Abstract
Hepatitis B virus (HBV) integration is strongly associated with hepatocellular carcinoma (HCC). However, the genetic alterations and pathogenesis mechanisms remain significantly unexplored, especially for HBV-inserted cancer-related genes. This study identified recurrent HBV DNA integration into the CCDC91 gene in HCC tissues (n = 17) using pooled analysis and HBV capture sequencing. CCDC91 expression was positively correlated with HBV DNA presence, and higher levels were linked to shorter overall survival in HCC patients. CCDC91 was upregulated in HCC and promoted HCC malignancy in vitro and in vivo. CCDC91 deficiency increased sensitivity to sorafenib treatment. By RNA sequencing and co-immunoprecipitation assays, we further demonstrated that the Ct-HBx upregulated CCDC91 and that CCDC91 induced aerobic glycolysis by activating LDHA to drive HCC progression. In conclusion, the HBV integrated gene CCDC91 is a novel HCC-related gene that functions through the Ct-HBx/CCDC91/LDHA axis. Our work sheds light on the mechanism in driving HCC progression and sorafenib resistance.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) was the sixth most common cancer worldwide in 2020, with approximately half of all new cases originating in China1. While surgical resection and liver transplantation are viable treatments for early-stage HCC, most patients eventually succumb to tumor recurrence and metastasis2. HCC is the third deadliest malignancy globally, and it surpasses other types of cancer in terms of mortality for men and women. Thus, preventing and curing this disease is highly challenging.
The etiology of HCC is complex and multifactorial, with chronic hepatitis B virus (HBV) infection being a primary cause, particularly in China3. Several major molecular mechanisms of hepatocarcinogenesis are caused by HBV infection. Recently, increasing evidence has indicated that HBV DNA integration plays a crucial role in the development of HCC4,5,6,7. One significant consequence of HBV DNA integration into the host genome is the truncation of the HBV genome, especially at the C-terminus of HBx (Ct-HBx). HBx functions as a transcriptional transactivator that is involved in the progression of HBV-related HCC8. Previous studies have shown that the integrated HBx gene can regulate WNT-5a expression through microRNA modifications9,10, which lead to increased cell proliferation in vitro and in vivo11. Recently, Zhang et al. reported that the Ct-HBx could mediate metabolic reprogramming from mitochondrial respiration to aerobic glycolysis via the NFACT2-TXNIP signaling pathway in hepatocarcinogenesis12. On the other hand, HBV DNA integration can cause genomic instability and insertional mutagenesis of various cancer-related genes. For instance, the human telomerase reverse transcriptase (TERT) gene is the most common site for HBV DNA integration in HCC. Li et al. demonstrated that the recurrence of HBV DNA integration in the TERT promoter region in HCC can activate TERT transcription in response to sex hormones13. Moreover, Sze et al. revealed that HBV integration at the TERT promoter was associated with increased TERT mRNA expression and more aggressive tumor behavior. They also demonstrated an ELF4-HBx-TERT promoter signal axis in hepatocarcinogenesis14. In addition to TERT, many HCC cases are therefore expected to have HBV integration in cyclin E1 (CCNE1). Doxycycline-induced expression of Ccnel in mice has demonstrated that Cyclin E1 overexpression leads to liver-specific molecular and cellular defects15. Because one third of HBV preferential target genes are cancer-related genes, it is believed that insertional mutagenesis may play a role in HCC development, and this proposal requires further exploration.
Our previous study revealed that the integration of viral DNA into the host genome was an important mechanism for HBV-related carcinogenesis in HCC and in non-Hodgkin lymphoma (NHL)16. Notably, the gene coiled-coil domain-containing protein 91 (CCDC91) was identified as a recurrent target of HBV DNA integration in HCC and NHL samples. The coiled-coil structural domain (CCD), consisting of 2–5 intertwined α-helices, is widely expressed in a variety of proteins, regulates cell motility, and is involved in cell signaling. Recently, abnormal activation of CCD-containing proteins has been observed in several tumors17, such as CCDC17818 and CCDC3419 in HCC, CCDC88A in pancreatic cancer20, and CCDC821 in lung cancer. In the field of cancer research, CCDC91 has not been studied as extensively as other CCD-containing proteins. CCDC91 is a 441 amino- acid polypeptide that belongs to the superfamily of coiled-coil domain-containing proteins and is involved in Golgi-to-lysosome transport. Variants at 12p11, including CCDC91, have been associated with a greater risk of breast cancer22. In NHL, we found that patients with high expression of CCDC91 had poor survival, with borderline significance16. However, the role of CCDC91 in HCC has not been studied.
Metabolic reprogramming affects the occurrence and development of various cancers, including HCC23. Intriguingly, in cancer cells, despite the presence of oxygen, a preference for glycolysis (Warburg effect) often results in increased lactate production and rapid adenosine triphosphates (ATP) generation. ATP and many molecules required for carcinogenesis are synthesized in cancer cells to regulate the tumor microenvironment, maintain cancer biology, and resist antitumor therapy. In this study, we identified aerobic glycolysis, particularly lactate dehydrogenase A (LDHA), as a downstream target of CCDC91 in regulating HCC proliferation and metastasis. LDHA is a key enzyme in the glycolytic pathway, playing a critical role in cellular energy metabolism. It catalyzes the conversion of pyruvate to lactate, a process particularly vital under hypoxic conditions where oxidative phosphorylation is limited. LDHA is frequently overexpressed in various cancers, driving this glycolytic phenotype and contributing to tumor progression, invasion, and metastasis24,25,26,27. For instance, previous study has shown that the higher acetylation level of LDHA enhances the proliferation and metastasis of head and neck squamous carcinoma24. In HCC, LDHA was found to interact with FOXK2-142aa and activate its phosphorylation28. Notably, no investigations have focused on the regulatory mechanisms of HCC energy metabolism by CCDC91 and LDHA.
In the present study, we investigated the role of the HBV integrated gene CCDC91 in HCC. Our results revealed that CCDC91 promoted HCC growth, metastasis, and resistance to sorafenib by enhancing aerobic glycolysis. These findings suggest that CCDC91 may serve as a novel molecular target for HCC intervention.
Methods
Patients and samples
Paraffin-embedded of four HCC tissues were obtained from the First Affiliated Hospital of the University of Science and Technology of China (USTC) for HBV capture sequencing between June 2019 and January 2021. All included HCC samples were positive for HBV serum markers and negative for the spirochete that causes syphilis and for other viruses, such as hepatitis C virus and human immunodeficiency virus. The clinicopathologic features of the patients are shown in Table S1. The study was approved by the institutional ethics review committee of the First Affiliated Hospital of USTC and conducted according to the principles of the Declaration of Helsinki. The ethics committee waived the need for written informed consent because the study was conducted using residual samples from previous clinical diagnoses, the risks of the study did not exceed minimal risks, and the waiver of informed consent would not adversely affect the subjects. All ethical regulations relevant to human research participants were followed.
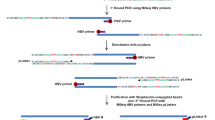
HBV capture sequencing
HBV capture sequencing was conducted by MyGenostics (Beijing, China). The detailed experimental procedure has been previously described29. Briefly, DNA from paraffin-embedded HCC tissues was extracted and prepared for sequencing. The capture sequencing targeted HBV DNA to identify integration sites within the host genome. The integration sites were identified via Clipping Reveals Structure (CREST) analysis. HBV breakpoints with a supporting read number ≥2 were regarded with high confidence as HBV integration sites. The University of California Santa Cruz (UCSC) Genome Browser (GRCh37/hg19) was employed to annotate genes near the breakpoints.
