Abstract
Cervical cancer (CC), a leading cause of cancer-related deaths among women worldwide, is primarily driven by high-risk human papillomavirus (HPV) infections and comprises two major histological subtypes: adenocarcinoma (AC) and squamous cell carcinoma (SCC). Despite advances in prevention and treatment, the molecular and cellular heterogeneity of these subtypes poses significant challenges to achieving optimal clinical outcomes. Here, we integrate single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) to dissect the cellular and spatial heterogeneity of AC and SCC, uncovering distinct tumor microenvironment (TME) dynamics that underlie their divergent clinical behaviors. Our scRNA-seq analysis reveals that AC is enriched in epithelial cells, while SCC exhibits a more immunogenic TME with elevated plasma cells and NK/T cells. Spatial transcriptomics further highlights robust interactions between CD8 + T cells and epithelial subtypes in SCC, contrasting with the stromal-rich, immune-cold phenotype of AC. We identify subtype-specific immune and stromal features, including ICOS+ Tregs, IDO1+ cancer-associated fibroblasts (CAFs), and PLVAP+ endothelial cells, which may drive immune evasion, angiogenesis, and metastasis. These findings provide a comprehensive framework for understanding CC heterogeneity and offer actionable insights for developing subtype-specific therapeutic strategies, such as combining immune checkpoint inhibitors with stromal-targeting agents. This study underscores the potential of spatial multi-omics technologies to advance precision oncology and improve outcomes for cervical cancer patients.
Similar content being viewed by others
Introduction
Cervical Cancer (CC) remains a significant global health challenge, ranking as one of the most common malignancies affecting women worldwide. It is the fourth leading cause of cancer-related deaths among women, particularly in low- and middle-income countries where access to screening and preventive measures is limited1,2. The disease is primarily caused by persistent infection with high-risk human papillomavirus (HPV) strains, which drive the transformation of cervical epithelial cells into malignant phenotypes3,4. Two primary histological subtypes, cervical adenocarcinoma (AC) and squamous cell carcinoma (SCC), account for the majority of cervical cancer cases5. While SCC is more prevalent, AC has been increasingly diagnosed in recent years and is often associated with poorer prognosis due to its deeper location within the cervix and resistance to conventional therapies6. Despite significant advances in diagnosis, prevention (e.g., HPV vaccination), and treatment modalities such as surgery, radiotherapy, and chemotherapy, the molecular and cellular heterogeneity of these subtypes poses a major challenge to achieving optimal clinical outcomes. Understanding the distinct molecular and cellular landscapes of AC and SCC is therefore essential for developing more effective, subtype-specific therapeutic strategies and improving patient survival rates.
Recent developments in single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) have revolutionized our ability to study tumor biology at an unprecedented resolution. These cutting-edge technologies provide detailed insights into the complex cellular ecosystems within tumors, enabling researchers to dissect the heterogeneity of the tumor microenvironment (TME) and uncover the mechanisms driving tumor progression, immune evasion, and therapy resistance7,8. scRNA-seq offers a powerful approach to characterize the heterogeneity within the TME by analyzing gene expression profiles at the single-cell level. This technology has revealed diverse cell states, rare cell populations, and dynamic interactions between tumor cells, immune cells, stromal cells, and other components of the TME.
Complementing scRNA-seq, spatial transcriptomics (ST) technology provides critical spatial context by mapping gene expression data to the physical locations of cells within the tissue architecture9,10. This technology allows researchers to visualize the spatial organization of cell types, identify cellular neighborhoods, and study the interactions between tumor cells and their surrounding microenvironment. By integrating scRNA-seq and ST, researchers can achieve a comprehensive understanding of both the cellular composition and spatial organization of tumors, uncovering critical insights into tumor architecture, cellular interactions, and potential therapeutic targets. Together, these technologies have the potential to transform our understanding of cervical cancer biology and pave the way for precision medicine. Additionally, they can elucidate mechanisms of therapy resistance and inform the development of combination therapies that target both tumor cells and their supportive microenvironment. As these technologies continue to evolve, they hold great promise for improving the diagnosis, treatment, and prognosis of cervical cancer, ultimately reducing its global burden and improving the lives of affected women.
In this study, we aim to compare cervical adenocarcinoma and squamous cell carcinoma combining scRNA-seq and ST, especially focusing on characterization of distinct cellular populations and states within AC and SCC, as well as deciphering the spatial distribution and interactions of these populations within distinct tumor microenvironment. Our analysis provides a comprehensive framework for understanding the cellular and spatial features that differentiate AC and SCC, offering a foundation for personalized therapeutic strategies and advancing the field of cervical cancer research.
Results
Single-cell and spatial transcriptomics profiling of different histological subtypes of cervical cancer
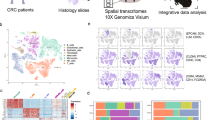
In this study we collected samples from three histological subtypes of cervical cancer patients with similar stages (Fig. 1a, Supplementary Data 1), including adenocarcinoma (AC), squamous cell carcinoma (SCC), and adenosquamous carcinoma (ASC), and then performed single-cell RNA sequencing (scRNA-seq) as well as Xenium spatial in situ hybridization assay on those samples to investigate the contribution of tumor microenvironment to different cancer types (Fig. 1b).
a, b Schematic illustrations of experimental workflows for single-cell transcriptomic profiling and spatial transcriptomics analysis performed on cervical cancer (CC) specimens. c Two-dimensional UMAP visualization of single-cell RNA sequencing (scRNA-seq) data depicting cell types within CC tissues. Colors in UMAP indicate different cell types. d Quantitative analysis of major cell type proportions derived from scRNA-seq between two histopathological classifications of CC: AC and SCC; Student’s t test was used to perform statistical analysis, and “*” means statistical p value < 0.05, “***” means statistical p value < 0.001. e Comparative heatmap visualization displaying expression levels of lineage-specific marker genes among identified cell clusters in scRNA-seq datasets. f Sample-specific distribution patterns of cellular subpopulations identified through scRNA-seq analysis. Colors in the histograms indicate the major cell types; triangles with different colors at the bottom indicate AC or SCC cancer subtypes. g Two-dimensional UMAP visualization of Xenium spatial transcriptomic data depicting major cell types within CC tissues, with different colors indicating specific cell types as in (c). h Histopathological subtype-dependent cellular composition quantification using Xenium in situ sequencing platforms in CC specimens, there was no statistical difference for any cell types between AC and SCC samples. i Comparative heatmap visualization displaying expression levels of lineage-specific marker genes among identified cell clusters in Xenium datasets. j Sample-specific distribution patterns of cellular subpopulations identified through Xenium data analysis. Colors in the histograms indicate the major cell types; triangles with different colors at the bottom indicate AC or SCC cancer subtypes.
The scRNA-seq was performed by using fresh tumor specimens from 16 patients, including 8 AC, 6 SCC, and 2 ASC cases. After removing low-quality cells and the batch correction process, we performed dimensionality reduction on the data including a total of 150,798 single cells (Fig. 1c), followed by cell annotation based on expression of cell-type-specific marker genes, leading to identification of different major cell types, including epithelial/tumor cells, endothelial cells, fibroblasts and so on (Fig. 1d). The proportions of three major cell types varied significantly between AC and SCC samples: AC samples exhibited a higher proportion of epithelial cells, while SCC group was enriched in plasma cells and NK/T cells (Fig. 1e). These differences indicated distinct cell compositions in the tumor microenvironment between AC and SCC. The hierarchical clustering based on the proportions of all cell types in different samples can distinguish the SCC samples from AC samples, indicating obvious difference in cell compositions between SCC and AC (Fig. 1f, Supplementary Data 2).
