Abstract
Arrhythmogenic cardiomyopathy (ACM) is an inherited heart disease marked by progressive fattyfibro replacement of the ventricular myocardium, life-threatening arrhythmias, and sudden cardiac death. To dissect epicardial contributions to ACM pathogenesis, we generated iPSC lines from patients carrying plakophilin 2 (PKP2) 1849C > T or PKP2 2013delC mutations, their CRISPR/Cas9–corrected isogenic controls, and a PKP2 knockout line. Epicardial cells (hPSC-EPCs) differentiated from mutant and knockout backgrounds exhibit enhanced epithelial-to-mesenchymal transition characteristics, increased lipid accumulation, and a pronounced fibrotic phenotype. RNA-seq performed on ACM hPSC-EPCs reveals dysregulation of Wnt, interferon, and Rho GTPase signaling, including an upregulation of insulin growth factor 2 (IGF2) and a key adipogenic transcription factor, CEBPA. Subsequent treatment of control and PKP2KO hPSC-EPCs with recombinant IGF2 enhances CEBPA expression, suggesting that insulin growth factor signaling contributes to ACM fattyfibro remodeling.
Similar content being viewed by others
Introduction
Arrhythmogenic cardiomyopathy (ACM) is a complex genetic disorder that is characterized by progressive fattyfibro remodeling of the ventricular myocardium that disrupts electrical conduction, leading to lethal arrhythmias1 and increasing the risk of sudden cardiac death2. Typically, the symptoms of ACM (e.g., palpitations, syncope, shortness of breath, and chest discomfort) arise during adolescence or early adulthood and become progressively worse over time3. To date, the majority of mutations associated with ACM have been found in genes coding for desmosome proteins, with almost half of the mutations occurring in the gene plakophilin-2 (PKP2)4,5,6.
The pathogenesis of ACM has been studied in multiple cell types and animal models7,8. However, studying fattyfibro remodeling using animal models has been challenging, given that little to no fattyfibro remodeling has been found in animal models9. Cardiac cell types derived from human pluripotent stem cells (hPSCs) provide a model to study ACM by eliminating species-specific differences between human and animal models and allowing one to study patient-specific disease characteristics. Human induced pluripotent stem cell (hiPSC) derived-cardiomyocytes (hiPSC-CMs) from ACM patients have been previously shown to recapitulate features of the ACM phenotype, including disrupted desmosomes and increased formation of lipid droplets upon treatment of cells with an adipogenic cocktail containing dexamethasone, insulin, and IBMX10. While cardiomyocytes express abundant desmosomes and have received much attention for their involvement in ACM, the complex pathophysiology of the disease suggests that epicardial cells (EPCs) play a significant role in fibrosis and fatty infiltrates into the myocardium. In addition, EPCs are multipotent and have been shown to differentiate into various cell types within the heart11. Specifically, during heart development, a subset of EPCs undergo epithelial-to-mesenchymal transition (EMT) and contribute to the cardiac fibroblast12, vascular smooth muscle cell13, pericyte14, and vascular endothelial cell populations15. The involvement of EPCs in the development of ACM is further supported by the observation that EPCs also express desmosomes and thus, would be impacted by the expression of mutant desmosome proteins16. This is supported by a previous study demonstrating that lowering Pkp2 levels in neonatal rat EPCs leads to an increase in lipid accumulation, increased migration potential, and increased expression of an EMT marker (α-SMA)16. Similarly, another study showed that human induced pluripotent stem cell derived-EPCs (hiPSC-EPCs) spontaneously undergo EMT and differentiate into fibroblasts and fat cells, suggested to be regulated by the transcription factor TFAP2A17. These observations support the involvement of EPCs in the pathogenesis of ACM.
While EPCs have been an area of discussion due to their role in ACM fattyfibro pathology, a complete understanding of the signaling mechanisms and growth factors promoting or repressing the pathologic remodeling of myocardial tissue is still needed. In addition, it is not known whether epicardial-derived fibroblasts are more susceptible to contributing to the fattyfibro pathology when compared to non-epicardial derived fibroblasts. Therefore, to investigate the signaling mechanisms driving epicardial-dependent fattyfibro pathology, we generated hiPSCs from two families harboring either a PKP2 2013delC/- or PKP2 1849C > T/-heterozygous ACM-causative truncation mutation and performed CRISPR/Cas9 genetic editing to generate isogenic corrected hiPSC lines. Furthermore, we generated a PKP2 knockout (KO) line (PKP2KO) to assess whether a total loss of PKP2 recapitulates similar ACM cellular phenotypes compared to hiPSCs harboring ACM mutations. Subsequently, we differentiated ACM hiPSC lines, PKP2KO lines, and isogenic control lines into epicardial cells (hPSC-EPCs). Differentiation of EPCs derived from ACM hiPSC lines and PKP2KO lines demonstrated perturbed PKP2 localization, enhanced cell migration, and increased lipid accumulation potential. Moreover, fibroblasts derived from epicardial cells demonstrated an enhanced fibrotic phenotype and fibrotic gene expression profile when compared to cardiac fibroblasts that did not undergo an epicardial state. Transcriptomic analysis revealed ACM hPSC-EPCs and ACM human heart tissue expressed higher levels of adipogenic transcription factors CEBPA and IGF2. Exogenous exposure of IGF2 onto hPSC-EPCs enhanced the expression of CEBPA and PPARG, suggesting that insulin growth factors drive ACM fattyfibro remodeling.
Results
Generation of ACM and PKP2KO hPSCs, isogenic control hPSCs, and hPSC-EPCs
Studying ACM fattyfibro pathology has been difficult given that some animal models do not fully recapitulate the fattyfibro pathology seen in ACM patients18. To better understand human ACM fattyfibro pathology and the molecular signaling mechanism leading to the fattyfibro pathology, we identified two families, Family 1025 and Family 398, carrying ACM-causative heterozygous PKP2 mutations, PKP2 1849C > T/- or PKP2 2013delC/-. Collectively, these patients presented with ventricular arrhythmias, inverted T-waves on EKG, and fattyfibro pathology in the myocardium (Supplementary Table 1). We reprogrammed blood from patients with ACM (398100, 398110, 1025100, 1025200), as well as blood from family members who did not have ACM (1025110, 1025201) into hiPSCs. To mitigate genetic background differences, we corrected the PKP2 1849C > T/- and PKP2 2013delC/- mutation in two lines (1025100, 398110) to generate isogenic control hiPSC cell lines using CRISPR/Cas9 gene editing (Fig. 1A). In addition, to understand whether cellular phenotypes could be recapitulated in cells without PKP2 expression, we generated a PKP2KO line via CRISPR/Cas9 editing within the H7 human embryonic stem cell (hESCs) line (H7 hESCs) (Fig. 1B). All hPSC lines expressed pluripotency markers (OCT4 and SSEA4), lacked large chromosomal abnormalities, and did not exhibit any tested CRISPR/Cas9 off-target effects, as predicted by COSMID19 (Supplementary Fig. 1). To investigate the role of epicardial cells in ACM pathogenesis, both hESC and hiPSC lines were differentiated into epicardial cells (hPSC-EPCs) (Fig. 2A–C). Immunocytochemistry performed on hPSC-EPCs demonstrated expression of the epicardial marker, WT1, and in addition, qPCR analysis identified WT1 and TBX18 in hPSC-EPCs (Fig. 2D–F). To determine whether hPSC-EPCs harboring PKP2 mutations affected the localization of PKP2, immunofluorescence (IF) (Fig. 2G) and Western blot analysis (Fig. 2H–J, Supplementary Fig. 3) were performed. PKP2 was observed to localize to the cell borders in control hiPSC-EPCs and hESC-EPCs while diffuse PKP2 labeling was observed in ACM hiPSC-EPCs and PKP2 was undetected in PKP2KO hESC-EPCs. Additionally, Western blot quantification showed reduced PKP2 expression in ACM hiPSC-EPCs and as expected, PKP2 expression was undetected in PKP2KO hESC-EPCs.
A Pedigrees and genotypes of ACM patient families 1025 and 398, showing inheritance of PKP2 mutations. Family 1025 carries a nonsense mutation (PKP2 c.1849C>T, p.Q617X), and Family 398 carries a frameshift mutation (PKP2 c.2013delC, p.P672fsX683). Squares represent males and circles represent females. Shaded symbols indicate individuals from whom iPSC lines were derived. Gene-corrected control lines were generated using CRISPR/Cas9 and are denoted by dashed lines. Predicted protein structures show the location and potential structural impact of the mutations. Sanger sequencing traces confirm the presence of the mutant allele (top) and successful correction (bottom) for both patient-derived lines (1025–100 and 398–110). B Schematic of the human PKP2 gene locus, including isoforms Pkp2a and Pkp2b. Arrows indicate transcriptional direction. The position of exon 2, which harbors the mutations used in this study, is highlighted in red. Generation of isogenic control, ACM, and PKP2KO human pluripotent stem cells (hPSCs).
