Abstract
Lung squamous cell carcinoma (LUSC) represents a major subtype of lung cancer, and it demonstrates limited treatment options and worse survival. Identifications of a prognostic model and chemoresistance mechanism can be helpful for improving stratification and guiding therapy decisions. The integrative development of machine learning-based models reveals a random survival forest (RSF) prognostic model for LUSC. The 12-gene RSF model exhibits high prognostic power in more than 1,000 LUSC patients. High-risk LUSC patients are associated with worse survival and the activation of the epithelial-mesenchymal transition pathway. Additionally, high-risk LUSC patients are resistant to docetaxel or vinorelbine treatment. In vitro and in vivo drug sensitivity experiments indicates that high-risk HCC15/H226 tumour cells and cell line-derived xenograft models are more resistant to vinorelbine treatment. Furthermore, the combination of chemotherapy with transforming growth factor-β inhibition augments antitumour responses in LUSC tumours. Our study provides valuable insights into prognosis stratification and the development of therapeutic strategies for LUSC.

Similar content being viewed by others
Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide. Lung squamous cell carcinoma (LUSC) is the second most common lung cancer subtype and is responsible for 20–30% of lung carcinoma deaths1. However, unlike lung adenocarcinoma (LUAD), for which targeted therapies (including EGFR and ALK inhibitors) have demonstrated significant efficacy2, the lack of active genetic alterations and corresponding targeted therapies that are approved for the treatment of LUSC has hindered the development of precision medicine3.
Chemotherapy is the standard treatment for LUSC4. Platinum doublet chemotherapy (which involves paclitaxel, gemcitabine, docetaxel and vinorelbine combined with platinum) is the first-line treatment for patients with advanced LUSC who are in good health (PS 0–1), and single-drug chemotherapy (which involves gemcitabine, paclitaxel, docetaxel, and vinorelbine) is recommended for patients with advanced LUSC (at PS = 2)5,6,7. However, chemoresistance is a significant obstacle for treatment and may lead to poor survival8. Therefore, identifications of a prognostic model and chemoresistance mechanism are urgently needed for LUSC.
Individual risk prediction demonstrates important benefits for clinical decision-making, as it can identify high-risk individuals and provide guidance for appropriate individual interventions and treatments. In recent years, machine learning algorithms have also been employed to predict the individual risk of disease development9,10. A pioneering machine learning method is the random survival forest (RSF), which estimates a hazard function via the ensemble prediction of many decision trees. Previous studies have demonstrated the ability of RSF to effectively predict survival and identify molecular biomarkers in colorectal cancer11, hepatocellular carcinoma12, and oesophageal carcinoma13.
In this study, we combined an integrated analysis of LUSC multiomics data and experimental validation to propose a RSF prognostic model for predicting patient clinical outcomes; importantly, this combined design may also suggest future directions for treatment strategies in LUSC.
Results
Identification of 12 core fitness genes in LUSC
A complete flowchart of this study is shown in Fig. 1. To identify LUSC fitness genes, we performed an integrative analysis of 22 LUSC cell lines from the DepMap database, which stores genome-wide CRISPR information on various cancer cells. Genes with a CERES score of less than -1.0 in at least 80% of 22 LUSC cell lines were defined as fitness genes. In addition, we identified tumour-upregulated genes and prognosis-related genes in the TCGA-LUSC dataset. We intersected these three gene lists, and 31 essential fitness genes were identified (Fig. 2A and Supplementary Table 1). Via the training cohort from TCGA, we employed the “randomForestSRC” package to further select the final 12 core fitness genes (Fig. 2A and Supplementary Table 2), which were uniquely identified in LUSC (Supplementary Fig. 1A).
A Core fitness gene selection in LUSC. B The 5-year AUCs and C-indices for seven prognostic models in the overall TCGA-LUSC dataset (n = 493). C Kaplan-Meier estimates of overall survival according to the RSF risk score in the training cohort from TCGA (n = 345) (Log-rank test). D Kaplan-Meier estimates of overall survival according to the RSF risk score in the test cohort from TCGA (n = 148) (Log-rank test). E Kaplan-Meier estimates of overall survival according to the RSF risk score in the GSE157011 cohort (n = 484) (Log-rank test). F Multivariate Cox regression analysis of the RSF and seven prognostic models in the overall TCGA-LUSC dataset (n = 493). G Multivariate Cox regression analysis of the RSF and seven prognostic models in the GSE157011 cohort (n = 484).
Integrative construction of the prognostic model
Some prognostic models using fitness genes have been reported in acute myeloid leukaemia14, bladder cancer15, and renal cell carcinoma16 but not in LUSC. With the training cohort from TCGA, seven prognostic models, including RSF, GBM, Lasso, Suprepc, Ridge, ElasticNet, and CoxPH, were established based on the 12 core fitness genes. In the training cohort, the predictive performance of the RSF model (0.798) was better than that of the other models, with C-indices of 0.734 for GBM, 0.591 for Lasso, 0.585 for Suprepc, 0.589 for Ridge, 0.588 for ElasticNet, and 0.582 for CoxPH. We also performed comparative analysis among these models in the overall TCGA-LUSC dataset (Fig. 2B). The results also revealed that the RSF (0.806) had higher 5-year AUCs than did the other models. After comprehensive consideration of the C-indices and 5-year AUCs, the RSF model was demonstrated to have better predictive performance than the other models and was chosen for further explorations.