The Xenium spatial in situ hybridization assay was performed using paraffin-embedded sections from 22 patients, including 9 AC, 10 SCC, and 3 ASC samples (Supplementary Data 1). Using the Xenium data only, we were also able to perform dimensionality reduction on the data including a total of 1,963,636 single cells (Fig. 1g), and then clustered different cell types based on the transcriptome profiles of the 430 genes included in our Xenium gene panel (Supplementary Data 3) and then annotated those cell types based on related marker genes. We were able to identify most major cell types using the 430 gene panel, such as epithelial/cancer cells, endothelial cells/pericytes, fibroblasts, B cells, plasmablasts, myeloid cells, mast cells and NK/T cells (Fig. 1h). However, there was no significant proportional difference of these cells between AC and SCC samples (Fig. 1i, j, Supplementary Data 4); the inconsistency with scRNA-seq data may be due to biased sampling of the spatial transcriptomic platform without homogenization of the materials. By integrating spatial coordinates for individual cells, we reconstructed a two-dimensional tissue map that precisely localized all identified cell types within their native architectural context (Supplementary Fig. 1).
We also integrated the scRNA-seq data and Xenium data and projected the Xenium single cells into the scRNA-seq data UMAP (Supplementary Fig. 2a, b). Annotation of majority of the cell types from both datasets was consistent, with epithelial and endothelial cells the highest consistency and mast cells the lowest consistency (Supplementary Fig. 2c), indicating that the Xenium data had similar sensitivity to identify those cell types although with a small set of gene panels.
Deciphering the tumor and immune cell heterogeneity between AC and SCC cervical cancer subtypes using scRNA-seq data
Next, to systematically explore the cellular heterogeneity underlying the distinct biological behaviors of AC and SCC, we subclustered the tumor cells and immune cells independently, as they exhibited marked compositional disparities between the two tumor subtypes. For epithelial tumor cell compartment analysis, we applied the same method, unsupervised dimensionality reduction combined with graph-based clustering algorithms to the epithelial cluster from the scRNA-seq data. This approach further resolved the epithelial cell population into 11 clusters (Fig. 2a). Three subclusters demonstrated statistically significant proportional imbalances between AC and SCC, with Cluster 1 exhibiting pronounced enrichment in SCC samples (Fig. 2b, Supplementary Data 5). This SCC-predominant cluster displayed robust expression of canonical squamous lineage markers, including KRT5 and KRT6A (Fig. 2c, Supplementary Data 6), which were hallmark genes for stratified epithelial differentiation11,12. Intriguingly, this cluster also showed elevated expression of S100A9 (Fig. 2c, Supplementary Data 6), a pro-inflammatory alarmin linked to myeloid cell recruitment and tumor-stroma crosstalk13,14,15, suggesting a potential interplay between SCC-specific tumor subpopulations and immune modulation. For the other two clusters which were more abundant in AC samples, cluster 0 overexpressed many histone family genes such as HIST1H3A and HIST1H4B (Supplementary Fig. 3a, Supplementary Data 7), indicating high DNA damage response in those cells16; cluster 2 overexpressed genes such as tetraspanin family genes TSPAN1/3/8 (Supplementary Fig. 3b, Supplementary Data 8), the roles of which were controversial in cancer11,17. These findings highlight the intrinsic molecular divergence between AC and SCC at the tumor cell level, with AC and SCC-enriched subclusters potentially driving subtype-specific microenvironmental compositions and clinical behaviors.
a Two-dimensional UMAP visualization of scRNA-seq depicting epithelial cell subclusters. b The boxplot indicating proportional difference of different epithelial cell subclusters across AC and SCC tumor subtypes in scRNA-seq datasets; three subclusters showed statistically significant proportional difference and marked with “*” indicating p values < 0.05 and “**” indicating p values < 0.01. c The dot plot showing marker genes of two epithelial cell subclusters which showed consistency with the Xenium data, including the subcluster 1 significantly more enriched in SCC samples compared with AC. d Proportional distribution of different epithelial cell subclusters in samples identified through scRNA-seq analysis; triangles with different colors at the bottom indicate AC or SCC cancer subtypes. e Two-dimensional UMAP visualization of scRNA-seq depicting immune cell subtypes within CC samples. f Dot plot showing RNA expression levels of marker genes across different immune cell populations. g Proportions distribution of different immune cells in samples identified through scRNA-seq analysis; triangles with different colors at the bottom indicate AC or SCC cancer subtypes. h Comparative distribution of immune cells across AC and SCC tumor subtypes in scRNA-seq datasets; Student’s t test was used to perform statistical analysis, and “**” means statistical p value < 0.01, “***” means statistical p value < 0.001. i Violin plot showing the expression of ICOS+ Th1-like Tregs related genes in immune cell subtypes. j Dot plot showing expression of ICOS ligand gene in immune cell subtypes, with the highest expression in pDC subtype.
We performed hierarchical clustering of the AC and SCC samples based on the frequency distribution of the 11 epithelial subclusters (Fig. 2d, Supplementary Data 5). The SCC samples did not form distinct clusters separate from the AC samples, suggesting that these tumor types could not be distinguished based on epithelial cell subclustering. Besides, we integrated the scRNA-seq data with Xenium spatial transcriptomic data to identify consistent marker genes of the subclusters for both datasets. However, only two subclusters from the scRNA-seq UMAP analysis—S100A9+ epithelial cells and MKI67+ epithelial cells—showed consistency between the two datasets (Fig. 2c, Supplementary Fig. 3c–e). The observed discrepancies may stem from the inherent genomic instability, heterogeneous transcriptomic profiles of tumor cells, and the diverse genetic backgrounds of individual patients, which could lead to artifactual clustering of many subpopulations. Consequently, it is unlikely that those subclusters represent biologically meaningful subtypes of tumor cells in cervical cancer.
We also performed sub-clustering of immune cells in the scRNA-seq data (Fig. 2e). Distinct immune cell populations, including CD4 + T cells, CD8 + T cells, plasmablasts, and others, were identified within specific clusters based on the expression of canonical marker genes (Fig. 2e, f). Some population of CD8 T cells were high in AP-1 transcription factors (such as ATF3 and FOSB) expression, and were clustered with NK cells, this subcluster was then annotated as “Activated CD8 T/NK cells”. The AP-1 transcription factors were important for the cytotoxic lymphocyte development and antitumor immunity18,19, implying this subcluster of cells may play important roles in the TME. Based on the sub-clustering results, our analysis revealed marked proportional differences in Activated CD8 T/NK cells, CD8 + T cells, regulatory T cells (Tregs), and plasmacytoid dendritic cells (pDCs) between AC and SCC (Fig. 2g, Supplementary Data 9). Statistical result revealed that the Treg cells and pDCs were more abundant in SCC samples compared with AC samples, indicating that these two immune cell types may play important roles in the tumor immune microenvironment. Moreover, the hierarchical clustering based on the proportions of different immune cells in different samples can distinguish the SCC samples from AC samples (Fig. 2h, Supplementary Data 9), highlighting distinct immune microenvironment profiles between these cervical cancer subtypes. We next performed deconvolution (CIBERSORTx) on TCGA-CESC bulk RNA-seq data, and deciphered the enrichment of plasma cells, CD8 T cells as well as NK cells in SCC samples, which was consistent with our results based on scRNA-seq data (Supplementary Fig. 3f).
We further investigated Tregs and pDCs in terms of other marker genes. Interestingly, we found that the ICOS gene was highly expressed in Tregs (Fig. 2i), while ICOSLG was highly expressed in pDCs (Fig. 2j), implying direct interactions between these ICOS+ Tregs and the ICOSL secreting pDCs, and the importance of ICOSL-ICOS pathway in the immune microenvironment in SCC. Apart from that, we also found the Th1 cells related marker genes were highly expressed in Treg populations, such as CXCR3, LTA and LTB (Fig. 2i), implying that many of the cells in that cluster were Th1-like Tregs20,21. Unlike common Tregs, these ICOS+ Th1-like Tregs act like the Th1 cells, promoting anti-tumor immunity and even inhibiting immunosuppression caused by common Tregs21,22.
In addition, we further compared the expression of MDSC (Myeloid Derived Suppressor Cells) marker genes S100A8/9 within all “myeloid cells” between AC and SCC samples. The result revealed significantly higher expression of S100A8/9 in AC samples (Supplementary Fig. 3g), suggesting their potential role in driving poor patient prognosis.
