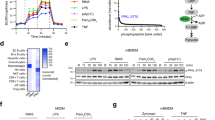
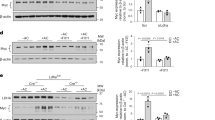
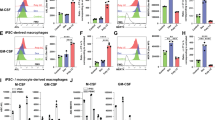
Abstract
Resolving-type macrophages prevent chronic inflammation by clearing apoptotic cells through efferocytosis. These macrophages are thought to rely mainly on oxidative phosphorylation, but emerging evidence suggests a possible link between efferocytosis and glycolysis. To gain further insight into this issue, we investigated molecular–cellular mechanisms involved in efferocytosis-induced macrophage glycolysis and its consequences. We found that efferocytosis promotes a transient increase in macrophage glycolysis that is dependent on rapid activation of the enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), which distinguishes this process from glycolysis in pro-inflammatory macrophages. Mice transplanted with activation-defective PFKFB2 bone marrow and then subjected to dexamethasone-induced thymocyte apoptosis exhibit impaired thymic efferocytosis, increased thymic necrosis, and lower expression of the efferocytosis receptors MerTK and LRP1 on thymic macrophages compared with wild-type control mice. In vitro mechanistic studies revealed that glycolysis stimulated by the uptake of a first apoptotic cell promotes continual efferocytosis through lactate-mediated upregulation of MerTK and LRP1. Thus, efferocytosis-induced macrophage glycolysis represents a unique metabolic process that sustains continual efferocytosis in a lactate-dependent manner. The differentiation of this process from inflammatory macrophage glycolysis raises the possibility that it could be therapeutically enhanced to promote efferocytosis and resolution in chronic inflammatory diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the present study are available within the manuscript and supplementary information files. Source data are provided within this paper. Source data are provided with this paper.
References
Raymond, M. H. et al. Live cell tracking of macrophage efferocytosis during Drosophila embryo development in vivo. Science 375, 1182–1187 (2022).
Morioka, S., Maueröder, C. & Ravichandran, K. S. Living on the edge: efferocytosis at the interface of homeostasis and pathology. Immunity 50, 1149–1162 (2019).
Doran, A. C., Yurdagul, A. Jr. & Tabas, I. Efferocytosis in health and disease. Nat. Rev. Immunol. 20, 254–267 (2020).
Vandivier, R. W., Henson, P. M. & Douglas, I. S. Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease. Chest 129, 1673–1682 (2006).
Yurdagul, A. Jr., Doran, A. C., Cai, B., Fredman, G. & Tabas, I. A. Mechanisms and consequences of defective efferocytosis in atherosclerosis. Front. Cardiovasc. Med. 4, 86 (2017).
Wu, Y., Singh, S., Georgescu, M. M. & Birge, R. B. A role for Mer tyrosine kinase in alphavbeta5 integrin-mediated phagocytosis of apoptotic cells. J. Cell Sci. 118, 539–553 (2005).
Gardai, S. J. et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123, 321–334 (2005).
Lemke, G. & Burstyn-Cohen, T. TAM receptors and the clearance of apoptotic cells. Ann. NY Acad. Sci. 1209, 23–29 (2010).
Yurdagul, A. Jr. et al. Macrophage metabolism of apoptotic cell-derived arginine promotes continual efferocytosis and resolution of injury. Cell Metab. 31, 518–533 (2020).
Mehrotra, P. & Ravichandran, K. S. Drugging the efferocytosis process: concepts and opportunities. Nat. Rev. Drug Discov. 21, 601–620 (2022).
Park, D. et al. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature 477, 220–224 (2011).
Wang, Y. et al. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell 171, 331–345 (2017).
Morioka, S. et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563, 714–718 (2018).
Liu, Y. et al. Metabolic reprogramming in macrophage responses. Biomark. Res. 9, 1 (2021).
Viola, A., Munari, F., Sánchez-Rodríguez, R., Scolaro, T. & Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019).
Freemerman, A. J. et al. Myeloid Slc2a1-deficient murine model revealed macrophage activation and metabolic phenotype are fueled by GLUT1. J. Immunol. 202, 1265–1286 (2019).
Nishizawa, T. et al. Testing the role of myeloid cell glucose flux in inflammation and atherosclerosis. Cell Rep. 7, 356–365 (2014).
Wu, N. et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol. Cell 49, 1167–1175 (2013).
Waldhart, A. N. et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep. 19, 2005–2013 (2017).
