Abstract
Metabolic coupling between neurons and glial cells plays a critical role in brain activity and myelin plasticity. Understanding its role in physiological and pathological contexts requires advanced methods to map metabolism and myelin in live tissue with high spatiotemporal resolution. Here, we present a label-free, multimodal, nonlinear optical microscopy platform integrated with an advanced image processing framework that simultaneously maps cellular metabolism and myelin distribution in organotypic cerebellar cultures. We combine third-harmonic generation microscopy for high-resolution myelin imaging with single axon precision with two-photon fluorescence lifetime microscopy of NAD(P)H metabolic biomarker to assess redox states with single-cell resolution. We introduce automated image analysis methods for cell segmentation and myelinated axon detection, enabling quantitative metabolic and myelin assessment in intact tissue during experimental myelination, demyelination and remyelination. Using this framework, we map the 3D myelin distribution in cerebellar folia and identify distinct metabolic signatures in neurons, oligodendrocytes, and microglia. Furthermore, we measure a metabolic shift in microglia along with myelin distribution changes during experimental demyelination. In conclusion, we establish label-free optical imaging as a powerful tool for the non-invasive characterization of neuro-glial metabolic coupling and myelin organization in living brain tissue, opening new perspectives for research in neuroinflammation and neurodegeneration.
Similar content being viewed by others
Introduction
Proper brain activity and myelin formation in the nervous system rely on the metabolic coupling between neurons and glial cells and axon-myelin interaction1,2,3. Glial cells encompass notably myelin-producing oligodendrocytes, astrocytes, and microglia, which are the innate immune cells of the central nervous system (CNS). Under physiological conditions, the brain is intrinsically characterized by metabolic heterogeneity at the cellular level, and the metabolic functions of neurons and glial cells are closely linked to support neuronal function4,5,6,7,8. Recent studies have uncovered a novel role for myelinating oligodendrocytes in delivering glycolytic metabolites such as lactate or pyruvate to fast-spiking axons1,7,9,10,11 and providing an energy reserve for white matter axons with their fatty acid metabolism12. As the strong metabolic cooperation between neurons and glia is essential for neuronal activity and survival, disruptions in this communication can lead to brain diseases5. Demyelination and impaired energy metabolism are indeed hallmarks of neurodegenerative diseases such as multiple sclerosis (MS)3,5,13 and Alzheimer14,15 leading to significant cognitive and behavioral effects. Growing evidence suggests that demyelination and neuronal energy deficiency are tightly connected10,16,17,18,19. Simultaneously, immunometabolism has emerged as a central player in maintaining tissue homeostasis and protection while driving the progression of neurodegenerative diseases20. Microglia can adopt either pro-inflammatory or pro-regenerative/promyelinating phenotypes6, with metabolic changes closely linked to their functional states21,22,23.
The neuron-glia metabolic coupling and its influence on demyelination and remyelination in the nervous tissue are still not well understood at high spatiotemporal resolution. This gap in knowledge hinders the development of improved treatments and therapeutic strategies for neurodegenerative diseases. A significant challenge is the lack of non-invasive imaging methods capable of probing metabolism at the cellular level and visualizing myelin at the single-fiber level in vivo. Traditional imaging techniques such as magnetic resonance imaging (MRI) and positron emission tomography (PET) are constrained by their spatial resolution, making it difficult to capture the intricate spatial complexity and heterogeneity of these processes at the micrometer scale13,24,25,26. Light microscopy and two-photon excited fluorescence (2PEF) can visualize myelinated axons with micrometer resolution using exogenous dyes and transgenic mice27 and can also monitor single-cell energy metabolism through the use of genetically encoded biosensors28. However, these methods often require labeling strategies that may interfere with normal tissue function. Label-free nonlinear optical (NLO) methods, such as 2PEF of endogenous fluorophores, second- and third-harmonic generation (SHG and THG), and coherent anti-Stokes Raman scattering microscopy, offer functional imaging of live tissues with various contrast mechanisms without the need for exogenous staining29,30,31,32,33,34. Additionally, NLO techniques allow for the study of the local environment in femtoliter volumes deep within tissue, thanks to their intrinsic three-dimensional resolution, high penetration depth, minimal out-of-focus photobleaching, and reduced photodamage.
2PEF of intrinsic biomarkers, combined with fluorescence lifetime imaging microscopy (FLIM), has demonstrated significant potential for non-invasive monitoring of metabolic processes in unstained living tissues34,35,36,37,38,39. The primary intracellular sources of endogenous fluorescence are reduced nicotinamide adenine dinucleotide (NADH), reduced nicotinamide adenine dinucleotide phosphate (NADPH) and flavin adenine dinucleotide (FAD), the major fluorescent cofactors of cellular redox (reduction/oxidation) reactions in the cell and central regulators of energy production and metabolism40. Since NADH and NADPH are not spectrally separable, the combined fluorescence is often denoted as NAD(P)H. The lifetimes of NAD(P)H and FAD are highly sensitive to enzymatic binding41, making them valuable indicators of various metabolic pathways, including oxidative phosphorylation (OXPHOS), glycolysis, fatty acid oxidation, and oxidative stress35,36,37,42,43,44. 2PEF lifetime imaging microscopy (2P-FLIM) of these intrinsic metabolites has demonstrated the spatial resolution necessary to characterize metabolic cellular heterogeneity and intracellular compartmentalization45,46,47. This technique has also demonstrated its ability to investigate metabolic patterns and changes in neurons48,49,50, immune cells47,51,52,53,54,55,56,57, and within the context of neuroinflammation and neurodegeneration13,50,51,58,59.
THG is another label-free NLO process in which three photons at the fundamental frequency are converted into one photon at the third-harmonic. THG microscopy is sensitive to optical heterogeneity at the sub-μm scale, such as lipid/water interfaces31, it therefore allows the visualization of the tissue morphology of tissues without staining32,60,61,62. THG has shown the capability to provide label-free, micron-resolution imaging of myelinated axons in both the CNS and peripheral nervous system (PNS) across ex vivo and in vivo mouse and zebrafish models63,64,65,66. THG is, however, not specific to myelin67,68, but we recently demonstrated that THG specificity and sensitivity to myelin can be enhanced using polarization-resolved THG contrast66,69. THG microscopy can visualize both large nerves64 and thin myelinated axons in the cortex in vivo65. It has shown potential for probing myelin damage and pathology63,66.
In this work, we develop and implement a novel label-free NLO imaging framework that simultaneously probes cellular metabolism and myelin distribution in living cerebellar slices, a live model for myelination, demyelination, and remyelination70. We also introduce advanced automated image analysis techniques to quantify single-cell metabolism and the distribution of myelinated axons. First, we present multimodal microscopy images of organotypic cerebellar slices from mice, integrating several contrast modalities: 2PEF lifetime imaging microscopy of NAD(P)H and THG. Using transgenic fluorescent mouse lines to identify different cell types, we demonstrate the visibility of sub-μm myelinated axons in the cerebellar folia and reveal their high metabolic heterogeneity. Next, we develop automated image analysis tools for single-cell segmentation and metabolic fingerprint measurements, as well as automated segmentation of myelinated axons to assess myelin content in nerve tissue. We conduct a systematic analysis of single-cell heterogeneity in cerebellar folia and demonstrate the effectiveness of 2P-FLIM in characterizing the metabolic states of various cell types, including neurons and glial cells. Finally, we showcase the potential of the FLIM-THG imaging platform for studying metabolic shifts in microglia at the single-cell level and monitoring changes in myelin distribution under demyelination and remyelination conditions using the organotypic culture of sagittal cerebellar slices.
Results
Label-free multimodal nonlinear microscopy to probe metabolism and myelin
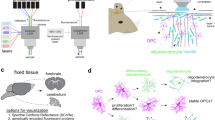
We implemented a label-free, multimodal, nonlinear microscopy platform that combines two-photon excitation, fluorescence lifetime microscopy (FLIM), and third-harmonic generation microscopy (THG). The setup is described in Fig. 1a. We integrated a custom-built microscope with a commercial femtosecond laser that can be tuned at different wavelengths to generate different NLO contrasts. We minimized the pulse width at the sample to optimize THG efficiency generation (See Methods). Fluorescence signals were epi-detected in three different spectral ranges (blue, green, and red), while the THG signal was forward-detected (Fig. 1a).
a The multiphoton microscope optimized for multimodal label-free THG and FLIM imaging with a dual-output laser and TCSPC electronics. b Jablonski diagrams depicting THG and 2PEF absorption at different wavelengths, along with the absorption and emission spectra of the exogenous fluorophores used in this study, including tdTomato, dsRed and NAD(P)H. c Sketch of key metabolic pathways relevant to this study: oxidative phosphorylation in the mitochondrion and glycolysis in the cytoplasm. d Simplified sketch of the mouse cerebellum with an enlarged view of a folium. White matter track and myelinated axons are represented in white, molecular layer in light gray, granule cell layer in intermediate gray and Purkinje cell bodies in dark gray. e Cell types relevant to this study in the cerebellum, including Purkinje cells, oligodendrocytes involved in neuron myelination, and microglia, the immune cells of the central nervous system. Sketches were created using BioRender. f Examples of multimodal imaging showing THG, 2PEF of labeled oligodendrocytes, and the intensity and lifetime of NAD(P)H. g Zoomed-in view of a region of interest of the granule cell layer of the folium.
Multi-contrast imaging was performed sequentially. Label-free imaging of myelin in organotypic cerebellar slices was first performed with THG with an excitation wavelength of 1150 nm to avoid three-photon resonance from blood66,68,71. At this wavelength, we were able to simultaneously excite red fluorescent proteins expressed in the cells of interest using two-photon excitation (see Fig. 1b)72. Emission filters were chosen to specifically select the THG signal (390/40) and the red fluorescence simultaneously (628/32). We then implemented two-photon excitation and FLIM of the intrinsic biomarker NAD(P)H involved in metabolism (Fig. 1c) to perform label-free metabolic imaging in live cerebellar slices (Fig. 1d) and to study the metabolic states of different cells of the nervous tissue, such as Purkinje neurons, oligodendrocytes and microglia (1e) (see Methods). We focused on the endogenous fluorescence of NAD(P)H because it is the most well-characterized metabolic biomarker73 and it enables reliable and robust measurements in the presence of a red fluorophore, due to its significant spectral separation47. We utilized specific transgenic mouse lines to express red fluorescent protein in myelin-producing oligodendrocytes or microglia (see Methods)74,75.
