CTNNA3 deficiency results in cell death through apoptosis, we first silenced fortilin and/or CTNNA3 in 239T cells by transfecting them with siRNA against fortilin (siRNAfortilin) and/or siRNACTNNA3. We validated the silencing of respective genes, using JESS™ (Fig. 4a; Supplementary Fig. S12) and observed as expected that silencing fortilin with siRNAfortilin (Fig. 4a, panel-B of the lanes 1–3 vs. lanes 10–12; Fig. 4b, columns 1 vs. 4) decreased CTNNA3 expression (Fig. 4a, panel-A of the lanes 1–3 vs. lanes 10–12; Fig. 4c, columns 1 vs. 4), while we found that silencing CTNNA3 with siRNACTNNA3 (Fig. 4a, panel-A of the lanes 1–3 vs 4–6; Fig. 4c, columns 1 vs. 2) did not alter fortilin expression (Fig. 4a, panel-B of the lanes 1–3 vs. lanes 4–6; Fig. 4b, columns 1 vs 2). The overall cell morphology post-transfection remained similar to its pre-transfection state, although cell density appeared less in the cells transfected with siRNAfortilin and/or siRNACTNNA3 (Supplementary Fig. S5a). We first subjected the transfected cells to trypan blue exclusion assay and found that the siRNACTNNA3-treated cells exhibited a significant (94%) increase in cell death (trypan blue positivity) compared with siRNAcontrol-treated cells (Fig. 4d, columns 1 vs. 2). As expected from the previous report1,4,5, the siRNAfortilin-treated cells exhibited a significant (154%) increase in cell death compared with the siRNAcontrol-treated cells (Fig. 4d, columns 1 vs. 4). The trypan blue positivity of the cells treated with both siRNAfortilin and siRNACTNNA3 was greater than that of the cells treated by siRNACTNNA3 or siRNAfortilin alone (Fig. 4d, column 3 vs. columns 2 and 4). To determine whether apoptosis contributed to the cell death observed in the trypan blue exclusion assay following CTNNA3 and/or fortilin silencing, we subjected the same four groups to both activated caspase-337 and DNA fragmentation9,10 assays. The siRNACTNNA3-treated cells showed a significant increase in both activated caspase-3 (86%) and DNA fragmentation (121%) levels compared with the siRNAcontrol-treated cells (Fig. 4e, f, columns 1 vs. 2), suggesting that CTNNA3 protects cells from apoptosis. The siRNAfortilin-treated cells also exhibited a significant increase in both the activated caspase-3 (174% increase) and DNA fragmentation (272%) levels, compared with the siRNAcontrol-treated cells (Fig. 4e, f, columns 1 vs. 4), confirming previous reports that fortilin protects cells against apoptosis1,4,5. Finally, both the activated caspase-3 and DNA fragmentation levels in cells treated with both siRNAfortilin and siRNACTNNA3 were significantly higher than in cells treated by siRNACTNNA3 or siRNAfortilin alone, although the effect was not fully additive (Fig. 4e, f, columns 3 vs. 2 and 4), suggesting that the fortilin deficiency induces apoptosis both by promoting the degradation of the anti-apoptotic protein CTNNA3 and through CTNNA3-independent pathways.
In summary, under normal conditions, fortilin binds to the pro-survival molecule CTNNA3 and prevents it from being phosphorylated, ubiquitinated, and proteasomally degraded, thereby protecting cells against apoptosis (Fig. 5, left panel). However, when fortilin levels decrease in the cell, CTNNA3 becomes phosphorylated, ubiquitinated, and proteasomally degraded, precipitating apoptotic cell death (Fig. 5, right panel).
FT fortilin, P phosphorus, Ub ubiquitin molecule. The figure represents a proposed model of the positive regulation by fortilin of CTNNA3. Under normal conditions, fortilin binds CTNNA3 and protects CTNNA3 against its phosphorylation, ubiquitination, and proteasome-mediated degradation, thereby maintaining cellular integrity (left panel, Fortilin (+)). Reduction of fortilin levels within the cell disrupts this protective mechanism, leading to increased phosphorylation, ubiquitination, and proteasome-mediated degradation of CTNNA3, resulting in increased apoptosis (right panel, Fortilin (–)).
Discussion
The innovation of this study lies in the discovery that fortilin physically and functionally interacts with CTNNA3 (Fig. 1 & Supplementary Fig. S1). Although fortilin has been shown to bind the cytoskeletal proteins actin and tubulin, its binding to a protein of the adherens junctional complex has not been reported previously. Despite the sequence homology of CTNNA3 to other catenins38, fortilin binds only to CTNNA3 and not to CTNNA1, CTNNA2, or CTNNB (Fig. 1a–f; Supplementary Fig. S1a, e, f, g), suggesting that fortilin binds CTNNA3 through the region(s) of CTNNA3 that is less homologous to that of the other catenins (e.g., the region spanning between Q149 and I296) (Supplementary Fig. S3a–c). It was widely accepted that CTNNA3 binds only a few proteins—CTNNB38, plakophilin-2 (PKP2)39, one of the desmosome proteins implicated in arrhythmogenic right ventricular dysplasia40, and SPATA3341, a protein that is specifically expressed in the testis and plays a role in mitophagy42,43,44. More recently, however, Huttlin et al. established a large-scale affinity purification-mass spectrometry platform in which 293T and HCT116 cells were stably transduced by lentiviral vectors containing the cDNAs of human genes fused to the human influenza hemagglutinin (HA)-epitope tag construct at the 3ʹ end (“bait”). After the cells were lysed, HA-tagged bait-proteins were pulled down along with their binding partners by anti-HA agarose resin and subjected to liquid chromatography mass spectrometry. The interactome data deposited in BioPlexExplorer (URL: https://bioplex.hms.harvard.edu) represent protein-protein interactions from more than 15,000 human affinity-purified or immunoprecipitated proteins45. In this database, CTNNA3 was found to interact with at least 35 proteins (Supplementary Fig. S5b). Although many of the proteins reported were cell structural proteins such as cadherins (CDH1, CDH2, CDH3, CDH4, CDH10, CDH12, CDH24) and catenins and their homologs (CTNNB, CTNND1, PKP4 (also known as P0071)46, and ARVCF47), CTNNA3 was also found to bind Wnt signaling pathway proteins (APC48 and AMER149), RNA processing proteins (AGO450 and INTS1251), and cell-growth-related protein kinases (FYN52 and PKN253) (Supplementary Fig. S5b). The biological significance of these interactions is unknown and needs to be experimentally evaluated in the future. Interestingly, a fortilin-CTNNA3 interaction is not listed in the database.
Herein, we also established the positive post-translational regulation of CTNNA3 by fortilin (Fig. 2f, g). CTNNA3 is an 895-amino acid polypeptide with a molecular weight of ~100 kDa, and it represents the newest member of the α-catenin family, which includes CTNNA1, CTNNA2, and CTNNB54. α-catenins plays a critical role in linking the cadherin-based adherens junction complex and the actin cytoskeleton54. CTNNA3 is abundantly expressed in cardiomyocytes, skeletal muscle cells, testis, and the brain54. Although it Vanpoucke et al. reported that CTNNA3 expression is regulated at the transcriptional level by transcriptional factors GATA-4 and MEF2C54, the post-translational regulation of the CTNNA3 protein remained unknown until now. Our results shed light on how CTNNA3 is post-translationally regulated and clarify the biological significance of the fortilin-CTNNA3 interaction. Specifically, they show that fortilin prevents unphosphorylated CTNNA3 from being phosphorylated (Fig. 3a, c), ubiquitinated (Fig. 3d, e), and proteasomally degraded (Fig. 3f, g). Fortilin binds more strongly to unphosphorylated CTNNA3 than to phosphorylated CTNNA3 (Fig. 3h, i).