Evaluation and validation of the RSF model
We calculated the RSF risk score for each patient and divided them into high-risk and low-risk groups. The high-risk group was associated with a significantly worse OS (HR: 6.06, 95% CI: 4.05–9.06; univariate Cox regression p < 0.01) (Fig. 2C) and remained a significant independent prognostic factor in the training cohort from TCGA (Supplementary Table 3). High-risk scores were also associated with worse survival (HR: 1.69, 95% CI: 1.03–2.78, univariate Cox regression p = 0.039) (Fig. 2D) and were an independent prognostic factor in the test cohort from TCGA (Supplementary Table 4).
To ensure that the constructed RSF model had prognostic value in the different cohorts, another two independent validation cohorts were used. In GSE157011, the high-risk score group exhibited worse survival (HR: 1.29, 95% CI: 1.01–1.65, univariate Cox regression p = 0.039) (Fig. 2E) and was determined to be an independent prognostic factor (Supplementary Table 5). Furthermore, LUSC patients with high risk scores also demonstrated worse survival in the CPTAC-LUSC cohort (Supplementary Fig. 1B).
To validate the ability of the RSF model to predict survival outcomes, we derived an unbiased prognostic signature score in the TCGA training dataset and compared these two prognostic indices via multivariate Cox regression analysis. The results revealed that the RSF risk score was independent of the unbiased prognostic signature across the TCGA training, test, and GSE157011 datasets (all p < 0.05; Supplementary Tables 6–8). Furthermore, compared with seven reported prognostic models for LUSC (Model 117, Model 217, Model 318, Model 419, Model 520, Model 621, and Model 722) via multivariate Cox regression analysis, the RSF model was an independent prognostic factor in the TCGA training, TCGA overall, and GSE157011 datasets (all p < 0.05; Supplementary Fig. 1C and Fig. 2F, G).
Subgroup analysis for the prognostic value of the RSF model in LUSC
In the overall dataset from TCGA, we found that patients with higher RSF risk scores exhibited significantly worse OS, disease-free survival (DFS), and progression-free survival (PFS) (Supplementary Fig. 2A, C, E). To further investigate the impacts of clinical characteristics on the prognostic value of the RSF model, we conducted stratified analysis according to the baseline characteristics of LUSC patients (including age, sex, and disease stage) and found that the RSF model was able to discriminate LUSC patients into significantly different prognostic groups for OS, DFS, and PFS (Supplementary Fig. 2B, D, F).
Genomic analysis in high-risk and low-risk groups of LUSC
To obtain more comprehensive insights into the mutational landscape associated with the RSF model, we depicted a landscape of commonly mutated genes in NSCLC between the high-risk and low-risk groups (Supplementary Fig. 3A). Moreover, we revealed that the B2M mutation was associated with a higher risk score and poor survival (Supplementary Fig. 3B, C). However, the CSMD3 mutation was associated with a lower risk score and good survival (Supplementary Fig. 3D, E). The tumour mutational burden (TMB) is reported as being a predictive biomarker in solid tumours. We found that low-risk patients demonstrated higher TMB and better OS (Supplementary Fig. 3F, G), which was consistent with the results of Riviere’s study23. These results indicate that there is tumour genomic heterogeneity between the high-risk and low-risk groups and that precise treatment is needed.
To gain a more comprehensive understanding of the molecular differences that exist between the low- and high-risk groups, we also analysed the copy number variation (CNV) data in the TCGA-LUSC dataset. Using GISTIC2, we detected aberrant CNV regions in the low/high-risk groups and identified several common and specific copy number alterations between these two groups. Both groups exhibited amplification in the cytoband of 8p11.23 and deletion in the cytoband of 2q22.1. In addition, specific deletions were detected in the cytoband 15q21.1 region in the low-risk group and in the cytoband 7q36.3 region in the high-risk group (Supplementary Fig. 4A, B). Moreover, there were specific amplifications observed in cytobands of 22q11.22 in the low-risk group and unique amplifications observed in cytobands of 2q31.2 in the high-risk group (Supplementary Fig. 4A, B). Notably, NFE2L2 is part of a recurrent focal amplicon at the 2q31.2 region in LUSC. Increased NFE2L2 accumulation in cancer tissues is strongly correlated with poor clinical outcomes in various cancer types24,25,26. Tumours with 2q31.2 amplification exhibited significantly increased NFE2L2 expression in the TCGA-LUSC cohort (Supplementary Fig. 4C), which may partly explain the poor survival that is observed in patients with high-risk tumours. HIBCH is a central enzyme of valine catabolism27, and recent investigations have demonstrated HIBCH as being a key gene involved in the development of various tumours, such as clear cell renal cell carcinoma28, colorectal cancer29, and prostate cancer30. LUSC tumours with 2q31.2 amplification exhibited significantly increased HIBCH expression (Supplementary Fig. 4D), and more studies are needed to explore whether HIBCH is functionally involved in LUSC pathogenesis.
A multivariate Cox regression model that included survival-associated genetic abnormalities and the RSF risk score revealed that the B2M mutation, CSMD3 mutation, and 22q11.22 amplification lost their prognostic significance. However, the RSF risk score and TMB remained significant independent prognostic factors (Supplementary Table 9).