Two-photon excitation of NAD(P)H of the same areas was performed at 760 nm excitation wavelength permitting the simultaneous excitation of NAD(P)H and red fluorescent proteins in the lower part of their absorption spectra29,72 (Fig. 1b). Appropriate emission filters (Fig. 1c) were chosen to select the endogenous fluorescence signal with a good compromise between fluorophore specificity and signal-to-noise ratio (SNR) (see Methods and Fig. 1b)36,43. Under these excitation and emission conditions, NAD(P)H is the dominant contributor to the blue channel signal and interference from other intrinsic fluorophores, such as FAD and lipofuscin, is considered negligible for NAD(P)H lifetime measurements29,34,37,50.
FLIM was implemented with a custom-made time-resolved electronics providing simultaneous channels with 24 temporal bins of 500 ps (see Methods)76. We determined the minimal optimal illumination conditions permitting 2P-FLIM of NAD(P)H with a good SNR and with negligible photoperturbation (see Methods) to create non-invasive metabolic maps of the metabolic coenzymes. The fluorescence lifetime of NAD(P)H varies depending on whether it is free or bound to enzymes. Specifically, protein-bound NAD(P)H exhibits a longer fluorescence lifetime compared to free NAD(P)H41 and NAD(P)H lifetime depends on the type of binding enzymes that influence the fate of cellular glucose carbon, such as pyruvate dehydrogenase (PDH) in mitochondria and Lactate dehydrogenase (LDH) in the cytoplasm77. This difference in fluorescence lifetimes can be exploited to estimate the relative contributions of glycolysis and OXPHOS in cellular metabolism35,37,42,43 as well as to estimate fatty acid synthesis and β-oxidation contribution37. It is important to note that due to the spectral overlap between NADH and NADPH, their collected fluorescence signal is a combination of the two, often referred to as NAD(P)H. As a result, the NAD(P)H signal can reflect changes in both energy production and oxidative stress within cells. Specifically, NADH is primarily associated with OXPHOS in the mitochondria and glycolysis in the cytoplasm, while NADPH is more involved in antioxidant defense mechanisms and biosynthetic pathways.
The fluorescence lifetime decays were analyzed using phasor analysis36,78, as previously described44 (see Methods) and using our custom-written open-source software79 (See Methods). The intensity decay is transformed with an FFT into g and s coordinates. To quantify subcellular metabolism, we calculated the fluorescence lifetime (τϕ) of NAD(P)H at each pixel (Fig. 1f). We also quantified the fraction of bound NAD(P)H by graphically calculating the distance between the multi-exponential experimental point on the phasor plot from the phasor location of free NAD(P)H, assumed to have a single exponential lifetime of 0.4 ns, while no assumption was made on the location of bound NAD(P)H (see Methods)44,47,79.
Organotypic sagittal cerebellar slices are a well-established and suitable model for studying myelination and demyelination, as they maintain axonal survival70,80. Brain slices from the cerebellum were extracted from the brains of 8 and 10-day-old mice (see Method). Representative multimodal NLO images of a live cerebellum slice are shown in Fig. 1f. We used two transgenic mouse lines expressing either dsRed or GFP protein associated with the proteolipid protein (PLP)74,81, enabling the identification of oligodendrocyte bodies and processes. High-resolution THG images of the cerebellar folium reveal myelinated fibers of Purkinje neurons branching out from the white matter tracts and colocalizing with the red processes of oligodendrocytes (Fig. 1g). At the end of each axon, the cell body of the Purkinje neuron is visible as a negative contrast in the THG image, with the Purkinje cell bodies arranged in a crown pattern, as schematized in Fig. 1d. The intensity map of NAD(P)H reveals significant heterogeneity among cells, with some displaying a star shape and very high intensity, while Purkinje cell bodies appear as dark nuclei surrounded by brighter mitochondria. The map of NAD(P)H lifetime reveals metabolic heterogeneity among cells and the subcellular metabolic compartmentalization. Purkinje cell bodies and dendrites display a long NAD(P)H lifetime, while the granule cell layer of the folium has a shorter lifetime. NAD(P)H lifetime maps show heterogeneity at the cellular level.
THG microscopy and automatic myelinated axon segmentation probe dynamic changes of myelin distribution
THG microscopy is emerging as an effective label-free contrast to visualize both large myelinated axons and nerves in different areas of the brain and spinal cord 63,64,66 and small (1–4 micrometers) diameter axons in the cortex of mouse brain65,66. However, automated detection of axons and myelin content in tissue images, as well as quantification of myelin content and distribution, remains challenging and unexplored in brain tissue. Thus, here we aim to: a) develop an analysis workflow to automatically detect axons and determine myelin score in THG images from nervous tissues; b) determine whether this method can detect and quantify changes in myelin distribution during myelination; c) determine the sensitivity to myelinated axons and explore the applicability to 3D THG volumetric imaging.
Recent studies showed that myelin is a primary source of THG contrast in the mouse cortex and corpus callosum66 when the excitation wavelength is set to ≈1100–1150 nm, i.e., far from hemoglobin resonance near 1250–1300 nm68. Here, we extend this observation to cerebellar tissue (Fig. 2a). High-resolution THG-2PEF images from a transgenic line with PLP-GFP in the membrane confirm that THG highlights myelinated axons both when axons are separated from each other (Fig. 2a top right panel) and when they are bundled in white matter tracts in the cerebellum folium (Fig. 2a top bottom panel). THG highlights axons oriented along all 3D directions65,66. Therefore, in 2D THG images out-of-plane axons appear as bright dots while in-plane axons appear as elongated fibers (Figs. 1g and 2a).
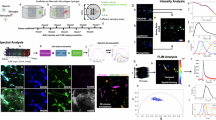
a THG and PLP-GFP in fixed cerebellar slices, along with excitation and emission spectra. b Automated myelin segmentation process and myelin score determination. The steps include preprocessing the raw image (ii), Hessian and Meijering filtering (iii), intermediate labeling (iv) and K-means filtering (v) for the final segmentation. c Application of the segmentation algorithm to 2D THG images during saggital cerebellar slices development shows an increase in myelinated axons density and myelin score as myelination occurs in cerebellar slices from 4 days in vitro (4 DIV) to 8 DIV. d Myelin score during tissue development, where each point represents an ROI from an entire cerebellar folium of the cerebellum slice including all the layers. For 4 DIV: N = 9 ROI from 1 animal (avg ± stdv = 0.0175 ± 0.005), for 5 DIV N = 4: ROI from 1 animal (avg ± stdv = 0.0203 ± 0.009), for 8 DIV: N = 5 ROI from three animals (avg ± stdv = 0.0348 ± 0.016). e Comparison of automated axons detection workflow from THG images and myelin manual tracing in the 3D dataset at the peak of myelination (6 DIV). The first row displays the sum of the images across all the optical sections for raw data (THG), manually segmented data (Manual), and automatically segmented data (Automated), while the second row displays a zoomed-in view of the sum of a few optical sections. A 3D display of the THG raw data and a 3D reconstruction of the manually traced myelinated fibers is shown. 3D reconstruction of the white matter tract outlier (in gray) and myelinated fibers (in blue); the intersections between fibers and white matter tract outlier were highlighted in red. Only the fiber portions outside of the white matter tract are shown. f Comparison between myelin score profiles calculated from automated myelin detection (purple) and manually segmented data (green) as a function of the optical depth at 4 DIV and 6 DIV stages. Myelin scores are calculated in ROIs that encompass the entire cerebellar folium, including all the layers. Myelin scores are normalized by dividing the values of the myelin score by the maximum value in each 3D stack.
We developed a fully automated workflow to detect and segment in-plane axons from 2D THG images (Fig. 2b) by using custom-written and open-source software82,83 (see Methods). The fiber detection workflow (Fig. 2b(i)) comprises four main steps: preprocessing (Fig. 2b(ii)), fiber segmentation (Fig. 2b(iii)), intermediate labeling (Fig. 2b(iv)) and K-means filtering (Fig. 2b(v)). We first optimized the preprocessing step to enhance the quality and reliability of the THG images by applying three filters to the raw THG image (see Methods). The fiber segmentation step was optimized and enhanced by using a combined filtering strategy that integrates Hessian and Meijering approaches (Fig. 2b(iii)). While Hessian-based filtering exhibits high sensitivity to tubular geometries, it is susceptible to noise-induced false positives; in contrast, Meijering filtering demonstrates greater efficacy in attenuating noise contributions82, thereby complementing the Hessian response. We chose a spatial-domain framework over frequency-domain methods (e.g., Gabor or steerable filters) for three reasons: it exploits local geometric properties without costly frequency transforms, reducing computations to simple matrix operations; Hessian-based strategies, unlike frequency approaches, are well validated for vessel and fiber detection (e.g., Frangi vesselness); and Hessian filtering better captures irregular, branching fibrillar architectures, whereas frequency methods are optimized for regular, periodic, globally consistent patterns for texture analysis. An intermediate labeling step was developed to identify single axons (Fig. 2b(iv)) and finally K-means filtering was applied as a post-processing method to separate true myelin structures from other segmented objects in our pipeline. K-means clustering was selected for discriminating genuine fibrils from imaging artifacts owing to its algorithmic simplicity and ease of implementation. While alternative clustering strategies, such as density-based spatial clustering of applications with noise or Gaussian mixture models, impose additional constraints, K-means clustering provides satisfactory performance with minimal parameterization and low computational overhead. Finally we obtained the final mask with pixels that we associate with in-plane axons (Fig. 2b(v)) (see Methods).
We then establish a score to quantify myelin content in a given region of interest (ROI), starting from myelin-associated pixels assigned to the binary mask (Fig. 2b(v)). We defined a “myelin score” as the ratio of the number of pixels in the mask (Fig. 2b(v)) corresponding to myelinated fibers to the total number of pixels of the image, i.e.,: Myelin score = Nfiber/Ntotal where Nfiber represents the number of pixels associated with detected fibers, while Ntotal represents the total number of pixels of the image. We note that the myelin score depends on both the distribution of myelin within the ROI and the ROI size; therefore, the ROI selection and dimensions should be reported, and quantitative comparisons should be made under similar conditions. We also note that the obtained myelin score does not account for out-of-plane myelinated fibers that appear as bright dots or small segments in the 2D THG images. As a result, the score may underestimate the density of axons with out-of-plane orientations. To assess the sensitivity of the developed automated workflow and scoring method, we quantified myelin content and scores in cerebellar folia during myelination occurring in development (Fig. 2c, d). Additionally, we compared automated and manual segmentation of myelin in a 3D THG dataset (Fig. 2e). We used organotypic sagittal cerebellar slices70 as a model to investigate different stages of myelination during development. Folia were imaged from the onset of the myelination process at 4 days in vitro (DIV) to the peak of myelination at 6 DIV, up to DIV 8 (Figs. 2c and S1). At the beginning of myelination, we observed less THG signal from myelinated axons, resulting in a smaller number of segmented fibers in the folium. In contrast, at the peak of myelination, when most axons are myelinated, more THG signal from fibers is observed, leading to a higher number of segmented fibers (Figs. 2c and S1). Following the automated segmentation of axons during development (Fig. S2), we measured an increasing myelin score as myelination progressed (Fig. 2d).
