Abstract
Background
Chronological age does not capture individual health or resilience. Advances in metabolomics have enabled development of molecular aging biomarkers that capture deviations between biological and chronological age, highlighting how genetics, environment, and lifestyle shape biological aging. Despite their promise, metabolomic biomarkers face challenges such as interpretability, non-linearity, and reproducibility.
Methods
We have developed a metabolomic predictor of biological age based on untargeted metabolomic profiling of individuals aged 45–85 years from the Canadian Longitudinal Study on Aging. To enhance interpretability, we first identified metabolites related to health based on variance heterogeneity. For metabolites with identifiable optimal levels, or “sweet spots”, we modeled non-linearity using deviations from these values. A penalized regression model was trained on the Frailty Index using sweet spot deviations as predictors.
Results
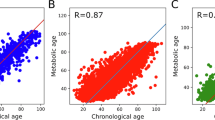
Here we show that the Sweet Spot Clock built on 178 health-related metabolites is strongly associated with all-cause mortality (HR = 1.08, p = 5.8×10−12, C-index=0.841) and age-related diseases. The biomarker outperforms models trained on chronological age and those using raw metabolite levels, underscoring the importance of modeling non-linearity. It remains predictive after adjusting for age, sex, lifestyle and socioeconomic factors, though its added value over standard health and demographic measures is modest. The model generalizes to an independent cohort of individuals aged 85 years and older.
Conclusions
The Sweet Spot Clock provides a reproducible and interpretable measure of biological age. By modeling deviations from optimal metabolite levels and training on health status rather than age, it offers a tool for understanding aging heterogeneity and identifying individuals at risk of health decline.
Plain Language Summary
Chronological age does not fully reflect a person’s health or resilience. We used data from Canadians aged 45–85 to develop a biomarker of biological age based on metabolites—small molecules in the blood that reflect body processes. We focused on 178 health-related metabolites and identified “sweet spots,” or optimal levels, for 74 of them. Our model, the Sweet Spot Clock, was strongly related to mortality and age-related diseases. These findings held up in an independent group of Canadians aged 85 and older. By focusing on health status and accounting for non-linear patterns, our approach offers a reproducible and interpretable way to measure biological aging and understand why some people age more healthfully than others.
Similar content being viewed by others
Data availability
Data are available from the Canadian Longitudinal Study on Aging (www.clsa-elcv.ca) for researchers who meet the criteria for access to de-identified CLSA data. The Super Seniors dataset is stored at Genome Sciences Center, BC Cancer, Canada. For inquiries about this dataset, or for requests to collaborate on projects involving this dataset, please contact Dr. Angela Brooks-Wilson at abrooks-wilson@bcgsc.ca. The list of metabolites used for analysis is provided in Supplementary Data 1. Source data for recreating figures are available at https://zenodo.org/records/1759410665 and in Supplementary Data Files 2–7: The source data for Fig. 2a, b is in Supplementary Data 2, the source data for Fig. 2c is in Supplementary Data 3, the source data for Fig. 3 is in Supplementary Data 4, the source data for Fig. 4d is in Supplementary Data 6, and the source data for Fig. 4e is in Supplementary Data 7. The list of predictors included in the Sweet Spot Clock model is in Supplementary Data 5.
Code availability
All code used in this study is available at https://zenodo.org/records/1759410665. The data analysis was performed in R v. 4.4.2.
References
Lowsky, D. J., Olshansky, S. J., Bhattacharya, J. & Goldman, D. P. Heterogeneity in healthy aging. J. Gerontol. A Biol. Sci. Med. Sci. 69, 640–649 (2014).
Rae, M. J. et al. The demographic and biomedical case for late-life interventions in aging. Sci. Transl. Med. 2, 1–6 (2010).
van den Berg, N. et al. Longevity defined as top 10% survivors and beyond is transmitted as a quantitative genetic trait. Nat. Commun. 10, 1–12 (2019).
Chiu, M. et al. Mortality risk associated with psychological distress and major depression: A population-based cohort study. J. Affect. Disord. 234, 117–123 (2018).
Stringhini, S. et al. Socioeconomic status, non-communicable disease risk factors, and walking speed in older adults: multi-cohort population based study. BMJ 360, 1–10 (2018).