A Differentiation protocol used to generate hPSC-EPCs. B Phase-contrast images of hPSC differentiating into hPSC-EPCs from day 0 up to day 12 with epicardial cobblestone morphology (scale bar= 100 µm). C Phase contrast images of ACM, PKP2KO, and isogenic control hPSC-EPCs (scale bar= 100 μm). D Immunofluorescence images of WT1 in ACM, PKP2KO, and isogenic control hPSC-EPCs (scale bar= 100 μm). E qPCR of the epicardial marker WT1 in ACM, PKP2KO, and isogenic control hPSC-EPCs relative to undifferentiated hPSCs (mean ± SEM, N = 6 of hPSCs, hPSC-EPCs and ACM/PKP2KO hPSC-EPCs). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. F qPCR of the epicardial marker TBX18 in ACM, PKP2KO, and isogenic control hPSC-EPCs relative to undifferentiated hPSCs (mean ± SEM, N = 4 of hPSCs, hPSC-EPCs and ACM/PKP2KO hPSC-EPCs). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. G Immunofluorescence images of PKP2 in ACM, PKP2KO, and isogenic control hPSC-EPCs (scale bar= 50 μm). H Representative western blot image of PKP2 in ACM, PKP2KO, and isogenic control hPSC-EPCs. I Densitometry analysis of PKP2 expression in ACM hiPSC-EPCs relative to the control (hiPSC-EPCs) (mean ± SEM, N = 8 of hiPSC-EPCs and ACM hiPSC-EPCs). Statistical significance was evaluated using an one sample t test (p = 0.0642). J Densitometry analysis of PKP2 expression in PKP2KO hESC-EPCs relative to hESC-EPCs (mean ± SEM, N = 4 of hESC-EPCs and PKP2KO hESC-EPCs). Each N value represents a separate epicardial differentiation from the hPSC state.
ACM and PKP2KO hPSC-EPCs demonstrate enhanced EMT characteristics compared to isogenic control hPSC-EPCs
To model the cellular processes driving fattyfibro remodeling, we first examined whether ACM and PKP2KO hPSC-EPCs demonstrated enhanced epithelial-to-mesenchymal transition (EMT) characteristics. During EMT, the expression of E-cadherin decreases while the expression of N-cadherin increases. Therefore, to investigate whether ACM/PKP2KO and isogenic control hPSC-EPCs demonstrate enhanced EMT characteristics, we measured the expression of CDH1 (E-cadherin) and CDH2 (N-cadherin) in hPSC-EPCs in the absence or presence of a growth factor shown to enhance EMT (TGFβ). In the absence of TGFβ, levels of E-cadherin were significantly reduced in both ACM hiPSC-EPCs and PKP2KO hESC-EPCs compared to their respective isogenic control. In contrast, transcript levels of N-cadherin were enhanced in both ACM hiPSC-EPCs and PKP2KO hESC-EPCs in the TGFβ treated group. Similarly, N-cadherin expression was significantly elevated in PKP2KO hESC-EPC under baseline conditions (i.e., no TGFβ treatment) (Fig. 3A). To further assess EMT characteristics, we performed an in vitro cell migration assay to evaluate the motility of ACM, PKP2KO, and control hPSC-EPCs. Epicardial cells were tested for their ability to migrate at 0, 4, 8, 24, and 48 hours (Fig. 3B, C) after a pipette was used to generate a gap in the cell monolayer. Compared to control hiPSC-EPCs, ACM hiPSC-EPCs demonstrated a significant increase in cell migration at 24 and 48 hours, whereas PKP2KO hESC-EPCs exhibited enhanced cell migration at 4, 8, 24, and 48 hours compared to control hESC-EPCs (Fig. 3D, E). These findings indicate that PKP2 deficiency promotes EMT and enhances epicardial cell migration.
A qPCR of the EMT marker CDH1 (E-cadherin) and CDH2 (N-cadherin) with or without TGFβ treatment relative to isogenic control hPSC-EPCs, ACM hiPSC-EPCs, and PKP2KO hESC-EPCs (mean ± SEM, N = 3). Statistical significance was evaluated using a one-way ANOVA with Bonferroni correction. B Representative images of 48-hour cell migration assay, showing the start of the migration of hiPSC-EPCs and ACM hiPSC-EPCs or C hESC-EPCs and PKP2KO hESC-EPCs from the introduced gap at hour 0 and the final migration results of hiPSC-EPCs and ACM hiPSC-EPCs at 48 hour (scale bar = 500 μm). D Gap closure quantification of ACM hiPSC-EPCs relative to control (hiPSC-EPCs) over 48 hours post introduction of the gap (mean ± SEM, N = 10 of hiPSC-EPCs and ACM hiPSC-EPCs with ** at 24 hour (p = 0.0071), with *** at 48 hour (p = 0.0072)). Statistical significance was evaluated using a two-way ANOVA test with Bonferroni correction. E Quantification gap closure of PKP2KO hESC-EPCs relative to control (hESC-EPCs) over 48 hours post introduction of the gap (mean ± SEM, N = 5 of hESC-EPCs and PKP2KO hESC-EPCs with * at 4 hour (p = 0.0408), ** at 8 hour (p = 0.0037), ** at 24 hour (p = 0.0079), with ** at 48 hour (p = 0.0011)). Statistical significance was evaluated using a two-way ANOVA test with Bonferroni correction. Each N value represents a separate epicardial differentiation from the hPSC state.
ACM and PKP2KO hPSC-EPCs exhibit increased lipid accumulation under adipogenic induction conditions
ACM fattyfibro pathology is initially formed subepicardially, suggesting that epicardial cells contribute to the formation of the fattyfibro pathology16,20,21. Given that ACM hPSC-EPCs expressed higher levels of transcription factors involved in adipogenesis (CEBPA and PPARG), we investigated whether PKP2 deficiency promotes epicardial lipid accumulation using an adipogenic induction protocol. We used Nile Red, a live fluorescence dye, to quantify lipid accumulation, enabling longitudinal tracking of lipid uptake across multiple time points in the same experiment (Fig. 4A, B). When assessing fluorescent intensity of ACM and control hiPSC-EPCs on days 0, 4, and 8, ACM hiPSC-EPCs showed increased levels of lipids at baseline (day 0) and at day 4 compared to control hiPSC-EPCs. Additionally, PKP2KO hESC-EPCs also demonstrated significant lipid accumulation at baseline (day 0) and on days 4, and 8 compared to the control hESC-EPCs (Fig. 4C). To assess whether key adipogenic transcription factors were changed during adipogenic induction, we performed qPCR analysis targeting the expression of CEBPA and PPARG in hPSC-EPCs under baseline and adipogenic induction conditions (Fig. 4D, E). Under adipogenic induction, hPSC-EPCs showed elevated CEBPA and PPARG expression compared to their baseline counterparts. However, ACM and PKP2KO hPSC-EPCs had significantly increased levels of CEBPA and PPARG compared to isogenic control hPSC-EPCs under adipogenic induction conditions.
A Representative fluorescence images of Nile Red staining of lipids in ACM hiPSC-EPCs and hiPSC-EPCs or B hESC-EPCs and hESC-EPCs during adipogenic induction at days 0, 4, and 8 (scale bar = 100 μm). C Quantification of Nile Red fluorescent intensity per cell in each field view of ACM hiPSC-EPCs and hiPSC-EPCs relative to the control (hiPSC-EPCs at day 0) at days 0, 4, and 8 (mean ± SEM, N = 10 of hiPSC-EPCs and ACM hiPSC-EPCs). Quantification of Nile Red fluorescent intensity per cell in each field view of PKP2KO hESC-EPCs and hESC-EPCs relative to the control (hESC-EPCs at day 0) at days 0, 4, and 8 (mean ± SEM, N = 6 of hESC-EPCs and PKP2KO hESC-EPCs). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. D qPCR of CEBPA in ACM hiPSC-EPCs and hiPSC-EPCs with or without adipogenic induction treatment relative to untreated hiPSC-EPCs (mean ± SEM, N = 6 of hiPSC-EPCs and ACM hiPSC-EPCs). qPCR of CEBPA in PKP2KO hESC-EPCs and hESC-EPCs with or without adipogenic induction treatment relative to untreated hESC-EPCs (mean ± SEM, N = 8 of hESC-EPCs and PKP2KO hESC-EPCs). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. E qPCR of PPARG in ACM hiPSC-EPCs and hiPSC-EPCs with or without adipogenic induction treatment relative to untreated hiPSC-EPC (mean ± SEM, N = 7 of hiPSC-EPCs and ACM hiPSC-EPCs). qPCR of PPARG in PKP2KO hESC-EPCs and hESC-EPCs, with or without adipogenic induction treatment, relative to untreated hESC-EPC (mean ± SEM, N = 8 of hESC-EPCs and PKP2KO hESC-EPCs). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. Each N value represents separate epicardial differentiation from the hPSC state.