Gerlach, B. D. et al. Efferocytosis induces macrophage proliferation to help resolve tissue injury. Cell Metab. 33, 2445–2463 (2021).
Nishi, C., Yanagihashi, Y., Segawa, K. & Nagata, S. MERTK tyrosine kinase receptor together with TIM4 phosphatidylserine receptor mediates distinct signal transduction pathways for efferocytosis and cell proliferation. J. Biol. Chem. 294, 7221–7230 (2019).
Yancey, P. G. et al. Macrophage LRP-1 controls plaque cellularity by regulating efferocytosis and Akt activation. Arterioscler. Thromb. Vasc. Biol. 30, 787–795 (2010).
Mor, I., Cheung, E. C. & Vousden, K. H. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb. Symp. Quant. Biol. 76, 211–216 (2011).
Novellasdemunt, L. et al. Akt-dependent activation of the heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) isoenzyme by amino acids. J. Biol. Chem. 288, 10640–10651 (2013).
Deprez, J., Vertommen, D., Alessi, D. R., Hue, L. & Rider, M. H. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J. Biol. Chem. 272, 17269–17275 (1997).
Rossi, D. C. et al. A metabolic inhibitor arms macrophages to kill intracellular fungal pathogens by manipulating zinc homeostasis. J. Clin. Invest. 131, e147268 (2021).
Finucane, O. M., Sugrue, J., Rubio-Araiz, A., Guillot-Sestier, M. V. & Lynch, M. A. The NLRP3 inflammasome modulates glycolysis by increasing PFKFB3 in an IL-1β-dependent manner in macrophages. Sci. Rep. 9, 4034 (2019).
Jiang, H. et al. PFKFB3-driven macrophage glycolytic metabolism is a crucial component of innate antiviral defense. J. Immunol. 197, 2880–2890 (2016).
Tawakol, A. et al. HIF-1α and PFKFB3 mediate a tight relationship between proinflammatory activation and anerobic metabolism in atherosclerotic macrophages. Arterioscler. Thromb. Vasc. Biol. 35, 1463–1471 (2015).
Thorp, E., Cui, D., Schrijvers, D. M., Kuriakose, G. & Tabas, I. Mertk receptor mutation reduces efferocytosis efficiency and promotes apoptotic cell accumulation and plaque necrosis in atherosclerotic lesions of Apoe–/– mice. Arterioscler. Thromb. Vasc. Biol. 28, 1421–1428 (2008).
Lee, M. et al. Mutation of regulatory phosphorylation sites in PFKFB2 worsens renal fibrosis. Sci. Rep. 10, 14531 (2020).
Gronski, M. A., Kinchen, J. M., Juncadella, I. J., Franc, N. C. & Ravichandran, K. S. An essential role for calcium flux in phagocytes for apoptotic cell engulfment and the anti-inflammatory response. Cell Death Differ. 16, 1323–1331 (2009).
Kasikara, C. et al. Deficiency of macrophage PHACTR1 impairs efferocytosis and promotes atherosclerotic plaque necrosis. J. Clin. Invest 131, e145275 (2021).
Ampomah, P. B. et al. Macrophages use apoptotic cell-derived methionine and DNMT3A during efferocytosis to promote tissue resolution. Nat. Metab. 4, 444–457 (2022).
Scott, R. S. et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411, 207–211 (2001).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470 (2016).
Murphy, M. P. & O’Neill, L. A. J. Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell 174, 780–784 (2018).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Nakayama, Y., Mukai, N., Kreitzer, G., Patwari, P. & Yoshioka, J. Interaction of ARRDC4 with GLUT1 mediates metabolic stress in the ischemic heart. Circ. Res. 131, 510–527 (2022).
Freemerman, A. J. et al. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289, 7884–7896 (2014).
Kumagai, S. et al. Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell 40, 201–218 (2022).
Choi, S. Y., Collins, C. C., Gout, P. W. & Wang, Y. Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J. Pathol. 230, 350–355 (2013).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014).
Wang, T. et al. HIF1α-induced glycolysis metabolism Is essential to the activation of inflammatory macrophages. Mediators Inflamm. 2017, 9029327 (2017).
Luo, W. et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 (2011).
Palsson-McDermott, E. M. et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 (2015).
Van den Bossche, J., O’Neill, L. A. & Menon, D. Macrophage immunometabolism: where are we (going)? Trends Immunol. 38, 395–406 (2017).
O’Neill, L. A. A broken krebs cycle in macrophages. Immunity 42, 393–394 (2015).