HPV infection was considered to be closely associated with immune microenvironment in cervical cancer23. In order to better understand how the HPV infection influences immune tumor microenvironment, we compared the proportions of different immune cells between HPV + AC and HPV- AC samples. Only CD8 T cells and NK cells showed statistically higher proportions in HPV + AC than HPV- AC, abundance of other immune cells such as plasma cells was not much different (Supplementary Fig. 4a). In addition, we also compared the HPV + AC samples and the SCC samples which were all HPV infected. Similar with the comparison between all AC and SCC samples, we found much higher abundance of CD8 T cells, NK cells, Tregs and pDCs in SCC samples than in HPV + AC samples (Supplementary Fig. 4b). The result indicated that the immunogenic tumor microenvironment may not be correlated with HPV infection, but more related to the cancer subtypes.
Although only three ASC samples were included in our study, with no statistically significant results found for ASC subtype, we further performed hierarchical clustering on all three cancer subtypes based on the frequency distribution of the major cell populations using scRNA-seq data (Supplementary Fig. 4c) or Xenium data (Supplementary Fig. 4d). Hierarchical clustering based on the scRNA-seq data indicated that the ASC subtype was more similar with the AC, which was consistent with their clinical similarities; while hierarchical clustering based on the Xenium data didn’t distinguish the three subtypes very well.
Subclutering of the major cell types in Xenium data to identify cell subtypes
To further resolve cellular heterogeneity within the Xenium dataset, we performed subclustering on major cell populations, including epithelial cells, immune cells, cancer-associated fibroblasts (CAFs), fibroblasts, and endothelial cells. Subclusters exhibiting strong marker gene expression patterns were annotated based on their molecular signatures, while those lacking definitive markers in the current Xenium panel were conservatively labeled as “other.” Among the epithelial cells, we identified and annotated 10 distinct subclusters based on their marker gene profiles (Fig. 3a, Supplementary Data 10). These marker genes included: ligand-encoding genes (e.g., CXCL1, CXCL10, CCL20), immune-related markers (e.g., IL6, S100A9), and proliferative markers (e.g., MKI67), suggesting diverse biological functions across these subtypes (Fig. 3a). Compared to scRNA-seq data, subclustering of epithelial cells using the Xenium data might provide a more refined approach to identify biologically relevant tumor cell subtypes, thereby deepening our understanding of the diverse functional roles these cells play in cancer development, progression, and treatment.
a Subclustering the epithelial/tumor cells and identifying epithelial subtypes according to respective marker gene expression profiles; the UMAP after clustering is shown on the left and heatmap (right) indicates marker genes expression in different subclusters. b Subclustering the immune cells in the Xenium data and identifying specific subtypes based on related markers genes expression; two-dimensional UMAP visualization is shown on the left and heatmap indicating respective marker genes expression is shown on the right. Two-dimensional UMAP visualization of Xenium data depicting fibroblasts (c), CAFs (d) and endothelial cells (e) subclusters and their respective marker gene expression profiles; the UMAP is on left side and heatmap showing respective marker gene expression profiles is on the right.
For the immune cell compartment, we successfully resolved 20 distinct subtypes, encompassing a diverse array of cell populations, including 3 plasma cell subtypes, 6 macrophage subtypes, 6 T cell subtypes, as well as NK cells, B cells, conventional dendritic cells (cDCs), plasmacytoid dendritic cells (pDCs), and mast cells (Fig. 3b). The plasma cells were further categorized into 4 subtypes based on their antibody secretion profiles: IGHM+ plasma cells representing early or IgM-secreting plasma cells; IGHG2+ plasma cells characterized by IgG2 antibody production, indicative of a more mature or class-switched state; and the IGKV2-30+ plasma cells suggesting a unique clonal population; the fourth cluster was unlike the plasma cells due to the expression of RGS5, VWF, NOTCH3 and NOTCH1 genes, it was therefore annotated as “NOTCH1+ pericytes” to discriminate from other pericytes populations (Supplementary Fig. 5a, b). These subtypes reflect the functional diversity of plasma cells in terms of their antibody repertoire and immune responses. Among the 6 macrophage subtypes, we identified two distinct populations with M2-like polarization, characterized by the overexpression of CD163, a well-known M2 marker24: MMP12 + M2 macrophages with overexpression of MMP12, and the M2 macrophages representing a general M2 population without additional specific markers (Supplementary Fig. 5c, d). Three other subtypes exhibited M1-like polarization, each marked by distinct functional genes including IL1B, CCL4 and G0S2. The final macrophage subtype was annotated as CCL21 macrophage, as their expression of CCL21 did not clearly align with either M1 or M2 polarization, implying a potentially unique or transitional state (Supplementary Fig. 5c). For the six T cell subtypes (Supplementary Fig. 5e, f), two of them were CD8 T cells, namely CCL4 + CTL and CD8 T cells (other CTLs), the other four were CD4 T cells, including CCL13+ Th, IL7R+ Th, Treg and CD4 T cells (other Th cells). This comprehensive classification of immune cell subtypes provides a detailed framework for understanding their functional diversity and roles in the immune microenvironment.
Within the stromal compartment, we identified 4 fibroblast subtypes marked by CSF3R, LIF, CXCL1, and CXCL14 (Fig. 3c), CAF subtypes expressing S100A9, PLVAP, and IDO1 (Figs. 3d, 2) endothelial subtypes characterized by CXCL12 and CCL2 expression (Fig. 3e). Pericytes, defined by RGS5 and VWF expression, were not further subclustered due to the low cell numbers. The “PLVAP + CAF” subcluster not only expressed ACTA2 and DCN, but also expressed the endothelial marker VWF; since PLVAP was well known as the tumor endothelial marker gene, there was no evidence that CAFs expressed PLVAP gene at all, we believed these cells were mixed up between CAFs and endothelial cells, therefore we renamed this cluster of cells as “PLVAP+ Endo” to discriminate with other tumor endothelial cells.
This annotation step is critical for downstream analysis, as it enables systematic measurement of spatial distances between distinct cell subtypes within these highly populated regions.
Determining specific cell-cell interactions in AC or SCC samples with CellChat
Our scRNA-seq data has identified specific cell types and subtypes enriched in AC and SCC samples. Next we tried to determine distinct cell-cell interactions in these tumor types, especially focusing on two epithelial cell subtypes (S100A9+ and MKI67 + ), which were also detected in the Xenium data, along with two immune cell types (Tregs and pDCs) that showed the most significant proportional differences between AC and SCC.
Using the CellChat tool25, we analyzed cell-cell interactions in the scRNA-seq data from each sample and employed t-tests to evaluate differences between AC and SCC groups. The results revealed significantly stronger cell-cell interactions in SCC samples for these four cell types (Fig. 4a). In addition to interactions within these four cell types, we also found enhanced interactions between them and other cell populations in the SCC microenvironment (Fig. 4a). For example, S100A9+ and MKI67+ epithelial cells, as well as pDCs, exhibited stronger interactions with activated CD8 T/NK cells, CD8 + T cells, CD163+ macrophages, and proliferating macrophages in SCC, while Tregs showed stronger interactions only with activated CD8 T/NK cells and CD8 + T cells, suggesting distinct functional roles for these cells in SCC. Then we focused on their interactions among those four cell types and activated CD8 T/NK cells and CD8 + T cells, in total six cell types/subtypes. We found more interaction events among these six cell identities in SCC samples compared with AC samples (Fig. 4b).
a Comparative heatmap illustrating the most divergent cell-cell interactions involving pDCs, Tregs, S100A9+ and MKI67+ epithelial/tumor cells-both with themselves and with other cell populations- between AC and SCC. b Network graph of cell communication analysis of six cell subtypes (pDC, Treg, activated CD8 T/NK, CD8 T cells, MKI67+ and S100A9+ epithelial cells) in AC and SCC tumors. Comparison of putative ligand-receptor pairs involving interactions between CD8 + T cells or activated CD8 + T/NK cells and Tregs (c), pDCs (d), S100A9+ epithelial cells (e), and MKI67+ epithelial cells (f) in AC and SCC.