An enhanced fibrotic phenotype is observed in fibroblast-derived cells from hPSC-EPCs when compared to non-epicardial-derived fibroblasts
Epicardial cells undergo EMT and have been shown to differentiate into fibroblasts in cardiac injury models22,23,24. Additionally, adipogenic induction performed on fibroblasts has been previously demonstrated to transdifferentiate fibroblasts into adipocytes25. Therefore, fibroblasts derived from epicardial cells may be a unique cell type demonstrating enhanced fattyfibro characteristics. At baseline, ACM hiPSC-EPCs and PKP2KO hESC-EPCs expressed higher levels of fibroblast markers (Col1α1 and PDGFRA) (Fig. 5A, B). Hence, we assessed whether cardiac fibroblasts derived from epicardial cells (hPSC-EpiCFs) were more amenable to fattyfibro characteristics when compared to non-epicardial derived fibroblasts. ACM, PKP2KO, and isogenic control hPSC-EPCs were differentiated into hPSC-EpiCFs using a fibroblast promoting medium26, and cardiac fibroblasts (hPSC-CFs) were generated using a separate protocol that was devoid of an epicardial transition cell state (Fig. 5C, D). The levels of Col1α1 and PDGFRA were higher in hPSC-EpiCFs when compared to hPSC-CFs with ACM and PKP2KO hPSC-EpiCFs expressing the highest levels of Col1α1 and PDGFRA in comparison to ACM and PKP2KO hPSC-CFs (Fig. 5E, F). To assess lipid accumulation in epicardial-derived cells, we performed Nile Red staining at baseline (Day 0) and after 8 days in culture (Day 8) in both hiPSC- and hESC-derived epicardial fibroblasts (EpiCFs) and cardiac fibroblasts (CFs). Representative fluorescence images demonstrated a marked increase in Nile Red signal in ACM hiPSC-EpiCFs compared to control hiPSC-EpiCFs by Day 8 (Fig. 5G). Similarly, PKP2KO hESC-EpiCFs exhibited enhanced lipid staining relative to control hESC-EpiCFs (Fig. 5H). Quantification of mean fluorescence intensity revealed that at Day 0, lipid content was comparable among groups. However, by Day 8, ACM hiPSC-EpiCFs displayed significantly elevated Nile Red fluorescence compared to both ACM CFs and control EpiCFs (Fig. 5I). A similar trend was observed in the hESC-derived cells, where PKP2KO hESC-EpiCFs exhibited the highest fluorescence intensity at Day 8, indicating increased lipid accumulation compared to all other groups (Fig. 5J). CEBPA and PPARG expression of isogenic control, ACM, and PKP2KO hPSC-CFs and hPSC-EpiCFs with and without adipogenic induction demonstrated enhanced CEBPA expression in ACM hPSC-EpiCFs compared to ACM hPSC-CFs (Fig. 5K, L). These data suggest that ACM-associated and PKP2-deficient epicardial-derived cells have a heightened propensity for lipid accumulation over time, potentially contributing to disease pathology.
A qPCR of fibrotic markers Col1α1 (mean ± SEM, N = 3) and B PDGFRA in hiPSC-EPCs and ACM hiPSC-EPCs (mean ± SEM, N = 4), and in hESC-EPCs and PKP2KO hESC-EPCs (mean ± SEM, N = 3). Statistical significance was evaluated using a one sample t test. C Phase contrast images of non-epicardial derived fibroblasts (ACM, PKP2KO, and isogenic control hPSC-CFs) and D fibroblasts derived from epicardial cells (ACM, PKP2KO, and isogenic control hPSC-EpiCFs). E qPCR of Col1α1 and F PDGFRA in hiPSC-CFs, ACM hiPSC-CFs, hiPSC-EpiCFs, ACM hiPSC-EpiCFs as well as in hESC-CFs, PKP2KO hESC-CFs, hESC-EpiCFs, and PKP2KO hESC-EpiCFs (mean ± SEM, N = 6). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. G, H Representative fluorescence images of Nile red staining of lipids in ACM, PKP2KO, and isogenic control hPSC-EpiCFs and hPSC-CFs during adipogenic induction at days 0 and 8 (scale bar = 100 μm). I Quantification of Nile red fluorescent intensity per cell in ACM hiPSC-CFs and hiPSC-CFs relative to control (hiPSC-CFs at day 0) and ACM hiPSC-EpiCFs and hiPSC-EpiCFs relative to control (hiPSC-EpiCFs at day 0) at day 8 (mean ± SEM, N = 3 of hiPSC-CFs, ACM hiPSC-CFs, hiPSC-EpiCFs, and ACM hiPSC-EpiCFs). J Quantification of Nile red fluorescent intensity per cell in each field of view of PKP2KO hESC-CFs and hESC-CFs relative to control (hESC-CFs at day 0) and PKP2KO hESC-EpiCFs and hESC-EpiCFs relative to control (hESC-EpiCFs at day 0) at day 8 (mean ± SEM, N = 4 of hESC-CFs, PKP2KO hESC-CFs, hESC-EpiCFs, and PKP2KO hESC-EpiCFs). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. K qPCR of CEBPA or L PPARG in hiPSC-CFs, ACM hiPSC-CFs, hiPSC-EpiCFs, ACM hiPSC-EpiCFs, hESC-CFs, PKP2KO hESC-CFs, hESC-EpiCFs, and PKP2KO hESC-EpiCFs (mean ± SEM, N = 6). Statistical significance was evaluated using a one-way ANOVA test with Bonferroni correction. Each N value represents separate hPSC-CFs and hPSC-EpiCFs differentiation from the hPSC state.
Transcriptomic profiling of ACM hiPSC-EPCs reveals dysregulated inflammatory, Wnt, and metabolic signaling pathways
To determine how PKP2 perturbations influence epicardial gene expression, RNA sequencing (RNA-seq) was performed on ACM, PKP2KO and isogenic control hPSC-EPCs (Fig. 6, Supplementary Fig. 2). A heatmap was used to display differentially expressed genes (DEGs) identified using DESeq2 (1202 genes, p < 0.05), with transcription factors and growth factors labeled on the right panel (Fig. 6A). Notably, IGF2 and CEBPA (a transcription factor regulating adipogenesis) were expressed at higher levels in ACM hPSC-EPCs. Reactome pathway enrichment analysis identified changes in the Rho, Interleukin, and in metabolic signaling pathways (Fig. 6B). To further understand the transcription factors regulating DEGs, motif analysis was performed using DNA sequences that are 2 kb upstream and downstream of the DEG start site. HOMER predicted an enrichment in the motif sequences for TEAD Family members (TEAD2 and TEAD4), which are associated with Hippo signaling, as well as Forkhead Family members (FOXA1 and FOXD3), which regulate metabolic and inflammatory processes (Fig. 6C). Lastly, we performed a STRING network analysis to predict distinct clusters of protein-protein interactions associated with biological pathways. Interferon alpha/beta signaling/ISG15-protein conjugation (FDR = 0.0063) and Metabolic pathways (FDR = 0.0263) were two significantly enriched pathways. Key hub proteins in this cluster included IL1B and RSAD2 (which are central to inflammatory and innate immune responses) and IGF2 (which is important to cellular growth, metabolic regulation, and tissue repair), thus indicating a link between metabolic dysregulation and inflammation in ACM. Lastly, Wnt signaling members (WNT5B, WNT16, and FZD1) were identified, reinforcing the role of Wnt signaling in ACM pathology. The presence of IGF2 and IL1B as prominent nodes suggests potential cross-talk between metabolic regulation and inflammatory responses.