Tissue microarray (TMA) and immunohistochemistry (IHC)
This study included 123 HCC tumor tissues (all pathologically confirmed) for immunohistochemical staining, derived from two sources: 80 matched pairs of tumor and corresponding paratumor tissues (i.e., 80 tumor + 80 paratumor samples) and 43 additional non-matched HCC tumor tissues (without paired paratumor samples). Those tissues were obtained by the Shanghai National Engineering Research Center from Taizhou Hospital from 2010 to 2014 (approved by the Research Ethics Committee of Taizhou Hospital). All ethical regulations relevant to human research participants were followed. Comprehensive CCDC91 expression and survival analyses were performed on all 123 HCC cases. LDHA was specifically evaluated in 80 matched tumor pairs. In addition, the tumor tissues from the orthotopic xenografts and human HCC samples in this study were resected, fixed in 4% phosphate-paraformaldehyde (for at least 72 h), embedded in paraffin and sectioned at 5 μm. The IHC procedures included deparaffinization, rehydration, antigen retrieval, and blocking as reported previously30. Sections were incubated with primary antibodies at 4 °C overnight and then treated with secondary antibodies for 30 min at room temperature. The antibodies used for IHC targeted CCDC91 (1:200, sc-514452, Santa Cruz Biotechnology, Inc., U.S.A.), LDHA (1:200, 380954, Zenbio, Beijing, China), and ki-67 (1:200, 27309-1-AP, Proteintech, Rosemont, IL, USA). The intensity of the staining was evaluated as follows: negative (0 points), weak (1 point), moderate (2 points), and strongly positive (3 points). The percentage of positive tumor cells was scored as 0 (<5%), 1 (<25%), 2 (25%-50%), 3 (51%-75%), and 4 (>75%). The final scores (0 to 12) were based on the percentage of positive cells and the staining intensity.
Cell lines and cell cultures
The human HCC cell lines SNU449, Huh-7, HepG2, and PLC/PRF/5, and normal liver cell line THLE-2 were obtained from the American Type Culture Collection (ATCC, United States). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Huh-7, HepG2, and PLC/PRF/5) or RPMI-1640 (SNU449) supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 at 37 °C. Cell lines were authenticated by Biowing Applied Biotechnology Co., Ltd. (Shanghai, China), and short tandem repeat (STR) profiling was used to confirm their identity. For functional inhibition experiments, the indicated HCC cells were treated with sorafenib (Meilunbio, Dalian, China) at a final concentration of 8 μΜ31. The cells were incubated with sorafenib under the same culture conditions for the duration of the experiments.
Gene overexpression and knockdown
Full-length cDNA encoding human CCDC91 was amplified via PCR and subsequently cloned and inserted into the pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro lentiviral vector system. The primers used for constructing the CCDC91 overexpression vector were as follows: CCDC91-F: 5’- TGACCTCCATAGAAGATtctaga; and CCDC91-R: 5’-gaattcGAATTTAAATCGGATCCG. The sequences of the short hairpin RNAs (shRNAs) used for CCDC91 knockdown were as follows: shCCDC91-1: 5’-GAAGAGCAGAAACGAAGUGAATT; shCCDC91-2: 5’-UUCACUUCGUUUCUGCUCUUCTT. Plasmids for lentiviral packaging were transfected into 293 T cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). HCC cells grown at 60%-70% confluence were infected with the viral particle mixture. Short interfering RNA (siRNA) oligonucleotides targeting CCDC91 and LDHA were designed and synthesized by RiboBio (Guangzhou, China). The siRNA target sequences for CCDC91 were as follows: siCCDC91-1: 5’-CAACAUCUCCUGCUAUUCCUUTT and 5’-AAGGAAUAGCAGGAGAUGUUGTT; siCCDC91 -2: 5’-GAAGAGCAGAAACGAAGUGAATT and 5’-UUCACUUCGUUUCUGCUCUUCTT; and siCCDC91-3: 5’-GGCUCCUUGAAAUGCUAGAUATT and 5’-UAUCUAGCAUUUCAAGGAGCCTT. The siRNA target sequences for LDHA were as follows: siLDHA-1: 5’-CCUUAGAACACCAAAGAUUTT and 5’-AAUCUUUGGUGUUCUAAGGTT; siLDHA-2: 5’-GCACCCAGAUUUAGGGACUTT and 5’-AGUCCCUAAAUCUGGGUGCTT; and siLDHA-3: 5’- GAUUACAGUUGUUGGGGUUTT and 5’- AACCCCAACAACUGUAAUCTT. Furthermore, the 3-end deleted X genes (encoding HBx-180, Ct-HBx-180) were cloned and transduced into HepG2 cells according to the manufacturer’s instructions. Transfection efficiency and gene expression levels were confirmed via qPCR and Western blot analysis.
CCK8 assay
Cell proliferation was measured with a Cell Counting Kit-8 (CCK8, Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. Equal numbers of HCC cells were seeded into 96-well culture plates and incubated for 3–5 days. Incubate the 96-well plates for 2 h in the incubator and measure the absorbance at 450 nm using a microplate reader (Tecan, Switzerland). Each experiment was performed in triplicate.
Cell migration and invasion assays
Cell migration or invasion was measured using Transwell inserts with an 8 μm pore size culture membrane (Corning, Ithaca, NY, USA) covered without or with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) before use. For cell migration, equal numbers of HCC cells were resuspended in serum-free medium, and 200 μL of cell suspension was added to the upper compartment and incubated at 37 °C, with 700 μL of medium containing 10% FBS in the lower chamber. For cell invasion, cells in a 200 μL, serum-free culture medium was added to the compartment with a Matrigel-coated membrane diluted with serum-free culture medium. The cells were stained and counted after being cultured for 8-24 h.
Wound-healing assays
For the cell motility assay, cells were seeded in 12-well plates to reach 90%-95% confluence. A single scratch wound was created with a 20 μL pipette tip. The cell debris was removed by washing with PBS and replaced with culture medium. The samples were immediately photographed at a suitable time after wounding.
Animal experiments
To construct a subcutaneous HCC model, CCDC91-overexpresion and -knockdown and control HCC cells (3 × 106) were injected into the right flanks of five-week-old male BALB/c nude mice (n = 5 per group). Sample sizes for animal studies were determined based on established laboratory experience with this xenograft model, with n = 5 mice per group providing consistent detection of biologically significant effects in prior experiments. All health mice were sourced from Shanghai SLAC Laboratory Animal Co. Ltd and randomly allocated to groups using the drawing lots. Cage location and order of procedures were balanced across groups to minimize environmental bias. All animals were maintained in specific-pathogen-free conditions. The tumor was excised prior to reaching a maximum volume of 2000 mm³. Tumor volumes were measured at the indicated time intervals, and the primary tumors were collected and weighed.
To determine the antitumor effect of combined treatment with shCCDC91 and sorafenib, CCDC91 -knockdown and control HCC cells were injected subcutaneously into the right flank of nude mice. The mice were randomly assigned to receive either sorafenib or control treatment (n = 5 per group). Vehicle and sorafenib (20 mg/kg/d) were given intraperitoneally five days per week. Tumor volumes were measured at the indicated time intervals. On day 28 of sorafenib treatment, the mice were sacrificed by cervical dislocation under deep anesthesia with CO2, and the primary tumors were collected and weighed.
All experiments involving mice were approved by the Animal Care and Use Committees of the First Affiliated Hospital of USTC, and were performed in accordance with the Institutional Animal Welfare Guidelines. We have complied with all relevant ethical regulations for animal use.
