Abstract
Ovarian cancer (OC) is one of the deadliest gynecological malignancies. As the prevalent post-transcriptional regulation, alternative polyadenylation (APA) plays a crucial role in various tumors. Here we identify that the APA regulator NUDT21 is upregulated in OC and promotes malignant progression. We further demonstrate that IGF2BP3 interacts with NUDT21, which suggests m6A modification could regulate APA processing. Mechanistically, IGF2BP3, recognizing the m6A-modified site in intron 32 of SPTBN1, recruits NUDT21 to promote the usage of the SPTBN1 proximal polyadenylation site (PAS), thus increasing the generation of short transcripts in OC cells. Intriguingly, the SPTBN1 long variant demonstrates tumor-suppressive properties, whereas the short variant enhances oncogenic activity in OC. Subsequently, we illustrate that the long isoform inhibits tumor growth and metastasis by binding to CDK1 and blocking the G2/M phase of the cell cycle. In conclusion, this study uncovers a previously unrecognized regulatory mechanism in OC, which could provide potential therapeutic strategies for OC.
Similar content being viewed by others
Introduction
Ovarian cancer (OC) is the most lethal gynecological malignancy among women, and its incidence and mortality rates have steadily increased over the years1. Owing to the absence of specific early clinical symptoms, ~70% of OC patients present at advanced stages, which results in a high likelihood of recurrence and metastasis and a dismal 5-year survival rate of <40%2. Thus, investigations of the mechanisms involved in the occurrence and development of OC are urgently needed. Recent studies have begun to explore the role of alternative polyadenylation (APA) in cancer biology, revealing its potential as a critical regulatory mechanism influencing mRNA stability, translation efficiency, and protein localization3. Molecular inhibitors can be independently developed for specific APA regulatory factors to exert antitumor effects.
Mammalian 3′end processing is mediated primarily by a set of multiprotein complexes, including cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CSTF), and cleavage factors I and II (CFIm and CFIIm)4. Approximately 70% of precursor mRNA (pre-mRNA) transcripts contain multiple PASs, allowing for the generation of diverse 3′ untranslated region (UTR) isoforms5. This diversity contributes significantly to the complexity of the transcriptome and proteome. Recent studies have highlighted the dysregulation of APA factors in various cancers, including lung cancer, liver cancer, gastric cancer, pancreatic cancer, esophageal cancer, bladder cancer, prostate cancer, breast cancer and glioblastoma, where CPSF, CSTF and CFIm components have been implicated in tumor progression6,7,8,9,10,11,12,13,14,15,16. In OC, our previous research reported that the upregulation of CSTF3 in platinum-resistant OC cells could increase the production of shorter NEAT1 isoforms to promote drug resistance17. However, the specific roles of APA regulators in the progression of OC have not been fully explored.
N6-methyladenosine (m6A) is the most prevalent mRNA modification in eukaryotes and is predominantly located near stop codons and 3′UTRs close to or overlapping with PASs18. Emerging evidence suggests that m6A modifications can interact with APA factors to modulate gene expression19. For example, YTHDC1, a m6A reader, has been shown to bind to the APA regulator FIP1L1 and influence the recruitment of CPSF4, thereby inhibiting the use of proximal PAS sites in target genes, as demonstrated in 293T and MCF-7 cells20. In lung cancer, YTHDC1 has been shown to recognize m6A modifications on the 3′UTR of the FSP1 gene and recruit CSTF3 to generate short 3′UTR transcripts, reducing the mRNA stability of FSP121. Zheng et al. reported that CSTF2 could regulate m6A installation through slowing the RNA Pol II elongation rate during gene transcription in pancreatic ductal adenocarcinoma22. The interplay between m6A and APA may be an innovative molecular mechanism for tumor growth.
In this study, we indicated that the m6A reader IGF2BP3 recruits the APA factor NUDT21 to facilitate the use of the proximal PAS in SPTBN1, leading to the production of shorter transcript that may drive malignant processes in OC. Furthermore, our findings indicate that the SPTBN1 long transcript functions as a tumor suppressor by binding to CDK1 to block the G2/M phase of the cell cycle. These findings highlight the potential of targeting m6A and APA interactions as an emerging therapeutic strategy for OC treatment.
Results
NUDT21 is upregulated in human OC and has a poor clinical prognosis
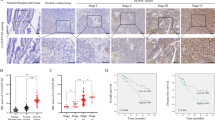
The dysregulation of APA is critical for cancer initiation and progression23. To systematically screen the expression patterns of key APA regulators involved in OC progression, we performed single-cell sequencing of two OC patients tissue samples and one normal ovarian tissue sample (Fig. 1A). After strict quality control, 29,331 cells were retained for subsequent analyses. We classified all the cells into 15 clusters via the Seurat software package (Supplementary Fig. 1A). On the basis of previously reported canonical cell markers, five main known cell types were identified: stromal fibroblasts, epithelial cells, myeloid cells, lymphocytes, and endothelial cells (Fig. 1B). Epithelial cells (EPCAM, CD24D, PROM1, PAX8, and KRT18 as markers; Supplementary Fig. 1B–F) represented the highest proportion of tumor samples, whereas stromal cells constituted the highest proportion of normal samples (Fig. 1C). We subsequently observed that almost all APA regulators factors were highly expressed in epithelial cells, with NUDT21 and CPSF6 being the most prominent (Fig. 1D). NUDT21 and CPSF6 are subunits of the CFIm complex, and NUDT21 binds directly to the UGUA sequence on RNA, influencing PAS selection24. NUDT21 has been implicated in human cancers, but its role in tumors is controversial. For example, in cervical cancer, NUDT21 has been shown to promote carcinogenesis25, while in bladder cancer, it functions as a tumor suppressor26. However, the precise role of NUDT21 in OC remains unclear. To further investigate the role of NUDT21 in OC cells, mRNA expression in OC cells and fallopian tube epithelial cells from the Gene Expression Omnibus (GEO) database revealed that NUDT21 was significantly upregulated in OC cells (Fig. 1E). Additionally, analysis from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) illustrated that NUDT21 protein levels were significantly elevated in OC tissues compared with normal fallopian tube epithelial tissues (Fig. 1F). To further validate these findings, we performed Western blot analysis on clinical tissue samples, which confirmed markedly increased NUDT21 expression in OC tissues (Fig. 1G). Kaplan–Meier plotter (KMplot) survival analysis further demonstrated that high NUDT21 expression was significantly associated with poor overall survival (OS) and progression-free survival (PFS) in OC patients (Fig. 1H).
A–D scRNA-seq analysis of two OC patients tissue samples and one normal ovarian tissue sample. A Composition of the three scRNA-seq datasets according to the UMAP plot. B UMAP plot of major cell types, with the cell number as well as the proportion of each type illustrated in (C). D Dot plot of key APA regulators in five major cell types. E Boxplot of key APA regulators in OC cells vs. fallopian tube epithelial cells. Data from the GEO database (GSE151316). F CPTAC database analysis of SPTBN1 protein expression (normal: n = 25, tumor: n = 100). G Validation of NUDT21 expression in clinical tissue samples (The upper panel represents 6 paired samples, while the lower panel represents 6 unpaired samples. F: Fallopian tube epithelial tissue, T: OC tissue). H Kaplan‒Meier curves of progression-free survival (n = 614) and overall survival (n = 655) in OC patients with high and low NUDT21 levels.