A Heatmap representing log2 fold changes in 1202 differentially expressed genes (p < 0.05) between ACM and isogenic control hiPSC-EPCs (N = 5, transcription factors and growth factors annotated on right panel). B Reactome pathways called using differentially expressed genes (Rho-GTPase, interleukins, and IGF signaling terms highlighted in red). C Motif sequences for TEAD2/4, FOXD3/FOXA1, and progesterone receptor (PR) were enriched within 2 kb of the differentially expressed gene transcriptional start sites. D STRING network analysis highlighting the interaction between members of the Wnt Pathway (WNT5B, WNT16, FZD1), IGF (IGF2), interleukin (IL1B), interferon, and metabolic genes.
Next, we performed RNA-seq on PKP2KO hPSC-EPCs and isogenic control H7 hPSC-EPCs. 339 genes were identified to be differentially expressed (p < 0.05) and plotted as a heatmap (transcription factors annotated on the right) (Supplementary Fig. 2A). Reactome pathway analysis revealed upregulation of interferon and interleukin signaling while lipid metabolism was downregulated (Supplementary Fig. 2B). HOMER analysis consistently identified Foxa3 as a potential binding motif within the DEGs along with an interferon regulatory factor motif (ISRE(IRF)/ThioMac-LPS-Expression, Tbox:Smad, and Brn1 motifs (Supplementary Fig. 2C). STRING network analysis consistently identified interferon alpha/beta signaling as a significantly enriched pathway (FDR = 0.0024) including genes within the Wnt family (WNT2, SFRP1, and FRZB) (Supplementary Fig. 2D). Integrated analyses of ACM and PKP2KO hPSC-EPCs revealed that perturbations in PKP2 influence key signaling pathways in epicardial cells, including interferon-mediated inflammation, metabolic regulation, and Wnt signaling, all of which may contribute to the fattyfibro remodeling characteristics of ACM.
Insulin growth factor 2 regulates hPSC-EPC CEBPA and PPARG expression
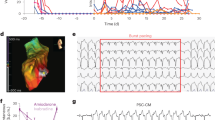
Insulin growth factors are key regulators of adipogenesis and lipid metabolism. Interestingly, we found that IGF2 was significantly elevated in ACM hPSC-EPCs compared to isogenic controls. To assess whether IGF2 is also elevated in the hearts of ACM patients, we performed qPCR for IGF2, PPARG, and CEBPA on control and ACM human right ventricle heart tissue. Tissue from ACM patients was found to have a significant increase in the levels of IGF2, PPARG, and CEBPA compared to control human heart tissue (Fig. 7A). To validate whether IGFs regulate markers involved in ACM fattyfibro pathology, we exogenously treated control hESC-EPC and PKP2KO hESC-EPC with IGF2 for six days (Fig. 7B). Both control and PKP2KO hESC-EPCs treated with IGF2 exhibited a significant increase in CEBPA and PPARG expression (Fig. 7C). To assess whether inhibition of IGF signaling could reduce the expression of pro-adipogenic transcription factors, we utilized an IGF-1R inhibitor (GSK-1904529), which has previously been shown to reduce CEBPA expression and adipocyte formation during adipocyte differentiation of human bone marrow stromal stem cells27 and reassessed lipid uptake upon adipogenic induction in control and PKP2KO hPSC-EPCs treated with GSK1904529A for eight days (Fig. 7D). While Nile Red staining was significantly reduced in control hPSC-EPCs with GSK1904529A treatment, no significant change was observed when PKP2KO hPSC-EPCs were treated with GSK1904529A (Fig. 7E). Furthermore, we evaluated whether IGF-1R inhibition could modulate the expression of CEBPA and PPARG by qPCR (Fig. 7F). While IGF-1R inhibition reduced the expression of CEBPA (p = <0.0001, N = 4) and PPARG (p = 0.0041, N = 4) in control hPSC-EPCs, no significant change in CEBPA and PPARG expression was observed in PKP2KO hPSC-EPCs treated with GSK1904529A.
A qPCR analysis of human heart tissue shows elevated expression of IGF2 (p = 0.0526, N = 4), PPARG (p = 0.0827, N = 3), and CEBPA (p = 0.0972, N = 3) in ACM patients compared to control hearts. B Phase-contrast images of control and PKP2KO hPSC-derived epicardial cells (hPSC-EPCs) after six days of adipogenic induction, with or without IGF2 treatment. IGF2 induces morphological changes consistent with adipogenic differentiation. C qPCR analysis of CEBPA and PPARG expression in control (gray) and PKP2KO (purple) hPSC-EPCs treated with IGF2. In control hPSC-EPCs, IGF2 treatment significantly increases CEBPA (p = 0.0082, N = 6) and PPARG (p = 0.0186, N = 5). In PKP2KO hPSC-EPCs, IGF2 also elevates CEBPA (p = 0.0112, N = 6) and PPARG (p = 0.0.0681, N = 5). D Nile Red staining reveals lipid accumulation after 8 days of adipogenic induction with or without IGF-1R inhibitor GSK1904529A. Lipid content is visibly reduced in control hPSC-EPCs following inhibitor treatment, with no obvious change in PKP2KO hPSC-EPCs. E Quantification of Nile Red fluorescence intensity confirms that GSK1904529A significantly reduces lipid accumulation in control hPSC-EPCs (p 0.0386, N = 5), but not in PKP2KO hPSC-EPCs (p = 0.3967, N = 5). F qPCR analysis of CEBPA and PPARG expression in hPSC-EPCs treated with GSK1904529A. In control cells, CEBPA (p < 0.0009, N = 4) and PPARG (p = 0.0205, N = 4) levels are significantly reduced. In contrast, GSK1904529A has no significant effect on CEBPA (p = 0.9106, N = 4) or PPARG (p = 0.4736, N = 4) expression in PKP2KO hPSC-EPCs. Data are presented as mean ± SEM. Statistical comparisons were made using a one sample t-test.
Discussion
The progressive accumulation of fat and fibrosis within the myocardium is a characteristic pathology seen in ACM, which can lead to cardiac dysfunction, ventricular arrhythmias, and sudden cardiac death. Understanding the molecular mechanisms underlying fattyfibro pathology is critical for developing targeted interventions to prevent or reverse disease progression. However, studying the mechanisms by which this pathology arises is limited, as many mouse models do not fully recapitulate the pathogenesis of ACM observed in human hearts. To address this limitation, we sought to utilize a human model to understand the molecular mechanisms regulating fattyfibro remodeling in ACM. Epicardial cells are predicted to contribute to the fattyfibro pathology given that: (1) a significant increase in WT1+ (an epicardial marker) mesenchymal cells was found in the fattyfibro subepicardial regions of ACM hearts17, (2) epicardial cells have the capacity to differentiate into various cell types within the heart upon injury, and (3) fattyfibro remodeling is initiated subepicardially21. Thus, we generated epicardial cells from ACM and PKP2KO patient hPSC lines to investigate the molecular mechanisms leading to ACM fattyfibro pathology.
An initiating step involved in epicardial activation is an epithelial-to-mesenchymal transition followed by cell migration, both of which facilitate epicardial-derived cell differentiation and pathological remodeling. Our findings demonstrate that ACM and PKP2KO hPSC-EPCs expressed lower levels of E-cadherin, higher levels of N-cadherin, and migrated faster than isogenic control hPSC-EPCs, suggesting that perturbations in desmosomal proteins promote an epithelial-to-mesenchymal transition. Given that cell migration involves the loss of cell-to-cell contacts and results in disassembly of junctional proteins28, it is not surprising that hPSC lines with desmosomal mutations display an inherently less adherent state, predisposing them to increased motility. Following cell migration, epicardial cells differentiate into various cell types, therefore we sought to understand the intrinsic potential of epicardial cells to differentiate into a more fibrotic or fatty phenotype when harboring PKP2 mutations. To model the ability of epicardial cells to develop a fatty phenotype, epicardial cells were subjected to an adipogenic induction protocol. Epicardial cells derived from ACM hPSC lines accumulated significantly larger quantities of lipids, suggesting that mutations in desmosome proteins lead to cellular metabolic changes that promote adipogenesis. This is consistent with a prior study demonstrating increased lipid accumulation in knock-out Pkp2 neonatal rat epicardial cells using an adipogenic induction protocol16. In addition, increased lipid accumulation was also found in other ACM cellular models, including fibro-adipocyte progenitors and cardiac mesenchymal stromal cells29. To model the extent of which epicardial cells in ACM contribute to a fibrotic phenotype, we assessed the expression levels of two fibroblast markers (PDGFRA, Col1α1) in ACM and PKP2KO hPSC-EPCs. At baseline, both ACM and PKP2KO hPSC-EPCs expressed higher PDGFRA and Col1α1, demonstrating that ACM hPSC-EPCs are primed to develop a fibrotic phenotype. Furthermore, given that fibroblasts derived from epicardial cells (hPSC-EpiCFs) expressed higher levels of PDGFRA and Col1α1 when compared to fibroblasts that did not go through an epicardial cell state (hPSC-CFs), hPSC-EpiCFs may also be a specific fibroblast population with enhanced fibrotic characteristics. However, ACM and PKP2KO hPSC-CFs also expressed elevated levels of Col1α1 and PDGFRA when compared to isogenic control hPSC-CFs, suggesting that hearts from ACM patients may also develop interstitial fibrosis.