Finally, we extended the automated fiber detection to 3D THG data, where myelinated axons were manually traced (Fig. 2e), and we tested the sensitivity of the method by comparing it to manual tracing in two datasets with different myelin content: at the beginning of myelination (4 DIV) and at the peak of myelination (6 DIV). Manual tracing was performed by identifying myelinated axons displaying an unambiguous THG signal to the human eye, with primary emphasis on large internodes of Purkinje cell axons that showed a high signal-to-background ratio (SBR) (see Fig. S3b and Methods). Following manual tracing, 3D reconstruction of axons was performed at both myelination stages (Fig. S3b), enabling the quantification of the number of myelinated axons, their orientation, length, and tortuosity (Figs. 2e and S3c–f; see Methods). The folium at 6 DIV exhibited a higher number of myelinated axons (Fig. S3c), longer fibers (Fig. S3e), and greater tortuosity (Fig. S3f) compared to the folium at 4 DIV. We then compared the performance of automated fiber detection with manual tracing (Fig. 2e) as well as the resulting myelin scores (Fig. 2f) to demonstrate the capability of the automated workflow to capture myelin fibers. Fig. 2e demonstrates that the automated segmentation (right column) using the developed workflow accurately detects all myelinated axons manually traced (center column) from a 3D THG dataset (left column), both at the level of the entire folium (top row) and within smaller tissue areas (bottom row). We note that automated fiber detection demonstrates greater sensitivity, identifying a larger number of myelinated segments compared to manual tracing, including internodes with weaker THG SBRs-likely representing either developing internodes or Purkinje cell axon collaterals. After normalization, to account for differences in fiber thicknesses between automated and manual detection, we compared myelin scores obtained using both methods across various tissue depths in the folium at both four DIV and six DIV developmental stages (Fig. 2f). The myelin scores derived from the two methods exhibited very similar profiles across tissue depths (Fig. 2f), showing consistency between automated and manual detection, and confirming the sensitivity of the automated method for detecting myelinated axons. Slight discrepancies between the myelin scores estimated from the two methods may result from the underestimation of out-of-plane axonal contributions in the automated detection, the exclusion of internodes with weaker THG SBRs in the manual segmentation, and the dependence of automated segmentation performance on SBR, which varies with imaging depth.
Single-cell endogenous fluorescence analysis reveals metabolic differences between neuronal, oligodendrocytes, and microglia populations
The brain exhibits significant metabolic heterogeneity (Fig. 1f), with cellular metabolic states closely linked to their specific functions. The metabolic interplay between neurons and glial cells plays a crucial role in supporting overall brain activity. Beyond the simple adaptation of energy supply to neuronal consumption, evidence suggests that energy delivery is not a passive response to neuronal demand but is dynamically regulated by an integrated neuron-glia network, actively shaping cerebral activity. Each cell type has distinct workloads and energy requirements, resulting in unique metabolic signatures5,84,85. Neurons are highly specialized for rapid electrical signaling and can exhibit variable spiking activity. This requires maintaining ion gradients through ATP-dependent pumps and supporting processes such as neurotransmitter release and synaptic activity. These energy-intensive tasks necessitate a highly efficient energy production system, primarily mitochondrial OXPHOS, which provides a high ATP yield per glucose molecule5,84,85. Glial cells, such as astrocytes, oligodendrocytes, and microglia, play supportive roles, including maintaining homeostasis, recycling neurotransmitters, myelinating, and modulating immune responses. These tasks are less dependent on rapid ATP generation. Instead, glial cells rely more on glycolysis with respect to neurons2,5,85,86, which is less efficient in ATP production but provides metabolic intermediates that are essential for biosynthesis and antioxidant defense. Of particular importance in MS is the energetic coupling between axons, myelin, and oligodendrocytes. By ensheathing axons, oligodendrocytes serve as a source of lactate for neurons through the myelin sheath. Lactate, a glycolytic product, is essential for neuronal survival and function and plays a critical role in the generation of metabolic energy1,9.
Therefore, we sought to determine whether differences in metabolic states of different cell types can be observed and characterized at the cellular level in living brain tissue, and whether cell-scale analysis can reveal metabolic heterogeneity across the tissue. We implemented single-cell metabolic analysis in three different cell types of cerebellum organotypic slices: (i) Purkinje cells, which are fast-spiking inhibitory neurons (ii) oligodendrocytes, and (iii) microglia. We first developed an automated workflow for cell segmentation as detailed in Figs. 3 and S4 (see Methods). We used the open-source segmentation software CellProfiler87 to segment individual cells from red fluorescence images, where cell bodies were visible with good contrast (Fig. 3 and Fig. S4a). The CX3CR1-CreErt2/Rosa-tdTomato line was used to highlight microglia75, while the PLP-dsRed line was used to highlight oligodendrocyte bodies81. From the single-cell mask, shape parameters—such as cell area and eccentricity were extracted for each cell (see Methods and Supplementary material). To isolate Purkinje neuron cell bodies visible with shadow contrast in the THG image, we first generated a probability map using the deep-learning-based ilastik software88, and then segmented individual cells using the CellProfiler software (Figs. 3 and S4b, see Methods). Once the masks were obtained, they were applied to the images of the intensity or fraction of bound NAD(P)H, as shown in Fig. 3a. The metabolic state of each cell was then estimated by measuring the mean lifetime and distance to free NAD(P)H within each cell’s mask. Higher levels of bound NAD(P)H are generally associated with increased OXPHOS activity, fatty acid synthesis or oxidative stress, whereas lower bound NADH fractions are associated with elevated glycolysis and fatty acid β-oxidation36,37. In healthy cerebellar brain tissue, we observe a different NAD(P)H lifetime (Fig. S5) and fraction of bound NAD(P)H in these different cell types (Fig. 3a, b). Purkinje neurons cell bodies show the longest lifetime and the highest fraction of bound NAD(P)H, possibly indicating the highest OXPHOS metabolism, whereas microglia show the shortest lifetime and the lowest fraction of bound NAD(P)H, indicating the most glycolytic phenotype or the most fatty acid β-oxidation metabolism among these cell types (Fig. 3c). Oligodendrocyte cells have an intermediate value of the fraction of bound NAD(P)H. Their state is therefore more glycolytic or relying more on fatty acid β-oxidation than neurons, but more OXPHOS or relying more on fatty acid synthesis than microglia. These results show that glial cells (microglia and oligodendrocytes) have an overall more glycolytic phenotype or fatty acid β-oxidation compared to neurons, in line with their different functions and metabolic requirements5. Our results are also in agreement with previous in vivo measurements in which microglia exhibited a glycolytic phenotype, characterized by a low NAD(P)H lifetime compared to the surrounding neuropil55 and in vitro measurements in which microglial cells exhibited a lower NAD(P)H lifetime compared to non-microglial cells54. We also find that endogenous fluorescence analysis with single-cell precision highlights metabolic heterogeneity among cells (Fig. 3c), revealing the highest metabolic heterogeneity in microglia.
a Measurement of the fraction of bound NAD(P)H of single cells in a cultured tissue slice. Each column represents a step in the analysis workflow, and each row corresponds to a different cell type. Either the THG map (for Purkinje cells) or the 2PEF map of the exogenous fluorophore (for oligodendrocytes and microglia) was used to segment single cells. These single-cell masks were then applied to the lifetime maps to provide information at the single-cell level. b The fraction of bound NAD(P)H of single-cell measurements reveals metabolic differences between the three different cell types of the cerebellum (Purkinje cells, oligodendrocytes and microglia) as well as metabolic heterogeneity among cells. Each point in the box plot represents a single cell. N = 12 oligodendrocytes from 2 animals (avg ± stdv = 0.443 ± 0.019), N = 17 Purkinje neurons from two animals (avg ± stdv=0.465 ± 0.011), and N = 115 microglia from five animals (avg ± stdv = 0.384 ± 0.034) at 8 DIV. c Purkinje neurons rely more on the oxidative phosphorylation pathway, which is linked to the abundance of bound NAD(P)H. In contrast, oligodendrocytes and microglia have shorter, more glycolytic lifetimes, with microglia having the shortest lifetime, which is linked to the abundance of free NAD(P)H. Sketches were created using BioRender. The asterisk * indicates a statistically significant difference in the data following a t-test analysis (p < 0.05).
Finally, we investigated whether NAD(P)H intensity measurement could provide quantitative and complementary measurements of the metabolic differences between different cell types in living tissues. In Fig. S6, we applied the same single-cell mask to the NAD(P)H intensity images (Fig. S6a) and measured the NAD(P)H intensity distribution in different cell types (Fig. S6b). Despite the very high heterogeneity in intensity values, we observed that both oligodendrocytes and microglia had higher NAD(P)H levels compared to neurons. The elevated levels of NAD(P)H intensity in glial cells are consistent with other label-free 2PEF imaging studies, both in hippocampal slice preparations86 and in engineered three-dimensional brain tissue model50. However, we do not observe a significant difference in NAD(P)H intensity between microglia and oligodendrocytes (Fig. S6b), while they show a difference in lifetime (Fig. 3b). In summary, these results indicate that NAD(P)H FLIM measurements are more sensitive and reliable than NAD(P)H intensity measurements because FLIM measurements are unaffected by fluorophore concentration, excitation variability, or tissue absorption and scattering.
Single-cell endogenous fluorescence analysis reveals metabolic changes in microglia during demyelination and remyelination
Recent studies suggest that, during demyelination and remyelination of neural tissue, microglia exhibit distinct phenotypes characterized by distinct molecular signatures, metabolic states and functions20,21,22. A commonly used yet oversimplified vision categorizes microglia into two states: pro-inflammatory (M1) and anti-inflammatory/pro-healing (M2)89,90. Pro-inflammatory stimuli are generally thought to trigger a metabolic shift from OXPHOS to glycolysis, while a shift to OXPHOS metabolism is associated with protective and regenerative microglial phenotypes that support remyelination91,92,93. However, a more plastic and dynamic perspective is emerging89,94, highlighting the spatial and temporal heterogeneity of microglial transcriptomes, metabolism and function and showing how microglial activation and function are in turn shaped by the local tissue environment, influencing regional differences in remyelination89,95,96. While regional differences in the rate of remyelination have been observed95 their correlation to microglia heterogeneity97,98 is unknown due to a lack of high-resolution imaging approaches in a living tissue.