RSF risk score is associated with the tumour microenvironment in LUSC
Tumour microenvironment differences were subsequently evaluated. First, we focused on several immune checkpoints and found that PDCD1, CTLA4, HAVCR2, TIGIT, and BTLA were highly expressed in the high-risk group (Supplementary Fig. 5A). In addition, the high-risk group demonstrated significantly higher stromal scores and immune scores (Supplementary Fig. 5B, C). Afterwards, we detected increased signatures of T cells, myeloid dendritic cells, and fibroblasts in the high-risk group (Supplementary Fig. 5D). Moreover, the high-risk group exhibited a higher TIDE score31, thus suggesting that high-risk patients may be resistant to ICB therapy (Supplementary Fig. 5E). Although high-risk patients exhibit high T-cell infiltration, most T cells may maintain an exhausted status and lack the practical ability to control tumour progression, thereby prolonging the survival times of these patients (Supplementary Fig. 5F). Moreover, we observed a greater expression level of TGFB1 in the high-risk group (Supplementary Fig. 5G), which is consistent with the findings of a previous study indicating that the cytokine TGF-β (encoded by TGFB1) plays a suppressive role during antitumour immunotherapy32. Moreover, multiple genes involved in TGF-β signalling are regulated via methylation of the promoters of these genes, thereby potentially contributing to mechanisms of immune evasion33. For example, the TGF-β1 complex is activated by MMP2, and the transduction of TGF-β signalling requires MMP2 activity34,35. As shown in Supplementary Fig. 5H-I, we found that MMP2 exhibited a lower methylation level and a higher gene expression level in the high-risk group. Furthermore, SMAD7, which can be activated by TGF-β to restrain immune cell activation36, exhibited a lower methylation level and greater expression in the high-risk group (Supplementary Fig. 5J, K). Therefore, our investigation revealed that high-risk tumours exhibit a unique methylation pattern with a potential mechanism for regulating the immunosuppressive TME.
A high risk score is associated with the EMT signalling pathway
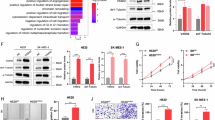
We further investigated the gene expression patterns related to the RSF score in the TCGA-LUSC cohort. We found that positively correlated genes were mainly enriched in the PI3K-AKT signalling, focal adhesion, and ECM interaction pathways (Fig. 3A). Genes involved in the EMT, PI3K-AKT signalling, focal adhesion, and TGF-β signalling pathways were extracted to generate a heatmap (Fig. 3B). The negatively correlated genes were mainly enriched in the ribosome, cell cycle, and DNA replication pathways (Fig. 3C). GSEA further revealed that the EMT, ECM, and TGF-β signalling pathways were significantly enriched in the high-risk group (Fig. 3D). Pathways enriched in the low-risk group were associated with E2F targets, DNA repair, and the cell cycle. Due to the fact that EMT has also been reported to be associated with poor survival37, we compared the prognostic value of the RSF risk score and EMT score in the TCGA-LUSC cohort. The results revealed that LUSC with higher EMT scores was associated with a worse prognosis (Supplementary Fig. 6). However, compared with the RSF risk score, the EMT score had an inferior ability to predict overall survival in the TCGA-LUSC cohort (5-year AUC: 0.56 vs. 0.81).
A GO enrichment analysis of genes that were positively correlated with the risk score. B Associations between gene expression in the EMT, PI3K-AKT signalling, focal adhesion, and TGF-β signalling pathways and the RSF risk score. C GO enrichment analysis of genes that were negatively correlated with the risk score. D Volcano plot showing normalized enrichment scores based on gene set enrichment analysis for significant pathways that were correlated (right) or anticorrelated (left) with the risk score.
A high risk score predicts resistance to chemotherapy in LUSC
In TCGA-LUSC chemotherapy-treated patients, the high-risk group also exhibited worse survival (Supplementary Fig. 7A), and patients who did not respond to chemotherapy had higher RSF risk scores (Supplementary Fig. 7B). Moreover, high-risk patients exhibited significantly worse survival after treatment with docetaxel, vinorelbine, navelbine, or cisplatin (Supplementary Fig. 7C–F).
We further applied the RSF model to 9 LUSC cell lines with proteomics data. We conducted correlation analysis between the risk score and protein expression and subsequently performed GSEA. The EMT, ECM and focal adhesion pathways were significantly associated with a high risk score (Fig. 4A), which was consistent with the results from the TCGA-LUSC cohort (Fig. 3D). Drug susceptibility analysis revealed a positive correlation between the risk score and the LN IC50 of vinorelbine from the GDSC database (Fig. 4B). In addition, the risk score was significantly associated with the IC50 and AUC values of docetaxel in both databases (Fig. 4C, D). These results suggest that the RSF risk score can predict the effectiveness of chemotherapy in LUSC and that high-risk LUSCs are more resistant to chemotherapy.
A GSEA revealed the upregulation of EMT signalling in the high-risk group. B The risk score was positively correlated with the LN IC50 of vinorelbine in the GDSC database (n = 7) (Pearson’s correlation test). C The risk score was positively correlated with the IC50 of docetaxel in the CTRP database (n = 8) (Pearson’s correlation test). D The risk score was positively correlated with the AUC of docetaxel in the GDSC database (n = 6) (Pearson’s correlation test).
LUSC tumour cells with high risk scores exhibit significant resistance to vinorelbine or docetaxel treatment
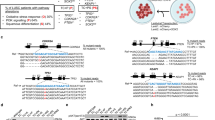
According to the abovementioned results, 9 LUSC cell lines were divided into high-risk (n = 4) and low-risk (n = 5) groups (Supplementary Table 10). HCC15 (high-risk), NCI-H226 (high-risk) and NCI-H520 (low-risk) cells were used for further experimental validation. We examined the IC50 values of vinorelbine and docetaxel in these three cell lines. Vinorelbine more efficiently inhibited the growth of NCI-H520 cells, with an IC50 value of 4.727 μM (95% CI: 4.092–5.467 μM), which was lower than the IC50 values for HCC15 (9.352 μM, 95% CI: 8.057–10.89 μM) and NCI-H226 (9.238 μM, 95% CI: 8.222–10.41 μM). Furthermore, the IC50 value of docetaxel was 12.47 μM (95% CI: 11.51–13.52 μM) in NCI-H520 cells, which was also lower than the IC50 values for HCC15 (19.57 μM, 95% CI: 17.84–21.58 μM) and NCI-H226 (25.34 μM, 95% CI: 23.44–27.48 μM) cells (Fig. 5A, B). Compared to NCI-H520 cells, the high-risk HCC15 and NCI-H226 cell lines were more resistant to vinorelbine and docetaxel treatment. Furthermore, the results of the cell colony formation assay suggested that high-risk cells were more resistant to vinorelbine or docetaxel (Fig. 5C, D). The difference in drug resistance between the high- and low-risk groups indicates that the RSF risk score can predict the efficacy of chemotherapy in LUSC patients.