Next, we examined potential ligand-receptor interactions of these four cell types with AP-1 activated CD8 T/NK cells and CD8 + T cells separately, and identified a greater number of interaction pairs in SCC compared to AC (Fig. 4c–f), further supporting the notion of more robust cell-cell communications in SCC. For instance, pDCs exhibited more extensive interactions with CD8 + T cells or activated CD8 + T/NK cells in SCC samples, characterized by a higher number of potential ligand-receptor pairs (Fig. 4c). These interactions included key pathways such as APP-CD74, CD80/CD86-CTLA4, as well as more robust engagements between various HLA class I molecules and the CD8 protein. The HLA class I molecules are responsible for presenting intracellular antigens to cytotoxic T cells and also activating the cytotoxic T cells26, the enhanced interactions indicate more activated antitumor immunity of cytotoxic lymphocytes in SCC samples compared with AC samples. The function of APP-CD74 pathway was usually considered to suppress immune response in cancer27,28, but in some other cases, it was shown to promote the antitumor immunity through activating the cytotoxic T cells29,30. The CTLA4 was proved to suppress T cell activation and proliferation and inhibit antitumor immunity31,32, while in our case, the CD80/CD86-CTLA4 interactions between pDCs and CD8 T cells may inhibit overactivation of the CD8 T cells in SCC. For Tregs interacting with CD8 + T cells or AP-1 activated CD8 T/NK cells (Fig. 4d), the major difference was interactions between HLA class I molecules from Tregs and the CD8 proteins from CTLs, which was considered to suppress CTL activity33,34, probably indicating a dynamic equilibrium between immune activation driven by larger numbers of CTLs and suppression caused by Tregs in SCC. For the other two epithelial subtypes, their interaction patterns were largely similar, with the most notable difference being that interaction pairs involving ICAM1-SPN/CD43 and CDH1-(ITGAE + ITGB7) were exclusively observed in SCC samples for both CD8 + T cells and activated CD8 + T/NK cells (Fig. 4e, f). The interplay between ICAM1 and SPN/CD43 remains poorly understood, with only a limited number of studies suggesting their role in tumor adhesion in vitro35. On the other hand, the interactions between CDH1 on epithelial cells and ITGAE/ITGB7 on CD8 + T/NK cells are known to promote the retention of cytotoxic lymphocytes at tumor sites, thereby enhancing local immune surveillance36,37.
In general, our results showed that SCC exhibited a larger variety of cell-cell interactions, with these cell types interacting through more ligand-receptor pairs, these interactions may be related to distinct characteristics of the SCC tumor type.
Determining tumor type specific Interactions in cervical cancer based on distance analysis
After annotating the cell types and subtypes in the spatial transcriptomic data, we performed permutation distance analysis38,39,40 on the data, and determined the pairs of cells showing significantly closer distance in AC or SCC samples. The principle of this method was to first fix an “index cell” and the surrounding cells, perform Voronoi tessellation after determining the cell centroids, and then use permutation methods to generate randomized cell distance data, followed by a permutation test to assess the spatial relationships between “index cell” and others (Fig. 5a). The closer distance would infer higher possibility of those pairs of cells interacting with each other, representing unique tumor microenvironment in AC or SCC. Each annotated cell type was independently designated as an ‘index cell’; using permutation distance analysis, we identified its most proximal cell types as potential interaction partners.
a Schematic illustration of permutation distance analysis for spatial transcriptomic data: first fix an “index cell” and the surrounding cells, perform Voronoi tessellation after determining the cell centroids, and then use permutation methods to generate randomized cell distance data, followed by a permutation test to determine interactions using a predefined p value cutoff; scale bar is 10 µm. b The top 20 cell-type pairs exhibiting the most significant spatial distribution divergence between AC and SCC tumors, ranked by mean distance fold-change values. c The top 20 immune & non-immune cell-type pairs exhibiting the most significant spatial distribution divergence between AC and SCC tumors, ranked by mean distance fold-change values. The top 20 cell-type pairs involving pDC (d) or Treg (e) with others which showed the most significant spatial distribution divergence between AC and SCC tumors, ranked by mean distance fold-change values. f Permutation distance analysis result showing the most significant spatial distribution divergence between AC and SCC tumors for S100A9+ epithelial cells (upper) and MKI67+ epithelial cells (lower) separately.
Our permutation distance analysis revealed several pairs of cell types with the most divergent spatial distributions between AC and SCC tumors (Fig. 5b). For instance, G0S2 + M2 macrophages exhibited significantly closer spatial proximity to two specific cell subtypes: IGHG2+ plasmablasts and VEGFA+ epithelial cells. Interestingly, the distance between G0S2 + M2 macrophages and IGHG2+ plasmablasts was markedly reduced in SCC samples, whereas their proximity to VEGFA+ epithelial cells was more pronounced in AC samples (Fig. 5b). The spatial proximity between M2-macrophages and IgG2+ plasmablasts in SCC may be due to the mechanism that CD163 + M2 macrophages induce differentiation and isotype switching of plasma cells41. These findings suggest that the same cell type can exhibit distinct interaction patterns within different tumor microenvironments, potentially contributing to the unique characteristics and progression of each tumor type.
It is also noteworthy that more than half of the top 20 most divergent interactions between AC and SCC samples occurred among the immune cell subpopulations, highlighting the distinct immune microenvironments associated with these two cancer types (Fig. 5b). These differences may underlie their contrasting clinical behaviors and outcomes. For example, CCL4 + M1 macrophages demonstrated close interactions with both IL1B + M1 macrophages and CD4 + T cells; however, the communication with IL1B + M1 macrophages was stronger in AC samples, while interactions with CD4 + T cells were more prominent in SCC samples (Supplementary Fig. 6). This divergence may reflect differences in the anti-tumor immune responses elicited in AC versus SCC, potentially influencing their respective therapeutic responses and prognoses.
The observed variability in immune cell behavior across different microenvironments underscores the need for tailored treatment strategies. For instance, the distinct interaction patterns of immune cells in AC and SCC may require different therapeutic approaches to effectively target their unique microenvironmental dynamics. By leveraging spatial transcriptomic techniques, we can gain deeper insights into these specific cellular behaviors, paving the way for more precise and personalized therapeutic interventions. Such an approach holds great promise for improving treatment efficacy and patient outcomes, ultimately leading to better prognosis and survival rates.
In addition, we extended our analysis to investigate the interactions between immune cells and non-immune cells, revealing that the most divergent interactions occurred between epithelial cell subtypes and various immune cell populations (Fig. 5c). For instance, CD8 + T cells were found to engage in significant interactions with multiple epithelial subtypes, including CXCL1 + , CXCL10 + , VEGFA + , and S100A9+ epithelial cells. Notably, all of these interactions were markedly stronger in SCC samples compared to AC samples. This observation suggests that SCC may exhibit a more robust anti-tumor immune response, potentially driven by enhanced CD8 + T cell activity and their closer spatial proximity to epithelial cells. These findings highlight the distinct immune microenvironment of SCC, which may be associated with its high link to HPV infections driving carcinogenesis and therapeutic responsiveness.
We also analyzed the spatial distances for those four specific cell types: pDCs, Tregs, S100A9+ epithelial cells, and MKI67+ epithelial cells. pDCs were found to be in close proximity to numerous immune cell types, and for most pairs, the distances were shorter in AC samples compared to SCC samples. However, the distance between pDCs and IgG+ plasma cells was notably shorter in SCC samples (Fig. 5d). This observation aligned with the established role of pDCs in promoting B cell maturation and plasma cell differentiation42,43, suggesting that the closer interaction between pDCs and plasma cells in SCC samples may reflect enhanced plasma cell maturation. For Tregs, the most striking difference between AC and SCC samples was observed in their proximity to CCL13+ Th cells, with significantly shorter distances in AC samples (Fig. 5e). The reason why Tregs preferentially interact with CCL13+ Th cells, rather than other Th cell subsets, remains unclear and warrants further investigation. Additionally, Tregs were found to be closer to CCL4+ CTLs and other CD8 + T cells in AC samples, suggesting a role for Tregs in suppressing cytotoxic T cell activity within the AC tumor microenvironment (TME). In contrast, the distance analysis for the two epithelial subtypes (S100A9+ and MKI67+ epithelial cells) revealed fewer interactions compared to pDCs and Tregs (Fig. 5f). Both epithelial subtypes were predominantly located near other epithelial cells, with two exceptions: both were also found in close proximity to CCL4+ CTLs and other CD8 + T cells, implying a cytotoxic role for these T cells in targeting and eliminating tumor cells.