Therefore, we sought to assess i) whether changes in microglial metabolic function during demyelination and remyelination can be observed at the single cell level in living tissue, simultaneously with changes in myelin distribution, ii) whether such changes are associated with microglial shape and their activation phenotypes (pro-inflammatory and pro-regenerative), and iii) whether these changes correlate with myelin distribution in the cerebellar brain tissue and with the presence of myelin debris within microglia.
For this purposes, we performed multimodal, label-free assessment of single-cell metabolism and myelin content in organotypic saggital cerebellar slices70,80, a model for demyelination and remyelination, by analyzing the NAD(P)H lifetime readout and comparing them with THG image changes (Fig. 4a). We used an overnight demyelinating treatment lysophosphatidylcholine (LPC) (see Methods) to induce demyelination in the cultures slices. This treatment is known to reach the peak of demyelination around 24 hours post treatment70. A spontaneous remyelination initiates three days following demyelination and is achieved within a week. In our experiments, the peak of demyelination in treated slices is observed at 8 DIV, and remyelination is proceeding at 11 DIV (Fig. 4b), which were the timepoints included in this study.
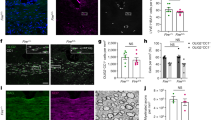
a Maps of THG, myelin segmentation, 2PEF, and distance from free NAD(P)H in the entire tissue, single cells, and a few zoomed-in cells in a region of interest for control (representing healthy condition), LPC at 8 DIV (representing the peak of demyelination), and LPC at 11 DIV (representing the peak of remyelination). b Experimental schedule: slices are kept in the incubator at 0 DIV, and the medium is changed every 2–3 days. LPC is added at 6 DIV to the treated slices to induce demyelination, and imaging is performed at 8 DIV and 11 DIV. c Box plots of the myelin score showing a decrease at 8 DIV and a recovery in remyelinated tissue at 11 DIV, with each point representing an ROI. There are N = 21 ROIs for the control in 10 animals (avg ± stdv = 0.068 ± 0.023), N = 5 ROIs for the demyelinating condition in 3 animals (avg ± stdv = 0.005 ± 0.003), and N = 13 ROIs for the remyelinating condition in five animals (avg ± stdv=0.051 ± 0.017). Myelin scores are calculated in ROIs within the granule cell layer of the cerebellar folium. d Box plots of eccentricity, which measures how circular (eccentricity=0) or elliptical (eccentricity 1) a shape is, for the shape of microglia in different conditions, namely pro-regenerative and pro-inflammatory. A significant difference is observed between pro-inflammatory microglia at 8 DIV compared to pro-regenerative microglia in control and 11 DIV conditions. e A decrease in the lifetime towards a more glycolytic phenotype is seen at 8 DIV, followed by a recovery in the metabolic phenotype at 11 DIV. d, e Each point represents a single cell. N = 256 cells from 10 animals in control (avg ± stdv for fraction of bound NAD(P)H = 0.384 ± 0.041) (avg ± stdv for eccentricity = 0.788 ± 0.145); N = 179 microglia from three animals in the demyelinating condition (avg ± stdv for fraction of bound NAD(P)H = 0.365 ± 0.048) (avg ± stdv for eccentricity = 0.683 ± 0.174) and N = 147 microglia from five animals in the remyelinating condition (avg ± stdv for fraction of bound NAD(P)H = 0.377 ± 0.071) (avg ± stdv for eccentricity=0.742 ± 0.154) with 2–3 ROIs for each animal. f A schematic representation of the microglia states ranging from pro-regenerative to pro-inflammatory, with both lifetime and eccentricity decreasing from the former to the latter. The asterisk * indicates a statistically significant difference in the data following a t-test analysis (p < 0.05).
To achieve high-quality imaging while preserving the physiological health of the tissue, we used the THG contrast modality to identify a myelinated folium (Figs. 1f and S7a). Representative THG images of the entire cerebellar folia for control, demyelination, and remyelination conditions are shown in Fig. S7a. We then performed multimodal THG and FLIM imaging in 2–3 smaller (≈250 × 250 μm) regions within the granule layer of the folium (Fig. 4a). First, we performed THG imaging exciting at 1150 nm and simultaneously collecting THG signals from myelin (Fig. 4a first column) and 2PEF signals from microglia. Subsequently, we performed FLIM imaging with an excitation wavelength of 760 nm, allowing the simultaneous acquisition of 2PEF signals emitted by tdTomato-tagged microglia (Fig. 4a, third column) and NAD(P)H (Fig. 4a, fourth column).
Automatic segmentation of myelinated axons in each region of interest (Fig. 4a second column) reveals a different myelin distribution in demyelinating and remyelinating conditions (Fig. 4a) and allows evaluation of the myelin score in the three different conditions (Fig. 4c). We note that, after the peak of myelination at DIV 6, i.e. between DIV 8 and DIV 11, myelin score of control slices that were not treated with the demyelinating drug LPC is not affected by time of culture (Fig. S8), therefore we merge the control data of the 2 days (Fig. 4c). Control slices consistently exhibited higher myelin scores. In contrast, demyelinating slices had the lowest myelin content and score, reflecting the extensive demyelination of fibers throughout the folia. During the transition to the remyelination phase, the myelin scores increased significantly. Consequently, at 11 DIV, the difference in myelin scores between control and LPC-treated slices became insignificant, indicating progress in remyelination.
In parallel, we performed automated single-cell analysis of the microglial population to assess their shape and metabolic fingerprint (See Methods). It is well-established in the literature that changes in microglial function are often accompanied by changes in cell shape, making morphological analysis a key method for identifying activation phenotypes89,90. Homeostatic and pro-regenerative microglia typically display a “branched” shape with elongated processes, whereas pro-inflammatory microglia exhibit a morphology characterized by round cell bodies and increased phagocytic activity. To quantitatively assess cell shape and morphological differences, we measured cell eccentricity99 (See Methods). In control folia, myelination appeared to be intact, as evidenced by well-defined myelinated fibers observed in THG images. Microglial cells in these samples exhibited elongated processes and high eccentricity values (closer to 1), suggesting active surveillance within the brain tissue (Figs. 4a top and S7a). In contrast, microglia in the demyelinated state displayed a more rounded shape with lower eccentricity values (closer to 0), indicating a polarization towards a pro-inflammatory phenotype Figs. 4a and S7a. During remyelination at 11 DIV, microglial morphology became more versatile and appeared to be influenced by their spatial location within the folia. Near remyelinated regions, microglia in LPC-treated slices tended to exhibit an elliptical shape with higher eccentricity values, resembling an elongated structure. These observations highlight dynamic changes in microglial morphology during the transition to a remyelination state, reflecting their role in tissue remodeling and repair processes. Cellular eccentricity of microglia was quantified across multiple folia and animals, revealing an overall decrease in eccentricity during demyelination, followed by a recovery in remyelinating slices (Fig. 4d). To evaluate the robustness of the microglial eccentricity metric (Fig. 4d), which is automatically assessed at the single-cell level, we compared it to the number of microglial processes (Fig. S7a, third row, and Fig. S7b) that were manually evaluated using Sholl analysis without tracing or reconstruction100, (https://imagej.net/plugins/sholl-analysis). Microglia with high eccentricity values typically exhibit a greater number of processes, whereas those with low eccentricity values have fewer processes (Fig. S7a, third row). This finding suggests a strong correlation between the automatically scored eccentricity and the manually scores number of processes (Fig. S7b).
We then investigated the metabolic patterns across conditions at the single-cell level. To verify the effect of time in culture on microglia metabolism, we first compared the controls samples that were handled identically but not treated with the demyelinating drug LPC. As microglia NAD(P)H lifetime is not affected by the time in culture (Fig. S8) we combined the control groups from 8 DIV and 11 DIV (Figs. 4d, e and S9) to focus our analysis on differences attributable to LPC-induced demyelination and subsequent remyelination. During demyelination, microglia generally exhibited a lower levels of the fraction of bound NAD(P)H (Fig. 4e) and NAD(P)H lifetime (Fig. S9) compared to controls, suggesting a shift toward a glycolytic phenotype or increase in fatty acid β-oxidation metabolism21,37. However, by 11 DIV, at the peak of remyelination, no significant difference with the control was observed, indicating a recovery of the metabolic phenotype in remyelinating slices (Fig. 4e). A general correlation was observed between the fraction of bound NAD(P)H and cell eccentricity (Fig. 4c–e). We also observed a higher value of NAD(P)H intensity in microglial cells during demyelination compared to control and remyelination (Fig. S10). Taken together, these findings suggest that during demyelination, microglia tend to adopt a pro-inflammatory, with a round shape, low eccentricity and low fraction of bound NAD(P)H (Fig. 4f) possibly indicating a metabolic phenotype relying more on glycolysis or fatty acid β-oxidation. In contrast, during remyelination, more cells display a pro-regenerative phenotype with a “branched” shape with processes and high eccentricity values and high fraction of bound NAD(P)H possibly indicating a metabolic phenotype relying more on OXPHOS or fatty acid synthesis. Our findings in living tissue are consistent with previous in vitro studies on macrophages, which reported a decrease in the fraction of bound NAD(P)H in classically activated macrophages (M1/pro-inflammatory) while an increase in the fraction of bound NAD(P)H was observed in alternatively activated macrophages (M2/pro-healing)52. We observed significant metabolic heterogeneity within the cell population, especially under demyelinating conditions (Figs. 4e and S9). This single-cell metabolic variability may reflect regional heterogeneity and dynamic temporal and spatial microglial polarization reflecting a spectrum of states ranging from pro-inflammatory to anti-inflammatory101.
Finally, we explored the feasibility of THG microscopy to detect myelin debris during demyelination. It is known that microglia support remyelination by clearing myelin debris through internalization and by expressing genes associated with phagocytosis and lysosomal pathways97,98,102. For this purpose, we performed automated single-cell analysis using the red fluorescence image acquired simultaneously with THG (Fig. S11a) to ensure perfect co-registration of the images, and we measured the THG intensity inside every cell. In demyelinating folia, we observed higher THG signals within the microglial cytoplasm (Figs. 4a, S7a and S11a), resembling lipid droplets or debris31. This signal possibly corresponds to myelin and lipid debris phagocytosed by active microglia. To quantify this, we defined the “microglia THG score”, a normalized metric defined as \(\,{{{\rm{Microglia}}}}\; {{{\rm{THG}}}}\; {{{\rm{score}}}}\,=\frac{{I}_{{{{\rm{avg}}}}}}{{I}_{\max }}\), where Iavg is the average THG intensity within a single microglial cell and \({I}_{\max }\) is the maximum intensity across all ROIs and conditions. Single-cell analysis revealed significantly higher microglial THG levels in demyelinating tissue (Fig. S11c), possibly indicating active phagocytosis of myelin debris during inflammation (Fig. S11d). In control regions, the lower microglial THG scores combined with higher myelin scores reflect a healthy state with intact myelination. Remyelinating slices showed microglial THG scores similar to controls, suggesting a return to a non-phagocytic phenotype. However, the large standard deviation indicates spatial heterogeneity, with some areas still showing active phagocytosis, particularly where remyelination is incomplete. This highlights the importance of microglial localization within the tissue. The scatter plot in Fig. S11e shows the anti-correlation between microglial THG score and myelin score in different regions of interest of the cerebellum. Demyelinating regions of interest with low myelin scores generally show microglia with high THG scores, possibly reflecting increased phagocytic activity and reduced myelin content. On the other hand, remyelinating regions showed microglial low THG scores, although with considerable variability, reflecting differences in microglial activity between regions.