Co-immunoprecipitation
The co-immunoprecipitation (co-IP) experiments were performed on HepG2 and PLC/PRF/5 cells via the Pierce® Classic Magnetic IP/Co-IP Kit (Thermo Fisher, Waltham, MA, USA). Briefly, the cells were harvested using IP lysis buffer and lysed for 5 min. To reduce nonspecific binding, the total lysate was incubated with 25 μl of Pierce Protein A/G Magnetic Beads for 1 h. Primary antibody (anti-CCDC91, sc-514452,1:200, Santa Cruz Biotechnology; anti-LDHA,1:200, 380954, Zenbio; anti-HBx antibody,1:200, bs-2147R, Bioss, Beijing, China) was added at 4 °C overnight. The supernatant was then subjected to Western blot analysis.
Immunofluorescent confocal imaging
Cells cultured on coverslips were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 for 10 min. After being washed 3 times with PBS, the cells were blocked with 5% BSA for 30 min and incubated with primary antibodies at 4 °C overnight. The glass slides were subsequently incubated with secondary antibodies (anti-CCDC91, Santa Cruz Biotechnology, 1:100, sc-514452; anti-HBx, 1:200, 22741, Genetex, Alton Pkwy Irvine, CA, USA; anti-LDHA, 380954, Zenbio, 1:200) for 30 min in the dark. For nuclear staining, cells were incubated with DAPI (4’,6-diamidino-2-phenylindole; Servicebio, Wuhan, China) at a concentration of 5 μg/ml for 15 min at room temperature. Images of the cells were captured with a Leica TCS SP8 confocal system.
Glucose and lactate assays
A glucose assay kit (Beyotime, Shanghai, China) and a lactate assay kit (Solarbio, Beijing, China) were used to determine glucose uptake and lactate production, respectively, according to the manufacturer’s protocols. For the glucose uptake colorimetric assay, cells were seeded at a density of 5 × 106 cells per well in a 6-well plate and starved by preincubating with 100 μL of Cell and Tissue Lysis Buffer (Beyotime). Dilute the Glucose standard (200 mg/ml) provided in this kit with distilled water to the required concentrations for the standard curve: 0, 5, 10, 20, 50, and 100 mg/dl. Add 5 μL of sample or standard to 96-well PCR plates and then add Glucose Assay Reagent (Beyotime) to a final volume of 190 μL. Mix by vortex and centrifuge briefly and place the tubes in a thermal cycler. Heat at 95 °C for 8 min and then cool down to 4 °C. For the lactate assay, 5 × 106 cells were homogenized in 100 μL of the assay buffer provided by the lactate assay kit, followed by centrifugation, and the supernatant was analyzed. The lactate standard (100 μmol/mL) provided in this kit with distilled water to the required concentrations for the standard curve: 0, 0.078, 0.15625, 0.3125, 0.625, 1.25, and 2.5 mg/dl. Absorbance was measured at 630 nm for the glucose assay and 570 nm for the lactate assay. For both assays, the protein concentration of the cells was measured to normalize the glucose and lactate levels.
Western blot
To analyze gene expression at the protein level, the cells were lysed in cold Radioimmunoprecipitation Assay (RIPA) buffer freshly supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). The protein concentrations were measured with a Pierce BCA protein assay kit (Thermo Fisher). The quantification of proteins was performed to ensure consistent analysis, with 30 µg of total protein loaded per lane for subsequent Western blotting. Whole-cell protein lysates were separated by SDS‒PAGE and then transferred to a PVDF membrane. The membrane was blocked with 5% nonfat milk and then incubated with one of the following primary antibodies: anti-CCDC91 (1:1000, Abcam, ab227558, Cambridge, MA, USA), anti-HBx (1:500, Genetex, 22741), or anti-LDHA (1:1000, Zenbio (380954 and R380954)). Equal protein sample loading was confirmed via an anti-β-actin antibody (1:5000, 66009-1-Ig, Proteintech). All procedures strictly followed the kit manufacturer’s protocol.
Quantitative real-time PCR (qPCR)
To detect gene expression at the mRNA level, total RNA samples were isolated from the HCC cell lines in this study with TRIzol reagent (Thermo Fisher) according to the manufacturer’s protocol. We used a FastStart Universal SYBR Green kit (Roche, Indianapolis, IN, Switzerland) on StepOne Plus Real-Time PCR System (Applied Biosystems) to preform quantitative real-time PCR (qPCR). Gene-specific primers were used at a final concentration of 10 µM each. The primers used are listed in Table S2. qPCR was performed using the following cycling conditions: initial denaturation at 95 °C for 30 s; 40 cycles of denaturation at 95 °C for 5 s and annealing/extension at 60 °C for 30 seconds with fluorescence acquisition; melting curve analysis from 60 to 95 °C with 0.5 °C increments; and hold at 4 °C. The relative expression of the target genes was normalized to that of β-actin.
RNA-seq and bioinformatics analysis
RNA-seq library preparation and sequencing of CCDC91-overexpressing and control HepG2 cells were performed on an Illumina NovaSeq 6000 platform. Clean reads were aligned to the human genome hg38. The expression of the transcript was calculated by fragments per kilobase of exon model per million mapped reads (FPKM) using Perl. The filter criteria of p ≤ 0.05 and a fold change ≥2 were used to determine the differential expression of genes. We used a web-accessible functional annotation tool from the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8 (https://david.abcc.ncifcrf.gov) to analyze the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment for differentially expressed genes. The transcript profiles from this study were uploaded to the NCBI database under accession number PRJNA1138405.
Public database
The mRNA expression of CCDC91 in HCC tissues compared with noncancerous tissues was analyzed using data from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) and the Gene Expression Omnibus (GEO, https:// www.ncbi.nlm.nih.gov/geo/). Gene set enrichment analysis (GSEA) is a computational approach to determine if a pre-defined gene set can show a significant consistent difference between two biological states. We use the local version of the GSEA analysis tool http://www.broadinstitute.org/gsea/index.jsp, Hallmark data set were used for GSEA independently. The overall survival (OS) curves of HCC patients from the TCGA dataset were generated by the online survival analysis tool Kaplan‒Meier plotter (http://kmplot.com/analysis/). The CCDC91 high expression and low expression groups were classified using the “Auto select best cutoff” function. Survival curves were calculated using the Kaplan–Meier method and analyzed with a log-rank Mantel‒Cox test.
Statistics and reproducibility
The significance of differences between groups was assessed with a Student’s t test (for normally distributed variables) or a Wilcoxon rank sum test (for nonnormally distributed variables). The experiments were conducted in triplicate. The results of the statistical analyses are presented as the mean ± standard error of the mean (SEM). Correlations between CCDC91 or LDHA expression levels and clinicopathological features were analyzed via multivariate logistic regression models. The independent effects of CCDC91 or LDHA expression on OS or progression-free survival were evaluated using multivariate Cox regression models from the TCGA dataset and our IHC cohort dataset. The level of significance was set at 0.05 for all the analyses. Statistical analyses were performed using SPSS software version 22.0 (SPSS Inc., United States) or GraphPad Prism (GraphPad Software, United States).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
CCDC91 is upregulated in HCC tissues and is correlated with a poor prognosis
Our previous research revealed that the gene CCDC91 was recurrently inserted by HBV DNA in HCC (n = 16) and NHL (n = 1) samples (Fig. 1A). Recurrent insertion was identified using pooled analysis and HBV capture sequencing, respectively, suggesting that CCDC91 may share some common mechanisms in driving cancer initiation and progression. In this study, we further obtained HBV capture sequencing data from four HCC patients positive for HBV serum markers. A total of 454 viral‒host junctions were identified in those tissues (Supplementary Data 2). Interesting, we found that CCDC91 (with a distance of ~100 kb) was inserted by HBV DNA once in HCC tissue A-2 (Fig. 1A and Supplementary Data 2). Thus, a total of 17 HCC samples were identified with HBV integration around CCDC91 (Supplementary Data 3). Moreover, the protein level of CCDC91 in tissue A-2 was much greater than that in the other three HCC tissues (Fig. 1B), suggesting that HBV DNA integration may induce CCDC91 expression in HCC tissues.