NUDT21 knockdown inhibits OC cell growth, proliferation, metastasis and invasion in vitro and in vivo
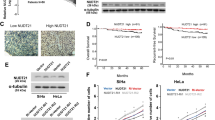
To explore whether NUDT21 has an impact on OC progression, we knocked down NUDT21 expression in A2780, OVCAR3, and SKOV3 cells (Fig. 2A). The results of the CCK8 assays indicated significant inhibition of tumor cell growth following NUDT21 knockdown (Fig. 2B). Consistently, colony formation assays demonstrated a marked reduction in cell proliferation upon NUDT21 knockdown (Fig. 2C). Additionally, we performed transwell assays and confirmed that NUDT21 knockdown significantly suppressed tumor cell migration and invasion (Fig. 2D). To evaluate the effect of NUDT21 on the proliferative ability of OC in vivo, xenograft tumor models were established via the subcutaneous injection of NUDT21-knockdown cells into nude mice. NUDT21 knockdown significantly inhibited the proliferative capacity of OC cells in vivo (Fig. 2E–G), which was characterized by a reduction in tumor volume and weight. We then investigated whether NUDT21 may impact the in vivo metastasis of OC cells. NUDT21-knockdown cells were intraperitoneally injected into nude mice. NUDT21 depletion inhibited the metastasis of OC cells (Fig. 2H–J). Subsequently, immunohistochemical staining revealed that the knockdown of NUDT21 led to a significant decrease in the expression of Ki-67, a marker of cell proliferation, and a corresponding increase in the expression of Caspase-3, a marker of apoptosis (Fig. 2K–M). Conversely, we next overexpressed NUDT21 in OC cells (Supplementary Fig. 2A), and by using a transwell assay, we observed that NUDT21 overexpression significantly enhanced the migration and invasive ability of tumor cells (Supplementary Fig. 2B–D). Taken together, these results indicate that the key APA regulator NUDT21 is highly expressed in OC and promotes the malignant process of tumors.
A Western blot analysis of NUDT21-knockdown cells. B-D CCK–8 (n = 4) (B), colony formation (n = 3) (C) and Transwell (n = 3) (D) assays of A2780, OVCAR3 and SKOV3 cells infected with NUDT21-knockdown or negative control cells. E–G Gross xenograft model anatomy (E), tumor volume (F) and tumor weight (G) after the subcutaneous injection of NUDT21-knockdown OVCAR3 cells into nude mice (n = 5). H and I Intraperitoneal tumor metastasis of NUDT21-knockdown OVCAR3 cells injected into nude mice. The graphs show the gross anatomy of the abdominal cavity (H), number of metastatic nodules (I) and metastatic tumor weight data (J) (n = 5). K–M IHC and HE staining demonstrating the expression of Ki-67 and Caspase-3 in tissues as indicated (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001.
The IGF2BP3-NUDT21 interaction in OC may regulate APA through m6A modification
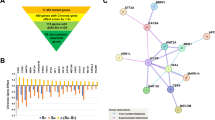
Phenotypic studies revealed that NUDT21 significantly promotes tumor progression in OC. Given these findings, we hypothesized that NUDT21 may exert its oncogenic effects through interactions with specific proteins. To elucidate the molecular mechanisms by which NUDT21 drives tumor progression, we employed mass spectrometry (MS) to analyze NUDT21 immunoprecipitation proteins in OVCAR3 cells (Supplementary Fig. 3A). Previous studies have shown that NUDT21 interacts with several RNA binding proteins (RBPs), such as PABPN1, VIRMA, SRSF3 and SRSF727,28. By subsequently integrating the Eukaryotic RBP Database (EuRBPDB) and the RBP Database for Tumorigenesis (RBPTD), we identified 66 regulatory factors that interact with NUDT21 (Tables S2 and S3). Among these, we selected splicing factors, protein modification factors, and post-transcriptional regulatory factors (Fig. 3A). To validate potential interactors, co-immunoprecipitation (Co-IP) assays were performed in OVCAR3 cells with IGF2BP3, RIM21, U2AF2, EIF3C, EIF3D, SRSF3, and SRSF6. Meanwhile, to avoid false positives, we used the protein CPSF6, which interacts with NUDT21, as a positive control (Supplementary Fig. 3B and C). Co-IP results showed that only the m6A reader protein IGF2BP3 interacted with NUDT21 (Fig. 3B). Given the existing functional crosstalk between IGF2BP3, an m6A reader protein, and NUDT21, a core alternative polyadenylation (APA) factor, we hypothesize that IGF2BP3 might collaborate with NUDT21 via m6A-mediated recognition mechanisms to jointly govern the selection of APA sites. To identify IGF2BP3 and NUDT21 as potential key regulatory factors that mediate this interaction, we further explored the interplay between m6A and APA in OC. To this end, we performed meRIP-seq, PAS-seq with NUDT21 knockdown, and RIP-seq for both IGF2BP3 and NUDT21 in OVCAR3 cells. Consequently, the PAS-seq results revealed that 2,464 genes presented APA events in response to NUDT21 knockdown (Table S4). Among these genes, 1,574 presented shortened APA events, whereas 890 presented lengthened APA events (Fig. 3C). NUDT21 knockdown tended to use the proximal PAS of target genes (Fig. 3D). Subsequent KEGG enrichment analysis suggested that the shortened genes were involved in various tumor-associated pathways (Fig. 3E, F). Additionally, meRIP-seq identified a total of 10,474 transcripts with m6A modifications (Supplementary Fig. 3D and Table S5), predominantly enriched in the GGAC motif (Supplementary Fig. 3E). Furthermore, RIP-seq analysis identified 3553 transcripts bound by IGF2BP3 and 1486 transcripts bound by NUDT21 (Fig. 3G, I, Tables S6 and S7), with KEGG enrichment revealing their involvement in multiple tumor-associated signaling pathways (Fig. 3H and J). To elucidate the role of IGF2BP3 and NUDT21 in coregulating APA target genes, we integrated meRIP-seq, PAS-seq, and RIP-seq data and identified 56 potential target genes (Fig. 3K and Table S8). Among these genes, two genes with shortened APA events, ICMT and ZEB1, and two with lengthened APA events, RNF213 and SPTBN1, were confirmed (Fig. 3L). Previous studies have primarily focused on the selection of proximal or distal PAS within the 3′UTR in APA, with limited research on the selection of intronic PAS29. When querying the UCSC database, we identified an isoform of RNF213 in intron 17 which aligned precisely with the proximal PAS (Fig. 3L). However, meRIP-seq revealed no detectable m6A modification sites in this intronic region (Table S5). In contrast, the UCSC database indicates that SPTBN1 generates two distinct transcript variants and that the PAS is located within the 32nd intron, which follows the proximal PAS according to PAS-seq (Fig. 3M). Notably, PAS-seq analysis revealed that NUDT21 knockdown decreases proximal PAS usage while increasing distal PAS usage in SPTBN1 (Fig. 3N). Furthermore, near the proximal PAS of SPTBN1, m6A modifications were detected via meRIP-seq (Fig. 3O).