Although we found that silencing the CTNNA3 gene significantly increased apoptosis in 293T cells (Fig. 4e, f), the biological activities of CTNNA3 remain somewhat controversial, as some research including ours suggested that it functions as a pro-survival, pro-proliferation protein35,55,56, whereas other studies suggested that it acts as a tumor suppressor57. On the one hand, Tyberghein et al. generated epiblast-specific CTNNA3 transgenic mice and found that overexpression of CTNNA3 resulted in expansion of the spongiotrophoblast population due to increased proliferation56. Li et al. generated constitutional CTNNA3 knockout mice and found that the CTNNA3-null mice, although viable, fertile, and lacking gross abnormalities, exhibited dilated heart with reduced LV systolic function at 3 months of age35. Although an apoptosis assay was not performed in the hearts of CTNNA3-null mice in that study, the hearts of cardiac-specific CTNNA1-null mice in another study exhibited extensive apoptosis36 where the protein sequences of CTNNA1 and CTNNA3 are 65.6% identical (Supplementary Fig. S3b), suggesting that cardiomyocyte apoptosis at least partially contributed to the LV dysfunction of the CTNNA3-null mice. On the other hand, both He et al.58 and Fanjul-Fernande et al.57 found that CTNNA3 decreased the proliferation, migration, and invasion of cells. Further investigation is needed to better elucidate the biological activity of CTNNA3 in the various microenvironmental contexts.
As a previously unreported observation, we demonstrate that phosphorylation of CTNNA3 facilitates its degradation via the ubiquitin-proteasome pathway (Fig. 3f, g). Reitz et al. performed a systematic, unbiased, phosphoproteomic analysis of myocardial tissue explants from heart failure patients and patients with non-failing hearts and found that phosphorylation of CTNNA3 was increased in the hearts of dilated cardiomyopathy (DCM) patients34. However, the causal relationship between hyperphosphorylated CTNNA3 and LV dysfunction was not entirely clear in the report. They found that phosphorylation of CTNNA3 was important, and not harmful, in maintaining the integrity of the cardiomyocyte intercalated disc, because AAV9-mediated overexpression of phospho-null CTNNA3 (CTNNA35A) in the heart resulted in LV dysfunction. Overexpression of phospho-mimetic CTNNA3 (CTNNA35D) was not evaluated in the study34. Based on the current work, we speculate that hyperphosphorylated CTNNA3 in DCM hearts is prone to ubiquitination and proteasomal degradation, leading to the loss of the pro-survival protein CTNNA3 in cardiomyocytes and the apoptotic loss of cardiomyocytes, which in turn manifests itself in the derangement of the cardiomyocyte intercalated disc and worsening of LV dysfunction as previously reported35. Further studies are needed to elucidate the impact of CTNNA3 phosphorylation on cardiomyocytes in vivo.
Our results support the idea that fortilin might function as a negative regulator of phosphorylation. Chattopadhyay et al. reported that fortilin interacts with PRX1, prevents it from being phosphorylated by Mst1, and protects it from proteasome-mediated degradation6. In addition, Pinkaew et al. showed that fortilin interacts with IRE1α and prevents it from being phosphorylated9. Herein, we demonstrated that fortilin interacts with CTNNA3 and prevents it from being phosphorylated (Fig. 3a, c). Further research is needed to identify additional proteins whose phosphorylation is inhibited by fortilin and to explore whether a unified mechanism underlies fortilin’s ability to prevent phosphorylation across a diverse group of proteins.
As we discussed previously9, one of the unique aspects of the anti-apoptotic activity of fortilin is that it exerts its anti-apoptotic activity by binding to various molecules that participate in cell survival and death and by enhancing the activity of anti-apoptotic molecules or mitigating the activity of pro-apoptotic molecules. As examples of the former function, fortilin (a) binds to and stabilizes MCL1, a BCL-2 family anti-apoptotic protein27, (b) binds to and sustains the activity of PRX1, an antioxidant, anti-apoptotic protein6, and (c) binds to and stabilizes CTNNA3, a structural protein that is also important for cell survival (Figs. 1–4). As examples of the latter function, fortilin (a) binds to and de-stabilizes transforming growth factor-β-stimulated clone-22, a pro-apoptotic molecule59, (b) binds to and sequesters Ca2+ to block Ca2+-dependent apoptosis60, (c) binds to and inhibits the tumor suppressor protein p5310, and (d) binds to and inhibits the activation of pro-apoptotic signaling of IRE1α9.
We still do not know the significance of the fortilin-CTNNA3 interaction in a whole organism. To date, a causal link has not been established between CTNNA3 deficiency and human diseases. Although van Hengel et al. identified CTNNA3 mutations in 2 patients among 76 arrhythmogenic right ventricular (RV) cardiomyopathy (ARVC) patients who were negative for other known ARVC genes—Plakophilin-2 (PKP2), Desmoplakin (DSP), Desmoglein-2 (DSG2), Desmocollin-2 (DSC2), and Junction Plakoglobin (JUP)55, constitutional knockout of the CTNNA3 gene in mice resulted in progressive dysfunction of the left ventricle (LV), not the RV35. Nevertheless, the cardiomyopathy phenotype seen in CTNNA3-null mice suggests that either disruption of the fortilin-CTNNA3 interaction or the simple lack of fortilin could result in less-than-appropriate levels of CTNNA3 in the heart, resulting in cardiomyopathy and heart failure. We previously reported that the cardiomyocyte-specific fortilin deficiency (fortilinKO-heart) leads to lethal heart failure in mice61. Although we did not determine the levels of CTNNA3 in the hearts of fortilinKO-heart mice in that study, it is possible that the reduction of CTNNA3 in cardiomyocytes in the absence of fortilin could have at least partially contributed to LV dysfunction of fortilinKO-heart mice.
Materials and methods
Reagents and materials
Chemicals
Cycloheximide (CHX, Catalog #: C7698), MG132 (Catalog #: M8699), and ML364 (Catalog #: SML1920) were purchased from Sigma-Aldrich (Burlington, MA, USA).
Recombinant proteins
We obtained recombinant human CTNNA1 (OriGene, Rockville, MD, USA; Catalog #: TP301766; cMYC-FLAG-tagged), CTNNA2 (OriGene, Rockville, MD, USA; Catalog #: TP308731; cMYC-FLAG-tagged), CTNNA3 (OriGene; Catalog #: TP313265; cMYC-FLAG-tagged), GFP-strep-tag (IBA LifeSciences, Gottingen, Germany; Catalog #: 2-1007-105), fortilin (OriGene, Catalog #: TP301664; cMYC-FLAG-tagged), His6-NQO2 (Abcam; Catalog #: ab93933), and p53 (OriGene; Catalog #: TP710022; cMYC-FLAG-tagged) from commercial sources. We generated recombinant human fortilin (rh-fortilin) as described previously16. Briefly, we transiently transfected 293T cells with the pESG-IBA5-human-fortilin mammalian expression vector (IBA LifeSciences), in which the human-fortilin cDNA was ligated in frame to the 3ʹ-terminal end of the Strep-tag II sequence using Lipofectamine™ 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA; Catalog #: L300015) and following the manufacturer’s instructions. After collecting the cells in Buffer W (IBA LifeSciences, Catalog #: 2-1003-100; containing 100 mM Tris HCl, pH8, 150 mM NaCl, 1 mM EDTA, and supplemented with protease inhibitor cocktail, 1 tablet/mL, ThermoFisher, Catalog #: 87786), we lysed the cells by repeated freeze-thaw and sonication, centrifuged the lysates, and passed the cleared cell lysates through a gravity flow Strep-Tactin® XT Superflow® high-capacity column (IBA LifeSciences, Catalog #: 2-4031-001). After washing the column five times with Buffer W, we eluted the Strep-tagged fortilin using Buffer BXT (Buffer W plus 50 mM biotin, Catalog #: 2-1042-025), concentrated the eluents using the ultra-centrifugal filter unit (Amicon™, Sigma, Catalog #: UFC501096), and dialyzed them in phosphate buffered saline (PBS) using a Slide-A-Lyzer™ MINI dialysis device (Thermo-Fisher Scientific, Waltham, MA, USA, Catalog #: 88401). We characterized the purified protein by western blot analysis using in-house rat anti-fortilin mAb (1:1000 dilution; BioXcell, Lebanon, NH, USA; Clone 8289).
Plasmid vectors
We obtained the following mammalian expression vectors from GeneCopoeia (Rockville, MD, USA):
-
(1)
pEZ-CTNNA3WT-FLAG: The pEZ-M39 containing human CTNNA3 cDNA construct (GenBank accession NM_001127384.2) fused at the 3ʹ-terminus to the FLAG-tag construct under the control of the cytomegalovirus promoter.