Moreover, we also performed the distance analysis among the six major cell identities used before to validate the results based on Cellchat. First, we cross-checked the consistency of cell annotations based on both omics dataset. Apart from the two epithelial subtypes showed before (Supplementary Fig. 3e), we found the consistency of Tregs, pDCs and CD8 T cells (Supplementary Fig. 7a); CCL4 CTLs (ST data) were most similar with activated CD8 T/NK (scRNA-seq data) based on our integrative analysis (Supplementary Fig. 7a). The results revealed stronger interactions among these cell types in SCC samples, especially pDC- Treg and aCD8 T/NK-Treg pairs (Supplementary Fig. 7b), which was consistent with the ligand-receptor based analysis.
Microenvironmental cell abundance analysis surrounding the target cells to differentiate AC and SCC
We also quantified the abundance of surrounding cell types within a 100-μm radius around the target cells (e.g., tumor cells, immune cells) to assess their spatial interactions and microenvironmental compositions. Here we still focused on pDCs, Tregs, S100A9+ and MKI67+ epithelial cells, which were regarded as “target” cells separately (Fig. 6a). We found three cell subtypes, including CXCL1+ Fib, CXCL6+ Epi and IgG+ plasmablasts were significantly more enriched in SCC samples than AC near the pDCs, while PLVAP+ Endothelial cells were more enriched in AC samples (Fig. 6b, c). It was well known that PLVAP+ Endothelial cells played important roles in tumor progression as well as angiogenesis44,45, and pDCs may enhance their functions in AC tumor type. Surrounding the Tregs, five cell subtypes, including CXCL12+ Endo, CXCL14+ Fib, MUC5AC+ Epi, Pericytes and PLVAP+ Endo were more enriched in AC samples, while two other subtypes, CXCL6+ Epi and IgG+ plasmablasts were more enriched in SCC samples (Fig. 6d, e). The most significant proportional difference was found surrounding the MKI67+ Epi, in total nine cell subtypes had proportional difference between AC and SCC samples, and S100A9+ Epi was the most divergent one, it was much more enriched in SCC samples, implying the importance of their interactions in the TME (Fig. 6f, g). Besides, among these 9 cell subtypes, there was only one immune cell subtype, cDCs which was more enriched in SCC samples. The cDCs play important roles in anti-tumor immunity by presenting tumor antigens, activating T cells, and modulating the tumor microenvironment; their ability to induce anti-tumor immune responses can directly or indirectly inhibit the proliferation of tumor cells46,47,48. Higher abundance of cDCs surrounding the proliferating tumor cells may imply enhanced antitumor immunity in SCC samples. For the S100A9+ Epi surrounded cells, only VEGFA+ Epi and MKI67+ Epi were significantly more enriched in SCC samples (Fig. 6h, i).
a Schematic illustration of quantified analysis on the abundance of surrounding cell types within specific area: for each target cell type, the spatial abundance of surrounding cell types was quantified within a 100-μm radial neighborhood of individual cells across all samples, followed by statistical analysis; scale bar is 50 mm. The boxplots showing cell types with statistically significant proportional difference (divided by 100-μm radial surrounding cell numbers) between AC and SCC samples surrounding pDC (b), Treg (d), MKI67+ epithelial cells (f) and S100A9+ epithelial cells (h). Representative images showing differential cell-type abundance around one target cell in AC and SCC samples, with the target cells pDC (c), Treg (e), MKI67+ epithelial cells (g) and S100A9+ epithelial cells (i).
Cervical cancer neighborhood analysis
To further explore the spatial organization of the tumor microenvironment in cervical cancer, we performed the neighborhood analysis on the spatial transcriptomic data of cervical cancer40. We determined 13 distinct neighborhoods based on local distributions of specific cell types within the set regions (Fig. 7a), and each neighborhood represented a specific clustering area of cells in space, reflecting the spatial structure of different cell types.
a The heatmap showing 13 cellular neighborhoods identified using an unsupervised method, each neighborhood is enriched with specific cell types or subtypes. b Representative image of the cluster 3 enriched with IDO1 + CAF and various kinds of immune cells. c Representative images of the spatial distributions and abundance of all cellular neighborhoods within the AC, SCC and ASC tumor samples. d The boxplot indicating proportions of different cellular neighborhoods (CNs) in AC and SCC samples; Student’s t-test was used to perform statistical analysis, and “*” means statistical p value < 0.05, “**” means statistical p value < 0.01.
In our neighborhood analysis, multiple tumor cell subpopulations were enriched in clusters 4, 10, and 12, indicating aggregation of these tumor cells in space and formation of distinct structures within the microenvironment (Fig. 7a). Additionally, cluster 3 was most abundant with various kinds of T cells, CCL21 macrophages, pDCs and IDO1+ CAFs (Fig. 7b). This implied strong interactions between IDO1+ CAFs with the immune cells within a small niche. IDO1+ CAFs was well known as pivotal metabolic-immune regulators in the TME, driving immune evasion and tumor progression via Trp metabolism, cytokine crosstalk, and stromal remodeling49,50. Based on our spatial transcriptomic data, we showed that they may interact with pDCs, CCL21 macrophages as well as various kinds of T cells to fulfill biological functions. The spatial distributions of different neighborhoods in the TME can be visualized (Fig. 7c).
Next we compared the abundance of different clusters in AC and SCC samples. The result showed that cluster 1 was significantly abundant in AC samples, while cluster 2 was most enriched in SCC samples (Fig. 7d). The cluster 1 was mainly composed of PLVAP+ epithelial cells, pericytes, PLVAP + , CCL2+ and CXCL12+ endothelial cells. This small niche may be responsible for tumor metastasis44,45. Since this cluster was more enriched in AC samples, implying that AC had higher risk of metastasis than SCC, which was consistent with the clinical experience. The cluster 2 was mainly composed of different subtypes of epithelial cells, with highest abundance S100A9+ and CXCL6+ epithelial cells. This reflected distinct tumor cell organizations in the SCC.
These results demonstrate that within the tumor immune microenvironment of cervical cancer, distinct spatial aggregation patterns exist among different cell types, particularly in regions where tumor cells and immune cells interact. These findings shed new light on the dynamics of cell-cell interactions in the tumor microenvironment and may lay a theoretical foundation for the development of future immunotherapy strategies. Furthermore, the identification of cellular neighborhoods will enhance our understanding of the functional roles played by specific cell types within the context of their surrounding microenvironment.
Discussion
Cervical cancer (CC) remains a significant global health burden, with adenocarcinoma (AC) and squamous cell carcinoma (SCC) representing distinct histological subtypes that exhibit divergent clinical behaviors and therapeutic responses. By integrating single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST), our study provides unprecedented insights into the cellular and spatial heterogeneity of these subtypes, revealing unique tumor microenvironment (TME) dynamics that may underlie their contrasting prognoses. Below, we contextualize our findings, discuss their implications for understanding CC biology, and propose future directions for therapeutic development.
Our scRNA-seq analysis revealed stark differences in the cellular composition of AC and SCC tumors. AC exhibited a higher proportion of epithelial cells, suggesting a tumor-cell-dominated TME, while SCC was enriched in immune cells such as plasma cells and NK/T cells. These findings aligned with clinical observations of SCC’s more immunogenic nature compared to AC, which often evaded immune detection due to its deeper anatomical location and stromal-rich microenvironment. The SCC-predominant epithelial subcluster expressing KRT5, KRT6A, and the pro-inflammatory alarmin gene S100A9 highlights a potential link between squamous differentiation, myeloid cell recruitment, and immune modulation. S100A9, a known driver of tumor-stroma crosstalk15,51, may foster an inflammatory niche in SCC, facilitating immune cell infiltration while paradoxically promoting immunosuppressive signaling. Conversely, AC-enriched subclusters overexpressing histone genes (e.g., HIST1H3A) and tetraspanins (e.g., TSPAN1/3/8) suggest distinct molecular pathways driving tumor progression, possibly linked to DNA damage repair and cell adhesion—mechanisms that warrant further exploration in the context of therapy resistance.