A, B NCI-H520, HCC15, and NCI-H226 cells were treated with vinorelbine or docetaxel in a dose-dependent manner. Cell viability was assessed to construct curve plots, after which the IC50 values were calculated (n = 3 per group; ∗∗, p < 0.01; ∗∗∗, p < 0.001; t test). C NCI-H520, HCC15, and NCI-H226 cells were plated into 6-well plates and then treated with DMSO, vinorelbine (3 μM), or docetaxel (10 μM). Fourteen days after implantation, the cells were stained with crystal violet and photographed with a camera (n = 5 per group). D NCI-H520, HCC15, and NCI-H226 cells were treated with DMSO, vinorelbine (3 μM), or docetaxel (10 μM). Following the colony formation assay, the number of colonies in each well was calculated by ImageJ (n = 5 per group). E NCI-H520, HCC15, and NCI-H226 cells were treated with DMSO or the TGF-β receptor I inhibitor SB525334 (5 μM) for 2 days. The cell lysates were immunoblotted with the indicated antibodies. F–G NCI-H520, HCC15, and NCI-H226 cells were treated with vinorelbine (3 μM) or docetaxel (10 μM) with either DMSO or SB525334 (5 μM) pretreatment. Cell viability was tested every day after cell implantation (n = 5 per group; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001, ANOVA). H NCI-H520, HCC15, and NCI-H226 cells were treated with multiple doses of the TGF-β receptor I inhibitor SB525334 combined with vinorelbine or docetaxel for 2 days. Synergy plots depicting Gaddum’s noninteraction model (HSA) for NCI-H520, HCC15, and NCI-H226 cells were separately drawn. An HSA value that exceeds 10 indicates a synergistic interaction.
High-risk LUSC patients may benefit from combined treatment with chemotherapy and TGF-β pathway inhibition
The EMT pathway was significantly enriched in the high-risk group in both the TCGA-LUSC dataset and the LUSC cell lines (Figs. 3D, 4A). TGF-β is a well-established and potent inducer of EMT38. The TGF-β receptor I inhibitor SB525334 has been reported to effectively inhibit the TGF-β pathway. We further investigated whether inhibition of the TGF-β pathway reverses chemotherapeutic resistance in high-risk HCC15 and NCI-H226 cells. To assess the efficacy of this inhibition, we examined the protein levels of downstream markers in the TGF-β pathway. After the inhibition of TGF-β receptor I, no significant changes in the expression levels of TGF-β receptor I itself were observed. Conversely, the protein levels of N-cadherin and vimentin were decreased, and these decreases were accompanied by an increase in E-cadherin expression (Fig. 5E). When combined with TGF-β pathway inhibition, both vinorelbine and docetaxel significantly suppressed the proliferation of high-risk HCC15 and NCI-H226 cell lines. However, for the low-risk NCI-H520 cell line, combined drug inhibition had a limited effect (Fig. 5F, G). To determine whether the inhibition of the TGF-β pathway enhances the effects of vinorelbine and docetaxel at multiple doses, we evaluated the synergistic effects by the highest single agent (HSA) synergy model. In high-risk HCC15 and NCI-H226 cells (but not in low-risk H520 cells), the HSA synergy scores far exceeded 10, thus indicating that SB525334 synergised with vinorelbine and docetaxel (Fig. 5H). These results suggest that vinorelbine or docetaxel combined with TGF-β inhibition (TGF-βi) may aid in the treatment of LUSC patients with high RSF risk scores.
Vinorelbine resistance is reversed via combined inhibition of the TGF-β pathway in mice
To further verify the efficacy of vinorelbine combined with TGF-β pathway inhibition in vivo, we conducted tumour implantation procedures in NOD-SCID mice. As shown in Fig. 6A, the mice were injected with NCI-H520, HCC15 or NCI-H226 cells and treated with vinorelbine, SB525334 or a combination of both drugs, respectively. Compared with the vehicle group, the growth of the NCI-H520-derived tumours was greater than that of the HCC15-derived and NCI-H226-derived tumours following vinorelbine treatment (Fig. 6B, C, D). Additionally, vinorelbine exerted minimal effects on the weights of the HCC15-derived and NCI-H226-derived tumours (Fig. 6E). Furthermore, HCC15- and NCI-H226-derived tumours exhibited increased sensitivity to vinorelbine after combination with SB525334, as did NCI-H520-derived tumours. Tumours derived from these LUSC cell lines were significantly inhibited after the combination treatment (Fig. 6D, E). These in vivo results provide valuable evidence for the development of therapeutic strategies for LUSC.