-
(2)
pEZ-CTNNA35A-FLAG: The pEZ-CTNNA3WT-FLAG vector in which the following five phosphosite amino acids are mutated so that they cannot be phosphorylated—S637A, S647A, T649A, S650A, and T653A—as described by Reitz et al.34.
-
(3)
pEZ-CTNNA35D-FLAG: The pEZ-CTNNA3WT-FLAG vector in which the following five phosphosite amino acids are mutated to mimic the constitutionally phosphorylated status (phosphomimetic)—S637D, S647D, T649D, S650D, and T653D—as described by Reitz et al.34.
-
(4)
A mammalian expression vector in which the His6-construct is fused to the 5ʹ-end of human ubiquitin (UBB) cDNA (pReceiver-His6-Ubiquitin3, Catalog #: EX-F0751-M01-50), where His6 represents the hexa-histidine tag.
Small interfering RNA (siRNA) for gene silencing assays
We obtained the predesigned Accell™ SMART siRNA pools against human fortilin (siRNAfortilin) and CTNNA3 (siRNACTNNA3) from Dharmacon Horizon Discovery (Lafayette, CO, USA). siRNAfortilin targeted the following four RNA sequences: GUGGCAAUUAUUUUGGAUC; GCAUGGUUGCUCUAUUGGA; UGACUGUGAUUUAUUUGGA; and CUUUAUUGGUGAAAACAUG. siRNACTNNA3 targeted the following four RNA sequences: CUCUCAUAAUCCAGGUUAC; CCAGGAUGCUGAUAAUUUA; CCAGGAAGCUACAGUUUUA; and GAAGCAACUUGGAAUUUAU. We used the non-targeting pool of siRNAs as control (siRNAcontrol). We then validated the ability of siRNAfortilin and siRNACTNNA3 to silence fortilin and CTNNA3, respectively, at the message and protein levels using real-time quantitative PCR (RT-qPCR) (Fig. 2c) and western blot analyses (Fig. 2d, e).
Cell culture and cell lines
HEK293T, U2OS, and THP1 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). HEK293T and U2OS cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (Corning, NY, USA; Catalog # 10-013-CV;) supplemented with 10% fetal bovine serum (Sigma, Catalog #: 21K475-A) and 1% antibiotic-antimycotic solution (GIBCO, Waltham, MA, USA; Catalog #: 15240062) at 37 °C in an atmosphere containing 5% CO2. THP1 cells were maintained in Roswell Park Memorial Institute 1640 medium (GIBCO, Catalog #: 61870127) supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic solution at 37 °C in an atmosphere containing 5% CO2. Cells are examined by Nikon ECLIPS Ts2 inverted microscope equipped with NIS-Elements image acquisition system (version 5.30.01, Nikon, Japan) and, when appropriate, photographed to document the overall cell morphology.
Fortilin-deficient THP1 cells and their control (THP1KO-fortilin and THP1WT-fortilin)
THP1 cells in which fortilin was knocked out (THP1KO-fortilin) by targeting exon 3 of the fortilin gene using the standard CRISPR-Cas9 system26,62 and the exon-targeting single guide RNA (TTTCGGTACCTTCGCCCTCG) were obtained from Synthego (Redwood City, CA, USA). The final clones were characterized using indel analysis, western blot analysis, and immunofluorescence staining. For the indel analysis, purified genomic DNA from THP1KO-fortilin clonal and homozygous KO cells were amplified with PCR using the following primers flanking the target cut site:
-
Forward PCR Primer: 5ʹ–TATACCCACTGCGAAAGAACCT–3ʹ
-
Reverse PCR Primer: 5ʹ–CGTGAACTATTGGCGTGGAAG–3ʹ
The gel-purified amplicons were subjected to Sanger sequencing using the forward PCR primer. The sequence around the target cut site was then evaluated using the ICE CRISPR Analysis Tool (Synthego), which showed the insertion of one nucleotide (+1) at the site. Mock-transfected pool THP1 cells were provided by Synthego and used as the control (THP1WT-fortilin).
Western blot analyses
SDS-PAGE and standard western blot analyses were performed as described previously9,16 with some modifications. Briefly, protein concentrations were measured using a Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA, USA; Catalog #: 5000006), and an appropriate amount of proteins was loaded into each lane of the 10% polyacrylamide gel for electrophoresis. When appropriate, 0.5% (v/v) 2,2,2-trichloroethanol (TCE) was added to a polyacrylamide gel before polymerization to quantify total proteins loaded on each lane of the gel. After standard SDS-PAGE, the TCE-containing gel was ultraviolet-irradiated on the Bio-Rad ChemiDoc MP Imaging System for 2 min. The image was electronically captured, and the cumulative band densities were calculated to assess loading conditions as described previously16. The proteins resolved on the gel were then blotted onto a nitrocellulose membrane (pore size = 0.45 µm; Bio-Rad; Catalog #: 1620115) using the Trans-Blot® Turbo™ semi-dry transfer device (Bio-Rad). The membrane was then blocked with 3% skim milk (Bio-Rad, Catalog #: 1704270) in tris-buffered saline with 0.1% Tween 20 overnight, incubated first with a primary antibody and then with an appropriate horseradish peroxidase (HRP)-labeled secondary antibody as described below. Developed images were electronically captured using the ChemiDoc MP Imaging System, and the signal intensities of protein bands were quantified using ImageJ software (National Institutes of Health (NIH), Bethesda, MD, USA).
To quantify fortilin expression (Fig. 2d), the signal intensity of the fortilin band was divided by that of the TCE bands in the same lane to derive a fortilin expression index, which was expressed as arbitrary units (A.U.). Similarly, a CTNNA3 expression index was calculated by dividing the signal intensity of the CTNNA3 band by that of the TCE bands in the same lane, with values in A.U. (Fig. 2f; Supplementary Fig. S4a, c). To quantify fortilin co-immunoprecipitated by CTNNA3 and its mutants (Fig. 3h), the signal intensity of the fortilin band was divided by that of the CTNNA3 band in the same lane, and the derived value was expressed as A.U. The following primary antibodies were used:
-
(1)
Mouse anti-fortilin monoclonal antibody (mAb) (1:2500 dilution; Abnova, Corporation, Taipei City, Taiwan; Catalog #: H00007178-M03);
-
(2)
Rabbit anti-CTNNA3 mAb (recombinant; 1:2500 dilution; Abcam, Waltham, MA, USA; Catalog #: ab184916);
-
(3)
Rabbit anti-CTNNA1 mAb (recombinant; 1:10,000 dilution; Abcam, Catalog #: ab51032);
-
(4)
Rabbit anti-CTNNA2 mAb (recombinant; 1:2000 dilution; Abcam, Catalog #: ab76015);
-
(5)
Rabbit anti-CTNNB polyclonal antibody (pAb) (1:2000 dilution; Abcam, Catalog #: ab16051);
-
(6)
Rabbit anti-fortilin mAb (recombinant; 1:3000 dilution; Abcam, Catalog #: ab133568);
-
(7)
Rabbit anti-FLAG pAb (1:1000 dilution; Sigma, Catalog #: F7425);
-
(8)
Rabbit anti-phospho (Ser/Thr) pAb (diluted to 2 µg/mL; Abcam, Catalog #: ab117253);
-
(9)
Rabbit anti-acetylated-lysine pAb (1:1000 dilution; Cell Signaling, Danvers, MA, USA; Catalog #: 9441S).
The following secondary antibodies were used:
-
(1)
IRDye 800CW goat anti-rabbit pAb (1:5000 dilution; LI-COR, Lincoln, NE, USA; Catalog #: D926-32211);
-
(2)
IRDye 680RD goat anti-mouse pAb (1:5000 dilution; LI-COR; Catalog #: 926-68070).