A Venn diagram of NUDT21 IP-MS data with or without RNaseA, the eukaryotic RBP database (EuRBPDB) and the RBP database for tumorigenesis (RBPTD). B Co-IP assay of NUDT21 and IGF2BP3 in A2780 OVCAR3 and SKOV3 cells. C–F PAS-seq analysis of NUDT21-knockdown OVCAR3 cells; density scatter plots of long and short genes (C); and boxplot of the proximal and distal PASs uses (D). KEGG enrichment analysis of long genes (E) and short genes (F). G and H Volcano plot of IGF2BP3-binding genes identified by RIP-seq (G) and KEGG enrichment analysis of these genes (H). I and J Volcano plot of NUDT21-binding genes identified by RIP-seq (I) and KEGG enrichment analysis of these genes (J). K Venn diagram of the candidate target genes screened via meRIP-seq, NUDT21 PAS-seq, NUDT21 RIP-seq, and IGF2BP3 RIP-seq data. L Information on four candidate target genes from 56 genes related to cancer and APA. M UCSC browser showing the long and short isoforms of SPTBN1, in which the proximal polyA site (pPAS) is in the 32 (35 total) introns. N PAS-seq data for SPTBN1 genes with NUDT21 knockdown. The proximal and distal PASs were marked by colored boxes, and the locations are labeled at the bottom. O MeRIP-seq results showing enrichment of m6A sites on intron 32 of SPTBN1.
IGF2BP3 regulates the APA of SPTBN1 by recruiting NUDT21 in a m6A-dependent manner
Subsequently, primers were designed for the long and short transcripts of ICMT, ZEB1, RNF213, and SPTBN1. METTL3 (Methyltransferase-like 3) is a key m6A “writer” enzyme that catalyzes the m6A modification30. RT‒qPCR confirmed that knockdown of IGF2BP3, NUDT21 and METTL3 (Supplementary Fig. 4A–C), the ratio of the SPTBN1 long transcript (NM_003128) to the short transcript (NM_178313) was significantly increased. This indicates that knockdown of these genes promoted the production of the longer isoform of SPTBN1, which aligns with the PAS-seq data (Fig. 4A–C). Moreover, we validated these findings via meRIP-qPCR and RIP-qPCR in OVCAR3 cells, which demonstrated that SPTBN1 transcripts, as well as the IGF2BP3 and NUDT21 proteins, were significantly enriched in immunoprecipitates with m6A antibodies (Fig. 4D). Following this, upon IGF2BP3 knockdown, NUDT21 RIP-qPCR (Fig. 4E) indicated a significant increase in its binding to the long SPTBN1 isoform and a significant decrease in its binding to the short isoform, with the differences being statistically significant (Fig. 4F). Similarly, METTL3 knockdown yielded comparable results in the NUDT21 RIP‒qPCR assay (Fig. 4G and H). To further investigate whether IGF2BP3 regulates the APA of SPTBN1 via m6A modification, we conducted a dual-luciferase reporter assay by constructing both wild-type (WT) and m6A sites mutant vectors containing the SPTBN1 intron 32. These vectors were cotransfected with IGF2BP3 or the control, and the results demonstrated that the relative luciferase activity was significantly increased in the WT group transfected with IGF2BP3 (Fig. 4I). These findings suggest that IGF2BP3 recognizes the m6A modification in SPTBN1 intron 32 and influences the selection of the PAS. It can therefore be hypothesized that the different SPTBN1 transcripts are coregulated by m6A modifications and APA processing in the intron 32 out of a total of 35 introns. In human cells, SPTBN1 (βII-spectrin) codes for two C-terminal splice variants, βII∑1 and βII∑231. Western blot assays revealed that the knockdown of IGF2BP3, NUDT21, and METTL3 in OC cells led to the upregulation of the long SPTBN1 isoform βII∑1 (Fig. 4J–L). Additionally, the ratio of βII∑1 to the short SPTBN1 isoform βII∑2 did increase. (Supplementary Fig. 4D and E). Moreover, the expression of total SPTBN1 was markedly lower in clinical OC samples than in normal fallopian tube epithelial tissue, as demonstrated by Western blot analysis (Supplementary Fig. 4G). In addition, the CPTAC database revealed that SPTBN1 protein levels were significantly lower in OC tissues than in normal fallopian tube epithelial tissues (Supplementary Fig. 4H). These findings suggest that the m6A regulators METTL3 and IGF2BP3 may work synergistically with the APA regulator NUDT21. Together, they modulate the APA of the target gene SPTBN1, generating two distinct transcripts that are translated into different protein isoforms, potentially driving the malignant progression of OC.
A–C RT‒qPCR was used to detect the relative changes in the lengths of the target genes ICMT, ZEB1, RNF213 and SPTBN1 in A2780, OVCAR3, and SKOV3 cells following NUDT21 (A), IGF2BP3 (B) and METTL3 (C) knockdown (n = 4). D MeRIP‒qPCR of SPTBN1 m6A modification; RIP‒qPCR of SPTBN1 interaction with IGF2BP3 and NUDT21 (n = 4). E and F RIP-qPCR was used to validate the relative fold change in SPTBN1 long and short isoform binding with NUDT21 in OVCAR3 cells (F) following IGF2BP3 knockdown (E) (n = 4). G and H RIP-qPCR was used to validate the relative fold change in SPTBN1 long and short isoform binding with NUDT21 in OVCAR3 cells (H) following METTL3 knockdown (G) (n = 3). I A dual-luciferase assay was performed on the 32nd intron of SPTBN1 with WT and m6A site mutations, which were both cotransfected with IGF2BP3 (n = 4). J–L SPTBN1 long and short isoform protein levels were detected by Western blot in A2780, OVCAR3, and SKOV3 cells after NUDT21 (J), IGF2BP3 (K), and METTL3 (L) were knocked down. *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown of SPTBN1 and its long isoform enhances the growth and metastasis of OC cells
To further investigate the distinct roles of SPTBN1 isoforms in OC progression, we selectively knocked down long isoform, short isoform and total SPTBN1 expression in OC cell lines (Fig. 5A). Functional assays were then performed to assess the effects of these knockdowns both in vitro and in vivo. In vitro, CCK-8 assays demonstrated that knockdown of the long SPTBN1 isoform significantly promoted OC cell proliferation (Fig. 5B), a finding that was further supported by colony formation assays, which showed increased proliferative capacity upon knockdown of the long isoform (Fig. 5C). Furthermore, transwell assays revealed increased migration and invasion of OC cells following the knockdown of the long isoform (Fig. 5D), whereas the knockdown of the short isoform yielded the opposite results (Fig. 5B–D). To assess in vivo effects, we established subcutaneous xenograft and intraperitoneal metastasis models in nude mice. Tumor growth measurements revealed that knockdown of the long isoform significantly accelerated tumor growth in vivo, whereas knockdown of the short isoform did not significantly affect tumor growth (Fig. 5E). In addition, intraperitoneal metastasis assays confirmed that knockdown of the long isoform enhanced OC cell metastasis (Fig. 5F), whereas knockdown of the short isoform reduced metastasis. Next, overexpression experiments were conducted in the same cell lines (Supplementary Fig. 5A), revealing that overexpression of the long isoform suppressed OC cell growth, proliferation, migration and invasion, while overexpression of the short isoform had the opposite effect (Supplementary Fig. 5B–D). To further validate the contribution of SPTBN1 to the function of NUDT21, rescue experiments were performed by overexpressing the short SPTBN1 isoform in NUDT21-knockdown OC cells (Fig. 5G). These experiments demonstrated that overexpression of the short isoform partially rescued the proliferative and metastatic capacity suppressed by NUDT21 knockdown (Fig. 5H–J). Taken together, these phenotypic results indicate that the long isoform of SPTBN1 inhibits OC cell growth and metastasis, whereas the short isoform promotes these processes.