Van den Bossche, J. et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 17, 684–696 (2016).
Crochet, R. B. et al. Crystal structure of heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) and the inhibitory influence of citrate on substrate binding. Proteins 85, 117–124 (2017).
May, P., Bock, H. H. & Nofer, J. R. Low density receptor-related protein 1 (LRP1) promotes anti-inflammatory phenotype in murine macrophages. Cell Tissue Res. 354, 887–889 (2013).
Doddapattar, P. et al. Myeloid cell PKM2 deletion enhances efferocytosis and reduces atherosclerosis. Circ. Res. 130, 1289–1305 (2022).
Gorovoy, M., Gaultier, A., Campana, W. M., Firestein, G. S. & Gonias, S. L. Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J. Leukoc. Biol. 88, 769–778 (2010).
Freire-de-Lima, C. G. et al. Apoptotic cells, through transforming growth factor-beta, coordinately induce anti-inflammatory and suppress pro-inflammatory eicosanoid and NO synthesis in murine macrophages. J. Biol. Chem. 281, 38376–38384 (2006).
Dalli, J. et al. Annexin A1 regulates neutrophil clearance by macrophages in the mouse bone marrow. FASEB J. 26, 387–396 (2012).
Matsuura, Y. et al. Diabetes suppresses glucose uptake and glycolysis in macrophages. Circ. Res. 130, 779–781 (2022).
Qu, J., Yang, J., Chen, M., Wei, R. & Tian, J. CircFLNA acts as a sponge of miR-646 to facilitate the proliferation, metastasis, glycolysis, and apoptosis inhibition of gastric cancer by targeting PFKFB2. Cancer Manag. Res. 12, 8093–8103 (2020).
Acknowledgements
This work was supported by an American Heart Association Postdoctoral Fellowship (900337; to M. S.); the Niels Stensen Fellowship (to M. S.) and NIH/NHLBI grants R35-HL145228 and P01-HL087123 (to I. T.). We thank H.-W. Snoeck for facilitating the Seahorse-based bioenergetic analyses, A. Yurdagul Jr (LSUHSC-Shreveport) for being a valuable advisor and consultant, and T. McGraw (Weill-Cornell Medical College) and N. Wu (Van Andel Institute) for helpful discussions related to TXNIP. We thank T. Swayne for assistance with confocal microscopy, which was conducted using the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grant P30CA013696. We thank C. Lu at the Columbia Center for Translational Immunology (CCTI) for assistance with flow cytometry experiments, which were conducted using the Herbert Irving Comprehensive Cancer Center Flow Cytometry Shared Resources funded in part through Center Grant P30CA013696. LC–MS/MS analysis of fructose-1,6-bisphosphate was conducted at the Albert Einstein College of Medicine Stable Isotope and Metabolomics Core, with the helpful advice of the Core’s director, I. Kirkland.
Author information
Authors and Affiliations
Contributions
M. S. and I. T. conceptualized the research and experimental design. D. N. provided intellectual contributions throughout the project. M. S. conducted the experiments, and D. N. assisted with the dexamethasone–thymus study. M. K. and D. A. P. provided bone marrow cells from the PFKFB2-mutant model and were important advisors. M. S. and I. T. wrote the manuscript, and all co-authors critically reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Vishwa Dixit and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editor: Alfredo Giménez-Cassina, in collaboration with the Nature Metabolism team
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Seahorse analysis of macrophages treated with either IFNγ and LPS or IL-4.
a, Naïve BMDMs were polarized towards a pro-inflammatory phenotype with IFNγ and LPS or a pro-resolving phenotype with IL-4 for 24 h followed by Seahorse analysis. The extracellular acidification rate (ECAR), a measure of glycolysis, was measured at baseline and after the addition of glucose (‘glycolysis’), oligomycin (‘glycolytic capacity’), and 2-DG. The oxygen consumption rate (OCR), a measure of oxidative phosphorylation, was measured at baseline (‘basal respiration’) and after the addition of oligomycin, FCCP (‘maximal respiration), and rotenone plus antimycin A (n = 15-16 wells/group). **P = 0.0017 (naïve vs. IFNγ and LPS), **P = 0.0048 (naïve vs. IL-4), ***P < 0.0001, as compared to the naïve groups. b, The same experiment as described under panel a was performed in HMDMs (n = 7-8 wells/group). *P = 0.046 for glycolysis, *P = 0.044 for glycolytic capacity, *P = 0.021 for basal respiration, as compared to the naïve groups. All values are means ± SEM, and significance was determined by one-way ANOVA with Fisher’s LSD post hoc analysis.