In cervical SCC, high plasma cell abundance may have dual functions: the IgG secreted plasma cells may promote antibody-dependent anti-tumor responses via viral antigen targeting (e.g., HPV E6/E7)52, these antibodies may neutralize the viral particles to inhibit infection of new cells, or opsonize tumor cells through phagocytosis (ADCP) or NK-cell killing (ADCC)53,54,55. Conversely, diffuse intratumoral plasma cells (IgA/IgG4-dominated) drive immunosuppression through M2 macrophage polarization, fibrosis, and T-cell exclusion56,57. Spatial distribution, antibody isotype, and HPV antigen specificity determine net impact.
The spatial transcriptomic data further illuminated the architectural divergence between subtypes. While SCC tumors displayed robust interactions between CD8 + T cells and epithelial subtypes (e.g., CXCL1 + , S100A9 + ), AC tumors were characterized by stromal-rich niches dominated by PLVAP+ endothelial cells and pericytes. PLVAP, a marker of vascular permeability and angiogenesis44,45, is associated with metastatic potential, consistent with AC’s clinical propensity for distant spread. These findings underscore the importance of spatially resolved analyses in dissecting subtype-specific TME features that bulk sequencing approaches may obscure.
The immune landscape of SCC was marked by heightened activity of cytotoxic lymphocytes (e.g., CCL4+ CTLs) alongside elevated immunosuppressive populations, including Tregs and pDCs, although part of the Tregs showed Th1-like activities with CXCR3 and LTA/LTB overexpression. The co-enrichment of these cells in SCC suggests a dynamic equilibrium between immune activation and suppression, potentially driven by bidirectional interactions. For instance, the ICOS-ICOSLG axis linking Tregs and pDCs may enhance immunosuppressive signaling, while the proximity of pDCs to IgG+ plasma cells in SCC points to their role in fostering plasma cell maturation. Intriguingly, SCC Tregs exhibited a Th1-like phenotype (e.g., CXCR3, LTA/LTB), a paradoxical feature that may reflect functional plasticity. Th1-like Tregs have been implicated in both suppressing effector T cells and secreting pro-inflammatory cytokines21,22, suggesting a dual role in SCC that merits further investigation. In contrast, AC’s immune microenvironment was less active but uniquely organized, with Tregs spatially proximal to CCL13+ Th cells and CCL4+ CTLs. This spatial arrangement may reflect AC-specific mechanisms of immune evasion, such as Treg-mediated suppression of cytotoxic lymphocytes. The scarcity of cDCs—critical antigen-presenting cells—in AC’s proliferative niches (MKI67+ Epi surrounded) further underscores its immune-cold phenotype, which may contribute to its resistance to checkpoint inhibitors.
Stromal cells emerged as key architects of subtype-specific TMEs, which can be potential therapeutic targets for CC. Both tumor types harbored IDO1+ cancer-associated fibroblasts (CAFs) that colocalized with immune cells (e.g., T cells, pDCs) in spatially defined neighborhoods. IDO1, a mediator of tryptophan metabolism, is a known driver of immune evasion and a promising therapeutic target58,59. Its association with CCL21+ macrophages—a population with ambiguous M1/M2 polarization—suggests a novel immunosuppressive axis in CC. Interestingly, AC’s stromal compartment was enriched in PLVAP+ endothelial cells and CXCL12+ fibroblasts, which may promote angiogenesis and metastatic dissemination. Targeting PLVAP or its downstream effectors (e.g., VEGF) could disrupt AC’s vascular niche, potentially mitigating metastasis.
In addition to decoding the molecular mechanisms of different CC subtypes, our findings also have several translational implications. For instance, the identification of spatial neighborhoods revealed coexistence of activated CD8 + T cells and immunosuppressive networks (e.g., ICOS+ Tregs, IDO1+ CAFs) in certain patients may reflect localized immunosuppressive niches. Combining checkpoint inhibitors (e.g., anti-PD-1) with ICOS/IDO1 blockade could potentially shift this balance towards immune activation, offering a strategy for precision medicine. The stromal and vascular dominance of AC highlights the need for therapies targeting PLVAP+ endothelial cells or histone-driven DNA repair pathways. Besides, the identification of neighborhoods enriched in metastatic (AC) or immune-active (SCC) cells could guide prognosis and treatment selection.
Limitations of this study include the small sample size of ASC cases and the restricted gene panel in Xenium spatial assays, which may have obscured some important cell populations such as γδ T cells. To improve this, customized panels and other single-cell level data (e.g., spatial proteomic data, scRNA-seq data) are needed to identify more cell types/subtypes which are biologically meaningful. Besides, future work should integrate multi-omics data (e.g., proteomics, epigenetics) to validate ligand-receptor interactions and explore the functional roles of Th1-like Tregs and CCL21+ macrophages.
In summary, by integrating scRNA-seq and spatial transcriptomic data, our study delineates the molecular and spatial landscapes of cervical adenocarcinoma and squamous cell carcinoma, revealing subtype-specific mechanisms of immune evasion, stromal remodeling, and metastasis. These insights not only advance our understanding of CC biology but also provide a roadmap for developing precision therapies tailored to the unique TME of each subtype. As spatial technologies evolve, their application in clinical trials could transform CC management, offering hope for improved outcomes in this globally significant malignancy.
Methods
Clinical samples collection
The cervical cancer samples, including 8 cases of adenocarcinoma, 6 cases of squamous cell carcinoma, and 2 cases of adenosquamous carcinoma, were obtained from multiple sites, including Sun Yat-sen Memorial Hospital of Sun Yat-sen University, the First Hospital of Jilin University and the Third Affiliated Hospital of Harbin Medical University. All these 16 fresh samples were subjected to single-cell RNA sequencing, and paraffin-embedded tumor tissues were prepared for 15 of them (Supplementary Data 1). Additionally, 7 more paraffin-embedded tissues collected from the Sun Yat-sen Memorial Hospital of Sun Yat-sen University together with 15 FFPE samples were used for Xenium spatial transcriptome analysis (n = 22, Supplementary Data 1). This study was reviewed and received approval from the Ethics Committee of Sun Yat-sen Memorial Hospital of Sun Yat-sen University, the First Hospital of Jilin University Ethics committee and the Ethics committee of Third Affiliated Hospital of Harbin Medical University. All ethical regulations relevant to human research participants were followed. All patient data were de-identified prior to analysis, as all data were fully anonymized and no individual can be identified, the requirement for informed consent for publication was waived by the Ethics Committees from the three hospitals. Relevant clinical information was collected from medical records.
Single-cell sample preparation
The fresh cervical cancer samples were collected and processed immediately with the procedure modified from the manufacturer’s protocol (Demonstrated Protocol, CG000147). Briefly, the fresh tumor tissues were cut into small pieces and incubated with digestion medium (RPMI1640 supplemented with 200 U/mL collagenase IV (C1889, Sigma), 10 U/mL DNAse II (D8764, Sigma), 2% heat-inactivated FCS) for 30 min at 37 °C. Then the single cell suspension was filtered through a prewetted 40 μm strainer to remove clumps and debris, followed by centrifuge and washing with RPMI1640 medium for three times. The cell pellet was resuspended with 1x chilled Red Blood Cell Lysis Buffer (ab204733, Abcam) and incubated on ice for 10 min, and then washed with cold PBS three times. The single cell suspension was then prepared at an appropriate concentration for the library preparation steps.
Single-cell library preparation and sequencing
The 5’ single-cell library was prepared strictly following the manufacturer’s instructions (User Guide CG000733, 10x Genomics) using the Chromium GEM-X Single Cell 5’ Kit v3 (cat# PN-1000699). The final libraries were pooled and sequenced at the Illumina NovaSeq6000 sequencer using the 150 bp PE kits (read 1 = 26 bp, i7 index = 10 bp, i5 index = 10 bp, read 2 = 90 bp read configuration) targeting an average of 20,000 reads per cell.
ScRNA-seq data pre-processing, dimension reduction and clustering
Single-cell RNA sequencing (scRNA-seq) data were processed using the 10x Genomics Cell Ranger pipeline (v7.1.0). Raw sequencing files (BCL format) were first demultiplexed into sample-specific FASTQ files using cellranger mkfastq, followed by alignment and feature quantification via cellranger count. To account for RNA content, reads were aligned to a pre-mRNA reference transcriptome (GRCh38-2020-A) that retains intronic regions, ensuring detection of unspliced nuclear transcripts.