JESS™ protein analyses
To assess the expression levels of FLAG-tagged CTNNA3s and His6-tagged ubiquitin in a highly quantitative fashion (Fig. 3d, f), we turned to the JESS™ Simple Western System (ProteinSimple®, San Jose, CA, USA) as described previously29,30 and following the manufacturer’s instructions. We first generated total cell lysates (TCL) in Lysis Buffer (0.1% NP-40, 100 mM Tris-HCl pH 6.8, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786)) and the PhosSTOP phosphatase inhibitor (Roche, Basel, Switzerland; Catalog #: 0490684500) at the concentration of 1 µg/µL. We then mixed the Fluorescent 5x Master Mix (ProteinSimple®, Catalog #: PS-FL01-8) with the sample at a ratio of 1:4, resulting in the final sample concentration of 0.8 µg/µL. We boiled the mixture at 95 °C for 5 min, centrifuged it at 2500 rpm for 5 min, and loaded it into an appropriate well within the pre-filled plate for the 12–230 kDa capillary system (ProteinSimple®, Catalog #: SM-W004-1). We used the following primary antibodies:
-
(1)
Rabbit anti-FLAG pAb (1:50 dilution; Sigma, Catalog #: F7425);
-
(2)
Mouse anti-His6 mAb (1:50 dilution; Sigma, Catalog #: 70796-M).
We used the following secondary antibodies:
-
(1)
anti-rabbit secondary HRP mAb (ProteinSimple®, Catalog #: 042-206)
-
(2)
anti-mouse secondary HRP mAb (ProteinSimple®, Catalog #: 042-205).
We used the Luminol-Peroxide Mix (Luminol-S, ProteinSimple®, Catalog #: 043-311; Peroxide, ProteinSimple®, Catalog #: 043-379) to visualize the chemiluminescent signals from bound antibodies. We then quantified total proteins loaded in each capillary using the total protein detection module (ProteinSimple®, Catalog #: DM-TP01).
We reviewed the digitized chemiluminescence signals for both specific and total proteins and quantified them using Compass Simple Western™ software (version 6.3.0). Specifically, we calculated the protein expression index (in A.U.) by dividing the area under the curve of a protein of interest by the total proteins loaded in the same capillary (“in-capillary normalization”).
We also used the JESS™ western blot system56,57 to quantitatively assess the impact of fortilin silencing on CTNNA3 expression levels, as well as the effect of CTNNA3 silencing on fortilin expression levels (Fig. 4a). Briefly, we plated 1 ×104 293T cells in each well of 24-well plates and transfected them with siRNAcontrol; siRNACTNNA3, siRNACTNNA3, and siRNAfortilin, or siRNAfortilin. We subjected 4 μg of the lysates to JESS™, employing the same methods described above and the following antibodies:
Primary antibodies
-
a.
Rabbit anti-fortilin (1:50 dilution; Abcam; Catalog #: ab133568)
-
b.
Rabbit anti-CTNNA3 (1:200 dilution; Abcam, Catalog #: ab184916)
Secondary antibodies: anti-rabbit secondary HRP mAb (ProteinSimple®, Catalog #: 042-206)
Finally, we used the JESS™ Simple Western System to further validate the pattern of CTNNA3 degradation in the presence and absence of MG132, a proteasome inhibitor, (Supplementary Fig. S2a, c). We plated 2 ×104 293T cells in each well of 24-well plates, incubated them overnight, treated them with either siRNAcontrol or siRNAfortilin in Opti-Mem (GIBCO, Catalog #: 31985070), incubated them again for 48 h, washed them with PBS, and exchanged the culture media for new media containing 100 µg/mL CHX with or without MG132, and incubated them at 37 °C in an atmosphere containing 5% CO2. We harvested the cells at times 0, 0.5, 1, 2, 4, 8, 12, 24, and 36 h after the media exchange into RIPA buffer (Chem Cruz, Catalog #: SC-24948; Chem Cruz, Santa Cruz, CA, USA) and subjected the 4 µg of lysates to JESS™56,57. We detected CTNNA3 in each capillary by anti-CTNNA3 mAb (1:200 dilution; Abcam; # ab184916) and anti-rabbit secondary HRP mAb (ProteinSimple®, Catalog #: 042-206). We also visualized total proteins and used them as a loading control. We calculated CTNNA expression index as the area under curve of the CTNNA3 peak divided by that of total proteins and expressed it as arbitrary unit (A.U.), using the Compass software (v6.3.0., ProteinSimple®) as described above in detail.
In vivo co-immunoprecipitation (Co-IP)-western blot analyses
To evaluate whether fortilin specifically interacts with CTNNA1, CTNNA2, CTNNA3, and CTNNB in the cell, we lysed 293T cells in 500 µL of Lysis Buffer (PBS, 0.001% NP-40, and protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786)) on ice, followed by brief sonication (Bioruptor®, Diagenode, Denville, NJ, USA). We then centrifuged the lysates at 14,000 rpm at 4 °C for 10 min and generated the total cell lysates. We used 10% of the total cell lysates (50 µL) for input analyses. For forward immunoprecipitation (IP), we incubated 200 μL of lysate with a rabbit anti-fortilin mAb (Abcam, #ab133568) overnight at 4 °C with rotation. The following day, we added 50 μL of Dynabeads® M-280 sheep anti-rabbit IgG suspension (Invitrogen, Catalog #: 01266125) to the mixture, which had been blocked with 3% bovine serum albumin in Wash Buffer (PBS, 0.0001% NP-40, and the protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786)). We incubated the mixture for 1 h at 4 °C, washed it three times with the same Wash Buffer, eluted the proteins from the beads in 2× SDS gel Loading Buffer (50 mM Tris·HCl, pH 6.8, 1% 2-mercaptoethanol, 1% SDS, 25 mM EDTA, 0.01% bromphenol blue, and 10% glycerol), and subjected the eluents to SDS-PAGE and western blot analyses using (a) anti-fortilin mAb to verify successful fortilin IP and (b) anti-CTNNA1 mAb, anti-CTNNA2 mAb, anti-CTNNA3 mAb, or anti-CTNNB pAb to evaluate the presence of respective proteins co-immunoprecipitated by fortilin (Fig. 1a–d). For reverse IP, we followed the same procedure, except we incubated the lysate with a rabbit anti-CTNNA3 mAb (Abcam, Catalog #: ab184916) and analyzed the eluates by western blot analyses using (a) anti-CTNNA3 mAb (Abcam, Catalog #: ab184916) to verify successful IP of CTNNA3 and (b) anti-fortilin mAb (Abcam, Catalog #: ab133568) to evaluate the presence of proteins co-immunoprecipitated by CTNNA3 (Fig. 1f). For the co-immunoprecipitation (Co-IP) experiment using recombinant proteins (Fig. 1e), we added human recombinant fortilin-FLAG (OriGene; TP301664), CTNNA3-FLAG (OriGene; Catalog #: TP313265), p53-FLAG (OriGene; Catalog #: TP710022), and His6-NQO2 (Abcam; Catalog #: ab93933) lysis buffer (PBS, 0.001% NP-40, and protease inhibitor cocktail [Thermo-Fisher, Catalog #87786]) and incubated the mixture overnight at 4 °C with rotation in the presence of rabbit anti-fortilin mAb (Clone: EPR5540, Abcam, Catalog #: ab133568). The following day, we added 50 μL of Dynabeads® M-280 sheep anti-rabbit IgG (Invitrogen, Catalog #01266125) and incubated for 1 h at 4 °C. After three washes with the same buffer, we eluted the proteins from the beads using 2x SDS gel loading buffer and subjected the eluents to SDS-PAGE, followed by western blotting. We probed with the mixture of α-FLAG mAb (1:1,000 dilution; Sigma, Catalog #: F1804) and α-His6 mAb (1:1,000 dilution; Abcam, Catalog #: ab18184) to verify both (a) successful immunoprecipitation of fortilin-FLAG and (b) the presence of co-immunoprecipitated CTNNA3-FLAG, with p53-FLAG as a positive control10 and His6-NQO2 as a negative control6,9.