Discussion
The study presented in this manuscript describes the implementation of a NLO microscopy framework for the investigation of cellular metabolism and myelin distribution in mouse nervous tissue during demyelination and remyelination processes (Fig. 1). The method is based on the combination of two complementary label-free contrast modalities: two-photon fluorescence lifetime of the cellular endogenous metabolic cofactor NAD(P)H is used to probe the metabolic state of intact tissue with single-cell resolution, while THG provides a label-free probe of myelin organization and distribution. We have also developed dedicated image analysis approaches, such as automated detection and scoring of myelinated axons based on THG contrast (Fig. 2) and measurement of metabolic signatures at the single cell level based on NAD(P)H lifetime (Fig. 3). A promising direction of this work would be to upscale the automated detection and segmentation of myelinated axons across brain regions, accounting for variations in axon diameter, myelin thickness, density, organization and orientation in 3D. We aim to adapt our analysis in 3D and combine spatial- and frequency-domain approaches to account for out-of-plane axons and myelinated axons. We used different mouse lines (PLP-dsRed, PLP-GFP, CX3CR1-CreErt2/Rosa-tdTomato) to specifically study oligodendrocytes and microglia. In addition, Purkinje neurons were identified on a morphological basis in organotypic cerebellar cultures. Combining FLIM measurement of intrinsic biomarkers and cell segmentation, we performed label-free measurements of metabolic signatures in single cells, revealing distinct metabolic phenotypes in different cell types (Fig. 3), possibly related to their function (Fig. 3b). Neurons have the longest NAD(P)H lifetime and the highest fraction of bound NAD(P)H, revealing a phenotype that relies primarily on OXPHOS, consistent with their electrical activity. Oligodendrocytes are found to have a shorter NAD(P)H lifetime, indicating a phenotype relying more on glycolysis or fatty acids β-oxidation, possibly related to their need to rapidly produce lipids for myelin synthesis and provide lactate to neurons for metabolic support. Microglia have the shortest NAD(P)H lifetime and the lowest fraction of bound NAD(P)H, suggesting that they are the most glycolytic cells among those we studied. As a future direction, it would be interesting to investigate whether different neuronal subtypes with different firing patterns and excitatory-inhibitory dynamics have different metabolic profiles that reflect their energy requirements. Interestingly, in the intact Drosophila brain, we recently demonstrated that neuronal subtypes display distinct basal metabolic states and that memory formation triggers a subtype-specific metabolic reprogramming in mid-term memory neurons103.
We then systematically compared the optical signatures recorded from demyelinatuing and remyelinating neural tissues from organotypic cultures of the mouse cerebellum (Fig. 4). Using automated axons detection we measured a decrease in myelin score during demyelination and its rescue during remyelination. At the same time, a shorter NAD(P)H lifetime and a lower fraction of bound NAD(P)H (phenotype relying on glycolysis or fatty acids β-oxidation) were measured in microglia with a pro-inflammatory profile (during demyelination), and a rescue of NAD(P)H lifetime and fraction of bound NAD(P)H (relying on OXPHOS, fatty acids synthesis or oxidative stress) was observed in the promyelinating (remyelination) states of microglia (Fig. 4). These findings are consistent with the emerging evidence that metabolic states are closely linked with immune cell function during homeostatic and inflammatory processes. During demyelination, we observed a change in microglial morphology towards a rounder shape, typically associated with pro-inflammatory polarization, along with an increase in THG intensity within microglial cells, which may be attributed to the presence of myelin debris within the microglia (Fig. S11).
This study is the first to use label-free optical imaging to simultaneously visualize myelin at the single-fiber level and metabolism at the single-cell level in living tissue. It highlights the potential of advanced microscopy to address key unanswered questions in neuroscience, such as how metabolic coupling between glial cells and neurons shapes brain activity, particularly in modulating myelin plasticity. Our approach provides a means to characterize spatial and temporal heterogeneity during the progression of demyelinating pathology. A promising direction for future research will be to further explore the spatial and temporal metabolic heterogeneity of individual neurons, microglia, oligodendrocytes and astrocytes during demyelination and remyelination, and to correlate their metabolic phenotypes with microglial phagocytosis activity and local myelination levels in different regions of the cerebellar folia. Such advanced analyses could help to quantify the metabolic heterogeneity and adaptability of immune cells during the remyelination process, shedding light on the dynamic and plastic nature of these mechanisms. Furthermore, the observed metabolic adaptability and heterogeneity of microglia highlight the potential of targeting microglial metabolism as a therapeutic strategy for demyelinating diseases15.
Since multiple metabolic pathways (glycolysis, OXPHOS, fatty acid β-oxidation and synthesis, as well as oxidative stress) modulate NAD(P)H lifetime, unambiguous quantification of the measured metabolic changes requires in vivo calibrations together with the readout of multiple optical parameters—such as FAD lifetime, optical intensity redox ratio or mitochondrial clustering to provide complementary information to NAD(P)H lifetime37,44,104. Therefore, a promising direction opened by this work would be to implement multiparametric metabolic imaging by incorporating FLIM measurements of the FAD biomarker alongside NAD(P)H, using wavelength mixing76, as well as mitochondrial clustering analysis based on NAD(P)H intensity signal37,44. Performing such multiparametric metabolic imaging across different cell types and tissue states could potentially identify distinct metabolic phenotypes associated with inflammation, oligodendrocyte dysfunction, neuronal metabolic failure and degeneration. In addition, studying metabolic changes in neurons during demyelination and remyelination would deepen our understanding of the metabolic interplay between different cell types. These label-free approaches could also be used to investigate how neuronal activity influences the microglial phenotypic switch to a pro-regenerative state during repair80,105, and how it affects the metabolic signatures of both neurons and glial cells. Finally, extending these microscopy tools to in vivo studies in zebrafish or mouse models of demyelination would allow longitudinal monitoring of the temporal and spatial dynamics of cellular metabolism in microglia and neurons during inflammation and demyelination, together with simultaneous visualization of myelination dynamics and repair using THG imaging. This will open up a new field of research in optical imaging of immune metabolism and increase the potential of therapeutic strategies for inflammatory diseases.
In conclusion, measuring cellular temporal and spatial metabolic patterns and myelin (dis)organization during de- and remyelination processes has the potential to improve therapeutic strategies to promote remyelination and ensure neuroprotection. Furthermore, disruption of the myelin sheath is also involved in several other neuropathies of the central and PNS, while oxidative stress and metabolic alterations are important hallmarks of several neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases. Therefore, the optical techniques developed in this work could be used in the future in several areas of basic and translational neuroscience.
Methods
Multimodal NLO setup and imaging conditions
We optimized a lab-built multiphoton laser scanning upright microscope, a sketch of which is shown in Fig. 1a. A dual-output femtosecond laser source (Insight X3, Spectra-Physics, Santa Clara, CA, USA) was used for excitation, offering a tunable output range from 680 to 1300 nm (120 fs pulses, 80 MHz) and a fixed output at 1045 nm (200 fs pulses). For THG and NAD(P)H imaging, the tunable output was set to 1150 nm and 760 nm, respectively. Beam power was adjusted using two independent motorized waveplates and polarizers (Semrock, Rochester, NY, USA). A typical power of 20 mW was used to excite NAD(P)H fluorescence at 760 nm. For THG at 1150 nm, the power was set to ~50 mW. Pulse width at the sample was minimized using the laser’s built-in dispersion pre-compensation, iteratively adjusted to maximize THG intensity at constant power. The absorption spectra of the red fluorescent proteins used in this work, along with imaging wavelengths and examples, are shown in Fig. 1b. GFP was excited at 900 nm.
To focus the beam on the sample, we used a water immersion objective (×25, 1.05 NA, XLPLN25XWMP, Olympus, Tokyo, Japan) with a working distance of 2 mm. The sample was positioned on an XYZ translation stage (Z-Deck, Prior Scientific), which can be controlled manually with a joystick or automatically. A conventional photon-counting photomultiplier tube (PMT) (SensTech, Langley, UK) was used to detect the THG signal in the forward direction by using a condenser of NA = 1.4 to collect the light. Fluorescence signals were epi-detected in three different spectral channels using GaAsP detectors (Hamamatsu H7422-40) for 2P-FLIM and green fluorescence and a standard PMT (SensTech, Langley, UK) for red fluorescence. Bandpass filters were placed in front of these detectors to collect THG (390/40), and 2PEF of NAD(P)H (450/70), GFP (525/50) and red biomarkers (628/32), respectively. This choice was based on the emission spectra of these fluorophores, as shown in Fig. 1b.
We used a fast discriminator (Hamamatsu C9744 photon-counting unit) to detect pulses from the detectors. FLIM data acquisition was performed simultaneously in two channels using custom counting electronics developed in our laboratory, allowing time-correlated single-photon-counting. The trigger for the laser pulse was derived from the Insight X3, and a time-to-digital converter (TDC) integrated on an electronics board using a Xilinx Spartan-6 FPGA (field-programmable gate array), was used to measure the arrival time between the photon and the laser pulse. For each channel, the TDC used two parallelized deserializers that were synchronized with the ×12 laser frequency and phase-shifted by 180° using a digital clock manager of the FPGA. The laser was operated with an interpulse interval of 12.48 ns, and the measured arrival times were histogrammed using 24 temporal bins with a width of 0.52 ns. To calibrate the FLIM system, we measured the lifetimes of HG at 0 ns and fluorescein at pH 9, which has a single exponential decay of 4 ns. The system included galvanometric mirrors (VM500S, GSI Lumonics, Bedford, MA, USA) for laser scanning, and the acquisition process was synchronized using custom LabVIEW software (National Instruments, USA) and a multichannel I/O board (PCI-6115, National Instruments, Austin, TX, USA). Typically, our FLIM image acquisitions consisted of 320 × 320 pixel images, accumulating between 400 and 1000 photons. The total acquisition time was ~120 s, during which four images with a pixel dwell time of 20 μs were accumulated. We note that, although broad time bins and instrument response function (IRF) of the FLIM microscope reduce the separability of fluorescence lifetimes, this loss can be compensated by increasing the number of detected photons106,107. We previously demonstrated that, given the instrument parameters of our microscope and FLIM card (24 bins and 0.52 bin width), a photon budget of ~500 photons is sufficient to reliably resolve physiologically relevant NAD(P)H lifetime differences in cells and tissues76.