A Map of the CCDC91 gene with HBV DNA insertion in HCC (n = 17) and NHL (n = 1) tissues. Black arrows indicate HBV integration sites identified through pooled-analysis in HCC. Red arrows represent HBV integration sites detected in HCC samples using our HBV capture sequencing approach. Blue arrows denote HBV integration sites identified in non-Hodgkin lymphoma (NHL) samples from our previous HBV capture sequencing study. B Immunohistochemistry staining showed the protein expression of CCDC91 in 4 HBV capture sequencing HCC tissues. C The mRNA levels of CCDC91 in HCC tissues and non-cancerous liver tissues from GSEA cohorts (GSE36376: n = 193 in non-tumor, n = 240 in tumor; GSE76427: n = 52 in non-tumor, n = 115 in tumor). D The mRNA levels of CCDC91 in HCC tissues and non-cancerous liver tissues from TCGA cohort (n = 50 in non-tumor, n = 371 in tumor). E Relative mRNA levels of CCDC91 in HCC cells as determined by qPCR (n = 3 independent biological repeats). F Representative IHC images of CCDC91 in HCC tissues and adjacent paratumor tissues (left) and IHC scores of CCDC91 staining in HCC tissues and non-cancerous liver tissues (right, n = 123). G The Kaplan–Meier analysis and log-rank test revealed the association of CCDC91 with the overall survival of HCC patients from TCGA dataset. H Multivariate logistic regression analyses for the associations of CCDC91 expression in HCC patients from TCGA dataset (n = 240). I Multivariate Cox regression analysis of the data from the TCGA dataset revealed that CCDC91 was an independent predictor of OS (n = 120). J The Kaplan–Meier analysis and log-rank test revealed the association of CCDC91 with the overall survival of HCC patients from TMA cohort (n = 123). K Multivariate logistic regression analyses for the associations of CCDC91 expression in HCC patients from TMA cohort (n = 80). Statistical analysis was performed using Student’s t test in (C, D, E, and F). Error bars represent the SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To determine the expression level of CCDC91 in HCC, we first analyzed data retrieved from the TCGA database, as well as microarray-based gene expression profiles (GSE36376 and GSE76427) from the GEO database (Fig. 1C, D). The results revealed that the CCDC91 mRNA level in HCC tissues was much higher than that in noncancerous liver tissues (p < 0.05, Fig. 1C, D). We also investigated the expression of CCDC91 in a panel of human HCC cell lines. CCDC91 expression was higher at the mRNA level (p < 0.05, Fig. 1E) in the 4 HCC cell lines. The protein expression of CCDC91 was subsequently examined in parallel via immunohistochemical staining of the TMA cohort. Consistently, the protein level of CCDC91 was markedly increased in HCC tissues compared with noncancerous tissues (p < 0.01, Fig. 1F).
To further explore the clinicopathological role of CCDC91 in HCC progression, we then analyzed data retrieved from the TCGA database and the Kaplan‒Meier survival analysis revealed that HCC patients with higher CCDC91 mRNA levels had shorter OS times than did patients with lower CCDC91 levels (p < 0.001, Fig. 1G). Multivariate logistic regression analyses for the TCGA cohort revealed that the CCDC91 mRNA level was positively correlated with TNM (tumor, lymph node, and metastasis) stage (p < 0.05, Fig. 1H and Table S3). Additionally, multivariate Cox regression analyses indicated that CCDC91 expression was an independent predictor of HCC aggressiveness (p < 0.05, Fig. 1I and Table S4). Based on our TMA cohort data, we also found that HCC patients with high CCDC91 protein expression had a significantly shorter OS time (p < 0.001, Fig. 1J). Interestingly, further multivariate logistic regression analysis revealed that CCDC91 expression in HCC tissues was positively associated with HBV DNA status (p < 0.05, Fig. 1K and Table S5). Collectively, these data suggest that CCDC91 plays a crucial role in HCC.
CCDC91 promotes the malignant behavior of HCC in vitro and in vivo
To investigate the functional role of CCDC91 in HCC, we chose HepG2 and PLC/PRF/5 cells for overexpression studies because they exhibit relatively low intrinsic expression of CCDC91, allowing us to more clearly observe the functional effects of CCDC91 upregulation. qPCR and Western blot confirmed the successful induction of CCDC91 overexpression in HCC cells (Fig. 2A, B). CCDC91 overexpression significantly increased the proliferation (p < 0.011; Fig. 2C), migration (p < 0.05; Fig. 2D–F), and invasion abilities (p < 0.01; Fig. 2G) of HepG2 and PLC/PRF/5 cells. Conversely, we selected Huh-7 and SNU449 cells for knockdown experiments due to their relatively high endogenous expression of CCDC91, which provides a robust model for studying the loss-of-function effects (Fig. S1). In contrast, knocking down CCDC91 significantly inhibited the proliferation, migration, and invasion of HCC cells (p < 0.05; Fig. 2H–L and Fig. S2).
A The mRNA level of CCDC91 in CCDC91-overexpressing HCC cells and control cells revealed by qPCR (n = 3). B The protein level of CCDC91 in CCDC91-overexpressing HCC cells and control cells detected by western blot. C Cell proliferation of CCDC91-overexpressing HCC cells and control cells detected by CCK8 assay (n = 3). D, E The migration abilities of CCDC91-overexpressing HCC cells and control cells measured by wound healing assay (n = 3). The migration (F) and invasion (G) abilities of CCDC91-overexpressing HCC cells and control cells measured by transwell assay (n = 3). H Cell proliferation of CCDC91 knockdown HCC cells and control cells detected by CCK8 assay (n = 3). I, J The migration abilities of CCDC91 knockdown HCC cells and control cells measured by wound healing assay (n = 3). K, L The migration and invasion abilities of CCDC91 knockdown HCC cells and control cells measured by transwell assay (n = 3). Statistical analysis was performed using Student’s t test in (A, C, E, F, G, H, J, and L). Data represents mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To investigate the in vivo role of CCDC91, Huh-7 cells infected with shCCDC91 or controls were subcutaneously inoculated into athymic nude mice and the growth of the tumors was monitored. Compared with control tumors, tumors derived from Huh-7-shCCDC91 cells grew significantly slower (p < 0.01; Fig. 3A). At 36 d postinoculation, consistent with the results of tumor growth, a significant decrease in tumor weight was observed in the CCDC91 knockdown group compared with the control group (0.656 ± 0.091 g vs. 0.466 ± 0.130 g, p < 0.05, Fig. 3B, C). Meanwhile, we also conducted in vivo experiments using subcutaneous mouse models of CCDC91 overexpressing HepG2 cells. Consistent with our CCDC91 knockdown findings in vivo, CCDC91 overexpression significantly increased the tumor burden in subcutaneous (0.133 ± 0.057 g vs. 0.377 ± 0.063 g, p < 0.05, Fig. 3D, E) mouse models of HCC. Ki-67 staining indicated that CCDC91 promoted cell growth (p < 0.01; Fig. 3F, G). Collectively, these findings suggest that CCDC91 plays a critical role in promoting the malignant behavior of HCC in vitro and in vivo.