A NCI-H520, HCC15, and NCI-H226 cells were subcutaneously injected into NOD-SCID mice. Once the average tumour volume reached 100 mm³, the mice were randomized to receive different treatments. At 21 days postimplantation, the tumours were collected and weighed (n = 5 mice per group). B Mice were euthanized at 21 days after implantation, and all of the collected tumours are displayed in the figure (n = 5 mice per group). C Tumour volumes of NCI-H520-, HCC15-, and NCI-H226-derived tumours were measured every three days following implantation. Curve plots were generated to illustrate tumour growth associated with different cell line-derived tumours (n = 5 mice per group; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001, ANOVA). D Tumour volumes of NCI-H520, HCC15, and NCI-H226-derived tumours on day 21 postimplantation were measured before the mice were euthanized (n = 5 mice per group; ns, no significance; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001, t test). E Tumour weights of NCI-H520, HCC15, and NCI-H226-derived tumours were measured after they were harvested from the mice (n = 5 mice per group; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001, t test).
Discussion
Treatment options and efficacy in LUSC remain limited. The lack of targetable mutations in LUSC has hindered progress in the development of more effective, personalized treatment approaches3. Since the implementation of the CRISPR-Cas9 system in cancer research, numerous candidate LUSC targets have been revealed via in vitro or in vivo CRISPR screens39,40. Given the critical importance of fitness genes for the functional phenotypes of cells, we speculated that these LUSC-specific fitness genes may also provide critical prognostic information. Thus, we established a powerful prognostic RSF model composed of 12 core fitness genes, which were validated in an internal cohort and two independent cohorts. Therefore, our study, which included more than 1,000 LUSC patients (representing the largest cohort that has ever been examined), proposed a potential prognostic indicator for LUSC.
To explore whether essential genes selected for the RSF were lineage-specific, we further derived essential genes in LUAD and SCLC and found that essential genes were predominantly specific for lung cancer histologic subtypes. GSEA revealed that the RSF risk score was associated with the EMT pathway; however, the RSF risk score performed much better than the EMT signature in survival prediction, thus highlighting the potential role of machine learning in enhancing the predictive power of prognosis41. Furthermore, when genomic prognostic alterations and RSF scores were adjusted via multivariate Cox regression analysis, both the RSF score and the TMB emerged as independent prognostic factors. These findings suggest that the combination of RSF and TMB may be recommended for further application.
Due to the lack of specific targeted drugs, chemotherapy remains the standard treatment for LUSC4. Our study serves as a proof-of-concept for the utility of the RSF model to identify potential therapeutic candidates for specific chemotherapy agents for patients with LUSC. In the drug sensitivity analysis of the TCGA-LUSC cohort, LUSC patients who received vinorelbine or docetaxel treatment were divided into significantly different prognosis groups according to the RSF risk score. The results of the LUSC cell line drug sensitivity analysis also suggest that a high RSF risk score can predict chemoresistance to vinorelbine and docetaxel.
Therefore, we performed in vitro and in vivo drug sensitivity validation to confirm the predictive value of the RSF model by using the LUSC H520 and HCC15/H226 cell lines and CDX models. We found that the H520 cell line or the CDX model with a low RSF risk score exhibited increased sensitivity to vinorelbine. Our pathway enrichment analysis revealed significant EMT activation in the high-risk group. TGF-β is a well-established potential inducer of EMT38. Although high-risk tumours are resistant to vinorelbine, combined treatment with a TGF-βi reversed resistance in our in vitro and in vivo models. These findings suggest that the RSF risk score may guide the rational use of vinorelbine in patients who are most likely to benefit, such as patients with low RSF risk scores. Furthermore, the combination of vinorelbine with a TGF-βi may provide potential benefits for LUSC patients.
A potential limitation of the current study is that we used only published datasets to validate the performance of our prognostic model. In addition, the differences in clinicopathological characteristics between the training and validation cohorts may influence the predictive power of the RSF model. Furthermore, a focus on LUSC fitness genes may overlook some superior prognostic genes that are not associated with cellular fitness. Moreover, the clinical utility of the RSF model needs to be further validated in further large-scale prospective studies in LUSC.
In summary, we established a robust 12-gene RSF prognostic model for the rapid risk assessment of newly diagnosed LUSC patients. The predictive power of the RSF model risk score for chemotherapy response indicates its considerable potential for clinical applications. Our study provides a roadmap for patient stratification and may help in developing strategies for personalized follow-up and individualized decision-making for patients with LUSC.
Methods
Data source and processing
RNA-seq expression data for TCGA-LUSC was downloaded from UCSC Xena (https://xenabrowser.net/). Mutation, copy number variation, DNA methylation and clinical data were downloaded from the Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/). External validation cohorts include GSE15701142 and CPTAC-LUSC43.
Dependence scores of LUSC cell lines were downloaded from DepMap (https://depmap.org/portal/download/); scores were obtained following a series of loss-of-function genomic screenings in different cell lines. The CERES algorithm44 was used to determine dependency scores: a negative score of a gene indicates that knocking out the gene inhibits the survival of a cell line, whereas a positive score indicates that knocking out the gene promotes survival and proliferation.
To evaluate the specificity of our screening flow, we applied the same criteria used in LUSC to identify essential genes in LUAD and small cell lung cancer (SCLC), the other two predominant subtypes of lung cancer. Differentially expressed genes and prognosis analysis were performed using the data from studies by Gillette et al. 45 (LUAD) and Liu et al. 46 (SCLC).
Construction and verification of a random survival forest model
For the LUSC dataset from TCGA, we performed random sampling at a ratio of 7:3 to split TCGA-LUSC patients into TCGA training cohort (n = 345) and TCGA test cohort (n = 148). In the training cohort, we further selected 12 genes using the ‘var.select’ function in the “randomForestSRC” package; next, we build prognostic models via seven different models, including RSF, GBM, Lasso, Suprepc, Ridge, Elasticnet, and CoxPH. The performance of these models was measured and compared with 5-year AUCs and C-indexes. The 5-year AUCs were calculated via “timeROC” R package. Comparison of time-dependent ROC curves was conducted via “compare” function in the “timeROC” R package.