To evaluate the interaction of fortilin with wild-type CTNNA3 (CTNNA3WT), phospho-null CTNNA3 (CTNNA35A), and phospho-mimetic CTNNA3 (CTNNA35D), we first transiently transfected 293T cells with the pEZ-M39-CTNNA3WT-FLAG, pEZ-M39-CTNNA35A-FLAG, or pEZ-M39-CTNNA35D-FLAG mammalian expression plasmid using Lipofectamine 3000 (Invitrogen; Catalog #: 3000-075). We then lysed the cells in Lysis Buffer (PBS, 0.001% NP-40, protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786)) and PhosSTOP phosphatase inhibitor (Roche, Catalog #: 04906845001), incubated the TCL with mouse anti-FLAG M2 Magnetic Beads (Sigma-Aldrich, Catalog #: SIAL-M8823) for 3 h at 4 °C, washed the beads with Wash Buffer (PBS, 0.0001% NP-40, and the protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786) and the PhosSTOP phosphatase inhibitor), and eluted the bound proteins in 2x SDS gel Loading Buffer. Finally, we subjected the proteins to SDS-PAGE and western blot analyses.
Proximity ligation assay (PLA)
To verify the presence of the fortilin-CTNNA3 interaction and also to determine whether fortilin interacts with CTNNA1, CTNNA2, and CTNNB in vivo, we performed the PLA6,9 using a commercially available kit (Sigma, Duolink® series) according to the manufacturer’s instructions. Briefly, we seeded U2OS cells into 2-chamber slides (Thermo-Fisher; Catalog #: 154461), incubated them for 24 h, fixed them with 4% paraformaldehyde, permeabilized them with 0.1% Triton-X-100 in PBS, blocked them with Duolink Blocking solution at 37 °C for 1 h, and incubated them with (a) mouse anti-CTNNA3 mAb (1:200 dilution; OriGene; Catalog #: BM6021P), mouse anti-CTNNA1 mAb (1:200 dilution; Abcam, Catalog #: ab231306), mouse anti-CTNNA2 pAb (1:200 dilution; Abnova (Taipei, Taiwan), Catalog #: H00001496-B01P), or mouse anti-CTNNB mAb (1:200 dilution; Sigma; Catalog #: C7207) and (b) rabbit anti-fortilin mAb (1:200 dilution; Abcam; Catalog #: ab133568) overnight at 4 °C. The next morning, after washing with Wash Buffer (Sigma, Duolink®, Catalog #: DUO82049), we incubated the cells with the Duolink PLA probes, which were anti-mouse and anti-rabbit mAbs conjugated to oligonucleotides (Sigma, Duolink® series; PLA probe anti-mouse MINUS, Catalog #: DUO92004 and PLA probe anti-rabbit PLUS, Catalog #: DUO92002) for 1 h at 37 °C. We then added ligase and two connector oligonucleotides to the solution so that the latter would hybridize to the two PLA probes and join them into a closed circle if they were in close proximity (30 nm) to each other. Finally, we added Texas Red®–labeled oligonucleotides that would hybridize to the rolling circle amplification products to the solution (Sigma, Duolink® series; PLA Detection Kit Red, Catalog #: DUO92008) and applied a coverslip using mounting media containing DAPI (Sigma, Duolink® series; Catalog #: DUO82040). We visualized the signals using a Nikon A1R confocal microscope controlled by NIS-Elements AR image acquisition software (version 5.11.01, Nikon, Tokyo, Japan) and analyzed the images using ImageJ software (NIH).
Microscale Thermophoresis (MST) analysis
For Microscale Thermophoresis (MST), we used the NanoTemper Monolith NT.115 Pico, Microscale Thermophoresis (MST) system (NanoTemper Technologies GmbH, München, Germany), following the manufacturer’s instructions22,23. We first labeled rh-fortilin [3] with the Monolith™ red-maleimide 2nd generation protein labeling kit (NanoTemper Technologies, Catalog #: MO-L014) according to the manufacturer’s protocol. We mixed 5 nM red-maleimide-labeled rh-fortilin with varying concentrations of CTNNA3 (range: 30 pM to 250 nM ; OriGene, Catalog #: TP313265) in PBS, loaded the mixture into Monolith NT.115 series premium glass capillaries (Catalog # MO-K025), placed the capillaries in the MST system, and measured the pattern of thermophoresis of rh-fortilin at 25 °C using the medium MST power and 1% LED excitation power. We first determined the normalized fluorescence value (Fnorml) by dividing the post-thermal signal (Fhot) by the pre-thermal signal (Finitial), where Finitial represented the fluorescence measured before the temperature gradient was applied, and Fhot represented the fluorescence measured after heating induced thermophoresis. We then calculated the dissociation constant (Kd) using the NanoTemper analysis software after conducting three independent experiments. We repeated the same experiments for CTNNA1 (OriGene; Catalog #: TP301766) and CTNNA2 (OriGene; Catalog #: TP308731).
Bio-layer interferometry (BLI) analysis
We performed BLI as previously described6,9,16. Briefly, we first dipped a streptavidin-coated biosensor attached to the BLItz system (ForteBio, Menlo Park, CA, USA) into biotinylated human recombinant fortilin protein in PBS at a concentration of 25 µg/mL for 600 s to immobilize the protein on the sensor tip. We then applied various concentrations of recombinant human CTNNA3 (0, 250, 500, 1000, 2000 nM) to the sensor tip for 180 s to evaluate the association between the two molecules. We then exchanged the CTNNA3 solution for PBS for 300 s to evaluate their dissociation. We calculated the dissociation constant (Kd) using BLItz analysis software. The data depicted in Supplementary Fig. S1g represent the results of three independent binding experiments.
Real-time reverse transcription quantitative PCR (RT-qPCR)
We performed real-time RT-qPCR assays as previously described6,9,16. Briefly, we harvested 293T cells that had been treated by either siRNAcontrol or siRNAfortilin, isolated total RNA using the GeneJET RNA Purification kit (Thermo-Fisher; Catalog #: K0731), and subjected 100 ng of the isolated total RNA to real-time RT-qPCR using the QuantiNova® Probe RT-PCR kit (QIAGEN, Hilden, Germany, Catalog #: 208354) and the following primer and probe sets (Integrated DNA Technologies, Coralville, IA, USA):
-
(1)
Human fortilin —Forward: 5′-ATGACTCGCTCATTGGTGGAA-3′, Reverse: 5′-TGCTTTCGGTACCTTCGCCC-3′, Probe 5′-FAM-TGCCTCCGC/ZEN/TGAAGGCCC-IBFQ-3′
-
(2)
Human 18S rRNA—Forward: 5′- CTGAGAAACGGCTACCACATC-3′, Reverse: 5′-GCCTCGAAAGAGTCCTGTATTG-3′, Probe 5′-JOEN-AAATTACCC/ZEN/ACTCCCGAC-IBFQ-3′
where FAM = carboxyfluorescein, ZEN™ = an internal quencher to enhance the quenching activity of the Iowa Black FQ 3’ Quencher (IBFQ) (IDT), and JOEN = 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein. We used the 2–ΔΔCT method to calculate the expression levels (in A.U.) of the fortilin gene relative to the 18S rRNA levels in the sample.
CTNNA3 degradation assays
Assay of native CTNNA3 degradation
We plated 2 ×104 293T cells in each well of 24-well plates, incubated them overnight, treated them with either siRNAcontrol or siRNAfortilin in Opti-Mem (GIBCO, Catalog #: 31985070), incubated them again for 48 h, washed them with PBS, and exchanged the culture medium for new medium containing 100 µg/mL CHX with or without MG132. We incubated the cells at 37 °C in an atmosphere containing 5% CO2. We harvested cells at 0, 0.5, 1, 2, 4, 8, 12, 24, and 36 h after the medium exchange and placed them in RIPA Buffer (Chem Cruz, Santa Cruz, CA, USA; Catalog #: SC-24948). We then subjected 20 µg of lysates to standard quantitative western blot analysis using anti-CTNNA3 mAb (1:2500 dilution; Abcam, #ab184916). We calculated the CTNNA3 expression index (in A.U.) as described above. To further validate the pattern of CTNNA3 degradation, we also subjected 4 μg of the lysates to JESS™ as described previously29,30 and as detailed above.