To understand the signaling pathways and molecular mechanisms leading to fattyfibro remodeling, we performed RNA-seq on isogenic control and ACM hPSC-EPCs. STRING network analysis identified an interaction between cytokines (IL1B, interferon signaling), regulators of Wnt signaling, Rho-GTPases, and genes involved in adipogenesis (IGF2, CEBPA). Additionally, motif analysis predicted an enrichment of genes regulated by TEAD transcription factors (Hippo pathway) and forkhead transcription factors. Therefore, it is likely that a cross-talk between insulin growth factors, Wnt signaling, and the Hippo pathway regulates fattyfibro remodeling in ACM. This is supported by prior studies demonstrating that IGF/PI3K-AKT signaling removes the suppressive effects of foxhead transcription factors on CEBPA and PPARG promoters and represses Wnt signaling. Furthermore, IGF signaling has been shown to suppress Hippo signaling by promoting YAP/TAZ degradation resulting in PKP2KO mouse hearts with reduced YAP expression. Finally, IGF-1R expression is well known to be regulated by Hippo-YAP/TAZ signaling30.
To support the involvement of insulin growth factor signaling in epicardial dependent fattyfibro remodeling, human heart tissue from ACM patients was found to express significantly elevated levels of IGF2, PPARG, and CEBPA. In addition, in a previous study, single-nuclei RNA-seq performed on ACM patient hearts reported an upregulation of insulin growth factor signaling in ACM hearts31. Furthermore, other studies17,32,33 have also reported that the expression of CEBPA and PPARG is upregulated in ACM. In our study, exogenous treatment with IGF2 performed on PKP2KO and control hPSC-EPCs promoted the expression of CEBPA and PPARG. Furthermore, IGF-1R inhibition by GSK1904529A decreased the expression of CEBPA and PPARG in control hPSC-EPCs but did not alter the levels of CEBPA and PPARG in PKP2KO hPSC-EPCs. The selective decrease in CEBPA and PPARG levels in control cells but not in PKP2KO cells may suggest that the loss of PKP2 alters the cell’s dependency on IGF-1R signaling. In addition, the effects of IGF2 may be IGF-1R independent. Therefore, while elevated insulin growth factor signaling may exacerbate fattyfibro remodeling in ACM patients, IGF-1R receptor inactivation alone may not be sufficient to prevent fattyfibro formation.
Our findings establish a human epicardial model of ACM that recapitulates key pathological features of fattyfibro remodeling and implicates IGF2 signaling, Wnt pathway modulation, and Hippo pathway suppression in the regulation of adipogenic and fibrotic differentiation. However, several critical questions remain. First, the mechanisms by which desmosomal dysfunction initiates or sensitizes epicardial cells to IGF2 signaling remain to be elucidated. Future studies should investigate whether altered mechanotransduction or junctional instability in PKP2-deficient cells primes transcriptional programs via YAP/TAZ dysregulation or epigenetic remodeling. Second, the relative contributions of IGF-1R-dependent versus independent mechanisms in mediating IGF2-driven remodeling warrant further investigation. This includes defining potential compensatory pathways or alternate receptors involved in IGF2 responsiveness, particularly in the context of PKP2 loss. Third, the distinct differentiation trajectories and phenotypic properties of hPSC-derived epicardial fibroblasts (EpiCFs) versus cardiac fibroblasts (CFs) raise the possibility that targeting epicardium-derived stromal subsets may offer greater therapeutic specificity. Single-cell transcriptomic and epigenomic analyses of epicardial derivatives in ACM hearts will be valuable to identify lineage-specific regulators of fattyfibro transformation. Finally, future in vitro studies utilizing engineered heart tissue or cardiac organoids incorporating epicardial components will be essential to validate therapeutic strategies that target IGF signaling or its downstream effectors. Together, these studies will help define new entry points for preventing or reversing fattyfibro remodeling in ACM and may offer broader insights into epicardial contributions to cardiac disease.
Methods
Reprogramming human peripheral blood mononuclear cells (PBMCs) from ACM family members into hiPSCs
ACM and family matched control blood was obtained through consent and IRB approval at Stanford and at John’s Hopkins University from two family cohorts (1025 and 398). All ethical regulations relevant to human research participants were followed. Individuals 1025–100, 398–100, and 398–110 were diagnosed with ACM, while individuals 1025–110, 1025–201 were derived from family-matched control individuals without ACM. PBMCs were reprogrammed into hiPSC lines as previously published34. hiPSCs were grown in E8 media (Thermo Fisher Scientific) on Matrigel (Corning) coated 6-well plates until passage 8–10 and deposited into the Stanford Cardiovascular Institute Biobank.
Sanger sequencing hiPSC lines
DNA was isolated from each hiPSC line using the JETFLEX Genomic DNA Purification Kit (Ca no: A30700, Thermo Fisher Scientific) and PCR was performed using PrimeSTAR GXL Master Mix (Ca no: R051A, Takara) containing one of the following primer sets: PKP2_1849C > T-F1: tcctccccttctccagctcaaca, PKP2_1849C > T-R1: agcagtacggttcatgggtagca, PKP2_1849C > T-F2: ccgcagtttaagcacacccagtt, PKP2_1849C > T-R2: atacccaaggccaaatatcaggg, PKP2_2013delC-F1: tctggtctcctggtttgagtgtga, PKP2_PKP2013delC-R1: tcacgacagaaagaaggcccacc. Sanger sequencing was performed at Eton Biosciences and analyzed using the ApE Plasmid editor software35.
CRISPR/Cas9 knock-in and isogenic control hiPSC lines
The PKP2 P672fsX740 2013delC or PKP2 Q617X 1849T > C mutations were corrected in two ACM hiPSC lines (1025–100, and 398–110) using CRISPR/Cas9 using the following protocol36. To correct the PKP2 Q617X 1849C > T mutation, hiPSC line 1025–100 was transfected with 1 µg of the pX458 (Addgene) plasmid expressing Cas9 and the guideRNA (gtcataacctctcctactagc) along with 3–4 µg of the single-stranded oligodeoxynucleotides (ssODN) (catccaatacttttgttgttgtcagtctggatattccggttttgaatatagatattctgggaatatttctctgggagctctgcctccagctggtaggagaggttatgaagaatgcacacacaattct). To generate the isogenic corrected PKP2 2013delC hiPSC line (C > X), line 398–110 was transfected with 1 µg of the pX458 plasmid expressing Cas9 and the guideRNA (gcggaggaaaagagcaaccca) along with 3–4 µg of the ssODN (gcttcttgtgtgtagttgcggacacttttggcgatcaaggacagatacatccttataacaatggaatgccacagccactccacgcccttggggttgctcttttcctccggcatcggcacgtcctggt). Transfections were performed using 5 µL lipofectamine 3000 and 10 µL 3000 reagent (Thermo Fisher Scientific) on 1 million cells. Two days after transfection, GFP+ cells were sorted using the FACS Aria (BD) and seeded at a cell density of 2000 cells per 6-well dish. Colonies were picked, expanded, and sequence verified using Sanger sequencing. Isogenic corrected hiPSC lines were deposited into the Stanford Cardiovascular Institute Biobank.