A–C The subcutaneous tumor model formed by Huh-7-shCCDC91 and control Huh-7-SC cells in BALB/c nude mice (n = 5 per group). A The tumor growth curves based on tumor sizes measured at a 5-day interval. B Representative images of tumors at the time of sacrifice 36 days post tumor cells inoculation. C The tumor weights at the time of sacrifice. D, E The subcutaneous tumor model formed by HepG2-CCDC91 and control HepG2-VC cells in BALB/c nude mice. Representative images were shown (D) and the tumors with xenografts were weighted (E) (n = 5, respectively). F Representative images of IHC staining of CCDC91 and Ki-67 in the HCC tumors. G Quantification of the percentage of Ki-67-positive cells and CCDC91-positive cells. Statistical analysis was performed using Student’s t test in (C, E, and G). Two-way ANOVA was used for tumor volume in (A). Data represents mean ± SEM.*, p < 0.05; **, p < 0.01; ***, p < 0.001.
CCDC91 exerts its protumor effects via the aerobic glycolysis pathway
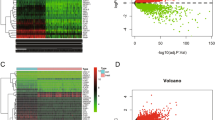
GSEA of the TCGA datasets was used to identify the molecular pathways that were significantly associated with CCDC91 expression. HCC patients were classified into two groups according to the mRNA expression level of CCDC91. The GSEA indicated that CCDC91 was associated mostly with genes involved in glycolysis (NES = 2.28, p < 0.001, Fig. 4A and Supplementary Data 4). Additionally, we compared the whole genome mRNA expression profiles of HepG2-VC and HepG2-CCDC91 cells using RNA-seq. A distinct transcriptome difference was observed (Fig. 4B) with 2625 genes significantly upregulated and 4033 genes downregulated by CCDC91 overexpression (|log2FC | ≥ 1, p ≤ 0.05). KEGG pathway enrichment analysis of the upregulated genes revealed that the glycolysis signaling pathway was also significantly enriched (p < 0.05, Fig. 4C).
A Upregulation of CCDC91 expression was positively correlated with activated glycolysis metabolic pathways in HCC as predicted by GSEA from TCGA dataset (GSEA, gene set enrichment analysis; NES, normalized enrichment score, p < 0.001). B The volcano graph showing the differentially expression genes (DEGs; |log2FC | ≥ 1, p ≤ 0.05) identified by RNA-seq between HepG2-CCDC91 and HepG2-VC cells. C The top 20 pathways revealed by KEGG enrichment analysis of DEGs. D Glucose levels were detected in CCDC91-overexpressing HCC cells and control cells (n = 3). E Lactate levels were detected in CCDC91-overexpressing HCC cells and control cells (n = 3). F The mRNA level of the key glycolysis differential expressed genes in CCDC91-overexpressing HCC cells and control cells by qPCR (n = 3). G The protein level of LDHA in CCDC91-overexpressing HCC cells and control cells by western blot (n = 3). H Correlation analysis of CCDC91 and LDHA levels in HCC tissues using the data from TCGA database. I Immunohistochemistry staining and analysis showed the protein expression of LDHA in HCC tissues and matched non-cancerous liver tissues (n = 80). J The Kaplan–Meier analysis revealed the association of LDHA with the overall survival of HCC patients from TMA cohort (n = 80). K Correlation analysis of CCDC91 and LDHA levels in HCC tissues using the data from TMA cohort (n = 80). Statistical analysis was performed using Student’s t test in (D, E, F, and I). Data represents mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We then investigated the potential role of aerobic glycolysis in the development of HCC following CCDC91 overexpression by examining the regulatory effects of CCDC91 on glucose uptake and lactate production. As anticipated, overexpression of CCDC91 resulted in increased lactate production in HepG2 and PLC/PRF/5 cell lines (p < 0.001, Fig. 4D, E). We also performed qPCR on key glycolysis-related genes, including GLUT1, HK2, MCT-1, MCT2, and LDHA, which were identified as differentially expressed in CCDC91-overexpressing cells via RNA-seq. The mRNA expression of LDHA was significantly greater in CCDC91-overexpressing cells than in control cells (p < 0.05, Fig. 4F). Western blot analysis also confirmed the increased protein expression of LDHA following CCDC91 overexpression (Fig. 4G). LDHA, a key enzyme in the glycolytic pathway, is highly expressed in various cancers. Clinical analysis of TCGA data clearly revealed that LDHA mRNA levels were positively correlated with CCDC91 levels (p < 0.001, R = 0.44; Fig. 4H). Consistent with previous studies32, in a TMA containing 80 HCC tissues, we found the expression of LDHA was markedly increased in HCC tissues compared with noncancerous tissues (p < 0.05, Fig. 4I). Moreover, we found that HCC patients with higher LDHA protein levels had a shorter OS time than did patients with lower levels of LDHA (p < 0.05, Fig. 4J). Interestingly, multivariate logistic regression analysis indicated a borderline significant positive association between LDHA expression and HBV DNA status in HCC tissues (p = 0.059, Table S6). Additionally, a significant correlation between LDHA and CCDC91 was confirmed (p < 0.001, R = 0.65; Fig. 4K) for protein levels. Collectively, these results indicate that CCDC91 and LDHA may be key regulatory factors involved in the HBV-related HCC and exert their effects through the glycolytic pathway.
LDHA is a downstream target of CCDC91 in CCDC91-overexpressing cells
Following the biosignature analysis that indicated that CCDC91 influenced glycolysis through LDHA, we investigated the potential interaction between the two proteins. As shown in Fig. 5A, co-IP demonstrated that CCDC91 and LDHA could interact with each other in PLC/PRF/5 and HepG2 cells. Additionally, immunofluorescence staining confirmed the significant colocalization of CCDC91 and LDHA in the cytoplasm of HCC cells (Fig. 5B). Notably, the immunofluorescence signals for both CCDC91 and LDHA were stronger in the CCDC91-overexpressing group than in the control group (Fig. 5B).
A Co-IP with antibody against CCDC91 followed by Western blot in HepG2 and PLC/PRF/5 cells (n = 3). B Immunofluorescence staining of CCDC91 and LDHA in CCDC91-overexpressing HCC cells and control cells. C The mRNA level of LDHA in CCDC91-overexpressing HCC cells with or without LDHA knockdown revealed by qPCR (n = 3). D The protein level of LDHA in CCDC91-overexpressing HCC cells with or without LDHA knockdown detected by western blot (n = 3). E Cell proliferation of CCDC91-overexpressing HCC cells with or without LDHA knockdown detected by CCK8 assay (n = 3). (F–I). F, G The migration abilities of CCDC91-overexpressing HCC cells with or without LDHA knockdown measured by transwell assay (n = 3). The migration (H, I) and invasion (J, K) abilities of CCDC91-overexpressing HCC cells with or without LDHA knockdown measured by wound healing and transwell assay, respectively (n = 3). L Glucose levels were detected in CCDC91-overexpressing HCC cells with or without LDHA knockdown (n = 3). M Lactate levels were detected in CCDC91-overexpressing HCC cells with or without LDHA knockdown (n = 3). Statistical analysis was performed using Student’s t test in (C, E, G, I, K, L, and M). Data represents mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To elucidate the biological function of LDHA in HCC, we transfected CCDC91-overexpressing cells with LDHA-siRNA and subsequently analyzed cell growth, migration, and invasion. qPCR and Western blot confirmed the successful reduction in LDHA in PLC/PRF/5 and HepG2 cells (p < 0.05, Fig. 5C, D). CCDC91 overexpression increased the proliferative, migratory, and invasive abilities of HCC cells, which were decreased by LDHA knockdown (p < 0.05, Fig. 5E–K). Moreover, LDHA knockdown weakened the capability of lactate production induced by CCDC91 in HCC cells (p < 0.01, Fig. 5L, M). In summary, these findings collectively suggest that CCDC91 promotes glycolysis by increasing LDHA expression in HCC.