Assay of the degradation of plasmid-derived, FLAG-tagged wild-type and mutant CTNNA3s
We plated 2 ×104 293T cells in each well of 24-well plates, incubated them overnight, and transiently transfected the cells using Lipofectamine 3000 (Invitrogen, Catalog #: 3000-075) with (a) pReceiver-His6-Ubiquitin3 and (b) pEZ-CTNNA3WT-FLAG, pEZ-CTNNA35A-FLAG, or pEZ-CTNNA35D-FLAG. We incubated the cells for 48 h, washed them with PBS, exchanged the culture medium for new medium containing 100 µg/mL CHX with or without MG132, and incubated them at 37 °C in an atmosphere containing 5% CO2. We harvested cells at 0, 4, 8, 12, and 24 h after the medium exchange and placed them in Lysis Buffer (0.1% NP-40, 100 mM Tris-HCl pH 6.8, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786)) and the PhosSTOP phosphatase inhibitor (Roche, Catalog #: 04906845001)). We then subjected 4 µg of lysates to JESS™ as previously described29,30. We detected plasmid-derived CTNNA3-FLAG in each capillary using anti-FLAG pAb (1:50 dilution; Sigma; Catalog # F7425). We also visualized total proteins and used them as a loading control. We used compass software (v6.3.0., ProteinSimple®) to calculate the CTNNA expression index as the area under the curve of the CTNNA3-FLAG peak divided by that of total proteins. Results were expressed in A.U.
Immunocytochemistry assays
To visualize the intracellular localization of CTNNA3 and fortilin, we performed immunocytochemical staining using THP1 human monocyte-macrophage cells as described previously6. In brief, we seeded THP1 cells on a 4-chamber slide (Thermo-Fisher, Catalog #: 154526), treated them with 100 ng/mL phorbol 12-myristate 13-acetate (Sigma, Catalog #: P8139) for 96 h, fixed them with 4% paraformaldehyde for 20 min, permeabilized them with 0.1% Triton-X-100 in PBS (PBST) for 15 min, and blocked them with 10% goat serum in PBST for 1 h. We then incubated the cells with both rabbit anti-fortilin mAb (1:500 dilution; Abcam; Catalog #: ab133568) and mouse anti-CTNNA3 mAb (1:500 dilution; OriGene; Catalog #: BM6021P) overnight at 4°C. The next day and after several washes, we incubated the cells with Alexa Fluor 488 goat anti-rabbit IgG (F + L) (1:2000 dilution; Invitrogen; Catalog #: 2179202) and Alexa Fluor 555 goat anti-mouse IgG (F + L) (1:2000 dilution; Invitrogen; Catalog #: A-21422), and then we counterstained the nucleus with DAPI. We visualized the fortilin and CTNNA3 signals using a Nikon A1R confocal microscope controlled by NIS-Elements AR image acquisition software (version 5.11.01, Nikon, Tokyo, Japan).
Ubiquitination assays
To evaluate the impact of CTNNA3 phosphorylation on its ubiquitination, we performed a standard ubiquitination assay as previously described63 with modifications. First, we transiently transfected 293 T cells using Lipofectamine 3000 (Invitrogen, Catalog #: 3000-075) with (a) pReceiver-His6-Ubiquitin and (b) pEZ-CTNNA3WT-FLAG, pEZ-CTNNA35A-FLAG, or pEZ-CTNNA35D-FLAG and incubated the cells for 48 h. We then treated the cells with 20 µM MG132, a proteasome inhibitor, and 10 µM ML364 (Sigma, Catalog #: SML1920), a deubiquitinase inhibitor, for 12 h. We lysed the cells in Denaturing Buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 10 mM imidazole at pH8.0), incubated the total cell lysates with Ni-NTA Superflow beads (QIAGEN, Catalog #: 30410) for 1 h, washed them four times with Wash Buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 50 mM imidazole), and eluted the bound proteins in Elution Buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 250 mM imidazole). We subjected the eluents to JESS™, using mouse anti-His6 mAb (1:50 dilution; Sigma, Catalog #: 70796-M) and rabbit anti-FLAG pAb (1:50 dilution; Sigma; Catalog #: F7425). Using Compass software (ProteinSimple®), we calculated the CTNNA3 ubiquitination index by dividing the total signal intensity of ubiquitinated CTNNA3 that was pulled down by Ni-NTA beads by the signal intensity of unmodified CTNNA3 present in the total cell lysates before pulldown. Results were expressed in A.U.
Phosphorylation assays
To evaluate the impact of fortilin on the phosphorylation of CTNNA3, we transiently transfected THP1WT-fortilin and THP1KO-fortilin cells with pEZ-M39-CTNNA3WT-FLAG using Lipofectamine 3000. We incubated the cells for 48 h and then lysed them in Lysis Buffer (PBS, 0.001% NP-40, protease inhibitor cocktail (Thermo-Fisher; Catalog #: 87786) and the PhosSTOP phosphatase inhibitor (Roche; Catalog #: 04906845001)). We incubated the total cell lysates with anti-FLAG M2 Magnetic Beads (Sigma, Catalog #: SIAL-M8823-1 ML) for 3 h at 4 °C, washed it with Wash Buffer (PBS, 0.0001% NP-40, and protease and phosphatase inhibitors), eluted the proteins in Elution Buffer (0.1 M Glycine-HCl, pH 2.5, protease inhibitor cocktail (Thermo-Fisher, Catalog #: 87786) and the PhosSTOP phosphatase inhibitor (Roche, Catalog #: 04906845001)), added the SDS Loading Buffer, and heated the eluates at 95 °C for 5 min. We performed SDS-PAGE on these samples along with PeppermintStick™ Phosphoprotein Molecular Weight Standards (Thermo-Fisher, Catalog #: P27167), fixed the gel in Fix Solution (50% methanol, 10% acetic acid), stained it with Pro-Q® Diamond (ThermoFisher, Catalog #: P33300) for 70 min to visualize phosphorylated CTNNA3, and captured the image of the stained gel using the ChemiDoc MP Imaging System (Illumination source: Epi-green, 520–545 nm excitation; Filter: 590/110 nm standard filter). We subsequently stained the gel overnight with SYPRO® Ruby (Thermo-Fisher, Catalog #: S12000) to visualize total CTNNA3, and we captured the image of the stained gel using the ChemiDoc MP Imaging System (Illumination source: Trans-UV, 302 nm excitation; Filter: 590/110 nm standard filter).
Trypan blue exclusion assay
We plated 1 ×104 293T cells in each well of 24-well plates; transfected them with siRNAcontrol; siRNACTNNA3, siRNACTNNA3 and siRNAfortilin, or siRNAfortilin. After 48 h of incubation, we assessed cell viability using the Trypan Blue exclusion method by mixing 5 μL of Trypan Blue dye (0.4%) with 5 μL of cell suspension and loading the mixture onto the Countess Cell Counting Chamber Slides (ThermoFisher, Catalog #:C10228). We then calculated the percentage of viable (unstained) and non-viable (stained) cells, using the Countess™ II automated cell counter (ThermoFisher).
Activated Caspase-3 assay
We quantified activated (cleaved) caspase-3 using the Human/Mouse Cleaved Caspase-3 (Asp175) DuoSet IC ELISA kit (R&D Systems; Minneapolis, MN, USA, Catalog #: DYC835-2), following the manufacturer’s instructions. Briefly, we plated 1 ×104 293 T cells in each well of 24-well plates and transfected them with siRNAcontrol, siRNACTNNA3, siRNACTNNA3 + siRNAfortilin, or siRNAfortilin. After 48 hours of incubation, we harvested and lysed the cells in a lysis buffer containing 0.5% Triton-X-100, followed by a 15-min incubation on ice. We then added 100 μL of the cell lysates or standards to wells coated with an anti-caspase-3 capturing antibody and incubated them for 2 h at room temperature. After washing, we added 100 μL of a biotinylated anti-cleaved caspase-3 detection antibody (150 ng/mL) to each well and incubated for another 2 h at room temperature. After additional washes, we added 100 μL of streptavidin conjugated to horseradish peroxidase (HRP) solution to each well and incubated for 20 min at room temperature, followed by the addition of 100 μL of 3,3’,5,5’-tetramethylbenzidine (TMB) substrate solution for a 20-min incubation at room temperature, protected from light. We stopped the reaction by adding 50 μL of 2 N H2SO4 stop solution and immediately measured the optical density (OD) using a microplate reader set to 450 nm, with wavelength correction at 570 nm. Activated caspase-3 activity was calculated as OD450 – OD570 and expressed in arbitrary units (A.U.).