A Western blot analysis of total, long, and short isoform SPTBN1-knockdown cells. B–D CCK-8 (B) (n = 4), colony formation (C) (n = 3) and Transwell (D) (n = 3) assays of A2780, OVCAR3, and SKOV3 cells infected with SPTBN1 total, long, or short isoform knockdown or negative control. E Gross xenograft model anatomy and tumor volume and tumor weight after subcutaneous injection of SPTBN1 total, long, and short isoform knockdown in OVCAR3 cells into nude mice (n = 5). F Intraperitoneal tumor metastasis of SPTBN1 total, long, and short isoform-knockdown OVCAR3 cells injected into nude mice; the graphs show the number of metastatic nodules (n = 5). G Western blot analysis of NUDT21-knockdown and SPTBN1 short isoform-overexpressing cells. H–J Overexpression of the SPTBN1 short isoform reversed the growth (H) (n = 4), proliferation (I) (n = 3) and metastasis (J) (n = 3) of OC cells in which NUDT21 was downregulated. *P < 0.05, **P < 0.01, ***P < 0.001.
The SPTBN1 long isoform regulates the cell cycle in OC cells via interaction with CDK1
According to previous studies, the long isoform of SPTBN1 contains a PH domain at its C-terminus, whereas the short isoform lacks this domain but contains phosphate groups31 (Fig. 6A). To further elucidate the molecular mechanisms by which SPTBN1 isoforms affect OC, we performed mass spectrometry (MS) analysis to identify proteins that interact with the short and long isoforms (Supplementary Fig. 6A). The results revealed that 617 proteins were commonly associated with both isoforms, 103 proteins interacted exclusively with the short isoform, and 1202 proteins were specific to the long isoform (Fig. 6B). KEGG pathway analysis of isoform-specific interacting proteins revealed that proteins uniquely binding to the long isoform were significantly enriched in pathways related to the cyclin-dependent kinase (CDK) gene family, multiple neurodegenerative diseases and DNA replication, with the cell cycle pathway being particularly prominent (Fig. 6C and D). To validate the interaction between SPTBN1 isoforms and the CDK family, we conducted Co-IP assays, which demonstrated that the long isoform of SPTBN1 interacted most strongly with CDK1 (Fig. 6E). Previous studies have shown that cyclins and CDK complexes play central regulatory roles in the cell cycle, including regulating interphase and mitotic processes32. CDK1 regulates the transition from the G2 phase to the M phase through interactions with CDK2, CyclinA, and CyclinB33. To further confirm whether the long isoform of SPTBN1 affects the interaction between CDK1 and its complex, we conducted subsequent Co-IP assays. The results revealed that knockdown of the long isoform of SPTBN1 enhanced the interaction between CDK1 and CyclinA2 (Fig. 6F). Conversely, overexpression of the long isoform of SPTBN1 significantly weakened the binding between CDK1 and CyclinA2 (Fig. 6F). Consequently, we speculate that the SPTBN1 long isoform inhibits the progression of OC by influencing the cell cycle. Cell cycle assays revealed an increased ratio of cells in the G2/M phase in OVCAR3 cells, which downregulated the expression of SPTBN1 and its long variants (Fig. 6G and Supplementary Fig. 6B). Similar results were observed after the knockdown of METTL3 (Supplementary Fig. 6D), IGF2BP3 (Supplementary Fig. 6E) and NUDT21 (Fig. 6H and Supplementary Fig. 6C).
A Structural diagram of the SPTBN1 long and short isoforms. B Venn diagram of SPTBN1 IP-MS data for long and short isoforms. C and D KEGG enrichment analysis of short unique genes (C) and long unique genes (D). E Co-IP assay of SPTBN1 long and short isoform binding to CDKs with RNase. F Co-IP was used to detect the interaction between CDK1 and its complex (CDK2, CyclinA2/B1) after total and long isoform SPTBN1 knockdown (upper panel) or SPTBN1 overexpression (lower panel). G and H Cell cycle assays were performed with knockdown of total SPTBN1 and the SPTBN1 long isoform (G), NUDT21 (H), and the statistics were presented for G2/M (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. I Diagram of the regulatory mechanism.
Taken together, these findings elucidated that IGF2BP3 recognizes the m6A modification of SPTBN1 pre-mRNA and recruits NUDT21 to bind the proximal PAS, which results in SPTBN1 generating two different variants with different functions. The long isoform of SPTBN1 blocks the G2/M phase of the cell cycle by interfering with the interaction of the CDK1-CyclinA2 complex, potentially suppressing the malignant progression of OC (Fig. 6I).
Discussion
The maturation of mRNAs is a critical process, particularly the cleavage and polyadenylation of precursor mRNAs (pre-mRNAs) at the PAS within the 3′UTR34. Most genes have more than one PAS, and different choices of these sites can generate transcript isoforms of varying lengths, known as APA35. On the basis of the location of the PAS, APA can be divided into two types: UTR-APA, which is located in the 3′UTR and affects the length of the 3′UTR and thus RNA stability and translation efficiency, and coding region APA (CR-APA), which is located in exons or introns and can lead to C-terminal truncated proteins with altered functions4. Dysregulation of APA factors has been implicated in various diseases, including cancer, where aberrant expression of these factors can lead to preferential selection of the PAS, contributing to tumorigenesis35.
The findings of our research underscore the role of the APA regulatory factor NUDT21 in OC, where its upregulation is correlated with poor prognosis. The NUDT21 subunit, a key component of the CFIm complex, selectively recognizes upstream sequence elements (USEs) of the PAS, influencing the selection and usage of the PAS36. Previous studies have documented the dual roles of NUDT21 in cancer, with evidence suggesting that both oncogenic and tumor-suppressive functions depend on the context37,38. In liver cancer, NUDT21 inhibited the expression of PSMB2 and CXXC5 through APA regulation, thereby suppressing tumor proliferation and metastasis34. In bladder cancer, NUDT21 inhibited tumor cell proliferation, migration, and invasion by regulating the APA processing of ANXA2 and LIMK239. In small-cell lung cancer, hypoxia-induced the downregulation of NUDT21 expression through HIF-1α, promoting the proliferation and inhibiting the apoptosis of tumor cells40. In cervical cancer, downregulation of NUDT21 expression also promoted tumor progression25. Conversely, in gastric cancer, NUDT21 promoted tumorigenesis by recognizing the UGUA sequence of the lncRNA ACVR1B 3′UTR (acv3UTR) to regulate its APA processing41. Zhou et al. reported that NUDT21 increased the use of the proximal PAS of MZT1 mRNA by recognizing the UGUA sequence of the proximal PAS, thereby promoting the progression of pancreatic cancer15. Additionally, Lin et al. confirmed that NUDT21 played an oncogenic role in prostate cancer and was involved in the process of enzalutamide resistance10. However, the potential function and mechanism of NUDT21 in OC remain unclear.