All THG images were acquired at depths between 20 and 70 μm in the tissue using laser powers of ~55 mW, with pixel sizes ranging from 0.5 to 1 μm, and a pixel dwell time of 20 μs accumulated twice, resulting in ~50–100 photons per image.
For live multimodal FLIM and THG imaging, the organotypic cerebellar slice preparation was positioned in a Petri dish filled with a non-fluorescent assay medium. The experimental setup was maintained in an incubation chamber at a temperature of 37 °C and 5% CO2 (Okolab, Pozzuoli, Italy).
Organotypic cerebellar slices preparation
We used mice with C57bl6 background. Mice were euthanized between postnatal days 8 and 10 (P8–P10, respectively), and both males and females were taken. Sagittal mouse cerebellar slices were prepared and then maintained in an incubator until the imaging days at 8 and 11 DIV (8 and 11 DIV, respectively) as described in ref. 70 and in Fig. 4b. Mouse cerebella at P8 to P10 were dissected in ice-cold Gey’s balanced salt solution supplemented with 4.5 mg/mL D-glucose and penicillin-streptomycin (100 IU/mL, Thermo Fisher Scientific). The cerebella were then cut into 250 μm parasagittal slices using a McIlwain tissue chopper, and the slices were placed on Millicell membranes (3–4 slices per membrane, 2 membranes per animal) (0.4 μmm Millicell, Merck Millipore). These slices were cultured in a medium consisting of 50% BME (Thermo Fisher Scientific), 25% Earle’s balanced salt solution (Sigma), and 25% heat-inactivated horse serum (Thermo Fisher Scientific), supplemented with GlutaMax (2 mM, Thermo Fisher Scientific), penicillin-streptomycin (100 IU/mL, Thermo Fisher Scientific), and D-glucose (4.5 mg/mL, Sigma). Organotypic cerebellar slices were maintained at 37 °C under 5% CO2 with medium changes every 2–3 days. To account for animal variability, organotypic cerebellar slices from each animal were cultured on two membranes-one treated with a demyelinating treatment and the other untreated as a control. This approach helped to minimize potential variability and allowed for paired experiments. To induce demyelination, one membrane from each animal was incubated overnight at 6 DIV in fresh culture medium containing 0.5 mg/mL LPC, while the other membrane was kept as a control70. To assess the demyelination process, imaging experiments were performed at 8 DIV to compare the peak of demyelination in LPC-treated samples with the peak of myelination in untreated samples. In order to assess the remyelination process, organotypic cerebellar slices were imaged at day 11, and LPC-treated slices were compared with untreated control slices. Three different transgenic mouse lines were used in order to identify different cell types. The first two mouse lines were PLP-dsRed81 and PLP-GFP to identify oligodendrocytes bodies74. To obtain the microglial expression of tdTomato, we crossed CX3CR1-creERT2 heterozygous mice (Jackson laboratory) with Rosa26-tdTomato Ai14 homozygous mice (Jackson laboratory). The activity of the Cre recombinase was then induced at postnatal day 3 by an intraperitoneal injection of 40 μL of Tamoxifen (T5648, Sigma, 15 mg/mL, diluted in 1:10 EtOH: Sunflower oil, S5007, Sigma), leading to the specific expression of tdtomato in the vast majority of microglial cells as previously described75. The care and use of mice conformed to institutional policies and guidelines (Sorbonne Université, INSERM, French and European Community Council Directive 86/609/EEC). The project underwent ethical validation (project number 20266).
Image analysis
We developed and implemented several advanced analysis workflows for single cell segmentation, FLIM analysis and automated myelinated axon detection. These workflows used different software tools such as CellProfiler87 and ilastik88 for cell segmentation, and Python GUIs FLUTE79 for phasor analysis of FLIM data. MATLAB was used for further processing of the single-cell analyses, while ImageJ, R, Origin, and MS Excel were used for visualization and statistical analysis.
Single-cell segmentation
To segment single oligodendrocytes and microglia, we processed the 2PEF images from the red fluorescent proteins using CellProfiler87 software (Fig. S1.a). First, the intensity of the 2PEF image is rescaled to enhance the contrasts by using the ’RescaleIntensity’ module (Fig. S1a(ii)). Next, the ’IdentifyPrimaryObjects’ module is employed to identify single cells (Fig. S1a(iii)). The identified objects are measured for their size and shape in CellProfiler software using the ’MeasureObjectSizeShape’ module. We extracted the eccentricity of each cell, defined as the ratio of the distance between the foci to the length of the major axis of an ellipse with the same second moments as the object99. A value of 0 corresponds to a perfect circle, while values approaching 1 indicate an increasingly elongated or elliptical shape. Subsequently, the outlines of the filtered cells are overlaid using the ’OverlayOutlines’ module to locate them within the tissue. The location of each cell in an image is exported to a spreadsheet via the ’ExportToSpreadsheet’ module. Finally, the image with the overlaid cells is saved using the ’SaveImages’ module.
To segment the cell bodies of Purkinje cell neurons, we used the THG image, as they are visible as a negative contrast. (Figs. 1f and S1b). We first performed a preprocessing step by stretching the contrast of the THG raw image (Fig. S1b(ii)) by using Fiji. We then trained ilastik software88 machine learning algorithm by annotating cell bodies with some THG images. We then used the same software to generate a probability map highlighting cell bodies (Fig. S1b(iii)). Finally, we used CellProfiler87 software to segment single cell masks from the probability map (Fig. S1b(iv)).
Analysis of FLIM data
FLIM Images acquired in the time domain were analyzed with Phasor analysis36,78. The phasor analysis of FLIM data was performed by using the open-source software FLUTE written in Python, in our lab79. The fluorescence decay of every pixel of the FLIM image was transformed with an FFT and converted into one pixel in the phasor plot, as previously described36,44,76 and detailed in the supplementary information (SI). The coordinates g and s in the phasor plot were calculated from the fluorescence intensity decay of each pixel using equations 1 and 2 of Supplementary Information. We applied a minimum intensity threshold of 20 photons to eliminate the background and pixels with low SNR. In addition, we applied a 3 × 3 median filter to the g and s matrices to enhance the SNR without compromising the spatial resolution. The matrices of g and s were extracted and then further processed with a custom-written Matlab code. For each pixel in the image, we computed the phase and modulation lifetimes (equations 7 and 8 of Supplementary Information) from g and s values. To estimate the fractional contributions of bound NAD(P)H, we estimated the distance from free NAD(P)H, by calculating the distance between the pixel in the phasor plot and the phasor location of the free coenzyme (equations 17 of Supplementary Information)79. We note that we disregard here the differences in quantum yield between the free and enzyme-bound metabolites.
Once the maps of NAD(P)H lifetime and distance from NAD(P)H are calculated, the single cell mask is applied (Fig. 3s), and the average values are calculated in single cell masks.
Automated myelinated axon segmentation workflow
We developed a fiber detection workflow to automatically extract myelinated axons from label-free THG images. This workflow involves four main steps: preprocessing (Fig. 2b(ii)), Hessian and Meijering filter for fiber segmentation (Fig. 2b(iii)), intermediate labeling (Fig. 2b(iv)) and K-means filtering (Fig. 2b(v)).
Preprocessig step (Fig. 2b(ii)) is performed with a custom-written software in python (https://github.com/laboratoryopticsbiosciences/thg-segmentation) that uses functions from the Scikit-image python library83. Preprocessing involves the application of three filters to the raw image to improve the quality and reliability of the THG images. First, we applied an inpainting filter with the function “skimage.restoration.inpaint()” to filter out Isolated bright pixels. Second, we applied the tophat transformation with “skimage.morphology.white.tophat()” function in order to enhance and extract the bright myelin structures from the dark background. Third, we removed the outliers by applying a median filter with the “skimage.filters.median()” function.
After preprocessing, segmentation of fiber structures (Fig. 2b(iii)) was performed using combining two methods: the Meijering neuriteness filter82 and a custom Hessian eigenvalue filter. For the custom Hessian filter, eigenvalues λ1 λ2 of the Hessian matrix were computed for each pixel in the preprocessed image. The Hessian matrix contains information about the second-order partial derivatives of an image and describes the curvatures of image intensity at each pixel. We select the largest absolute eigenvalue for each pixel, defined as λlarge = max(∣λ1∣, ∣λ2∣), and set the positive λlarge values to zero. The remaining non-zero λlarge values correspond to ridge-like structures within the image. We then threshold ∣λlarge∣ by keeping only the pixels with values above the 84.5th percentile for large ROI images and above the 80th percentile for small ROI images. This percentile-based thresholding helps identify the strongest ridge-like structures while adaptively accounting for variations in image intensity across different ROI sizes. The segmentation from both filters is combined using an intersection operation, preserving only those pixels identified by both the Meijering filter and the custom Hessian eigenvalue filter. This eliminates outliers and improves segmentation accuracy by emphasizing regions consistently identified by both methods. To refine the obtained segmentation and remove artifacts, such as folium borders and other non-myelin structures incorrectly captured, we apply K-means clustering to each object within the segmented masks. Initially, every object in each mask is assigned a distinct label (Fig. 2b(iv)). The labeled objects were created using skimage.measure.label() function. We input a binary segmentation (fiber mask) into the function, and the algorithm finds each separate object (connected pixels) and assigns a unique number to all the pixels. Next, using the “measure.regionprops” function from scikit-image version 0.24.083, the following features, as defined in Supplementary material, are measured and computed for each object: Equivalent diameter, Perimeter, Area, Bbox-area, Convex-area, Solidity, Minor-axis-length, Major-axis-length, Extent, Orientation, Eccentricity, Filled-area. K-means clustering (Fig. 2b(v)) with k = 10 is applied taking in account all parameters, and the clusters representing the objects of interest are visually selected. Each object is assigned to one of 10 clusters based on the similarity of these features. Then, by visual inspection of the properties of objects in each cluster, we select those clusters that contain predominantly myelin structures, while excluding clusters containing mostly artifacts (like folium borders or non-myelin structures). Typically, 3–4 clusters representing the fibers are retained. The objects within these selected clusters for each image are aggregated to form the final segmentation mask. In some cases, manual adjustments were made to correct regions where artifacts such as folium borders were erroneously identified as myelin in large ROIs.