CCDC91 deficiency confers sensitivity to sorafenib in HCC cell lines
Sorafenib, a multikinase inhibitor, is approved as the standard therapy for advanced HCC patients but provides only limited survival benefits, particularly for Asia–Pacific patients. Given that glycolysis signaling is implicated in a poor response to sorafenib33, we explored the effect of CCDC91 on sorafenib sensitivity. Based on our TMA cohort, we observed that HCC patients with stronger CCDC91 expression had a significantly shorter survival than those with weak CCDC91 expression when they were treated with sorafenib (Fig. 6A, B). Consistently, in PLC/PRF/5 cells, CCDC91 overexpression significantly counteracted the inhibitory effects of sorafenib on cell growth, migration, and invasion (p < 0.05, Fig. 6C–E). In HepG2 cells, a similar resistance phenotype was observed, with CCDC91 overexpression significantly diminishing sorafenib’s suppression of cell growth and migration (p < 0.05, Fig. 6C, D). However, for invasion, the effect was less pronounced, showing only a non-significant trend (Fig. 6E).
A The Kaplan–Meier analysis revealed the association of CCDC91 with the progression free survival of HCC patients that treated with sorafenib from TMA cohort (n = 77). B The Kaplan–Meier analysis revealed the association of CCDC91 with the overall survival of HCC patients that treated with sorafenib from TMA cohort (n = 80). C Cell proliferation of CCDC91-overexpressing HCC cells and control cells with or without sorafenib detected by CCK8 assay for 24 h, 48 h and 72 h separately (n = 3). The migration (D) and invasion (E) abilities of CCDC91-overexpressing HCC cells with or without sorafenib measured by transwell assay for 18 h (n = 3). F–H Huh-7 cells stably expressing SC or shCCDC91 were injected subcutaneously into the flanks of nude mice administrated without or with sorafenib (n = 5). Representative images of dissected tumors at the end of the experiment were shown. G Tumor growth curves of mice during sorafenib treatment were analyzed (n = 5). And xenografts were weighted (n = 5, H). All control cells were treated with an equivalent volume of DMSO to account for potential solvent effects, while the experimental groups were treated with sorafenib dissolved in DMSO. Statistical analysis was performed using Student’s t test in (C, D, E, and H). Two-way ANOVA was used for tumor volume in G Data represents mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To validate these findings in vivo, we employed a xenograft model. Our results revealed that either CCDC91 knockdown or sorafenib (20 mg/kg/day) alone inhibited tumor growth in vivo (Fig. 6F). Importantly, knockdown of CCDC91 expression in Huh-7 cells significantly increased the efficacy of tumor growth inhibition by sorafenib (p < 0.05, Fig. 6F–H). These results provide persuasive evidence that targeting CCDC91 signaling is an efficient strategy to increase the antitumor efficacy of sorafenib.
Ct-HBx mediates HBV integrated CCDC91 overexpression in HCC cells
Previously, we found that HBV DNA integration in HCC tissues may induce CCDC91 overexpression. While the Ct-HBx is crucial in the pathogenic mechanism of HBV DNA integration, we wanted to explore the interaction between the Ct-HBx and CCDC91. We transfected PLC/PRF/5 and HepG2 cells with Ct-HBx plasmid. Western blot confirmed the successful overexpression in HBx in PLC/PRF/5 and HepG2 cells (p < 0.05, Fig. 7A). To first determine whether the expression of CCDC91 mRNA is regulated by the Ct-HBx, qPCR was performed to compare the CCDC91 levels among the Ct-HBx infected cells as well as the control cells. The results revealed that CCDC91 was significantly upregulated in the Ct-HBx infected cells (p < 0.001, Fig. 7B), indicating that the Ct-HBx mediated CCDC91 overexpression and activation. To investigate the molecular mechanisms involved in regulating CCDC91 expression, we performed co-IP by transfecting the Ct-HBx plasmids into HepG2 and PLC/PRF/5 cells. As shown in Fig. 7C, co-IP demonstrated that HBx could bind to CCDC91. Additionally, immunofluorescence verified that CCDC91 and HBx significantly colocalized in the cytoplasm of HepG2 and PLC/PRF/5 cells (Fig. 7D), and when the cells were infected with the Ct-HBx plasmid, the immunofluorescence intensity of CCDC91 and HBx in the overexpression group was greater than that in the control group. Consistent with the function of CCDC91 overexpression in HCC, transfection of the Ct-HBx plasmid promoted the proliferation and metastasis of HepG2 and PLC/PRF/5 cells in vitro (p < 0.05, Fig. 7E–G). Moreover, by western blot we found that transfection of Ct-HBx promoted CCDC91-induced upregulation of LDHA (Fig. 7G) in HepG2 and PLC/PRF/5 cells.
A The protein level of HBx in HepG2 and PLC/PRF/5 cells with or without Ct-HBx transfection detected by western blot (n = 3). B The mRNA level of CCDC91 in HepG2 cells with or without Ct-HBx transfection revealed by qPCR (n = 3). C Co-IP with antibody against HBx followed by Western blot in HepG2 and PLC/PRF/5 cells (n = 3). D Immunofluorescence staining of CCDC91 and HBx in HepG2 and PLC/PRF/5 cells with or without Ct-HBx transfection. E Cell proliferation of HepG2 and PLC/PRF/5 cells with or without Ct-HBx transfection detected by CCK8 assay (n = 3). F, G) The migration and invasion abilities of HepG2 and PLC/PRF/5 cells with or without Ct-HBx transfection measured by transwell assay (n = 3). H The protein level of HBx, CCDC91, and LDHA in HepG2 and PLC/PRF/5 cells with or without Ct-HBx transfection revealed by western blot (n = 3). I Schematic diagram illustrating the proposed mechanisms of the effects of the HBV-integrated gene CCDC91 on HCC. Statistical analysis was performed using Student’s t test in (B, E, F, and G). Data represents mean ± SEM.*, p < 0.05; **, p < 0.01; ***, p < 0.001.
Overall, we propose that HBV DNA integration causes aberrant expression of CCDC91 in HCC, which leads to the activation of LDHA via the aerobic glycolysis pathway. CCDC91 deficiency also increased sensitivity to sorafenib treatment (Fig. 7H).
Discussion
Increasingly researchers agree that HBV DNA integration plays a crucial role in hepatocarcinogenesis34. On the basis of a sizeable cohort of clinical HBV-related HCC cases analyzed through pooled-analysis35 and our recent HBV capture sequencing data, we identified preferential HBV–human chimeric fusions with breakpoints at the CCDC91 gene. Moreover, we found a trend that HBV integration at CCDC91 was associated with increased CCDC91 protein expression, and HCC patients with higher CCDC91 expression had poorer survival. To date, these findings represent the first demonstration of a relationship between HBV integration and CCDC91.