This study elucidated the mechanisms by which NUDT21 promotes tumor growth and metastasis in OC, particularly through its interaction with the RNA-binding protein IGF2BP3, which modulates m6A methylation and APA processing. IGF2BP3, a m6A reader protein, is widely involved in the occurrence and development of various tumors, including OC, by regulating the RNA stability of target genes42. We further screened APA target genes that carried m6A modifications and were bound by IGF2BP3 and NUDT21. Strikingly, previous studies have shown that m6A modifications predominantly regulate APA in the 3′UTR, which influenced the expression of target genes but not the protein structure or function43. However, we discovered that this regulation occurs in the intronic region of the target gene SPTBN1. The m6A modification in intron 32 of SPTBN1 was recognized by IGF2BP3, which facilitated the recruitment of NUDT21 to the proximal PAS, thereby increasing the generation of the shorter isoform of SPTBN1. Mutation or downregulation of the m6A modification in the 32nd intron of SPTBN1 decreased the binding of the proximal PAS by NUDT21 and the expression of the short SPTBN1 variant.
The SPTBN1 gene, encoding βII spectrin, is a member of the spectrin family of proteins, which plays crucial roles in the organization and rearrangement of the actin cytoskeleton44. This protein is essential for maintaining cell shape, the arrangement of transmembrane proteins, and the organization of organelles45. Mutation of SPTBN1 can lead to cellular instability and mislocalization of membrane receptors and channels and is associated with severe neurodevelopmental disorders and neurodegenerative diseases46. In addition, SPTBN1 is involved in regulating various cellular functions, including apoptosis, adhesion, migration, and the cell cycle47. There are few studies on the SPTBN1 gene in cancers, with a focus on liver, kidney, colon, and breast cancers, where it generally exhibits tumor-suppressive properties48. Notably, βII spectrin exists in two isoforms, βII∑1 and βII∑2, which differ in their C-terminal structures, leading to distinct functional roles31. However, it remains unclear why there are two variants with different structures, and no studies have specifically investigated the unique functions of the long and short variants in tumors.
This study revealed an interesting molecular mechanism that underlies the generation of two functionally distinct SPTBN1 isoforms in OC. The interplay between IGF2BP3 and NUDT21 governs APA processing, resulting in the production of these isoforms. Innovative findings reveal a dualistic role of SPTBN1 isoforms in tumor biology. The long isoform performs tumor-suppressive functions, whereas the short isoform promotes oncogenic activities, counteracting the suppressive effects of NUDT21 depletion on cell proliferation and metastasis. Additionally, mechanistic studies identified a key role for the long isoform in regulating the cell cycle by binding to CDK1, which arrests OC cells in the G2/M phase and inhibits proliferation. This discovery elucidates the existence of these isoforms and provides critical insights into their opposing roles in cancer progression, suggesting potential avenues for therapeutic intervention.
In summary, our research demonstrated that NUDT21 acted as an oncogene and was recruited to the proximal PAS by interacting with the m6A reader IGF2BP3 to facilitate the production of a shorter SPTBN1 isoform that promoted the malignancy of OC. This regulation not only affected the expression levels of SPTBN1 isoforms but also highlighted the potential for differential functional outcomes on the basis of the isoform produced. The long isoform of SPTBN1 had an antitumor effect by inhibiting cell cycle progression. In the future, we will identify other specific target genes regulated by m6A and APA and delineate the unique functions of these regulatory mechanisms to provide a more comprehensive understanding of their roles in OC progression.
Methods
Clinical tissue sample collection
Patients diagnosed with OC were enrolled in this study, and all of the cancer and control samples were collected from OC patients. The clinical samples consist of 12 cases of high-grade serous ovarian cancer (HGSOC) and 12 cases of normal fallopian tube epithelium (6 paired samples and 6 unpaired samples). The histological subtypes of the samples were determined on the basis of the pathological results. The single-cell sequencing samples include 2 cases of HGSOC and 1 case of normal ovarian epithelium. Freshly excised biopsy tissues were stored in a tissue preservation solution and processed for scRNA-seq research within 48 h. Other samples were placed in liquid nitrogen for further examination. Written informed consent was obtained from all patients who provided samples. All procedures involving human participants were reviewed and approved by the ethical standards of the Institutional Review Board of the Third Affiliated Hospital of Chongqing Medical University (Approval No. 202293) and followed the 2013 revision of the Helsinki Declaration. All ethical regulations relevant to human research participants were followed.
Public bioinformatics analysis
To evaluate the clinical significance of NUDT21 in ovarian cancer, RNA-seq and protein expression data from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/, GSE151316) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) database (https://pdc.cancer.gov/pdc/) were downloaded. We compared the mRNA and protein expression levels between normal tissues and OC tissues. To assess the prognostic value of NUDT21, Kaplan‒Meier survival curves were generated via the Kaplan‒Meier Plotter program (www.kmplot.com) with probe 224830_at to analyze overall survival (OS) and progression-free survival (PFS).
Cell culture
The human OC cell lines A2780, OVCAR3, SKOV3, and the human embryonic kidney 293T cells were obtained from the Cell Bank of the National Infrastructure of Cell Line Resource (Beijing, China). Cell lines were authenticated by STR profiling. All cell lines were tested for mycoplasma and were mycoplasma negative. A2780, OVCAR3, and SKOV3 cells were cultured in RPMI 1640 (Gibco) medium, and 293T cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS; VivaCell Biology). All the cells were maintained at 37 °C with 5% CO2. 293T cells were used to generate lentiviruses.
Plasmid construction and cell transfection
To produce shRNAs for lentiviruses, complementary sense and antisense oligonucleotides targeting METTL3, IGF2BP3, NUDT21, and SPTBN1 were synthesized (Tsing Ke Biotechnology, China), annealed, cloned, and inserted into the pLKO.1 vector (Addgene). The sequences of the shRNAs are shown in Table S1. The NUDT21 and SPTBN1 overexpression plasmids were purchased from YouBio (Hunan, China). For the knockdown of target genes, 293T cells were transfected with the encoding shRNA psPAX2 and pMD 2. G plasmids via jetPRIME reagent (Polyplus Transfection, France). Stable cell lines were generated via lentiviral infection and infection for ectopic expression. All of the corresponding empty vector plasmids served as negative controls.
Western blot analysis
All protein samples were separated via SDS‒PAGE. After electrophoresis, the proteins were transferred to PVDF membranes (Millipore, USA). Each membrane was then incubated in a solution of TBS (Tris-buffered saline, pH 7.4) containing 5% skim milk powder for 1 h at room temperature. After that, the membrane was incubated with primary antibodies overnight at 4 °C. All primary antibodies were diluted to 1:1000 concentration and secondary antibodies were diluted to 1:10,000. The primary antibodies used were as follows: NUDT21 (Proteintech, 10322-1-AP), METTL3 (Proteintech, 15073-1-AP), IGF2BP3 (Proteintech, 14642-1-AP), CPSF6 (Proteintech, 15489-1-AP), TRIM21 (Proteintech, 12108-1-AP), EIF3C (Proteintech, 12733-1-AP), EIF3D (Proteintech, 10219-1-AP), U2AF2 (Proteintech, 15624-1-AP), SRSF3 (Proteintech, 10916-1-AP), SRSF6 (Proteintech, 11772-1-AP), FLAG-tag (MBL, PM020), SPTBN1 (Proteintech, 25681-1-AP), CDK1 (Proteintech, 19532-1-AP), CDK2 (Proteintech, 10122-1-AP), CDK4 (Proteintech, 11026-1-AP), CDK6 (Proteintech, 14052-1-AP), CDK7 (Proteintech, 27027-1-AP), CDK9 (Proteintech, 11705-1-AP), Cyclin B1 (Proteintech, 55004-1-AP), Cyclin A2 (Proteintech, 18202-1-AP) and GAPDH (Proteintech, 60004-1-Ig). The membrane was incubated with the corresponding secondary antibody at room temperature for 1 h on the following day. Protein signals were detected with an enhanced chemiluminescence (ECL) Western blot detection kit (Biosharp, China) after washing with TBST. Densitometry was used to quantify the intensity of each band relative to that of GAPDH with ImageJ software.