RSF model presented with the best performance and was screened out. Based on the median RSF risk score, patients in the training, test, and GSE157011 cohorts were divided into low- or high-risk groups. For CPTAC-LUSC, the cutoff was generated based on the association between risk score and survival data using the “survminer” package because of the short follow-up time. We evaluated and compared the survival differences between the high- and low-risk groups using Kaplan-Meier plots.
Unbiased prognostic signature in LUSC
We performed univariate cox analysis in the TCGA-training cohort and identified 132 prognostic genes (Univariate cox regression P < 0.01) which was used to construct an unbiased prognostic signature. The unbiased prognostic signature score was evaluated via single-sample gene set enrichment analysis.
Somatic mutation, copy number variation, DNA methylation, immune microenvironment, and functional enrichment analyses
Mutation data were processed using the “maftools” R package47 and we calculated the TMB for the TCGA-LUSC dataset. GISTIC248 was applied to detect aberrant CNV regions in the low-risk and high-risk groups.
MCPcounter49 evaluated the infiltration of different cell types. TIDE algorithm31 was used to estimate the likelihood of an immunotherapy response. The immune score of each sample was calculated via the “ESTIMATE” R package50.
Correlation analysis between the RSF risk score and gene expression was conducted via the Pearson correlation method. Gene set enrichment analysis (GSEA), biological functions and pathways were conducted using “clusterProfifiler4.0” package in R51. “Hallmark” and “KEGG” gene sets were acquired from MSigDb.
EMT score
We calculated the EMT score using the EMT signatures identified in the study by Mak et.al. 52 via single-sample gene set enrichment analysis.
Drug sensitivity analysis
Proteome data for 9 LUSC cell lines was acquired from Nusinow’s study53. Drug response data was acquired from Genomics of Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/) and Cancer Therapeutics Response Portal (CTRP) (https://portals.broadinstitute.org/ctrp). These two databases provide the IC50 (half the maximal inhibitory concentration) and/or the area under the dose-response curve (AUC) measures of drug sensitivity. A lower IC50 or AUC value indicates a higher sensitivity to an agent.
Cell Culture
NCI-H520 cell line (human lung squamous cell carcinoma, male), HCC15 cell line (human lung squamous cell carcinoma, male) and NCI-H226 cell line (human lung squamous cell carcinoma, male) were obtained from American Type Culture Collection (ATCC). All human lung squamous cells were maintained at 37 °C with 5% CO2 in RPMI-1640 (Gibco) medium supplemented with 10% foetal bovine serum (FBS, Sigma) and penicillin (100 units/ml)/streptomycin (0.1 mg/ml) (Gibco). All cell lines used were routinely confirmed as pathogen-free by PCR.
Cell viability and proliferation
Cell viability and proliferation were measured by using cell counting kit-8 (CCK-8) (Biomake, B34304). For IC50 determination, NCI-H520, HCC15 and NCI-H226 cells were plated in 96-well plates at 1 × 104 per well and treated with vinorelbine (Selleck, S4269) or docetaxel (Selleck, S1148) next day at different concentrations for 24 h. For cell proliferation measurement, cells were seeded into 96-well plates at 2 × 103 per well and treated with 5 μM transforming growth factor-β receptor I inhibitor SB525334 (Selleck, S1476) next day. After 2 days of medication, 3 μM vinorelbine or 10 μM docetaxel was added into plates. For HSA synergy score estimation, cells were seeded into 96-well plates at 1 × 104 per well and treated with vinorelbine or docetaxel at multiple doses for 24 h. Then, 10 μl CCK-8 reagent was added to each well at the time of harvest. The cells were incubated at 37 °C for 1.5 hours and then measured at 450 nm absorbance to determine the cell viability or proliferation using the microplate reader (AMR-100, ALLSHENG).
Clone formation
NCI-H520, HCC15 and NCI-H226 cells were plated in 6-well plates at 800 per well. The medium was changed every 3 days. After 12 days of culture, 3 μM vinorelbine or 10 μM docetaxel were added and the cells were continuously maintained for 2 days. Afterward, cells in plates were fixed with methanol and stained with crystal violet. Pictures of stained cells were taken by camera. Counts of colonies in each well were calculated by ImageJ software.
Immunoblotting assay
All the cell samples were lysed with RIPA buffer (Beyotime, P0013B) and then the supernatant was boiled in 5× loading buffer (Beyotime, P0013L) for 10 minutes. Each sample was electrophoresed on 10% PAGE gels, then transferred to PVDF membranes and immunoblotted with the antibodies targeting N-cadherin (Abclonal, A19083), E-cadherin (CST, 3195S), VIM (CST, 5741S), TGFβR1 (Abclonal, A0708) and β-Actin (Abclonal, AC004). Immunoblots were visualized by a chemiluminescence imaging system (Tanon).
Synergy analysis
CCK-8 assays were conducted to evaluate the synergistic interactions between TGF-β receptor I inhibitor (0, 0.5, 1, 2, 4, 8 μM) and vinorelbine (0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16 μM)/docetaxel (0, 0.25, 0.5, 1, 2, 4, 8, 16, 32 μM). The suppression rates of cell proliferation were calculated after normalized to day 0 measurement. Synergy scores were calculated using Gaddum’s non-interaction model - Highest Single Agent (HSA) with SynergyFinder (https://synergyfinder.fimm.fi/synergy)54. An HSA value that exceeds 10 indicates a synergistic interaction.