Manual myelinated axon segmentation and analysis
The myelinated fibers and the white matter tracts were first manually annotated using Imaris software. The white matter tract outliers were then approximated by using a convex hull in Meshlab108. Myelin fibers with a clearly distinguishable THG signal to the human eye were manually selected, primarily focusing on large internodes of Purkinje cell axons that exhibited a strong SBR in the THG images. Internodes with weaker SBRs–potentially corresponding to developing segments or Purkinje cell axon collaterals—were not systematically traced. The fibers coordinates and the white matter tract outliers were imported into GeNePy3D109 to extract the fibers portions staying outside of the white matter tracts. We then count them, compute their orientation distribution, lengths and tortuosity (Fig. S2). The source code for analyzing the myelinated fibers is available in a repository (https://github.com/laboratoryopticsbiosciences/thg-folium).
Microglia morphometry by Sholl analysis
To quantify microglia morphometry from 2D images by bypassing reconstruction and tracing of microglia processed, we performed Sholl analysis with an open-source program for ImageJ/Fiji100, (https://imagej.net/plugins/sholl-analysis). Sholl analysis of microglial morphology (n = 4 microglia per group) was performed by manually binarizing each cell, and the number of intersections was then calculated through the algorithm. The analysis measured intersections from the center of the microglia extending outwards to ~12 μm.
Statistical analysis and reproducibility
To perform statistical analysis, we utilized various software tools, including Microsoft Excel (2013), Origin (Version 2019, Northampton, MA: OriginLab Corporation), and R (Vienna, Austria: R Foundation for Statistical Computing). To ensure robust statistical comparisons and account for animal-to-animal variability, we employed a mixed-effect model t test. This method enabled us to assess the impact of different experimental conditions while considering inherent variations across animals. Numbers of samples for each experiment are specified in the figure legends. For the CX3CR1-tdTomato line, we used a total of 10 animals: 10 for the control condition, 3 for the demyelination condition, and 5 for the remyelination condition. For the PLP-dsRed line, we had 3 animals, resulting in 2 for the control and 2 for the demyelination conditions, with 1 animal serving both conditions. A summary has been provided in the following table.
Condition | #Cells | #Animals |
|---|---|---|
Microglia in control slices @8 DIV | 115 | 5 |
Microglia in control slices @11 DIV | 141 | 5 |
Microglia during demyelination @8 DIV | 179 | 3 |
Microglia during remyelination @11 DIV | 147 | 5 |
Oligodendrocytes in control slices @8 DIV | 12 | 2 |
Purkinje neurons in control slices @8 DIV | 17 | 2 |
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of the study are openly available in the following repository: https://zenodo.org/records/17105359.
Code availability
All the code and software used and developed in this publication are open source and available at the following ref. 79, (https://github.com/laboratoryopticsbiosciences/thg-segmentation, https://github.com/laboratoryopticsbiosciences/thg-folium).
References
Fünfschilling, U. et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–21 (2012).
Magistretti, P. & Allaman, I. A cellular perspective on brain energy metabolism and functional imaging. Trends Neurosci. 43, 854–869 (2015).
Stadelmann, C., Timmler, S., Barrantes-Freer, A. & Simons, M. Myelin in the central nervous system: structure, function, and pathology. Physiol. Rev. 99, 1381–1431 (2019).
Ronzano, R., Thetiot, M., Lubetzki, C. & Desmazieres, A. Myelin plasticity and repair: neuro-glial choir sets the tuning. Front. Cell Neurosci. 14, 42 (2020).
Bonvento, G. & Bolaños, J. Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33, 1546–1564 (2021).
Lloyd, A. & Miron, V. The pro-remyelination properties of microglia in the central nervous system. Nat. Rev. Neurol. 15, 447–458 (2019).
Saab, A. & Nave, K. Myelin dynamics: protecting and shaping neuronal functions. Curr. Opin. Neurobiol. 47, 104–112 (2017).
Osso, L. & Hughes, E. Dynamics of mature myelin. Nat. Neurosci. 27, 1449–1461 (2024).
Nave, K. Myelination and the trophic support of long axons. Nat. Rev. Neurosci. 11, 275–83 (2010).
Lee, Y. et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 485, 443–8 (2012).
Saab, A. et al. Oligodendroglial NMDA receptors regulate glucose import and axonal energy metabolism. Neuron 91, 119–32 (2016).
Asadollahi, E. et al. Oligodendroglial fatty acid metabolism as a central nervous system energy reserve. Nat. Neurosci. 27, 1934–1944 (2024).
Cleland, N., Al-Juboori, S., Dobrinskikh, E. & Bruce, K. Altered substrate metabolism in neurodegenerative disease: new insights from metabolic imaging. Curr. Opin. Neurobiol. 18, 248 (2021).
Jackson, J. et al. White matter tauopathy: transient functional loss and novel myelin remodeling. Glia 66, 813–827 (2018).
Cunnane, S. et al. Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 19, 609–633 (2020).
Calabrese, M., Filippi, M. & Gallo, P. Cortical lesions in multiple sclerosis. Nat. Rev. Neurol. 6, 438–444 (2010).
Haider, L. et al. Oxidative damage in multiple sclerosis lesions. Brain 134, 1914–24 (2011).
Franklin, R., ffrench Constant, C., Edgar, J. & Smith, K. Neuroprotection and repair in multiple sclerosis. Nat. Rev. Neurol. 8, 624–34 (2012).
Tepavcevic, V. Oligodendroglial energy metabolism and (re)myelination. Life 11, 238 (2021).
Bernier, L. P., York, E. M. & MacVicar, B. A. Immunometabolism in the brain: How metabolism shapes microglial function. Trends Neurosci. 43, 854–869 (2020).
O’Neill, L., Kishton, R. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–65 (2016).
Locatelli, G. et al. Mononuclear phagocytes locally specify and adapt their phenotype in a multiple sclerosis model. Nat. Neurosci. 21, 1196–1208 (2018).
Caputa, G., Castoldi, A. & Pearce, E. Metabolic adaptations of tissue-resident immune cells. Nat. Immunol. 20, 793–801 (2019).
Moll, N. et al. Multiple sclerosis normal-appearing white matter: pathology-imaging correlations. Ann. Neurol. 70, 764–73 (2011).
Petiet, A., Aigrot, M. & Stankoff, B. Gray and white matter demyelination and remyelination detected with multimodal quantitative mri analysis at 11.7t in a chronic mouse model of multiple sclerosis. Front. Neurosci. 27, 491 (2016).
Bonvento, G., Valette, J., Flament, J., Mochel, F. & Brouillet, E. Imaging and spectroscopic approaches to probe brain energy metabolism dysregulation in neurodegenerative diseases. J. Cereb. Blood Flow. Metab. 37, 1927–1943 (2016).
Romanelli, E. et al. Cellular, subcellular and functional in vivo labeling of the spinal cord using vital dyes. Nat. Protoc. 8, 481–90 (2013).
Zhang, Z., Cheng, X., Zhao, Y. & Yang, Y. Lighting up live-cell and in vivo central carbon metabolism with genetically encoded fluorescent sensors. Annu. Rev. Anal. Chem. 13, 293–314 (2020).
Zipfel, W. et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl. Acad. Sci. USA 100, 7075–80 (2003).
Wang, H., Fu, Y., Zickmund, P., Shi, R. & Cheng, J. Coherent anti-stokes raman scattering imaging of axonal myelin in live spinal tissues. Biophys. J. 89, 581–91 (2005).
Débarre, D. et al. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat. methods 3, 47–53 (2006).
Weigelin, B., Bakker, G.-J. & Friedl, P. Third harmonic generation microscopy of cells and tissue organization. J. Cell Sci. 129, 245–255 (2016).
James, D. & Campagnola, P. Recent advancements in optical harmonic generation microscopy: applications and perspectives. BME Front. 2021, 3973857 (2021).
Georgakoudi, I. & Quinn, K. Label-free optical metabolic imaging in cells and tissues. Annu. Rev. Biomed. Eng. 8, 413–443 (2023).
Skala, M. et al. In vivo multiphoton microscopy of nadh and fad redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA 104, 19494–19499 (2007).
Stringari, C. et al. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA 108, ar11 (2011).
Liu, Z. et al. Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci. Adv. 7, 3 (2013).
Datta, R., Heaster, T., Sharick, J., Gillette, A. & Skala, M. C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 25, 1–43 (2020).
Barroso, M., Monaghan, M., Niesner, R. & Dmitriev, R. Probing organoid metabolism using fluorescence lifetime imaging microscopy (flim): the next frontier of drug discovery and disease understanding. Adv. Drug Deliv. Rev. 201, 115081 (2023).
Huang, S., Heikal, A. & Webb, W. Two-photon fluorescence spectroscopy and microscopy of nad(p)h and flavoprotein. Biophys. J. 82, 2811–25 (2002).
Lakowicz, J. R. et al. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. USA 89, 1271–1275 (1992).
Bird, D. K. et al. Metabolic mapping of mcf10a human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme nadh. Cancer Res. 65, 8766–8773 (2005).
Stringari, C., Nourse, J., Flanagan, L. & Gratton, E. Phasor fluorescence lifetime microscopy of free and protein-bound nadh reveals neural stem cell differentiation potential. PLoS One 7, e48014 (2012).
Ung, T. et al. Simultaneous nad(p)h and fad fluorescence lifetime microscopy of long uva-induced metabolic stress in reconstructed human skin. Sci. Rep. 11, 22171 (2021).
Lorena, A.-A. et al. Spatial dynamics of sirt1 and the subnuclear distribution of nadh species. Proc. Natl. Acad. Sci. USA 113, 12715–12720 (2016).
Sánchez-Ramírez, E. et al. Coordinated metabolic transitions and gene expression by nad+ during adipogenesis. J. Cell Biol. 221, e202111137 (2022).
Paillon, N., Ung, T., Dogniaux, S., Stringari, C. & Hivroz, C. Label-free single-cell live imaging reveals fast metabolic switch in t lymphocytes. Mol. Biol. Cell. 35, ar11 (2024).
Chia, T. H. et al. Multiphoton fluorescence lifetime imaging of intrinsic fluorescence in human and rat brain tissue reveals spatially distinct nadh binding. Biomed. Opt. Express 16, 4237–49 (2008).
Yaseen, M. A. et al. Fluorescence lifetime microscopy of nadh distinguishes alterations in cerebral metabolism in vivo. Biomed. Opt. Express 8, 2368–2385 (2017).
Zhang, Y. et al. Synergistic label-free fluorescence imaging and mirna studies reveal dynamic human neuron-glial metabolic interactions following injury. Sci. Adv. 10, eadp1980 (2024).