Generation of a PKP2 knockout (PKP2KO) hESC line
To generate a human embryogenic stem cell (hESC) PKP2 knockout (KO) line (PKP2KO), H7 hESCs (WiCell) were passaged using Accutase, and 125,000 cells were plated into one well of a 24-well plate in E8 media plus 10 µM Y-27632 overnight. The following day, two guideRNAs (Synthego) designed to target a common exon (ENST00000070846.11: exon 2) encoding PKP2 (guideRNA1: UCCUACCUUUAGCAUGUCAU, guideRNA2: ACCUAUGACAUGCUAAAGGU) were resuspended in Tris-acetate EDTA (TAE) to a final concentration of 3 μM. In one 0.5 µL Eppendorf tube, 2 µL of each 3 μM guideRNAs was combined with 4 µL of 3 μM Cas9 protein (IDT) in 40 µL OptiMEM. In a separate 0.5 µL Eppendorf tube, 4 µL of RNAiMAX (Thermo Fisher Scientific) was added to 37 µL OptiMEM. Both tubes were combined and incubated at room temperature for 10 minutes. The combined RNP/RNAiMAX complex was added to the well of H7 cells with fresh E8 media. After four days, H7 cells were passaged using Cell Dissociation Buffer (Thermo Fisher Scientific) and plated at a density of 2000 cells per 6-well plate. After culturing for twelve days, H7 colonies were picked and sequenced at Eton Biosciences. Clones with deleted nucleotides within exon 2 of PKP2 were further propagated. To verify PKP2 was not expressed at the protein level, clones were differentiated into cardiomyocytes (hiPSC-CMs) or epicardial cells (hiPSC-EPCs), and Western blots were performed using antibodies against PKP2 (Cat no: PA5-53144, Thermo Fisher Scientific). The PKP2KO hESC line was deposited into the University of Arizona iPSC Core.
Characterization of iPSC lines
Chromosomal abnormality assessment, CRISPR-off target, and expression of pluripotent makers were previously performed on the PKP2 2013delC hiPSC line17. To confirm hPSC lines expressed pluripotent markers (OCT4 and SSEA4), immunofluorescence was performed as previously described37. To detect for chromosomal abnormalities such as duplications and deletions that may have occur during iPSC reprogramming, culture, or during CRISPR gene editing, DNA was isolated from the PKP2KO, H7 parental control line, PKP2 1849C > T/- and isogenic control line using the JETFLEX Genomic DNA Purification Kit (Ca no: A30700, Thermo Fisher Scientific). DNA samples were submitted to the UCLA Stem Cell and Genome Engineering Center for an NGS-based CNV analysis at 0.3X coverage. CRISPR off-target analysis was determined by Sanger sequencing. To predict off-target sites, default setting using the COSMID (CRISPR Off-target Sites with Mismatches, Insertions, and Deletions) tool was used19. Sanger sequencing was performed on each predicted site from each guideRNA used in generating the PKP2KO and PKP2 1849T > C isogenic control line and compared to the unedited/parental line (H7, 1849C > T/-).
Epicardial cells derived from hPSCs
Human PKP2 1849C > T/-, PKP2 2013delC/-, PKP2KO and isogenic control hPSC lines were differentiated into hPSC-EPCs as previously described38 with minor changes. hPSCs were cultured until cells were 80% confluent in E8 medium on Matrigel-coated 6-well cell culturing plate. On day 0, E8 medium was replaced by Dulbecco’s Modified Eagle Medium (DMEM) (Cat no: 10-092-CM, Fisher Scientific) that was supplemented with 128 μg/mL ascorbic acid-2-phosphate with 6 µM CHIR99021 (Cat no: C-6556, LC Laboratories), an agonist of Wnt signaling pathway. After twenty-four hours, the medium was replaced with fresh DMEM with 128 μg/mL ascorbic acid-2-phosphate for the following two days. On day 3, cells were treated for forty-eight hours in DMEM with 128 μg/mL ascorbic acid-2-phosphate supplied with 5 µM IWR-1-endo (Cat no: S7086, Selleckchem), an inhibitor of Wnt signaling pathway. On day 5, IWR-1-endo was replaced with fresh DMEM with 128 μg/mL ascorbic acid-2-phosphate. On day 6, the progenitor cells were dissociated using Accutase for five minutes at 37°C and centrifuged (200 × g for three minutes) to remove the Accutase. Cells were resuspended in epicardial maintenance medium (DMEM medium supplemented with 128 μg/mL ascorbic acid-2-phosphate and 10 μg/mL insulin) containing 10 μM Y27632. Cells were placed in a new Matrigel-coated 6-well culturing plate at a seeding density of 40,000 cells per cm2. On days 7 and 8, the medium was replaced with fresh epicardial maintenance medium supplied with 3 µM CHIR99021. Days 9, 10, and 11, the medium was replaced with fresh epicardial maintenance medium. From day 12 onwards, the cells were treated with 2 μM SB431542 (Cat no: S1067, Selleckchem), a TGF-β inhibitor for long-term maintenance and split when confluent. To split hPSC-EPCs, Accutase was added to the culture well for five minutes, cells were dislodged from the culture well with a pipette, and Accutase was removed by centrifugation (200 × g for 3 minutes). After 2–3 passages (approximately 30 day old hPSC-EPCs), SB431542 was removed from the epicardial maintenance medium three days before the start of experimentation.
Fibroblasts derived from hPSC-EPCs (hPSC-EpiCFs) and cardiac fibroblasts (hPSC-CFs)
To generate fibroblasts from hPSC-EPCs, hPSC-EPCs were treated using an adapted protocol26. hPSC-EPCs from days 10–12 of differentiation were placed in fibroblast medium containing 75 ng/ml bFGF, HLL supplement [500 µg/ml human serum albumin, 0.6 µM linoleic acid, 0.6 µg/ml lecithin], 1 µg/ml hydrocortisone hemisuccinate, 7.5 mM glutamine, and 5 µg/ml rh insulin to promote a profibroblast state. Once showing fibrotic characteristics, cells were maintained and split with the same treatment alongside 5% fetal bovine serum (FBS), allowing the cells to mature into epicardial derived cardiac fibroblasts (hPSC-EpiCFs). Cardiac fibroblasts (hPSC-CFs) that did not pass through an epicardial state were generated using an additional protocol39. In brief, hiPSCs were subjected to 6 µM CHIR99021 to upregulate the Wnt signaling pathway during mesendoderm induction. The medium was replaced after 24 hours with DMEM supplemented with 128 μg/mL ascorbic acid-2-phosphate. The following day, cells were placed in fibroblast medium to promote fibroblast differentiation for 20 days. Cells were split with this medium along 5% FBS and maintained within this medium.
Immunofluorescent imaging
hPSC-EPCs were passed onto Matrigel-coated glass coverslips and were allowed to grow until fully confluent. The media was removed and rinsed with phosphate-buffered saline (PBS) (Cat no: 14190-144, Thermo Fisher Scientific). For cell fixation and permeabilization, room temperature cells were treated for five minutes with 100% methanol that was chilled at −20°C, washed with PBS five times for five minutes, and then blocked using 2% bovine serum albumin (BSA) (Cat no: BP1600-100, Fisher Scientific) in PBS with 0.1% Tween-20 (Cat no: 85113, Thermo Fisher Scientific) (PBST) for one hour at room temperature. After blocking, primary antibodies (Supplementary Table 2) were diluted in 2% BSA in PBST and incubated for one hour at room temperature or overnight at 4°C. The cells were washed with PBST five times for three minutes before the cells were subsequently stained with the corresponding secondary antibodies (Supplementary Table 2) for one hour at room temperature. Cells were washed in a repeated manner and treated with 1:10,000 4’,6-diamidino-2-phenylindole (DAPI) (Cat no: D1306, Thermo Fisher Scientific) for five minutes. Cells were washed three times for three minutes each with PBS and cover-slipped using VECTASHIELD Vibrance® Antifade Mounting Medium (Cat no: H-1700, Vector Laboratories). Imaging was performed using a Nikon Eclipse Ti2 microscope equipped with a pco.edge camera.
Cell migration assay
hPSC-EPCs were seeded with a density of 300,000 cells per well in a 12-well cell culturing plate and were grown until a fully confluent monolayer was formed. hPSC-EPCs were treated with 10 μM cytosine β-D-arabinofuranoside (Cat no: 02100071-CF, MP Biomedicals), a selective DNA synthesis inhibitor to prevent undergoing cell division during the cell migration assay. The cell monolayer was scratched with a sterile P200 pipette tip to form a gap. The cell monolayer was imaged at hours 0, 4, 8, 24, and 48 using a Nikon Eclipse Ti2 microscope equipped with a pco.edge camera. The gap area of each image was quantified using ImageJ software with the wound size tool plugin installed40.