Following Cell Ranger processing, the data were imported into R via Seurat::Read10X (v5.1.0)60. Nuclei were filtered using thresholds of ≥500 genes, <10,000 genes, <20,000 UMIs, or ≥20% mitochondrial content per nucleus. To mitigate ambient RNA, single cells with UMIs <3 X the median droplet count (approximating empty droplets, as most droplets lack cells) were excluded. The data underwent doublet detection using DoubletFinder (PCs 1–20; parameters: nExp=0.076×(nCells²/10⁴), pN=0.25, pK optimized via paramSweep_v3).
All samples were merged into a unified Seurat object and processed via Seurat’s standard workflow. Data were log-normalized (scale factor=10,000), and 2000 variable features were selected using FindVariableFeatures (vst method). Scaling and PCA were applied to all genes. To mitigate donor-specific batch effects, Harmony integration (RunHarmony, grouped by donor) was performed prior to clustering, followed by UMAP dimensionality reduction. Expression of marker genes in the resulting clusters was then used to label clusters for downstream analysis.
Cell-cell communication analysis with the scRNA-seq data
The CellChat package (version 2.1.2)25 was used to perform cell-cell communication analysis in our cervical cancer dataset. First the data with normalized gene expression matrix and cell type annotations was used as input and incorporated with the built-in ligand-receptor interaction database to infer biologically significant cell-cell communications with the truncated mean (trim = 0.1) to reduce outlier effects. Then the inferred networks were aggregated at the cell type level and visualized using heatmaps, networks and circle plots independently. For multi-group comparisons (AC vs SCC in our case), the differential interactions were quantified using a permutation test and then used for statistical analysis.
Spatial transcriptomics using Xenium platform
The FFPE tissues were sectioned at a thickness of 5 µm and placed on the Xenium slides (cat# PN-1000659, 10x Genomics) according to the manufacturer’s instructions (CG000578, 10x Genomics). After baking at 42 °C for 3 h, slides were stored in 50 ml centrifuge tubes containing desiccant at room temperature until subsequent experiments. Next, the tissue sections were dewaxed and de-crosslinked following the manufacturer’s instructions (CG000580, 10x Genomics), and then treated according to the user guide for “Xenium In Situ Gene Expression” (CG000760, 10x Genomics). The whole process included a series of manual experiments, including Priming Hybridization, RNase Treatment & Polishing, Probe Hybridization, Post Hybridization Wash, Ligation, Amplification, Cell Segmentation Staining, and Autofluorescence Quenching. After completing these steps, the slides were processed using the user guide “Xenium Analyzer” (CG000584, Rev A, 10x Genomics). The pre-designed “Human Immuno-Oncology” gene panel (cat# PN-1000654) including 380 genes together with customized 50 genes was used in the assay (Supplementary Data 3). The 50 custom genes were designed to target specific cell subtypes based on the scRNA-seq data.
Spatial transcriptomic data processing
The raw count matrix generated by the Xenium Analyzer platform was pre-processed using the Seurat package (version 5.1.0) in R (version 4.4.1). Cells expressing at least three gene (min.freatures >=3) were retained. The raw count data were normalized using the SCTransform function. After PCA analysis using the RunPCA function, the data was further integrated by RPCA integration to minimize the batch effect, followed by clustering and visualization through UMAP dimensionality reduction, leading to identification of the major cell populations, including epithelial/cancer cells, endothelial cells, pericytes, cancer-associated fibroblasts (CAFs), fibroblasts, B cells, plasmablasts, myeloid cells, mast cells and NK/T cells.
For subclustering of each major cell identity, the process was repeated starting from the SCTransform step. A specific cell subtype was annotated only if the expression level of its marker gene was at least 4 times higher than in other subclusters (Log2FC > 2) and if more than 60% of the cells within that cluster expressed the gene. Subclusters that could not be annotated based on these criteria were labeled as “other” for downstream analysis.
Cell-cell interactions analysis
The permutation-based distance analysis was used to systematically evaluate cell-cell spatial interactions under different experimental/pathological conditions40. First, one annotated cell type was independently designated as an ‘index cell’, the observed spatial distances between cell types were computed using Delaunay triangulation within biologically independent regions to establish nearest-neighbor relationships. For each cell pair (index cell → celltype2), the mean triangulation distance was calculated across all regions. Statistical analysis was performed through implementation of a permutation approach by randomly shuffling cell type labels within each region while preserving spatial coordinates and tissue architecture. This randomization process was repeated 100 times to generate a null distribution of expected distances. The average distances for each field in each permutation cycle were calculated and compared with the observed distances using a Mann-Whitney U Test. The fold enrichment of the observed distance to the mean distance derived from the permutation test was determined for each pair of cells. The whole process was repeated each time switching the “index cell”.
Neighborhood enrichment analysis with Spatial Transcriptomic data
The Neighborhood enrichment analysis was performed as previously described40. A window size of 20–100 nearest neighbors was selected for analysis. Within each window, different cell types and subtypes were quantified, and a vector containing cell count information was generated. These vectors were then clustered to identify recurrently composed neighborhoods, revealing distinct spatial niches enriched with specific cell populations. Through extensive parameter optimization, we determined the optimal number of cellular neighborhoods that best captured tissue spatial organization and heterogeneity. Finally, we identified the most highly enriched cell identities within each neighborhood.
Statistics and reproducibility
For quantitative assessment of cellular microenvironment composition, we developed a spatial neighborhood frequency analysis pipeline. Four functionally relevant cell subtypes (pDC, MKI67+ epithelial cells, S100A9+ epithelial cells, and regulatory T cells) were selected as spatial anchors. For each anchor subtype, we first extracted all cellular coordinates and cross-joined them with coordinates of 47 other cell subtypes within the same tissue section using Cartesian product generation. Euclidean distances between anchor cells (x₁,y₁) and neighboring cells (x₂,y₂) were computed as d = sqrt((x₂−x₁)² + (y₂−y₁)²), with spatial proximity defined as d < 100 (equivalent to 100-unit radius). Neighborhood frequencies were normalized as percentages relative to total proximal interactions per sample.
Comparative analysis between AC and SCC groups was performed using two-tailed Student’s t tests with Welch’s correction. Statistical significance was hierarchically annotated (*p < 0.05, **p < 0.01, ***p < 0.001). The analytical workflow was implemented in R 4.4.1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq data for the cervical cancer patients have been deposited at ArrayExpress (EMBL-EBI) with accession number E-MTAB-15983, as well as OMIX with accession number OMIX012298 with controlled access; the spatial transcriptomic data have been deposited at BioImage Archive (EMBL-EBI) with accession number S-BIAD2378, and also OMIX with accession number OMIX012586 with controlled access.
Code availability
Details of publicly available software used in the study are given in the “Methods”. No custom code or mathematical algorithm that is deemed central to the conclusions was used.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Arbyn, M. et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob. Health 8, e191–e203 (2020).
Burmeister, C. A. et al. Cervical cancer therapies: Current challenges and future perspectives. Tumour Virus Res 13, 200238 (2022).
Chidebe, R. C. W., Osayi, A. & Torode, J. S. The global fund, cervical cancer, and HPV infections: what can low- and middle-income countries do to accelerate progress by 2030?. EClinicalMedicine 81, 103127 (2025).
Cohen, P. A., Jhingran, A., Oaknin, A. & Denny, L. Cervical cancer. Lancet 393, 169–182 (2019).
Wang, S. S., Sherman, M. E., Hildesheim, A., Lacey, J. V. Jr. & Devesa, S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976–2000. Cancer 100, 1035–1044 (2004).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e1624 (2017).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e1236 (2018).
Wang, Y. et al. Spatial transcriptomics: Technologies, applications and experimental considerations. Genomics 115, 110671 (2023).
Rao, A., Barkley, D., Franca, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Hemler, M. E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 (2005).
Moll, R., Divo, M. & Langbein, L. The human keratins:Biology and pathology. Histochem Cell Biol. 129, 705–733 (2008).