Mouse model
For animal studies, 5-week-old female NOD-SCID mice were chosen to perform tumour implantation. 1×106 NCI-H520, HCC15 or NCI-H226 LUSC cells were injected into the right flank of each mouse. Tumours were measured at serial time points after implantation by digital calipers and tumour volume was calculated as the 0.5×length×width×width. When average tumour volume reached 100 mm3, mice were randomized to treatment groups. For vinorelbine treatment, mice were injected intraperitoneally at 0, 2 and 4 days after grouping at a concentration of 5 mg/kg. SB525334 was used for TGF-β pathway inhibition through oral gavage at 10 mg/kg every other day. Both drugs were dissolved in DMSO, and then we prepareda solution in 10% DMSO suspension, 40% PEG300 (S6704, Selleck), 5% Tween-80 (A600562, Sangon Biotech), and 45% saline. Twenty-one days after tumour implantation, all mice were executed and dissected to collect tumour tissues and weigh tumours by digital electronic scales.
Animal care, housing, and procedures were performed following the Guide for the Care and Use of Laboratory Animals of the Shanghai Pulmonary Hospital Ethics Committee. Animals were randomized for this study and kept under specific pathogen-free (SPF) conditions at the Laboratory Animal Centre of Shanghai Pulmonary Hospital (SYXK (Shanghai) 2022-0013).
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Shanghai Pulmonary Hospital (K24-311).
Statistics and Reproducibility
Statistical analysis was performed using GraphPad Prism (Version 9) and R Studio software (version 4.3.1). Student’s t-test or Wilcoxon-rank test was used to compare the statistical differences between sample groups. One-way anova test was used to compare the effects of drugs in mouse model. All p-values were two-tailed with a significance level of 0.05.
Data availability
TCGA LUSC FPKM data were downloaded from the TCGA database via the UCSC Xena (https://xena.ucsc.edu/). GSE157011 was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds/). The expression data and clinic information for CPTAC-LUSC were acquired from Satpathy’s study43. Dependence scores of LUSC cell lines were downloaded from DepMap (https://depmap.org/portal/download/). Drug response data for human cancer cell lines (CCLs) from the GDSC website (https://www.cancerrxgene.org/, access date: September 11, 2022) and CTRP; https://portals.broadinstitute.org/ctrp). The source data for graphs can be found in the Supplementary Data 1-5. Uncropped blot for Fig. 5E can be reviewed in the Supplementary Fig. 8. All other data are available from the corresponding author on reasonable request.
References
Gandara, D. R. et al. Squamous cell lung cancer: From tumor genomics to cancer therapeutics. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 21, 2236–2243 (2015).
Yuan, M. et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 4, 61 (2019).
Thomas, A. et al. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 12, 511–526 (2015).
Chaft, J. E. et al. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat. Rev. Clin. Oncol. 18, 547–557 (2021).
Liao, B. C. et al. Comparative effectiveness of first-line platinum-based chemotherapy regimens for advanced lung squamous cell carcinoma. Clin. Lung cancer 16, 137–143 (2015).
Lu, S. et al. Nedaplatin plus docetaxel versus cisplatin plus docetaxel as first-line chemotherapy for advanced squamous cell carcinoma of the lung - a multicenter, open-label, randomized, phase III Trial. J. Thorac. Oncol. Publ. Int. Assoc. Study Lung Cancer 13, 1743–1749 (2018).
Shukuya, T. et al. Nedaplatin plus docetaxel versus cisplatin plus docetaxel for advanced or relapsed squamous cell carcinoma of the lung (WJOG5208L): a randomised, open-label, phase 3 trial. Lancet Oncol. 16, 1630–1638 (2015).
Min, H. Y. & Lee, H. Y. Mechanisms of resistance to chemotherapy in non-small cell lung cancer. Arch. Pharmacal Res. 44, 146–164 (2021).
Placido, D. et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 29, 1113–1122 (2023).
Shaikh, F. J. & Rao, D. S. Prediction of cancer disease using machine learning approach. Mater. Today.: Proc. 50, 40–47 (2022).
Nakatsu, G. et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–541.e525 (2018).
Villanueva, A. et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatol. (Baltim., Md.) 61, 1945–1956 (2015).
Mao, Y. et al. A seven-lncRNA signature predicts overall survival in esophageal squamous cell carcinoma. Sci. Rep. 8, 8823 (2018).
Jin, P. et al. Large-Scale In Vitro and In Vivo CRISPR-Cas9 Knockout Screens Identify a 16-Gene Fitness Score for Improved Risk Assessment in Acute Myeloid Leukemia. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 28, 4033–4044 (2022).
Sun, J. X. et al. A four-cell-senescence-regulator-gene prognostic index verified by genome-wide CRISPR can depict the tumor microenvironment and guide clinical treatment of bladder cancer. Front. Immunol. 13, 908068 (2022).
Liu, C. et al. Novel molecular classification and prognosis of papillary renal cell carcinoma based on a large-scale CRISPR-Cas9 screening and machine learning. Heliyon 10, e23184 (2024).
Ma, Z. et al. Construction of the metabolism-related models for predicting prognosis and infiltrating immune phenotype in lung squamous cell carcinoma. J. Cancer 14, 3539–3549 (2023).
Zhai, W. Y. et al. An aging-related gene signature-based model for risk stratification and prognosis prediction in lung squamous carcinoma. Front. Cell Dev. Biol. 10, 770550 (2022).
Kadasah, S. F. Prognostic significance of glycolysis-related genes in lung squamous cell carcinoma. Int. J. Mole. Sci. 25, 1143 (2024).