Extended Data Fig. 2 Seahorse analysis of macrophages at various times following incubation with apoptotic cells, and in apoptotic cells alone.
a, BMDMs (were incubated with apoptotic Jurkat cells (ACs) for 45 min (pulse), followed by rinsing to remove unengulfed ACs, and then subjected to Seahorse analysis 1, 6, or 24 h later (chase). The oxygen consumption rate (OCR), a measure of oxidative phosphorylation, was measured at baseline (‘basal respiration’) and after the addition of oligomycin, FCCP (‘maximal respiration), and rotenone plus antimycin A (n = 11-12 wells/group). *P = 0.0027, **P = 0.0011 (basal 1 h chase), **P = 0.0031 (basal 6 h chase), **P = 0.0036 (ATP production 1 h chase), as compared to the -AC groups. b, The same experiment as described under panel a was performed in HMDMs (n = 8 wells/group). ***P < 0.0001, as compared to the -AC group. c, A glycolysis stress test was performed in HMDMs differentiated with M-CSF instead of GM-CSF, after a 1 h chase without or with ACs (n = 11-12 wells/group). *P < 0.05, ***P < 0.001. d,e, A glycolysis stress test was performed in BMDMs after a 1 h chase without or with ACs (n = 14-15 wells/group; 20,000 macrophages per well), or in ACs alone (n = 3 wells; 100,000 ACs/well). The ECAR and OCR were measured to evaluate glycolysis (d) and oxidative phosphorylation (e), respectively. All values are means ± SEM, and significance was determined by one-way ANOVA with Fisher’s LSD post hoc analysis (a-b) or multiple two-tailed Student’s t-tests (c).
Extended Data Fig. 3 Flow cytometry analysis of cell-surface GLUT1 expression in efferocytic macrophages.
BMDMs were incubated with PKH67-labeled apoptotic cells (ACs) for 45 min, rinsed and harvested 1 h later for flow cytometric analysis of cell-surface GLUT1 using an APC-conjugated GLUT1 antibody. a, Gating strategy for AC− and AC+ macrophages, with PKH67 detected in the FITC channel. b, The mean fluorescent intensity (MFI) of cell-surface GLUT1 based on the flow cytometric data.
Extended Data Fig. 4 Analysis of siRNA silencing efficiency, efferocytosis-induced gene expression, and 2-NBDG uptake.
a, BMDMs were transfected with scrambled RNA or siSlc2a1, and Slc2a1 expression was measured by RT-qPCR (n = 2 wells/group). b, BMDMs (Mφs) pretreated with the Akt inhibitor MK-2205 (5 µM) were incubated with fluorescently labeled apoptotic Jurkat cells (ACs) for 45 min, followed by rinsing and addition of glucose-free medium containing 2-NBDG without or with MK-2205. After 1 h, macrophages were fixed and the mean fluorescent intensity (MFI) of 2-NBDG was quantified in AC− and AC+ macrophages by fluorescent microscopy (n = 3 images/group). **P = 0.0047. c,d, BMDMs were transfected with scrambled RNA or siTxnip, and knockdown was validated by RT-qPCR (n = 2 wells/group) and immunoblotting (n = 3 samples/group). e, BMDMs were incubated in the absence or presence of ACs for 45 min, rinsed, and Pfkfb2 expression was measured 1 h later (n = 4 wells/group). *P = 0.018 (pTXNIP), *P = 0.015 (tTXNIP). f, A 2-NBDG assay as described under panel B was performed in Mφs after 1 or 6 h of incubation with ACs (n = 3 images/group). *P = 0.011 (1 h ACchase), *P = 0.028 (6 h ACchase). g, BMDMs were transfected with scrambled RNA or siPfkfb2, and Pfkfb2 expression was measured by RT-qPCR (n = 2 wells/group). h, A 2-NBDG assay as described under panel B was performed in Mφs transfected with scrambled RNA or siPfkfb2 (n = 3 images/group). *P = 0.035 (Scrambled), *P = 0.027 (siPfkfb2) All values are means ± SEM, and significance was determined by two-way ANOVA with Fisher’s LSD post hoc analysis. ns, not significant (P > 0.05).