Adipogenic induction and lipid uptake quantification
hPSC-EPCs were seeded at a density of 150,000 cells per well in a 24-well cell culturing plate and grown to 90% confluency. Cells were treated with adipogenic induction medium [DMEM with 100 μM indomethacin, 500 μM 3-isobutyl-1-methylxanthine (IBMX), 700 nM dexamethasone, 1 μM insulin, and chemically defined lipid concentrate] for eight days with the media being replaced every other day41. For IFNγ treatment, cells were subjected to 50 ng/ml IFNγ (Ca no: 300-02, Peprotech) in adipogenic induction medium. Lipid staining was performed on days 0, 4, and 8 during adipogenic induction by incubating 20 ng/ml Nile red stain (Cat no: 300-02, Peprotech) for 20 minutes at 37°C. hPSC-EPCs were live imaged using the Nikon Eclipse Ti2 microscope equipped with an mCherry filter, which has an excitation range of 550–590 nm and an emission range of 608–683 nm at 50 milliseconds of exposure. Fluorescence intensity of the acquired images were saved in TIF format and then quantified using ImageJ software by measuring the mean grey value of each acquired image42.
Protein quantification using western blots
Protein was isolated from hPSC-EPCs and collected using a lysis buffer [50 mM tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, and protease inhibitor (Cat no: 11836153001, Sigma Aldrich) and sodium orthovanadate (Cat no: P0758S, New England BioLabs)]. Protein concentration was determined using the Pierce BCA Protein Assay. Sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and Western blot were performed using Mini-PROTEAN Tetra Vertical Electrophoresis Cell with Mini Trans-Blot (Ca no: 1703935, Bio-Rad). For each lysate, 30–40 μg of protein was loaded onto a 1.5 mm, 10–12% polyacrylamide gel and transferred to a polyvinylidene fluoride (PVDF) membrane using transfer buffer [25 mM Tris, 192 mM glycine, 20% methanol] overnight at 4°C. The next day, the membrane was blocked for an hour using tris-buffered saline (TBS) [50 mM Tris-Cl, (pH 7.5), 150 mM NaCl] with 0.1% Tween-20 (TBST) and incubated overnight at 4°C with the primary antibodies (Supplementary Table 3) diluted in 5% BSA in TBST. On the final day, the membranes were washed five times with TBST for ten minutes and incubated with the corresponding secondary antibodies (Supplementary Table 3) for one hour at room temperature. Membranes were subsequently washed again five times for ten minutes with TBST, and protein bands were visualized using the LI-COR Odyssey CLx imaging system (LI-COR Biosciences, Lincoln, NE). Protein densitometry was quantified and analyzed using Image Studio software43 and GraphPad Prism 9.5.1.
RNA isolation, RNA sequencing, and quantitative polymerase chain reaction (qPCR)
hPSC-EPCs were seeded with a density of 600,000 cells per well in a 6-well cell culturing plate and were grown until confluent. Cells were either subjected to either adipogenic induction, epicardial maintenance medium with 1 ng/ml TGFβ (Cat no: 100–21 C, Peprotech), or left untreated. Cells were treated for eight days and replaced with fresh medium every other day. Subsequently, cells were lysed with TRIzol (Cat no: R2050-1-200, Zymo Research), and total RNA was extracted and purified from the TRIzol lysates using the Direct-zol RNA MiniPrep kit and protocol (Cat no: R2052, Zymo Research). RNA-sequencing (RNA-seq) was performed targeting >20 million, 2×150 paired-end reads (Novogene). Reads were mapped to the hg38 genome using STAR, and a count matrix was produced using DESeq2. A string network was generated using STRING44, motif analysis using HOMER45, and gene enrichment analysis was performed using GSEA46. For quantitative polymerase chain reaction (qPCR), the total RNA was converted to single stranded complementary DNA (cDNA) using the High-Capacity cDNA Reverse Transcription kit (Cat no: 4368814, Thermo Fisher Scientific) or the EpiScript RNase H-Reverse Transcriptase kit (Cat no: ERT12925K, Biosearch Technologies) following the manufacturer’s instructions. cDNA samples were used for qPCR analysis using the GoldBio SYBR green master mix following the manufacturer’s protocol. Transcript quantities were compared using the relative Ct method, where the amount of each target gene is normalized to the amount of endogenous control gene (18S) and relative to the control sample as 2−∆∆Ct. Values were plotted and analyzed using GraphPad Prism 9.5.1 The sequences of primers used for qPCR are included in a table (Supplementary Table 4).
PKP2 mutant molecular modeling
The full-length structure of PKP2 isoform 2 is not available in the Protein Data Bank. Therefore, a computational method was used to model the full-length three-dimensional (3D) structure. Initially, the sequence of PKP2 isoform 2 was retrieved from Uniprot database47 and submitted to Robetta webserver48. A total of five models were retrieved from the Robetta server, and their geometry consistency/reliability were evaluated using SAVES and ProSA49 webservers. Based on these consensus model validation results, the most reliable PKP2 isoform 2 structure was chosen for further study. The most reliable PKP2 isoform 2 structure was then used as a homology template to model the PKP2 isoform 1 using MODELLER software50. After modeling, the predicted PKP2 isoform 1 structures geometry consistency/reliability was validated as described above. Finally, the mutant protein (i.e., PKP2 isoform 1) structure was generated using Discovery Studio Visualizer.
IGF2 and IGF-1R inhibitor treatment
hPSC-EPCs were seeded with a density of 600,000 cells per well in a 6-well cell culturing plate and were grown until fully confluent. The untreated cells only contained DMEM. Treated cells were subjected to DMEM medium with 10–50 ng/mL IGF2. Cells were treated for six days replaced with fresh treatment solutions every other day. Phase images were taken on days 0, 3, and 6 during the treatment period. On day 6, the cells were lysed, and total RNA was extracted to perform qPCR. To measure the effects of IGF-1R inhibition during adipogenic induction, control and PKP2KO hPSC-EPCs were seeded with a density of 150,000 cells per well in a 24-well culture plate. hPSC-EPCs were grown until confluent and subjected to the adipogenic induction described above. Control wells (untreated) were only exposed to the adipogenic induction medium, while IGF-1R inhibitor wells were treated with 1 µM GSK1904529A (S1093, Selleckchem) in adipogenic induction medium. Cells were treated for eight days with the medium and replaced with fresh treatment solutions every other day. Nile Red fluorescent intensity was quantified on day 8.
Human ACM and control heart tissue
Human ACM heart tissue and unaffected control heart tissue (10–20 mg) was obtained under IRB approval from the Bruce McManus Cardiovascular Biobank at St. Paul’s Hospital affiliated with the University of British Colombia (H23-03278). All ethical regulations relevant to human research participants were followed. All tissue samples were dissected from the same anatomical position (mid-RVFW). Heart tissue was minced using a sterile blade and placed in TRIzol. RNA was isolated and converted to cDNA using the same method described above. Samples were processed for qPCR using primers for CEBPA, PPARG, and IGF2.
Statistics and reproducibility
All experiments were performed at least in biological triplicate (n ≥ 3 independent differentiations or cell preparations), with each differentiation or preparation treated as a biological unit. No data points were excluded. Data are presented as mean ± standard error of the mean (SEM) unless otherwise noted. Statistical comparisons between two groups were made using unpaired two-tailed Student’s t-tests with Welch’s correction when variances were unequal, or one-sample t-tests when one group was normalized to a fixed reference. Multiple group comparisons were analyzed by one-way or two-way analysis of variance (ANOVA) with Bonferroni (for one-way) or Tukey’s (for two-way) post-hoc correction for multiple testing. A p-value < 0.05 was considered significant.
RNA-seq experiments were conducted on at least four independent differentiations per condition. Raw read counts were normalized and tested for differential expression using DESeq2. Due to high inter-line variance, a p < 0.05 threshold was used. Gene set enrichment and network analyses were performed using GSEA and STRING, respectively, with default settings.
Ethics declarations
All ethical regulations relevant to human research participants were followed. Written informed consent was obtained from all participants prior to enrollment. The study protocol was approved by the Institutional Review Boards of Stanford University and Johns Hopkins University, and all data were handled in a HIPAA-compliant, de-identified manner.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included in this published article [and its supplemental information files], and all relevant data are available from the corresponding author on reasonable request. Unprocessed gels/scans and numerical source data provided as Supplementary Fig. 3 and Supplementary Data 1. RNA-seq data are deposited under the GEO accession number GSE301005.
References
Marcus, F. I. et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur. Heart J. 31, 806–814 (2010).
Thiene, G., Nava, A., Corrado, D., Rossi, L. & Pennelli, N. Right ventricular cardiomyopathy and sudden death in young people. N. Engl. J. Med. 318, 129–133 (1988).
Nava, A. et al. Clinical profile and long-term follow-up of 37 families with arrhythmogenic right ventricular cardiomyopathy. J. Am. Coll. Cardiol. 36, 2226–2233 (2000).