Li, G. et al. Immunogenic cell death-related genes predict prognosis and response to immunotherapy in lung squamous cell carcinoma. Biotechnol. Appl. Biochem.72, https://doi.org/10.1002/bab.2652 (2025).
Zhou, Y. et al. Construction and validation of a tumor mutational burden and immune-related prognostic model for predicting the prognosis of patients with lung squamous cell carcinoma. J. Thorac. Dis. 15, 1319–1334 (2023).
Ding, Y. et al. A new risk model for CSTA, FAM83A, and MYCT1 predicts poor prognosis and is related to immune infiltration in lung squamous cell carcinoma. Am. J. Transl. Res. 14, 7705–7725 (2022).
Riviere, P. et al. High tumor mutational burden correlates with longer survival in immunotherapy-naïve patients with diverse cancers. Mol. Cancer Ther. 19, 2139–2145 (2020).
Inoue, D. et al. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci. 103, 760–766 (2012).
Ooi, A. et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer cell 20, 511–523 (2011).
Onodera, Y. et al. NRF2 immunolocalization in human breast cancer patients as a prognostic factor. Endocr. -Relat. cancer 21, 241–252 (2014).
Pan, Y. et al. 3-Hydroxyisobutyryl-CoA hydrolase involved in isoleucine catabolism regulates triacylglycerol accumulation in Phaeodactylum tricornutum. Philos. Trans. R. Soc. London. Series B, Biol. Sci. 372, 20160409 (2017).
Yang, L. et al. Prognostic and tumor microenvironmental feature of clear cell renal cell carcinoma revealed by m6A and lactylation modification-related genes. Front. Immunol. 14, 1225023 (2023).
Shan, Y. et al. Targeting HIBCH to reprogram valine metabolism for the treatment of colorectal cancer. Cell Death Dis. 10, 618 (2019).
Graff, J. N. et al. Sustained complete response to CTLA-4 blockade in a patient with metastatic, castration-resistant prostate cancer. Cancer Immunol. Res. 2, 399–403 (2014).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
de Streel, G. & Lucas, S. Targeting immunosuppression by TGF-β1 for cancer immunotherapy. Biochem. Pharmacol. 192, 114697 (2021).
Batlle, E. & Massagué, J. Transforming growth factor-β signaling in immunity and cancer. Immunity 50, 924–940 (2019).
Sharma, P. et al. Biphasic role of Tgf-β signaling during Müller glia reprogramming and retinal regeneration in zebrafish. iScience 23, 100817 (2020).
Li, X. et al. Different roles of matrix metalloproteinase 2 in osteolysis of skeletal dysplasia and bone metastasis (Review). Mol. Med. Rep. 23, 70 (2021).
Derynck, R. & Zhang, Y. E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577–584 (2003).
Pastushenko, I. & Blanpain, C. EMT transition states during tumor progression and metastasis. Trends cell Biol. 29, 212–226 (2019).
Su, J. et al. TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 577, 566–571 (2020).
Ma, Y. et al. CRISPR-mediated MECOM depletion retards tumor growth by reducing cancer stem cell properties in lung squamous cell carcinoma. Mol. Ther. : J. Am. Soc. Gene Ther. 30, 3341–3357 (2022).
Zhang, P. et al. Genomic sequencing and editing revealed the GRM8 signaling pathway as potential therapeutic targets of squamous cell lung cancer. Cancer Lett. 442, 53–67 (2019).
Swanson, K. et al. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 186, 1772–1791 (2023).
Bueno, R. et al. Multi-institutional prospective validation of prognostic mrna signatures in early stage squamous lung cancer (alliance). J. Thorac. Oncol. Publ. Int. Assoc. Study Lung Cancer 15, 1748–1757 (2020).
Satpathy, S. et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell 184, 4348–4371.e4340 (2021).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Gillette, M. A. et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 182, 200–225.e235 (2020).
Liu, Q. et al. Proteogenomic characterization of small cell lung cancer identifies biological insights and subtype-specific therapeutic strategies. Cell 187, 184–203.e128 (2024).
Mayakonda, A. et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756 (2018).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb. (Mass.)) 2, 100141 (2021).
Mak, M. P. et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 22, 609–620 (2016).
Nusinow, D. P. et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell 180, 387–402.e316 (2020).
Ianevski, A. et al. SynergyFinder 3.0: An interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic acids Res. 50, W739–w743 (2022).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grant No. 82125001, 82201948 and 82173393), the Innovation Programme of Shanghai Municipal Education Commission (Grant No. 2023ZKZD33), and Clinical Research Foundation of Shanghai Pulmonary Hospital (Grant No. FKLY20004 and FKYQ2308). We thank Prof. Haiyang Hu for his valuable opinions.
Author information
Authors and Affiliations
Contributions
L.Z., P.Z. and M.D. had the idea for and designed the study, had full access to all data in the study, and took responsibility for the data’s integrity and the data analysis’s accuracy. L.S., L.Z. and J.W. contributed to the statistical analysis and writing of the report. L.Z., J.W. contributed to experimental analysis. L.S., J.W., H.Y., Xin Z., J.Z., J.H., Y.Y., Xun Z., Y.Z. and G.J. contributed to critical revision of the report. All authors reviewed and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks John Minna and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaliya Georgieva.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, L., Wang, J., Yu, H. et al. Selective inhibition of TGF-β-induced epithelial-mesenchymal transition overcomes chemotherapy resistance in high-risk lung squamous cell carcinoma. Commun Biol 8, 152 (2025). https://doi.org/10.1038/s42003-025-07595-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07595-x