Extended Data Fig. 5 Additional efferocytosis assays and validation of siRNA silencing efficiency.
a, BMDMs were pretreated without or with 10 mM 2-DG for 1 h, followed by incubation with PKH26-labeled apoptotic Jurkat cells (ACs) for 45 min. Unengulfed ACs were then rinsed away, and the number of PKH26+ macrophages were quantified as a measure of single efferocytosis (n = 4 wells/group). b, BMDMs were transfected with scrambled RNA or siPfkfb2, and Pfkfb2 gene expression was measured by RT-qPCR (n = 3 wells/group). ****P < 0.0001. c, BMDMs were transfected with scrambled RNA or siLdha, and Ldha gene expression was measured by RT-qPCR (n = 3 wells/group). ****P < 0.0001. d, BMDMs were transfected with scrambled RNA, siPfkfb2 or siLdha, followed by a single efferocytosis assay as described for panel A (n = 3 wells/group). e, HMDMs were transfected with scrambled RNA or siPfkfb2, and PFKFB2 gene expression was measured by RT-qPCR (n = 3 wells/group). ****P < 0.0001. All values are means ± SEM, and significance was determined by the two-tailed Student’s t-test (a-c & e) or two-way ANOVA with Fisher’s LSD post hoc analysis in panel d. ns, not significant (P > 0.05).
Extended Data Fig. 6 Pfkfb3 and Pfkfb4 are upregulated by IFNγ and LPS, and partial silencing of Pfkfb3 and Pfkfb4 does not affect efferocytosis.
a,b, Expression levels of Pfkfb3 and Pfkfb4 were measured by RT-qPCR in naïve macrophages and macrophages polarized towards a pro-inflammatory phenotype with IFNγ and LPS (n = 4 wells/group). **P = 0.0011, ****P < 0.0001. c, BMDMs were transfected with scrambled RNA or siPfkfb3, and Pfkfb3 expression was measured (n = 3 wells/group). **P = 0.0018. d, BMDMs were transfected with scrambled RNA or siPfkfb4, and Pfkfb4 expression was measured (n = 3 wells/group). **P = 0.0018. e, BMDMs transfected with scrambled RNA, siPfkfb3, or siPfkfb4 were incubated with PKH26-labeled apoptotic Jurkat cells (ACs) for 45 min. Unengulfed ACs were removed by rinsing, and the number of PKH26+ macrophages were quantified (n = 4 wells/group). f, BMDMs transfected with scrambled RNA, siPfkfb3, or siPfkfb4 were first incubated with PKH67-labeled ACs for 45 min, rinsed, and 2 h later incubated with PKH26-labeled ACs for 45 min. The number of PKH67+ PKH26+ Mφs relative to PKH67+ Mφs was quantified as a measure of continual efferocytosis (n = 4 wells/group). All values are means ± SEM, and significance was determined by the two-tailed Student’s t-test (a-d) or one-way ANOVA with Fisher’s LSD post hoc analysis (e-f). ns, not significant (P > 0.05).
Extended Data Fig. 7 Macrophages from PFKFB2 mutant mice show attenuated efferocytosis-induced glycolysis.
BMDMs from PFKFB2 mutant mice (Mut) and wild-type littermates (WT) were incubated without or with apoptotic Jurkat cells (ACs) for 45 min, followed by rinsing to remove unengulfed ACs, and then subjected to Seahorse analysis 1 h later. a, The extracellular acidification rate (ECAR), a measure of glycolysis, was measured at baseline and after the addition of glucose (‘glycolysis’), oligomycin (‘glycolytic capacity’), and 2-DG (n = 8-9 wells/group). **P = 0.0028 (WT+AC vs. Mut+AC), **P = 0.0070 (Mut-AC vs. Mut+AC), ****P < 0.0001 for glycolysis, *P = 0.031, **P = 0.0057, ****P < 0.0001 for glycolytic capacity. b, The oxygen consumption rate (OCR), a measure of oxidative phosphorylation, was measured at baseline (‘basal respiration’) and after the addition of oligomycin, FCCP (‘maximal respiration), and rotenone plus antimycin A (n = 8-9 wells/group). ***P = 0.0001, ****P < 0.0001. All values are means ± SEM, and significance was determined by two-way ANOVA with Fisher’s LSD post hoc analysis. ns, not significant (P > 0.05).