Alcalai, R., Metzger, S., Rosenheck, S., Meiner, V. & Chajek-Shaul, T. A recessive mutation in desmoplakin causes arrhythmogenic right ventricular dysplasia, skin disorder, and woolly hair. J. Am. Coll. Cardiol. 42, 319–327 (2003).
Garcia-Gras, E. et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J. Clin. Invest. 116, 2012–2021 (2006).
Fressart, V. et al. Desmosomal gene analysis in arrhythmogenic right ventricular dysplasia/cardiomyopathy: spectrum of mutations and clinical impact in practice. Europace 12, 861–868 (2010).
Sommariva, E., Stadiotti, I., Perrucci, G. L., Tondo, C. & Pompilio, G. Cell models of arrhythmogenic cardiomyopathy: advances and opportunities. Dis. Models Mech. 10, 823–835 (2017).
Gerull, B. & Brodehl, A. Genetic Animal Models for Arrhythmogenic Cardiomyopathy. Front. Physiol. 11, 624 (2020).
Yamaguchi, Y. et al. Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPARgamma activation. Proc. Natl. Acad. Sci. USA 112, 2070–2075 (2015).
Ma, D. et al. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 34, 1122–1133 (2013).
Quijada, P., Trembley, M. A. & Small, E. M. The role of the epicardium during heart development and repair. Circ. Res 126, 377–394 (2020).
Lighthouse, J. K. & Small, E. M. Transcriptional control of cardiac fibroblast plasticity. J. Mol. Cell. Cardiol. 91, 52–60 (2016).
Christoffels, V. M. et al. Tbx18 and the fate of epicardial progenitors. Nature 458, E8–E9 (2009).
Volz, K. S. et al. Pericytes are progenitors for coronary artery smooth muscle. eLife 4, e10036 (2015).
Pérez-Pomares, J.-M. et al. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int. J. Dev. Biol. 46, 1005–1013 (2002).
Matthes, S. A., Taffet, S. & Delmar, M. Plakophilin-2 and the migration, differentiation and transformation of cells derived from the epicardium of neonatal rat hearts. Cell Commun. Adhes. 18, 73–84 (2011).
Kohela, A. et al. Epicardial differentiation drives fibro-fatty remodeling in arrhythmogenic cardiomyopathy. Sci. Transl. Med. 13, eabf2750 (2021).
Stroud, M. J. et al. Luma is not essential for murine cardiac development and function. Cardiovasc. Res. 114, 378–388 (2018).
Cradick, T. J., Qiu, P., Lee, C. M., Fine, E. J. & Bao, G. COSMID: a web-based tool for identifying and validating CRISPR/cas off-target sites. Mol. Ther. Nucleic Acids 3, e214 (2014).
Aliyari Ghasabeh, M. et al. Epicardial fat distribution assessed with cardiac CT in arrhythmogenic right ventricular dysplasia/cardiomyopathy. Radiology 289, 641–648 (2018).
Pilichou, K., Bezzina, C. R., Thiene, G. & Basso, C. Arrhythmogenic cardiomyopathy: transgenic animal models provide novel insights into disease pathobiology. Circ. Cardiovasc. Genet. 4, 318–326 (2011).
Shinde, A. V. & Frangogiannis, N. G. Fibroblasts in myocardial infarction: a role in inflammation and repair. J. Mol. Cell Cardiol. 70, 74–82 (2014).
Duan, J. et al. Wnt1/betacatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 31, 429–442 (2012).
Russell, J. L. et al. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ. Res. 108, 51–59 (2011).
Sowa, Y. et al. Direct conversion of human fibroblasts into adipocytes using a novel small molecular compound: implications for regenerative therapy for adipose tissue defects. Cells 10, 605 (2021).
Floy, M. E., Shabnam, F. & Palecek, S. P. Directed differentiation of human pluripotent stem cells to epicardial-derived fibroblasts. STAR Protoc. 3, 101275 (2022).
Ali, D. et al. Multiple intracellular signaling pathways orchestrate adipocytic differentiation of human bone marrow stromal stem cells. Biosci. Rep. 38, BSR20171252 (2018).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Sommariva, E. et al. Cardiac mesenchymal stromal cells are a source of adipocytes in arrhythmogenic cardiomyopathy. Eur. Heart J. 37, 1835–1846 (2016).
Xin, M. et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839–13844 (2013).
Reichart, D. et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science 377, eabo1984 (2022).
Lombardi, R. et al. Cardiac fibro-adipocyte progenitors express desmosome proteins and preferentially differentiate to adipocytes upon deletion of the desmoplakin gene. Circ. Res. 119, 41–54 (2016).
Yuan, P. et al. Single-cell RNA sequencing uncovers paracrine functions of the epicardial-derived cells in arrhythmogenic cardiomyopathy. Circulation 143, 2169–2187 (2021).
Churko, J. M., Burridge, P. W. & Wu, J. C. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol. Biol. 1036, 81–88 (2013).
Davis, M. W. & Jorgensen, E. M. ApE, a plasmid editor: a freely available DNA manipulation and visualization program. Front. Bioinform. 2, 818619 (2022).
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L. & Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344 (2016).
Stansfield, B. N., Rangasamy, S., Ramsey, K., Khanna, M. & Churko, J. M. Generation of an iPSC line from a Pontocerebellar Hypoplasia 1B patient harboring a homozygous c.395 A > C mutation in EXOSC3 along with a family matched control. Stem Cell Res. 65, 102944 (2022).
Bao, X. et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat. Biomed. Eng. 1, 0003 (2016).
Zhang, J. et al. Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat. Commun. 10, 2238 (2019).
Suarez-Arnedo, A. et al. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS One 15, e0232565 (2020).
Wen, J. Y. et al. Maturation-based model of arrhythmogenic right ventricular dysplasia using patient-specific induced pluripotent stem cells. Circ. J. 79, 1402–1408 (2015).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Pillai-Kastoori, L., Schutz-Geschwender, A. R. & Harford, J. A. A systematic approach to quantitative western blot analysis. Anal. Biochem. 593, 113608 (2020).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
UniProt, C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Kim, D. E., Chivian, D. & Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004).
Wiederstein, M. & Sippl, M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 (2007).
Webb, B. & Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2199, 239–255 (2021).
Acknowledgements
Research reported in this publication was supported by the Sarver Heart Center Investigator Awards, Gootter-Jensen Foundation, the BIO5 Institute, and the NHLBI (T32HL007249). The Johns Hopkins ARVC Program is supported by the Leonie-Wild Foundation, the Leyla Erkan Family Fund for ARVD Research, The Hugh Calkins, Marvin H. Weiner, and Jacqueline J. Bernstein Cardiac Arrhythmia Center, the Dr. Francis P. Chiramonte Private Foundation, the Dr. Satish, Rupal, and Robin Shah ARVD Fund at Johns Hopkins, the Bogle Foundation, the Campanella family, the Patrick J. Harrison Family, the Peter French Memorial Foundation, and the Wilmerding Endowments.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.M.C.; methodology, J.M.C., S.G.K., J.B.I., S.S., N.M., and S.L.F.; validation, J.M.C., S.L.F., J.B.I., N.M., and S.G.K.; formal analysis, J.M.C., S.L.F., S.S., and S.G.K.; investigation, J.M.C., S.G.K., R.L.G., N.M., and S.L.F.; resources, J.M.C., S.G.K., Z.L., P.H., E.J., T.M., L.R., H.C., C.A.J., and S.L.F.; writing—original draft, S.L.F., J.M.C.; writing—review & editing, J.M.C., S.L.F., J.B.I., B.E.B, and S.G.K.; visualization, J.M.C., J.B. I., S.G.K., and S.L.F.; supervision, J.M.C.; funding acquisition, J.M.C.
Corresponding author
Ethics declarations
Competing interests
C.A.J. has received research funding from Tenaya Therapeutics, Lexeo Therapeutics, Rocket Pharmaceuticals, ARVADA Therapeutics, StrideBio Inc., and EicOsis, Inc., and has served as a past consultant for Lexeo Therapeutics. HC has received research support from Tenaya Therapeutics, EicOsis Inc., and Stride Bio. All other authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Ngan Huang and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Falana, S.L., Kazmouz, S.G., Iwanski, J.B. et al. Modelling arrhythmogenic cardiomyopathy fattyfibro pathology with PKP2-deficient epicardial cells derived from human iPSCs. Commun Biol 8, 1502 (2025). https://doi.org/10.1038/s42003-025-08921-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08921-z