Quantitative real-time polymerase chain reaction (RT‒qPCR)
Total RNA was extracted from the transfected cells via TRIzol reagent (Invitrogen). For the RT‒qPCR assay, total RNA was reverse-transcribed using a cDNA synthesis kit (Vazyme, China). The cDNA quantity was subsequently detected with Universal SYBR qPCR Master Mix (Vazyme, Q711-02), and the reaction was performed on a 7500 Real-Time PCR system (ABI) according to the manufacturer′s instructions. The primers used in this study were synthesized by Tsing Ke (Chongqing, China), and their details can be found in Table S1. To calculate the relative change in gene expression, the 2−ΔΔCt method was employed, with GAPDH used as the reference gene.
Immunohistochemistry (IHC) and HE staining
IHC staining was used to analyze the levels of specific proteins in the mouse xenograft samples. The samples were fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 4 μm-thick sections. Following deparaffinization and rehydration, the tissue sections were subjected to antigen retrieval by heating in citrate buffer (pH 6.0) at 100 °C for 15 min. The sections were subsequently blocked with 5% bovine serum albumin (BSA) for 1 h and then incubated overnight at 4 °C with specific anti-Ki-67 antibody (Servicebio, GB111141-100, 1:200) and anti-Caspase-3 antibody (Servicebio, GB115600-100, 1:200). For the negative control group, phosphate-buffered saline (PBS) was used as the primary antibody. The following day, the sections were incubated with HRP-conjugated anti-rabbit IgG for 30 min at room temperature. The sections were subsequently stained with diaminobenzidine (DAB) and hematoxylin and mounted with resin. The xenograft sections were also stained with hematoxylin and eosin (HE) to evaluate immune reactivity and microscopy images were acquired.
Co-immunoprecipitation (Co-IP) and mass spectrometry
The cells were lysed via IP buffer (20 mM HEPES, 150 mM NaCl, 10 mM KCl, 3.75 mM MgCl2, 0.5 mM EDTA, 10% glycerol, and 0.5% Nonidet P-40) supplemented with 1 mM dithiothreitol (DTT) and 1× protease inhibitor cocktail (PIC). An aliquot of 5% of the lysate was reserved for input analysis. The cleared lysate supernatant was incubated overnight at 4 °C with gentle agitation using 3 μg of specific antibody or IgG (negative control, Millipore), followed by the addition of 50 μl of prewashed protein G beads (Invitrogen, 10004D) and further incubation for 3 h. The beads were collected via a magnet, washed five times with IP buffer, and resuspended in 2× SDS sample buffer. After boiling for 10 min at 75 °C, the immunoprecipitates were resolved via SDS‒PAGE and subjected to Western blot. For mass spectrometry analysis, OVCAR3 cells were lysed in an IP buffer with or without 200 ng/ml RNase A (Invitrogen, EN0531). IP was performed using an anti-NUDT21 antibody (Proteintech, 10322-1-AP), with rabbit IgG (Millipore, 12-370) serving as a negative control. Ultimately, the proteins were subjected to silver staining, and subsequent MS analysis was conducted by BGI (BGI-Shenzhen, China).
Cell counting kit-8 (CCK-8) assay
A CCK-8 assay was conducted to assess the relative cell growth rate. The cells from the different experimental groups were seeded at a density of 2 × 103 cells per well in 96-well plates containing 10% FBS and incubated at 37 °C. To evaluate cell proliferation, 10 μl of CCK-8 solution (APExBIO, USA) was added to each well at 0, 24, 48, 72, and 96 h of incubation. The plates were subsequently incubated for 2 h at 37 °C. The number of viable cells was determined by measuring the absorbance at 450 nm via a microplate reader.
Colony formation assay
A colony formation assay was used to determine cell viability in vitro. Briefly, 2 × 103 cells were seeded in a six-well plate. After 10 days of growth, the cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Finally, the colonies were washed with PBS and counted via ImageJ software.
Transwell assay
The transwell assay is an in vitro assay used to study the ability of cells to migrate and invade. The transfected cells, at a density of 1 × 105 per well, were seeded into the upper chamber (Corning, 353097) containing 200 μl of serum-free medium. The lower chamber of a 24-well plate, which was equipped with an 8 μm pore polycarbonate membrane either coated with Matrigel or uncoated (1:5, BD Biosciences), was filled with 600 μl of medium supplemented with 10% FBS. After 36 h of incubation, the cells remaining in the upper chamber were removed with a cotton swab. The migrated or invaded cells in the lower chamber were then fixed with 4% paraformaldehyde, stained with 0.5% crystal violet, and counted under a microscope (Nikon, Japan).
Cell cycle assay
The transfected cells were harvested by centrifugation at 1000 rpm for 5 min and fixed in 70% ethanol at 4 °C overnight. The next day, the cells were washed three times with PBS and stained with propidium iodide (PI) (DOJINDO, C543) and RNase solution for 30 min at room temperature in the dark. The cell cycle distribution was subsequently assessed via flow cytometry (Beckman, USA) and analyzed via ModFit LT software.
scRNA-seq
Raw reads in the fastq files were processed in the Cell Ranger Software Suite (10x Genomics Cell Ranger 4.0.0) to map reads on the human genome (GRCh38/hg38) and generate the unique molecular identifier (UMI) matrices. The raw gene expression matrices for each experimental condition were imported into R software (version 3.6.3) via the Seurat R package (version 3.2.2). We excluded cells with the following criteria: fewer than 200 unique genes expressed, more than 6000 unique genes expressed, or more than 25% of reads mapped to mitochondria. After quality filtering, 29,331 cells from three subjects were selected for the following analysis. The top 30 principal components were selected for UMAP construction. Harmony was employed to remove batch effects between different datasets. Clusters of the same cell type were selected for re-UMAP analysis, graph-based clustering, and marker analysis to identify specific cell types. Neighboring clusters with high similarity were merged.