Mossakowski, A. et al. Tracking cns and systemic sources of oxidative stress during the course of chronic neuroinflammation. Acta Neuropathol. 130, 799–814 (2015).
Alfonso-Garcia, A. et al. Label-free identification of macrophage phenotype by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 30, 46005 (2016).
Smokelin, I., Mizzoni, C., Erndt-Marino, J., Kaplan, D. & Georgakoudi, I. Optical changes in thp-1 macrophage metabolism in response to pro- and anti-inflammatory stimuli reported by label-free two-photon imaging. J. Biomed. Opt. 25, 1–14 (2020).
Sagar, M. et al. Microglia activation visualization via fluorescence lifetime imaging microscopy of intrinsically fluorescent metabolic cofactors. Neurophotonics 7, 035003 (2020).
Bernier, L. et al. Microglial metabolic flexibility supports immune surveillance of the brain parenchyma. Nat. Commun. 11, 1559 (2020).
Miskolci, V. et al. In vivo fluorescence lifetime imaging of macrophage intracellular metabolism during wound responses in zebrafish. Elife 11, e66080 (2022).
Fiedler, A. et al. Flimb: fluorescence lifetime microendoscopy for metabolic and functional imaging of femoral marrow at subcellular resolution. Biomed. Opt. Express 16, 1711–1731 (2025).
Plotegher, N. et al. Nadh fluorescence lifetime is an endogenous reporter of alpha-synuclein aggregation in live cells. FASEB J. 29, 2484–2494 (2021).
Chakraborty, S., Nian, F., Tsai, J., Karmenyan, A. & Chiou, A. Quantification of the metabolic state in cell-model of parkinson’s disease by fluorescence lifetime imaging microscopy. Sci. Rep. 13, 19145 (2016).
Sun, C.-K. et al. Higher harmonic generation microscopy for developmental biology. J. Struct. Biol. 147, 19–30 (2004).
Olivier, N. et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329, 967–971 (2010).
Aptel, F. et al. Multimodal nonlinear imaging of the human cornea. Investig. Ophthalmol. Vis. Sci. 51, 2459–2465 (2010).
Farrar, M. J., Wise, F. W., Fetcho, J. R. & Schaffer, C. B. In vivo imaging of myelin in the vertebrate central nervous system using third harmonic generation microscopy. Biophys. J. 100, 1362–1371 (2011).
Lim, H. et al. Label-free imaging of schwann cell myelination by third harmonic generation microscopy. Proc. Natl. Acad. Sci. 111, 18025–18030 (2014).
Redlich, M. & Lim, H. A method to measure myeloarchitecture of the murine cerebral cortex in vivo and ex vivo by intrinsic third-harmonic generation. Front. Neuroanat. 13, 65 (2019).
Morizet, J. et al. Multiscale characterization of myelin distribution with polarized THG microscopy. Optica 1122–1138 (2025).
Witte, S. et al. Label-free live brain imaging and targeted patching with third-harmonic generation microscopy. Proc. Natl. Acad. Sci. 108, 5970–5975 (2011).
Ferrer Ortas, J. et al. Label-free imaging of red blood cells and oxygenation with color third-order sum-frequency generation microscopy. Light. Sci. Appl. 12, 29 (2023).
Gleeson, M. et al. Multiplexed orthogonal polarizations harmonic imaging for probing dynamic biological processes. ACS Photonics 11, 3259–3266 (2024).
Thetiot, M., Ronzano, R., Aigrot, M., Lubetzki, C. & A., D. Preparation and immunostaining of myelinating organotypic cerebellar slice cultures. J Vis. Exp. 20, 145 (2019).
Chang, C.-F., Yu, C.-H. & Sun, C.-K. Multi-photon resonance enhancement of third harmonic generation in human oxyhemoglobin and deoxyhemoglobin. J. Biophotonics 3, 678–685 (2010).
Drobizhev, M., Makarov, N., Tillo, S., Hughes, T. & Rebane, A. Two-photon absorption properties of fluorescent proteins. Nat. Methods 8, 393–399 (2011).
Heikal, A. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark. Med. 4, 241–2463 (2010).
Spassky, N. et al. The early steps of oligodendrogenesis: insights from the study of the plp lineage in the brain of chicks and rodents. Dev. Neurosci. 23, 318–326 (2001).
Bedolla, A. et al. A comparative evaluation of the strengths and potential caveats of the microglial inducible creer mouse models. Cell Rep. 43, 113660 (2013).
Stringari, C. et al. Multicolor two-photon imaging of endogenous fluorophores in living tissues by wavelength mixing. Sci. Rep. 7, 3792 (2017).
Sharick, J. et al. Protein-bound nad(p)h lifetime is sensitive to multiple fates of glucose carbon. Sci. Rep. 8, 5456 (2018).
Digman, M., Caiolfa, V., Zamai, M. & Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, L14–L16 (2008).
Gottlieb, D., Asadipour, P., Kostina, B., Ung, T. & Stringari, C. Flute: a python gui for interactive phasor analysis of flim data. Biol. Imaging 6, e21 (2023).
Ronzano, R. et al. Microglia-neuron interaction at nodes of ranvier depends on neuronal activity through potassium release and contributes to remyelination. Nat. Commun. 12, 5219 (2021).
Fuss, B. et al. Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Dev. Biol. 218, 259–27 (2000).
Meijering, E. et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. A. 58, 167–76 (2004).
van der Walt, S. et al. scikit-image contributors. scikit-image: image processing in Python. Peer J. e453 (2014).
Padamsey, Z. & Rochefort, N. Paying the brain’s energy bill. Curr. Opin. Neurobiol. 78, 102668 (2022).
Vander Heiden, M., Cantley, L. & Thompson, C. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–33 (2009).
Kasischke, K., Vishwasrao, H., Fisher, P., Zipfel, W. & Webb, W. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science 305, 99–103 (2004).
Stirling, D. et al. Cellprofiler 4: improvements in speed, utility and usability. BMC Bioinformatics 22, 433 (2021).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Paolicelli, R. Microglia states and nomenclature: a field at its crossroads. Neuron 110, 3458–3483 (2022).
McWhorter, F., Wang, T., Nguyen, P., Chung, T. & Liu, W. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 110, 17253–17258 (2013).
Ghosh, S., Castillo, E., Frias, E. & Swanson, R. Bioenergetic regulation of microglia. Physiol. Rev. 66, 1200–1212 (2011).
Lauro, C. & Limatola, C. Metabolic reprograming of microglia in the regulation of the innate inflammatory response. Front. Immunol. 20, 493 (2020).
Bielanin, J. & Sun, D. Significance of microglial energy metabolism in maintaining brain homeostasis. Transl. Stroke Res. 14, 435–437 (2023).
Mosser, D. & Edwards, J. Exploring the full spectrum of macrophage activation. J. Biomed. Opt. 8, 958–969 (2016).
Skripuletz, T. et al. Cerebellar cortical demyelination in the murine cuprizone model. Neuron 95, 341–356.e6 (2010).
De Biase, L. et al. Local cues establish and maintain region-specific phenotypes of basal ganglia microglia. Neuron 95, 341–356.e6 (2017).
Rawji, K. et al. Deficient surveillance and phagocytic activity of myeloid cells within demyelinated lesions in aging mice visualized by ex vivo live multiphoton imaging. J. Neurosci. 28, 1973–1988 (2018).
Olah, M. et al. Identification of a microglia phenotype supportive of remyelination. Glia 60, 306–321 (2012).
Carpenter, A. et al. Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Ferreira, T., Blackman, A., Oyrer, J. et al. Neuronal morphometry directly from bitmap images. Nat. Methods 11, 982–984 (2014).
Hu, X. et al. Microglial and macrophage polarization—new prospects for brain repair. Nat. Rev. Neurol. 11, 1196–1208 (2015).
Cunha, M. et al. Pro-inflammatory activation following demyelination is required for myelin clearance and oligodendrogenesis. J. Exp. Med. 217, e20191390 (2020).
Roussel, P. et al. In vivo autofluorescence lifetime imaging of the drosophila brain captures metabolic shifts associated with memory formation. eLife 14, RP106040 (2025).
Walsh, A. et al. Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res. 73, 6164–6174 (2013).
Gibson, E. et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 344, 1252304 (2014).
Kollner, M. & Wolfrum, J. How many photons are necessary for fluorescence-lifetime measurements. Chem. Phys. Lett. 200, 199–204 (1992).
Trinh, A. L. & Esposito, A. Biochemical resolving power of fluorescence lifetime imaging: untangling the roles of the instrument response function and photon-statistics. Biomed. Opt. Express. 12, 3775–3788 (2021).
Phan, M.-S. & Chessel, A. Genepy3d. a quantitative geometry python toolbox for bioimaging. F1000Research 9, 1374 (2021).
Cignoni, P. et al. Meshlab: an open-source mesh processing tool. In Sixth Eurographics Italian Chapter Conference, 129–136 https://diglib.eg.org/items/cafa9acd-c34e-4247-be5c-e4b7beb14a46 (2008).
Acknowledgements
We thank Clement Perrot for help with the Sholl analysis and Arthur Boutillon and Nicolas David for advice on statistical analysis. We thank members of the LOB Advanced Microscopies group for discussions on tissue microscopy. This work was supported by the Agence Nationale de la Recherche (ANR) (contracts ANR-10-INBS-04 France BioImaging, ANR-11-EQPX-0029 Morphoscope2 and ANR-15-CE11-0012-01 NLOMMIT), by the Fondation Bettencourt-Schueller (Brain Harmonics), by the European Research Council (Horizon 2020 program, grant No 951330 HOPE), by Marie Skłodowska-Curie Actions (OpticMyeliMet project), by France SEP, Paris Brain Institute (ICM) and INSERM.
Author information
Authors and Affiliations
Contributions
B.A., C.S., E.B., B.S., and A.D. conceived the experiments and designed the research. C.S., E.B., P.M., and X.S. implemented the microscopy setup. M.S.A., R.R., and A.D. prepared samples. B.A., J.M., and C.S. performed experiments. B.A, J.M., and C.S. analyzed the data. B.A., X.Z., M.S.P., and A.C. developed advanced image analysis techniques. B.A. and C.S. drafted the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Catherine Best-Popescu, Pieter Vanden Berghe, and Raluca Niesner, who co-reviewed this paper with Anne Bias, for their contribution to the peer review of this work. Primary Handling Editors: Periklis Pantazis and Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Asadipour, B., Morizet, J., Ronzano, R. et al. Label-free nonlinear microscopy probes cellular metabolism and myelin dynamics in live tissue. Commun Biol 8, 1811 (2025). https://doi.org/10.1038/s42003-025-09192-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09192-4