Ehrchen, J. M., Sunderkötter, C., Foell, D., Vogl, T. & Roth, J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 86, 557–566 (2009).
Gebhardt, C., Nemeth, J., Angel, P. & Hess, J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharm. 72, 1622–1631 (2006).
Shabani, F., Farasat, A., Mahdavi, M. & Gheibi, N. Calprotectin (S100A8/S100A9): a key protein between inflammation and cancer. Inflamm. Res. 67, 801–812 (2018).
Chakraborty, U., Shen, Z. J. & Tyler, J. Chaperoning histones at the DNA repair dance. DNA Repair (Amst.) 108, 103240 (2021).
Zoller, M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40–55 (2009).
Schnoegl, D., Hiesinger, A., Huntington, N. D. & Gotthardt, D. AP-1 transcription factors in cytotoxic lymphocyte development and antitumor immunity. Curr. Opin. Immunol. 85, 102397 (2023).
Lynn, R. C. et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019).
Overacre-Delgoffe, A. E. et al. Interferon-gamma drives T(reg) fragility to promote anti-tumor immunity. Cell 169, 1130–1141.e1111 (2017).
Li, Y. et al. Potential anti-tumor effects of regulatory T cells in the tumor microenvironment: a review. J. Transl. Med 22, 293 (2024).
Liu, X. et al. FOXP3(+) regulatory T cell perturbation mediated by the IFNgamma-STAT1-IFITM3 feedback loop is essential for anti-tumor immunity. Nat. Commun. 15, 122 (2024).
Yuan, Y., Cai, X., Shen, F. & Ma, F. HPV post-infection microenvironment and cervical cancer. Cancer Lett. 497, 243–254 (2021).
Gordon, S. & Martinez, F. O. Alternative activation of macrophages: Mechanism and functions. Immunity 32, 593–604 (2010).
Jin, S., Plikus, M. V. & Nie, Q. CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat. Protoc. 20, 180–219 (2025).
Nguyen, A. T., Szeto, C. & Gras, S. The pockets guide to HLA class I molecules. Biochem Soc. Trans. 49, 2319–2331 (2021).
Chen, G. et al. Single-cell transcriptomic analysis reveals that the APP-CD74 axis promotes immunosuppression and progression of testicular tumors. J. Pathol. 264, 250–269 (2024).
Li, R. Q., Yan, L., Zhang, L., Zhao, Y. & Lian, J. CD74 as a prognostic and M1 macrophage infiltration marker in a comprehensive pan-cancer analysis. Sci. Rep. 14, 8125 (2024).
Xiao, N. et al. CD74(+) macrophages are associated with favorable prognosis and immune contexture in hepatocellular carcinoma. Cancer Immunol. Immunother. 71, 57–69 (2022).
Cheng, J. et al. CD74 facilitates immunotherapy response by shaping the tumor microenvironment of hepatocellular carcinoma. Mol. Med. 30, 116 (2024).
Rowshanravan, B., Halliday, N. & Sansom, D. M. CTLA-4: A moving target in immunotherapy. Blood 131, 58–67 (2018).
Vandenborre, K. et al. Interaction of CTLA-4 (CD152) with CD80 or CD86 inhibits human T-cell activation. Immunology 98, 413–421 (1999).
Cao, X. Regulatory T cells and immune tolerance to tumors. Immunol. Res 46, 79–93 (2010).
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Alkhamesi, N. A., Roberts, G., Ziprin, P. & Peck, D. H. Induction of Proteases in Peritoneal Carcinomatosis, the Role of ICAM-1/CD43 Interaction. Biomark. Insights 2, 377–384 (2007).
Webb, J. R., Milne, K., Watson, P., Deleeuw, R. J. & Nelson, B. H. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin. Cancer Res 20, 434–444 (2014).
Franciszkiewicz, K. et al. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res. 73, 617–628 (2013).
Edsgard, D., Johnsson, P. & Sandberg, R. Identification of spatial expression trends in single-cell gene expression data. Nat. Methods 15, 339–342 (2018).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Methods 15, 343–346 (2018).
Tan, Y. et al. SPACEc: A streamlined, interactive python workflow for multiplexed image processing and analysis. bioRxiv, https://doi.org/10.1101/2024.06.29.601349 (2024).
Xu, W. et al. Macrophages induce differentiation of plasma cells through CXCL10/IP-10. J. Exp. Med 209, 1813–1823 (2012).
Jego, G. et al. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity 19, 225–234 (2003).
Swiecki, M. & Colonna, M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 15, 471–485 (2015).
Carson-Walter, E. B. et al. Plasmalemmal vesicle associated protein-1 is a novel marker implicated in brain tumor angiogenesis. Clin. Cancer Res. 11, 7643–7650 (2005).
Denzer, L., Muranyi, W., Schroten, H. & Schwerk, C. The role of PLVAP in endothelial cells. Cell Tissue Res 392, 393–412 (2023).
Singer, M. et al. Promising cellular immunotherapy for colorectal cancer using classical dendritic cells and natural Killer T cells. Cells 14 https://doi.org/10.3390/cells14030166 (2025).
Marciscano, A. E. & Anandasabapathy, N. The role of dendritic cells in cancer and anti-tumor immunity. Semin Immunol. 52, 101481 (2021).
Del Prete, A. et al. Dendritic cell subsets in cancer immunity and tumor antigen sensing. Cell Mol. Immunol. 20, 432–447 (2023).
Kraman, M. et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 330, 827–830 (2010).
Liao, Z., Tan, Z. W., Zhu, P. & Tan, N. S. Cancer-associated fibroblasts in tumor microenvironment - Accomplices in tumor malignancy. Cell Immunol. 343, 103729 (2019).
Hiratsuka, S., Watanabe, A., Aburatani, H. & Maru, Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 8, 1369–1375 (2006).
Hung, C. F., Wu, T. C., Monie, A. & Roden, R. Antigen-specific immunotherapy of cervical and ovarian cancer. Immunol. Rev. 222, 43–69 (2008).
Wang, W., Erbe, A. K., Hank, J. A., Morris, Z. S. & Sondel, P. M. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol. 6, 368 (2015).
Su, S. et al. Immune checkpoint inhibition overcomes ADCP-induced immunosuppression by macrophages. Cell 175, 442–457.e423 (2018).
Clynes, R. A., Towers, T. L., Presta, L. G. & Ravetch, J. V. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat. Med. 6, 443–446 (2000).
Zhang, W. et al. Fc-Fc interactions and immune inhibitory effects of IgG4: implications for anti-PD-1 immunotherapies. J. Immunother. Cancer 12 https://doi.org/10.1136/jitc-2024-009034 (2024).
Jordakieva, G. et al. IgG4 induces tolerogenic M2-like macrophages and correlates with disease progression in colon cancer. Oncoimmunology 10, 1880687 (2021).
Platten, M., Nollen, E. A. A., Röhrig, U. F., Fallarino, F. & Opitz, C. A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 18, 379–401 (2019).
Liu, X. Q. & Wang, X. Indoleamine 2,3-dioxygenase in tumor induced tolerance. Chin. Med J. (Engl.) 122, 3072–3077 (2009).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Acknowledgements
The results published here are partly based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author information
Authors and Affiliations
Contributions
B.W., H.L. and H.S. designed and supervised the study. B.W. and Y.W. performed the data analysis and wrote the manuscript. Y.G. coordinated the samples collection, performed the single-cell RNA-sequencing experiments and the Xenium spatial transcriptomic assays. Z.W. collected the clinical samples and evaluated the pathological stages of the patients. H.W., Y.L., D.X. and L.G. helped collection of tumor specimens and FFPE sample preparation. X. L. helped perform spatial data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Benjamin Wolf and Yi-Qi Li for their contribution to the peer review of this work. Primary Handling Editors: Bibekanand Mallick and Johannes Stortz. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Y., Wei, B., Wei, Z. et al. Single-cell and spatial transcriptomic profiling reveals distinct tumor microenvironment dynamics in cervical adenocarcinoma and squamous cell carcinoma. Commun Biol 9, 46 (2026). https://doi.org/10.1038/s42003-025-09310-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09310-2