PAS-seq
The RNA libraries were constructed via the QuantSeq 3′ RNA-Seq Library Prep Kit (Lexogen, 016.24) according to the manufacturer′s protocol. Briefly, 500 ng of total RNA was used for first-strand cDNA synthesis in a PCR plate via oligodT primers containing Illumina-compatible sequences. Briefly, first-strand cDNA was synthesized from 500 ng of total RNA in a PCR plate via oligo-dT primers containing Illumina-compatible sequences. Second-strand cDNA was synthesized via random primers with Illumina-compatible adapter sequences at their 5′ ends, and the library was subsequently converted into double-stranded DNA. The library was purified via magnetic beads and amplified with complete adapter sequences and unique multiple indices. The final library was purified again from the PCR products. The library was subsequently sent to Annoroad Gene Technology (Beijing, China) for sequencing and analysis. Trimmed reads were mapped to the human genome (GRCh38.p5) using the Bowtie2 aligner. We evaluated differential PAS usage with PolyA miner to identify changes in APA and variations in transcript length49.
RIP-seq
RIP-seq is a technique used to study the binding of RNA to proteins in cells. The cells were lysed in IP buffer supplemented with DTT, PIC, and 40 U/µl RNase inhibitor (Invitrogen, EO0382), and 5% of the supernatant was collected as input samples. Anti-IGF2BP3 antibody (Proteintech, 14642-1-AP), anti-NUDT21 antibody (Proteintech, 10322-1-AP), or rabbit IgG was added to the lysate, which was subsequently incubated overnight at 4 °C for RIP. On the subsequent day, 50 μl of washed G-protein beads (Invitrogen, 10004D) were added to the mixture, which was subsequently rotated for 4 h at 4 °C. After washing 3 times, 10% of the IP samples were collected for quality control via Western blot. Co-precipitated RNA and input RNA within the beads were extracted with TRIzol reagent (Invitrogen), and isopropanol was precipitated with glycogen (Invitrogen). For sequencing, rRNA was removed via the NEBNext rRNA depletion kit (New England Biolabs). Library construction and sequencing were performed by Novogene Bioinformatics Technology Co., Ltd. Data analysis was performed in R v4.3.0. Precipitated RNAs were identified via the DESeq2 package with the criteria of a P value < 0.05 and a log2-fold change > 1.5.
meRIP-seq
OVCAR3 cells were prepared for methylated RNA immunoprecipitation (meRIP). Sequencing was performed by LC Bio-Technology (Hangzhou, China). Briefly, methylated RNA was precipitated with a Dynabeads Antibody Coupling Kit (Thermo Fisher) and m6A antibody in IP buffer (50 mM Tris–HCl, 750 mM NaCl, and 0.5% Igepal CA-630). Input and m6A IP samples were reverse-transcribed via SuperScriptTM II reverse transcriptase (Invitrogen, 1896649) and purified with AMPure XP beads. Following treatment of U-labeled second-strand DNA with heat-labeled UDG enzyme (NEB, M0280), the ligated products were amplified via PCR. Finally, paired-end sequencing (PE150) was conducted on the Illumina NovaSeq™ 6000 platform following the supplier′s recommended protocol. Data analysis was performed in the Python and R programming environments.
Dual-luciferase reporter assays
Wild-type (WT) and mutant SPTBN1 constructs targeting the m6A sites in intron 32 of 35 were generated. The mutant plasmid contained base substitutions at positions 55 (A>C), 81 (A>C), and 4046 (A>G) of SPTBN1 intron 32. Both the WT and mutant constructs were cloned and inserted into pMIR-Reporter luciferase vectors. The cells were cotransfected with either WT or mutant plasmids and control or IGF2BP3-expressing vectors via jetPRIME reagent (Polyplus Transfection). After 48 h, luciferase activity was measured with the Dual-Luciferase Reporter Assay Kit (TransGen, China) and normalized to Renilla luciferase activity to account for the transfection efficiency. Statistical analyses were performed to compare differences in luciferase activity between the experimental groups.
In vivo growth and metastasis assays
All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of Chongqing Medical University (Approval No. IACUC-CQMU-2023-0153). We have complied with all relevant ethical regulations for animal use. The female BALB/c-nude mice (3–4 weeks old) were purchased from Tenxin Co., Ltd. (Chongqing, China), and all the mice were randomly divided into treatment or control groups. For the tumor growth assay, the mice were injected subcutaneously with 5 × 106 OVCAR3 cells in 100 µl of PBS. Tumor sizes were measured via calipers, and the tumor volume was calculated via the formula (L × W2)/2. The final tumor sizes did not exceed the permissible limits set by the Institutional Animal Care and Use Committee. When the tumors reached a volume of 1000 mm3, the mice were sacrificed, and the tumors were collected, weighed, and analyzed. IHC and HE staining were performed on the xenograft tumors. In a separate experiment, a peritoneal metastasis model of OC was established via the intraperitoneal injection of 1 × 106 transfected OVCAR3 cells in 300 µl of PBS. Four weeks later, all the mice were necropsied and dissected, and the metastatic tumors were collected, counted, weighed, and analyzed.
Statistics and reproducibility
All the statistical analyses were conducted via GraphPad Prism 6. The data are presented as the means ± SDs from independent experiments. The results were analyzed using the χ2 test, Student′s t-test, and one-way analysis of variance (ANOVA) where appropriate. Cumulative survival was analyzed via the Kaplan‒Meier method with the optimal cutoff point. A P-value of <0.05 was considered statistically significant for all tests. All statistical analysis data can be found in Supplementary Data 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw sequencing data has been deposited in the NCBI SRA database (SRA accession: PRJNA1247180). The uncropped and unedited blot images are contained within the Supplementary Information, and the numerical source data can be found in the Supplementary Data files. Datasets associated with this work can be accessed from the corresponding author upon reasonable request.
References
Yokoi, A. et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 9, 4319 (2018).
Lo Riso, P. et al. A cell-of-origin epigenetic tracer reveals clinically distinct subtypes of high-grade serous ovarian cancer. Genome Med. 12, 94 (2020).
Levin, M. et al. Gene expression dynamics are a proxy for selective pressures on alternatively polyadenylated isoforms. Nucleic Acids Res. 48, 5926–5938 (2020).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017).
Sun, Y., Hamilton, K. & Tong, L. Recent molecular insights into canonical pre-mRNA 3′-end processing. Transcription 11, 83–96 (2020).
Ding, J. et al. The role of CSTF2 in cancer: from technology to clinical application. Cell Cycle 22, 2622–2636 (2023).
Guo, Q. et al. An alternatively spliced p62 isoform confers resistance to chemotherapy in breast cancer. Cancer Res. 82, 4001–4015 (2022).
Kang, W., Yang, Y., Chen, C. & Yu, C. CPSF1 positively regulates NSDHL by alternative polyadenylation and promotes gastric cancer progression. Am. J. Cancer Res. 12, 4566–4583 (2022).
Lin, P. et al. RBBP6 maintains glioblastoma stem cells through CPSF3-dependent alternative polyadenylation. Cell Discov. 10, 32 (2024).
Lin, S. C. et al. Un-methylation of NUDT21 represses docosahexaenoic acid biosynthesis contributing to enzalutamide resistance in prostate cancer. Drug Resist. Updat. 77, 101144 (2024).
Tan, S. et al. CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 40, 85 (2021).
Tan, Y. et al. Alternative polyadenylation reprogramming of MORC2 induced by NUDT21 loss promotes KIRC carcinogenesis. JCI Insight 8, e162893 (2023).
Zhang, N. et al. LINC00921 reduces lung cancer radiosensitivity by destabilizing NUDT21 and driving aberrant MED23 alternative polyadenylation. Cell Rep. 42, 113479 (2023).