DNA fragmentation apoptosis assay
We assessed the amount of DNA fragmentation, which is a marker of apoptosis, as described previously6,9 with modifications. We plated 1 ×104 293T cells in each well of 24-well plates and transfected them with siRNAcontrol; siRNACTNNA3, siRNACTNNA3, and siRNAfortilin, or siRNAfortilin. We incubated the mixtures for 48 h, harvested and lysed the cells in 200 µL Lysis Buffer provided in the Cell Death Detection ELISA PLUS kit (Roche; Catalog #: 11774425001), and added 20 μL each of cell lysates and 80 μL each of Assay Buffer containing anti-histone-biotin and anti-DNA-peroxidase to the streptavidin-coated wells of a 96-well plate in triplicate. After incubation and wash, we detected captured nucleosomes using 2,2’-azino-di (3-ethylbenzthiazoline-6-sulphonic acid), whose signal intensity was measured at 405 nm (A405). We calculated the DNA fragmentation index in A.U. using the following formula:
where A.U. A405study is the A405 of the study cell sample, A405medium is the A405 of the medium only sample, and A405positive control is the A405 of medium containing DNA-histone-complex provided in the kit.
Phylogram generation, homology and domain analyses, and sequence alignment
We used the following protein sequences from the database at the National Center for Biotechnology Information for the analyses:
-
(1)
CTNNA1, NP_001277238.1
-
(2)
CTNNA2, NP_001269526.1
-
(3)
CTNNA3, NP_001120856.1
-
(4)
CTNNB, NP_001895.1
We first determined percent (%) identify matrices using the Simple Phylogeny platform available on the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) website (URL: https://www.ebi.ac.uk/jdispatcher/phylogeny/simple_phylogeny) and by entering the protein sequences of these four catenins. We then visualized them by generating a phylogenetic tree using the same platform and calculated the evolutionary distances among these four catenins. Next, we evaluated the domain structures of the catenins using the InterProScan tool available on the EMBL-EBI web site (URL: https://www.ebi.ac.uk/interpro/). We found no vinculin head domain in CTNNB. Finally, we used SnapGene (version 7.2; Dotmatics, Boston, MA, USA) to align the protein sequences of CTNNA1, CTNNA2, and CTNNA3.
Identification of CTNNA3 interacting proteins using the BioPlex Interactome database
We used the BioPlex Interactome website (URL: https://bioplex.hms.harvard.edu/explorer/home)45 and entered CTNNA3 in the search field to identify the proteins that were identified as CTNNA3-interacting molecules by Huttlin et al.45.
Statistics and reproducibility
The degree of the spread of data was expressed by the standard deviation (±s.d.). Two-tailed unpaired t-tests were used to compare the means of two groups. To compare the means of three or more groups, we used one-way analysis of variance (ANOVA) with Fisher or Tukey–Kramer’s pairwise comparison. P < 0.05 was considered to be statistically significant. P < 0.10 was considered to show a trend toward statistical significance. Prism Software 10.1.2 (GraphPad Software, San Diego, CA, USA) and Minitab 21 (State College, PA, USA) were used for statistical analyses. The numbers of biological replicates used in each experiment and the statistical test used to analyze the particular experiment are provided in the manuscript.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. The numerical source data can be found in Supplementary Data 1. All relevant data are available from the authors upon request.
References
Li, F., Zhang, D. & Fujise, K. Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem. 276, 47542–47549 (2001).
Sinthujaroen, P. et al. Elevation of serum fortilin levels is specific for apoptosis and signifies cell death in vivo. BBA Clin. 2, 103–111 (2014).
Gross, B., Gaestel, M., Bohm, H. & Bielka, H. cDNA sequence coding for a translationally controlled human tumor protein. Nucleic Acids Res. 17, 8367 (1989).
Tuynder, M. et al. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc. Natl Acad. Sci. USA 99, 14976–14981 (2002).
Yang, Y. et al. An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 24, 4778–4788 (2005).
Chattopadhyay, A. et al. Fortilin potentiates the peroxidase activity of Peroxiredoxin-1 and protects against alcohol-induced liver damage in mice. Sci. Rep. 6, 18701 (2016).
Graidist, P., Phongdara, A. & Fujise, K. Antiapoptotic protein partners fortilin and MCL1 independently protect cells from 5-fluorouracil-induced cytotoxicity. J. Biol. Chem. 279, 40868–40875 (2004).
Zhang, D., Li, F., Weidner, D., Mnjoyan, Z. H. & Fujise, K. Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and Fortilin. The potential role of MCL1 as a fortilin chaperone. J. Biol. Chem. 277, 37430–37438 (2002).
Pinkaew, D. et al. Fortilin binds IRE1α and prevents ER stress from signaling apoptotic cell death. Nat. Commun. 8, 1–16 (2017).
Chen, Y. et al. Physical and functional antagonism between tumor suppressor protein p53 and fortilin, an anti-apoptotic protein. J. Biol. Chem. 286, 32575–32585 (2011).
Amson, R. et al. Reciprocal repression between P53 and TCTP. Nat. Med. 18, 91–99 (2011).
Yarm, F. R. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell Biol. 22, 6209–6221 (2002).
Kashiwakura, J. C. et al. Histamine-releasing factor has a proinflammatory role in mouse models of asthma and allergy. J. Clin. Invest. 122, 218–228 (2012).
MacDonald, S. M., Rafnar, T., Langdon, J. & Lichtenstein, L. M. Molecular identification of an IgE-dependent histamine-releasing factor. Science 269, 688–690 (1995).
Amzallag, N. et al. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J. Biol. Chem. 279, 46104–46112 (2004).
Pinkaew, D. et al. Fortilin interacts with TGF-beta1 and prevents TGF-beta receptor activation. Commun. Biol. 5, 157 (2022).
Gachet, Y. et al. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell Sci. 112, 1257–1271 (1999).
Tsarova, K., Yarmola, E. G. & Bubb, M. R. Identification of a cofilin-like actin-binding site on translationally controlled tumor protein (TCTP). FEBS Lett. 584, 4756–4760 (2010).
Kobielak, A. & Fuchs, E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat. Rev. Mol. Cell Biol. 5, 614–625 (2004).
Gielata, M., Karpinska, K., Pieczonka, T. & Kobielak, A. Emerging Roles of the alpha-Catenin Family Member alpha-Catulin in Development, Homeostasis and Cancer Progression. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms231911962 (2022).
Soderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Rainard, J. M., Pandarakalam, G. C. & McElroy, S. P. Using Microscale Thermophoresis to Characterize Hits from High-Throughput Screening: A European Lead Factory Perspective. SLAS Discov. 23, 225–241 (2018).
Romain, M., Thiroux, B., Tardy, M., Quesnel, B. & Thuru, X. Measurement of Protein-Protein Interactions through Microscale Thermophoresis (MST). Bio Protoc. 10, e3574 (2020).
Frenzel, D. & Willbold, D. Kinetic titration series with biolayer interferometry. PLoS One 9, e106882 (2014).
Shah, N. B. & Duncan, T. M. Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J. Vis Exper., e51383, https://doi.org/10.3791/51383 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Liu, H., Peng, H. W., Cheng, Y. S., Yuan, H. S. & Yang-Yen, H. F. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell Biol. 25, 3117–3126 (2005).
Aberle, H., Bauer, A., Stappert, J., Kispert, A. & Kemler, R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16, 3797–3804 (1997).
Campos, M. L. et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7, 12570 (2016).
DeRamus, M. L. et al. GARP2 accelerates retinal degeneration in rod cGMP-gated cation channel beta-subunit knockout mice. Sci. Rep. 7, 42545 (2017).
Orford, K., Crockett, C., Jensen, J. P., Weissman, A. M. & Byers, S. W. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J. Biol. Chem. 272, 24735–24738 (1997).
Schulenberg, B., Goodman, T. N., Aggeler, R., Capaldi, R. A. & Patton, W. F. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis 25, 2526–2532 (2004).