The characterization of integrated HBV DNA segments in the host genome provides a better understanding of the pathogenic mechanism and role of HBV in HCC. Previous studies have shown that the integrated DNA of HBV in HCC usually contains the viral basal core promoter (BCP)/enhancer II (Enh II), and the sequence encoding the C-terminal truncated HBx which is a pleiotropic transactivator36. Recently, Zhang et al. reported that the Ct-HBx mediated metabolic reprogramming from mitochondrial respiration to aerobic glycolysis via the NFACT2-TXNIP signaling pathway, indicating that the Ct-HBx plays a critical role in hepatocarcinogenesis12. In addition, full-length HBx also has pleiotropic properties that support viral gene expression and replication and contribute significantly to HCC development23,36. One study reported that HBx could interact with NF-κBp65 at both the transcriptional and protein levels, upregulate the NF-κBp65/HK2 axis and increase cell proliferation23. In this study, we found that HBx, which is induced by the Ct-HBx, could upregulate the CCDC91 in HepG2 cells. To investigate the molecular mechanisms involved in regulating CCDC91 expression in HBV-related HCC, we performed co-IP and found that HBx and CCDC91 interacted with each other and enhanced cell proliferation, migration, and invasion in HepG2 cells. Whether Ct-HBx affects CCDC91 in HCC cells at the transcriptional level requires further investigation.
The detailed function and mechanism of CCDC91 in cancer development remain unclear. Previously, we reported that patients with high expression of CCDC91 had poor survival, with borderline significance, for NHL29. In addition to cancers, some genome-wide association studies have reported that CCDC91 polymorphisms may be associated with lung function37, ossification of the posterior longitudinal ligament38, and bone remodeling39. Recently, Zhu et al. revealed that the CCDC91 gene is a pathogenic gene involved in the development of acrokeratoelastoidosis by using linkage analysis and whole-exome sequencing. Additionally, the authors reported that the CCDC91 gene plays a crucial role in elastin transport40. In this study, based on the loss-of-function and gain-of-function studies, CCDC91 was shown to contribute to the proliferation and metastasis of HCC. To date, the role of CCDC91 in HCC had not been studied. Our functional findings were also supported by the clinical observation that HCC patients with high tumor expression of CCDC91 had a poorer TNM stage and poorer survival. More interestingly, multivariate logistic regression analysis revealed that CCDC91 expression in HCC tissues was positively associated with HBV DNA status. High copy numbers of HBV DNA in HCC patients have been associated with a poor prognosis41. These data suggest a vital role for CCDC91 in HBV-related HCC. Furthermore, we discovered that CCDC91 modulates aerobic glycolysis levels by interacting with LDHA, a metabolite that promotes tumor growth and metastasis in HCC. LDHA is one of the key regulators involved in glycolytic metabolism and is highly expressed in various tumors, such as head and neck squamous carcinoma24, colorectal cancer25, and HCC26,28, and LDHA influences tumor proliferation, migration, and nuclear regulation. By performing an integrated analysis of data from HCC patients from the TCGA database and our RNA-sequencing data, we demonstrated that CCDC91 could upregulate the expression of LDHA. Moreover, the reduced proliferation, migration, and invasion ability induced by overexpression of CCDC91 in HCC cells was largely abolished by the introduction of siLDHA, indicating that therapeutic strategies that disrupt the interaction between CCDC91 and LDHA have the potential to prevent cancer growth and metastasis, especially in patients with high CCDC91 expression.
Although HCC is a highly aggressive type that remains difficult to treat, efforts to exploit effective therapeutic approaches (such as immune checkpoint inhibitors42, antiangiogenic agents43, and CDK4/6 inhibitors44) for treating this disease are ongoing. Among these, the multikinase inhibitor sorafenib is an attractive therapeutic option. Sorafenib is the standard of care for the treatment of advanced HCC, but it is associated with the development of resistance45. Several factors including HIF-1α46, EGFR47, MEK signaling48, and aerobic glycolysis33 limit the efficiency of sorafenib. Aerobic glycolysis is a major resistance mechanism induced by first-line treatment of HCC with sorafenib. In this study, HCC patients with stronger CCDC91 expression had significantly shorter survival than those with weak CCDC91 expression when they treated with sorafenib. By performing an in vivo study, we also found that the knockdown of CCDC91 in HCC cells resulted in increased sensitivity to sorafenib. These data indicate that CCDC91 may affect sorafenib sensitivity via aerobic glycolysis signaling, further investigation of this pathway might be useful for overcoming resistance to sorafenib in HCC.
There are several limitations to our study. First, we detected only 4 HBV-associated HCCs with HBV capture sequencing in this study. Most of the data on HBV DNA integration at the CCDC91 locus were obtained from our previous pooled-analyses. Therefore, the real pattern of HBV DNA integration at the CCDC91 locus awaits further larger-scale investigations. Second, combined with the corresponding in-vitro data, we highlight that the expression of CCDC91 may be induced by Ct-HBx and that HBx and CCDC91 bind with each other in HCC cells. However, we acknowledge that we did not provide direct in vitro evidence linking HBV DNA integration to CCDC91 upregulation. Further investigation is needed to fully understand the direct mechanism induced by the HBV-integrated sequence. Finally, understanding the detailed mechanism by which CCDC91 affects sorafenib sensitivity requires further study.
Overall, our work revealed that HBV DNA integration caused aberrant expression of CCDC91 in HCC, which led to the activation of LDHA via the aerobic glycolysis pathway. Furthermore, CCDC91 deficiency increased sensitivity to sorafenib treatment. The results strongly demonstrated that CCDC91 could serve as a potential biomarker and promising therapeutic target in HBV-induced HCC.
Data availability
The sequencing data were uploaded to the NCBI database (accession number: PRJNA1138405). All relevant data are included in the paper and/ or its supplementary information files (the source data was listed in Supplementary Data 1).
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249 (2021).
Yang, X. et al. Precision treatment in advanced hepatocellular carcinoma. Cancer Cell 42, 180–197 (2024).
Mak, L. Y. et al. Occult hepatitis B infection and hepatocellular carcinoma: Epidemiology, virology, hepatocarcinogenesis and clinical significance. J. Hepatol. 73, 952–964 (2020).
Yeh, S. H. et al. Hepatitis B virus DNA integration drives carcinogenesis and provides a new biomarker for HBV-related HCC. Cell. Mol. Gastroenterol Hepatol. 15, 921–929 (2023).
Peneau, C. et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut 71, 616–626 (2022).
Chen, X. P. et al. Viral integration drives multifocal HCC during the occult HBV infection. J. Exp. Clin. Cancer Res 38, 261 (2019).
Lin, S. Y. et al. Recurrent HBV integration targets as potential drivers in hepatocellular carcinoma. Cells 10, 1294 (2021).
Tu, H. et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 61, 7803–7810 (2001).
Yip, W. K. et al. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS One 6, e22888 (2011).
Ma, N. F. et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin. Cancer Res. 14, 5061–5068 (2008).
Huo, T. I. et al. Hepatitis B virus X mutants derived from human hepatocellular carcinoma retain the ability to abrogate p53-induced apoptosis. Oncogene 20, 3620–3628 (2001).
Zhang, Y. et al. C-terminal truncated HBx initiates hepatocarcinogenesis by downregulating TXNIP and reprogramming glucose metabolism. Oncogene 40, 1147–1161 (2021).
Li, C. L. et al. Androgen receptor enhances hepatic telomerase reverse transcriptase gene transcription after hepatitis B virus integration or point mutation in promoter region. Hepatology 69, 498–512 (2019).
Sze, K. M. et al. Hepatitis B virus-telomerase reverse transcriptase promoter integration harnesses host ELF4, resulting in telomerase reverse transcriptase gene transcription in hepatocellular carcinoma. Hepatology 73, 23–40 (2021).
Aziz, K. et al. Ccne1 overexpression causes chromosome instability in liver cells and liver tumor development in mice. Gastroenterology 157, 210–226.e12 (2019).
Li, M. et al. Characterization of hepatitis B virus infection and viral DNA integration in non-Hodgkin lymphoma. Int J. Cancer 147, 2199–2209 (2020).