Zhang, Y. et al. CPSF3 regulates alternative polyadenylation of CNIH2 to promote esophageal squamous cell carcinoma progression. Cancer Lett. 593, 216925 (2024).
Zhou, Y. et al. Nudt21-mediated alternative polyadenylation of MZT1 3′UTR contributes to pancreatic cancer progression. iScience 27, 108822 (2024).
Zhu, C. et al. CPSF6-mediated XBP1 3′UTR shortening attenuates cisplatin-induced ER stress and elevates chemo-resistance in lung adenocarcinoma. Drug Resist. Updat. 68, 100933 (2023).
Luo, X. et al. CSTF3 contributes to platinum resistance in ovarian cancer through alternative polyadenylation of lncRNA NEAT1 and generating the short isoform NEAT1_1. Cell Death Dis. 15, 432 (2024).
Shen, S. et al. Comprehensive analyses of m6A regulators and interactive coding and non-coding RNAs across 32 cancer types. Mol. Cancer 20, 67 (2021).
Tang, P. et al. Nuclear retention coupled with sequential polyadenylation dictates post-transcriptional m(6)A modification in the nucleus. Mol. Cell 84, 3758–3774.e10 (2024).
Chen, L. et al. Nuclear m(6) A reader YTHDC1 suppresses proximal alternative polyadenylation sites by interfering with the 3′ processing machinery. EMBO Rep. 23, e54686 (2022).
Yuan, S. et al. YTHDC1 as a tumor progression suppressor through modulating FSP1-dependent ferroptosis suppression in lung cancer. Cell Death Differ. 30, 2477–2490 (2023).
Zheng, Y. et al. CSTF2 mediated mRNA N(6)-methyladenosine modification drives pancreatic ductal adenocarcinoma m(6)A subtypes. Nat. Commun. 14, 6334 (2023).
Gallicchio, L., Olivares, G. H., Berry, C. W. & Fuller, M. T. Regulation and function of alternative polyadenylation in development and differentiation. RNA Biol. 20, 908–925 (2023).
Yuan, F. et al. Alternative polyadenylation of mRNA and its role in cancer. Genes Dis. 8, 61–72 (2021).
Xing, Y. et al. Downregulation of NUDT21 contributes to cervical cancer progression through alternative polyadenylation. Oncogene 40, 2051–2064 (2021).
Xiong, M. et al. NUDT21 inhibits bladder cancer progression through ANXA2 and LIMK2 by alternative polyadenylation. Theranostics 9, 7156–7167 (2019).
Dettwiler, S. et al. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 279, 35788–35797 (2004).
Yue, Y. et al. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10 (2018).
de Prisco, N. et al. Alternative polyadenylation alters protein dosage by switching between intronic and 3′UTR sites. Sci. Adv. 9, eade4814 (2023).
Jiang, X. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6, 74 (2021).
Hayes, N. V. et al. Identification of a novel C-terminal variant of beta II spectrin: two isoforms of beta II spectrin have distinct intracellular locations and activities. J. Cell Sci. 113, 2023–2034 (2000).
Bergman, M. T. et al. Binding modalities and phase-specific regulation of cyclin/cyclin-dependent kinase complexes in the cell cycle. J. Phys. Chem. B 128, 9315–9326 (2024).
Jones, M. J. & Jones, M. C. Cell cycle control by cell–matrix interactions. Curr. Opin. Cell Biol. 86, 102288 (2024).
Boreikaitė, V. & Passmore, L. A. 3′-End processing of eukaryotic mRNA: machinery, regulation, and impact on gene expression. Annu. Rev. Biochem. 92, 199–225 (2023).
Mitschka, S. & Mayr, C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 23, 779–796 (2022).
Brumbaugh, J. et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell 172, 106–120.e121 (2018).
Jafari Najaf Abadi, M. H. et al. CFIm25 and alternative polyadenylation: conflicting roles in cancer. Cancer Lett. 459, 112–121 (2019).
Xiao, S. et al. Relationship between NUDT21 mediated alternative polyadenylation process and tumor. Front. Oncol. 13, 1052012 (2023).
Tan, S. et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene 37, 4887–4900 (2018).
Gao, C. C. et al. NUDT21 suppresses the growth of small cell lung cancer by modulating GLS1 splicing. Biochem. Biophys. Res. Commun. 526, 431–438 (2020).
Liu, K. et al. CFIm25-regulated lncRNA acv3UTR promotes gastric tumorigenesis via miR-590-5p/YAP1 axis. Oncogene 39, 3075–3088 (2020).
Wang, J., Zheng, F., Wang, D. & Yang, Q. Regulation of ULK1 by WTAP/IGF2BP3 axis enhances mitophagy and progression in epithelial ovarian cancer. Cell Death Dis. 15, 97 (2024).
Chan, J. J., Tabatabaeian, H. & Tay, Y. 3′UTR heterogeneity and cancer progression. Trends Cell Biol. 33, 568–582 (2023).
Dai, L. P. et al. SPTBN1 attenuates rheumatoid arthritis synovial cell proliferation, invasion, migration and inflammatory response by binding to PIK3R2. Immun. Inflamm. Dis. 10, e724 (2022).
Rao, S. et al. β2-spectrin (SPTBN1) as a therapeutic target for diet-induced liver disease and preventing cancer development. Sci. Transl. Med. 13, eabk2267 (2021).
Cousin, M. A. et al. Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nat. Genet. 53, 1006–1021 (2021).
Yang, P. et al. βII spectrin (SPTBN1): biological function and clinical potential in cancer and other diseases. Int. J. Biol. Sci. 17, 32–49 (2021).
Chen, S., Li, J., Zhou, P. & Zhi, X. SPTBN1 and cancer, which links?. J. Cell Physiol. 235, 17–25 (2020).
Yalamanchili, H. K. et al. PolyA-miner: accurate assessment of differential alternative poly-adenylation from 3′Seq data using vector projections and non-negative matrix factorization. Nucleic Acids Res. 48, e69 (2020).
Acknowledgements
This work was funded by the National Natural Science Foundation of China [82172778]; the Chongqing Graduate Research Innovation Project [CYS23375] and the Incubation project with Third Affiliated Hospital of Chongqing Medical University [KY23039].
Author information
Authors and Affiliations
Contributions
Conception and design: X.L., P.Y., and J.X. Development of methodology: X.L., L.C.X., N.X.C., J.N.X., Y.Y., T.L., P.Y., and J.X. Acquisition of data: L.C.X., N.X.C., X.Y.J., X.Y.L., and X.H.M. Analysis and interpretation of data: X.L., L.C.X., N.X.C., and J.N.X. Writing, review, and/or revision of the manuscript: X.L., L.C.X., and J.X.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The research was conducted in accordance with the principles and guidelines of the Ethics Committee of The Third Affiliated Hospital of Chongqing Medical University. All animal experiments were approved by the Institutional Animal Care and Use Committee of Chongqing Medical University. The authors confirm that all methods were performed in accordance with the relevant guidelines and regulations.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Derrick Ong and Johannes Stortz.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, X., Xie, L., Ma, X. et al. IGF2BP3 recruits NUDT21 to regulate SPTBN1 alternative polyadenylation and drive ovarian cancer progression. Commun Biol 8, 680 (2025). https://doi.org/10.1038/s42003-025-08097-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08097-6