Berggren, K. et al. A luminescent ruthenium complex for ultrasensitive detection of proteins immobilized on membrane supports. Anal. Biochem. 276, 129–143 (1999).
Reitz, C. J. et al. Proteomics and phosphoproteomics of failing human left ventricle identifies dilated cardiomyopathy-associated phosphorylation of CTNNA3. Proc. Natl Acad. Sci. USA 120, e2212118120 (2023).
Li, J. et al. Loss of alphaT-catenin alters the hybrid adhering junctions in the heart and leads to dilated cardiomyopathy and ventricular arrhythmia following acute ischemia. J. Cell Sci. 125, 1058–1067 (2012).
Sheikh, F. et al. alpha-E-catenin inactivation disrupts the cardiomyocyte adherens junction, resulting in cardiomyopathy and susceptibility to wall rupture. Circulation 114, 1046–1055 (2006).
Roodveldt, C. et al. Preconditioning of microglia by alpha-synuclein strongly affects the response induced by toll-like receptor (TLR) stimulation. PLoS One 8, e79160 (2013).
Janssens, B. et al. alphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J. Cell Sci. 114, 3177–3188 (2001).
Goossens, S. et al. A unique and specific interaction between alphaT-catenin and plakophilin-2 in the area composita, the mixed-type junctional structure of cardiac intercalated discs. J. Cell Sci. 120, 2126–2136 (2007).
Gerull, B. et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet 36, 1162–1164 (2004).
Zhang, Y. SPATA33 affects the formation of cell adhesion complex by interacting with CTNNA3 in TM4 cells. Cell Tissue Res. 389, 145–157 (2022).
Miyata, H. et al. SPATA33 localizes calcineurin to the mitochondria and regulates sperm motility in mice. Proc. Natl Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2106673118 (2021).
Zhang, Y. et al. SPATA33 is an autophagy mediator for cargo selectivity in germline mitophagy. Cell Death Differ. 28, 1076–1090 (2021).
Chen, H., Yi, M., Sheng, Y., Cheng, H. & Zhou, R. A novel testis-enriched gene Spata33 is expressed during spermatogenesis. PLoS One 8, e67882 (2013).
Huttlin, E. L. et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184, 3022–3040 e3028 (2021).
Hatzfeld, M. & Nachtsheim, C. Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: evidence for a subfamily of closely related proteins. J. Cell Sci. 109, 2767–2778 (1996).
Sirotkin, H. et al. Identification of a new human catenin gene family member (ARVCF) from the region deleted in velo-cardio-facial syndrome. Genomics 41, 75–83 (1997).
Choi, S. H., Estaras, C., Moresco, J. J., Yates, J. R. 3rd & Jones, K. A. alpha-Catenin interacts with APC to regulate beta-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev. 27, 2473–2488 (2013).
Grohmann, A., Tanneberger, K., Alzner, A., Schneikert, J. & Behrens, J. AMER1 regulates the distribution of the tumor suppressor APC between microtubules and the plasma membrane. J. Cell Sci. 120, 3738–3747 (2007).
Modzelewski, A. J., Holmes, R. J., Hilz, S., Grimson, A. & Cohen, P. E. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev. Cell 23, 251–264 (2012).
Chen, J., Waltenspiel, B., Warren, W. D. & Wagner, E. J. Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex. J. Biol. Chem. 288, 4867–4877 (2013).
Kim, J. E. et al. Fyn is a redox sensor involved in solar ultraviolet light-induced signal transduction in skin carcinogenesis. Oncogene 35, 4091–4101 (2016).
Schmidt, A., Durgan, J., Magalhaes, A. & Hall, A. Rho GTPases regulate PRK2/PKN2 to control entry into mitosis and exit from cytokinesis. EMBO J. 26, 1624–1636 (2007).
Vanpoucke, G. et al. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 32, 4155–4165 (2004).
van Hengel, J. et al. Mutations in the area composita protein alphaT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 34, 201–210 (2013).
Tyberghein, K., Goossens, S., Haigh, J. J., van Roy, F. & van Hengel, J. Tissue-wide overexpression of alpha-T-catenin results in aberrant trophoblast invasion but does not cause embryonic mortality in mice. Placenta 33, 554–560 (2012).
Fanjul-Fernandez, M. et al. Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun. 4, 2531 (2013).
He, B. et al. CTNNA3 is a tumor suppressor in hepatocellular carcinomas and is inhibited by miR-425. Oncotarget 7, 8078–8089 (2016).
Lee, J. H., Rho, S. B., Park, S. Y. & Chun, T. Interaction between fortilin and transforming growth factor-beta stimulated clone-22 (TSC-22) prevents apoptosis via the destabilization of TSC-22. FEBS Lett. 582, 1210–1218 (2008).
Graidist, P. et al. Fortilin binds Ca2+ and blocks Ca2+-dependent apoptosis in vivo. Biochem. J. 408, 181–191 (2007).
Chunhacha, P., Pinkaew, D., Sinthujaroen, P., Bowles, D. E. & Fujise, K. Fortilin inhibits p53, halts cardiomyocyte apoptosis, and protects the heart against heart failure. Cell Death Discov. 7, 310 (2021).
Giuliano, C. J., Lin, A., Girish, V. & Sheltzer, J. M. Generating Single Cell-Derived Knockout Clones in Mammalian Cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol. 128, e100 (2019).
Li, M., Rong, Y., Chuang, Y. S., Peng, D. & Emr, S. D. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol. Cell 57, 467–478 (2015).
Acknowledgements
M.N. expresses heartfelt gratitude to Dr. Yasunori Miyamoto, Professor at the Organization for Human Life Innovation and Development, Institute for Human Life Sciences, Ochanomizu University, Tokyo, Japan, for his invaluable guidance and mentorship during her undergraduate and graduate studies, which served as the cornerstone of her research journey. The authors thank all members of the Fujise Laboratory for their collaborative efforts and Dr. Sandipan Mukherjee for critical review of and suggestion on the data. K.F. wishes to honor the memory of Dr. Hiroshi Fujise. The project was supported in part by grants from the National Heart, Blood, and Lung Institute within the National Institutes of Health (HL138992, HL117247, HL152723, and HL15283 to K.F.) and by the Locke Research Awards, the Dolsen Family Research Award, and the Harold T. Dodge-John L. Locke Chair in Cardiovascular Medicine at the Division of Cardiology, Department of Internal Medicine, University of Washington (to K.F.).
Author information
Authors and Affiliations
Contributions
K.F. conceived the general idea and framework of the project and oversaw the project to its completion. M.N. and K.F. designed the majority of the experiments. M.N. performed the majority of the experiments. D.P. performed biolayer interferometry (BLI) assays and one of the co-immunoprecipitation assays, using recombinant proteins. U.P. performed microscale thermophoresis (MST) to determine the dissociation constant of fortilin-CTNNA3 interaction. F.M. performed one of the co-immunoprecipitation assays, using recombinant proteins. L.T. performed a western blot analysis of THP1 cells lacking fortilin. H.H. performed a CTNNA3 acetylation assay. M.N. and K.F. analyzed data and created figures. M.N. and K.F. wrote the manuscript. M.N. and K.F. proofread the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Satarupa Bhaduri, Neha Sarodaya, and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nakashima, M., Pinkaew, D., Pal, U. et al. Fortilin binds CTNNA3 and protects it against phosphorylation, ubiquitination, and proteasomal degradation to guard cells against apoptosis. Commun Biol 8, 1 (2025). https://doi.org/10.1038/s42003-024-07399-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07399-5
This article is cited by
-
Parkinson’s disease-specific α-Synuclein variants potentially drive Lewy body formation by engaging in promiscuous and non-functional interactions
Communications Biology (2026)
-
Pluripotent stem cells-based neural organoids for modelling human brain development and diseases
Cell & Bioscience (2025)
-
GWM-HFN, a Gray-White Matter heterogeneous fusion network for functional connectomes
Communications Biology (2025)
-
Cognitive and neural mechanisms of mental imagery supporting creative cognition
Communications Biology (2025)
-
Fortilin deficiency induces anti-atherosclerotic phenotypes in macrophages and protects hypercholesterolemic mice against atherosclerosis
Communications Biology (2025)