Li, J. et al. Identification of potential prognostic biomarkers among gene models for coiled-coil domain-containing family members in hepatocellular carcinoma elucidates their influence on the hypoxia pathway and immune microenvironment. J. Gastrointest. Oncol. 14, 2559–2573 (2023).
Hu, X. et al. CCDC178 promotes hepatocellular carcinoma metastasis through modulation of anoikis. Oncogene 36, 4047–4059 (2017).
Lin, Z. et al. Up-regulated CCDC34 contributes to the proliferation and metastasis of hepatocellular carcinoma. Onco Targets Ther. 13, 51–60 (2020).
Tanouchi, A. et al. CCDC88A, a prognostic factor for human pancreatic cancers, promotes the motility and invasiveness of pancreatic cancer cells. J. Exp. Clin. Cancer Res. 35, 190 (2016).
Jiang, G. Y. et al. Coiled-coil domain-containing protein 8 inhibits the invasiveness and migration of non-small cell lung cancer cells. Hum. Pathol. 56, 64–73 (2016).
Zeng, C. et al. Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus. Breast Cancer Res. 18, 64 (2016).
Chen, L. et al. Aerobic glycolysis enhances HBx-initiated hepatocellular carcinogenesis via NF-kappaBp65/HK2 signalling. J. Exp. Clin. Cancer Res. 41, 329 (2022).
Lu, Y. et al. KAT7 enhances the proliferation and metastasis of head and neck squamous carcinoma by promoting the acetylation level of LDHA. Cancer Lett. 590, 216869 (2024).
Lin, J. et al. Circular RNA circTATDN3 promotes the Warburg effect and proliferation in colorectal cancer. Cancer Lett. 589, 216825 (2024).
Chen, S. et al. TAB182 regulates glycolytic metabolism by controlling LDHA transcription to impact tumor radiosensitivity. Cell Death Dis. 15, 209 (2024).
Feng, Y. et al. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 7, 6124–6136 (2018).
Zheng, J. et al. CircFOXK2 promotes hepatocellular carcinoma progression and leads to a poor clinical prognosis via regulating the Warburg effect. J. Exp. Clin. Cancer Res. 42, 63 (2023).
Li, M. et al. Characterization of hepatitis B virus infection and viral DNA integration in non-Hodgkin lymphoma. Int. J. Cancer. 147, 2199–2209 (2020).
Jin, Y. et al. Identification and impact of hepatitis B virus DNA and antigens in pancreatic cancer tissues and adjacent non-cancerous tissues. Cancer Lett. 335, 447–454 (2013).
Wang, L. et al. A novel multitarget kinase inhibitor BZG with potent anticancer activity in vitro and vivo enhances efficacy of sorafenib through PI3K pathways in hepatocellular carcinoma cells. Biomed. Pharmacother. 125, 110033 (2020).
Sheng, S. L. et al. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J. 279, 3898–3910 (2012).
Guo, S. et al. Glycolysis maintains AMPK activation in sorafenib-induced Warburg effect. Mol. Metab. 77, 101796 (2023).
Jiang, Y., Han, Q., Zhao, H. & Zhang, J. The mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatocell. Carcinoma 8, 435–450 (2021).
Li, M. et al. Characterization of hepatitis B virus DNA integration patterns in intrahepatic cholangiocarcinoma. Hepatol. Res. 51, 102–115 (2021).
Feitelson, M. A., Arzumanyan, A., Spector, I. & Medhat, A. Hepatitis B x (HBx) as a component of a functional cure for chronic hepatitis B. Biomedicines 10, 2210 (2022).
Soler Artigas, M. et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat. Commun. 6, 8658 (2015).
Nakajima, M. et al. A genome-wide association study identifies susceptibility loci for ossification of the posterior longitudinal ligament of the spine. Nat. Genet. 46, 1012–1016 (2014).
Sethi, A., Ruby, J. G., Veras, M. A., Telis, N. & Melamud, E. Genetics implicates overactive osteogenesis in the development of diffuse idiopathic skeletal hyperostosis. Nat. Commun. 14, 2644 (2023).
Zhu, Y. et al. A mutation in CCDC91, Homo sapiens coiled-coil domain containing 91 protein, cause autosomal-dominant acrokeratoelastoidosis. Eur. J. Hum. Genet. 32, 647–655 (2024).
Huang, J. T. et al. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clin. Chem. 61, 290–296 (2015).
Qin, S. et al. Camrelizumab plus rivoceranib versus sorafenib as first-line therapy for unresectable hepatocellular carcinoma (CARES-310): a randomised, open-label, international phase 3 study. Lancet 402, 1133–1146 (2023).
Chuma, M. et al. Clinical significance of circulating biomarkers of immune-checkpoint molecules with atezolizumab plus bevacizumab therapy in unresectable hepatocellular carcinoma. Hepatol Int. 18, 1472–1485 (2024).
Sheng, J. et al. Treatment of retinoblastoma 1-intact hepatocellular carcinoma with cyclin-dependent kinase 4/6 inhibitor combination therapy. Hepatology 74, 1971–1993 (2021).
Wan, S. et al. SPARC stabilizes ApoE to induce cholesterol-dependent invasion and sorafenib resistance in hepatocellular carcinoma. Cancer Res. 84, 1872–1888 (2024).
Song, Z. et al. HIF-1alpha-induced RIT1 promotes liver cancer growth and metastasis and its deficiency increases sensitivity to sorafenib. Cancer Lett. 460, 96–107 (2019).
Wang, C. et al. LARP4B promotes hepatocellular carcinoma progression and impairs sorafenib efficacy by activating SPINK1-mediated EGFR pathway. Cell Death Discov. 10, 208 (2024).
Ewald, F. et al. Vertical targeting of AKT and mTOR as well as dual targeting of AKT and MEK signaling is synergistic in hepatocellular carcinoma. J. Cancer 6, 1195–1205 (2015).
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (82203225 to M.L.), Fundamental Research Funds for the Central Universities (WK9110000172 to M.L.), Clinical Key Specialty Development Program of Anhui Province (2023silczdzk02 to Y.H.) and Health Commission of Anhui Province Scientific Research Project (AHWJ2023A30183 to S.W. and AHWJ2021b105 to Y.H.).
Author information
Authors and Affiliations
Contributions
Y.H. was responsible for the study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and obtaining funding. M.L. and S.W. reviewed clinical data, performed statistical analyses, and revised the manuscript. M.L. performed experiments, collected and analyzed data, and wrote the manuscript. H.L., J.N., Y.F., L.C., and W.C. collected samples and analyzed data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
HCC samples used in HBV sequencing were approved by the institutional ethics review committee of the First Affiliated Hospital of USTC and conducted according to the principles of the Declaration of Helsinki. The ethics committee waived the need for written informed consent because the study was conducted using residual samples from previous clinical diagnoses, the risks of the study did not exceed minimal risks, and the waiver of informed consent would not adversely affect the subjects. The IHC tissues were collected by Shanghai National Engineering Research Center from Taizhou Hospital (Approved by Research Ethics Committee of Taizhou Hospital). We confirmed that all studies are conducted following relevant guidelines/regulations. All ethical regulations relevant to human research participants were followed. All experiments involving mice were approved by the Animal Care and Use Committees of the First Affiliated Hospital of USTC, and were performed in accordance with the Institutional Animal Welfare Guidelines.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Isabela Pedroza-Pacheco, Johannes Stortz and Manuel Breuer.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, M., Wu, S., Luo, H. et al. HBV DNA integration gene CCDC91 is oncogenic and a potential therapeutic target for hepatocellular carcinoma. Commun Biol 8, 1079 (2025). https://doi.org/10.1038/s42003-025-08369-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08369-1