Extended Data Fig. 8 Immunoblotting for MerTK and LRP1, validation of siRNA silencing efficiency, and experiments with siSlc16a1.
a, BMDMs from PFKFB2 mutant mice and wild-type littermates were incubated without or with apoptotic Jurkat cells (ACs) for 45 min, rinsed, and harvested 2 h later for immunoblotting of MerTK, LRP1 and β-actin. The relative level of MerTK/LRP1 vs. β-actin was quantified by band densitometry (n = 3 samples/group). b, BMDMs were transfected with scrambled RNA or siPfkfb2, and Pfkfb2 gene expression was measured by RT-qPCR (n = 3 wells/group). ***P = 0.0002. c, BMDMs were transfected with scrambled RNA or siSlc16a1, and Slc16a1 expression was measured (n = 3 wells/group). ****P < 0.0001 d, BMDMs transfected with scrambled RNA or siSlc16a1 were incubated with ACs for 45 min, rinsed and 1 h later harvested for lactate measurement in the media (extracellular) and the cells (intracellular) (n = 3 wells/group). *P = 0.029. e,f, A continual efferocytosis assay (f) and single efferocytosis assay (g), as described in the legend of Extended Data Fig. 6, were performed in macrophages treated with scrambled RNA or siSlc16a1 and incubated with vehicle or lactate (10 mM) before adding the first or second round of ACs, respectively (n = 4 wells/group). *P = 0.031, **P = 0.0028. All values are means ± SEM, and significance was determined by two-way ANOVA with Fisher’s LSD post hoc analysis (a, e-f) or the one- or two-tailed Student’s t-test (b-d). ns, not significant (P > 0.05).
Extended Data Fig. 9 Additional analyses and representative images of the dexamethasone-thymus experiment.
a, Thymus weight and b, cellularity of PBS-injected hematopoietic wild-type (wt) and PFKFB2 mutant (mut) mice (n = 2-3 mice/group). c, Representative images of thymus sections from PBS-injected (n = 2) and dexamethasone (dex)-injected (n = 7) wild-type mice stained for Mac2 (macrophages) and TUNEL (apoptotic cells). d, Representative images of H&E-stained thymus sections of hematopoietic wild-type and PFKFB2 mutant mice at 10x magnification, and from the thymic cortex at 40x magnification, as indicated by the magnified inset (n = 2-3 mice in the PBS-injected groups, n=7 mice in the dex-injected groups). e, The mean fluorescent intensity (MFI) of phospho-PFKFB2 was measured in Mac2+ AC− (yellow arrowheads) and Mac2+ AC+ (that is, TUNEL+; white arrowheads) macrophages in the thymus of dex-injected wt mice by IFM (n = 7 mice/group). **P = 0.0011. All values are means ± SEM, and significance determined by the two-tailed Student’s t-test.
Extended Data Fig. 10 Flow cytometric analysis of apoptotic thymocytes from the dexamethasone-thymus experiment.
All cells were isolated from the thymi of hematopoietic PFKFB2 mutant and wild-type mice of the dexamethasone-thymus experiment (see Fig. 5 of the main manuscript) and subjected to flow cytometric analysis. The cells were immunostained for the apoptotic cell marker annexin V (AnnV; FITC) and, using the depicted gating strategy, quantified for the percentages of AnnV− (live) and AnnV+ (apoptotic) thymocytes.
Supplementary information
Supplementary Information
Supplementary Table 1 Primer sequences used for quantitative RT-PCR analysis
Source data
Source Data
Unprocessed western blots for Figs. 2, 3 and 6 and Extended Data Figs. 4 and 8.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schilperoort, M., Ngai, D., Katerelos, M. et al. PFKFB2-mediated glycolysis promotes lactate-driven continual efferocytosis by macrophages. Nat Metab 5, 431–444 (2023). https://doi.org/10.1038/s42255-023-00736-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-023-00736-8
This article is cited by
-
Immunomodulatory effect of efferocytosis at the maternal–fetal interface
Cell Communication and Signaling (2025)
-
Integrated multi-omics reveals glycolytic gene signatures of lung adenocarcinoma brain metastasis and the impact of Rac2 lactylation on immunosuppressive microenvironment
Journal of Translational Medicine (2025)
-
Neospora caninum hijacks host PFKFB3-driven glycolysis to facilitate intracellular propagation of parasites
Veterinary Research (2025)
-
LGR6 modulates intervertebral disc degeneration through regulation of macrophage efferocytosis
Journal of Translational Medicine (2025)
-
The potential role of 1,25(OH)2D3 (Active vitamin D3) in modulating macrophage function; implications for chronic obstructive pulmonary disease (COPD)
Journal of Inflammation (2025)