Moqri, M. et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758–3775 (2023).
Lohman, T., Bains, G., Berk, L. & Lohman, E. Predictors of biological age: the implications for wellness and aging research. Gerontol. Geriatr. Med. 7, 1–13 (2021).
Jylhävä, J., Pedersen, N. L. & Hägg, S. Biological age predictors. EBioMedicine 21, 29–36 (2017).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23, 715–727 (2022).
Lehallier, B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 25, 1843–1850 (2019).
Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 1–14 (2015).
Tanaka, T. et al. Plasma proteomic signature of age in healthy humans. Aging Cell 17, 1–13 (2018).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, 1–20 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Menni, C. et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 42, 1111 (2013).
Van Den Akker, E. B. et al. Metabolic age based on the BBMRI-NL 1H-NMR metabolomics repository as biomarker of age-related disease. Circ. Genom. Precis. Med. 13, 541–547 (2020).
Robinson, O. et al. Determinants of accelerated metabolomic and epigenetic aging in a UK cohort. Aging Cell 19, 1–13 (2020).
Deelen, J. et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 10, 1–8 (2019).
Galkin, F. et al. Human gut microbiome aging clock based on taxonomic profiling and deep learning. iScience 23, 1–10 (2020).
Sayed, N. et al. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nat. Aging 1, 598–615 (2021).
Kuiper, L. M. et al. Epigenetic and metabolomic biomarkers for biological age: a comparative analysis of mortality and frailty risk. J. Gerontol. A Biol. Sci. Med. Sci. 78, 1753–1762 (2023).
Okada, D., Cheng, J. H., Zheng, C., Kumaki, T. & Yamada, R. Data-driven identification and classification of nonlinear aging patterns reveals the landscape of associations between DNA methylation and aging. Hum. Genomics 17, 1–11 (2023).
Tanaka, T. et al. Plasma proteomic biomarker signature of age predicts health and life span. eLife 9, 1–24 (2020).
Robinson, O. & Lau, C. H. E. Measuring biological age using metabolomics. Aging 12, 22352–22353 (2020).
Rattray, N. J. W. et al. Metabolic dysregulation in vitamin E and carnitine shuttle energy mechanisms associate with human frailty. Nat. Commun. 10, 11 (2019).
Adav, S. S. & Wang, Y. Metabolomics signatures of aging: recent advances. Aging Dis. 12, 646–661 (2021).
Yu, Y. et al. Correcting batch effects in large-scale multiomics studies using a reference-material-based ratio method. Genome Biol. 24, 1–26 (2023).
Vishnyakova, O., Song, X., Rockwood, K., Elliott, L. T. & Brooks-Wilson, A. Physiological phenotypes have optimal values relevant to healthy aging: sweet spots deduced from the Canadian Longitudinal Study on Aging. Geroscience 46, 1589–1605 (2024).
Shen, X. et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat. Aging 4, 1619–1634 (2024).
Raina, P. et al. Erratum: Cohort profile: the Canadian Longitudinal Study on Aging (CLSA)(International Journal of Epidemiology (2019) 2019 DOI: 10.1093/ije/dyz173). Int. J. Epidemiol. 48, 2066 (2019).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Forgetta, V. et al. Cohort profile: genomic data for 26 622 individuals from the Canadian Longitudinal Study on Aging (CLSA). BMJ Open 12, 1–9 (2022).
Forgetta, V. et al. The Canadian Longitudinal Study on Aging Genome-wide Genetic Data Release (version 3). CLSA Data Support Document v20201207.1851 https://www.clsa-elcv.ca/wp-content/uploads/2023/06/clsa_gwas_v3.pdf (2023).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Wang, J., Wen, S., Fraser Symmans, W., Pusztai, L. & Coombes, K. R. The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inf. 7, 199–216 (2009).
Halaschek-Wiener, J. et al. The super-seniors study: phenotypic characterization of a healthy 85+ population. PLoS ONE 13, 1–11 (2018).
Darst, B. F., Koscik, R. L., Hogan, K. J., Johnson, S. C. & Engelman, C. D. Longitudinal plasma metabolomics of aging and sex. Aging 11, 1262–1282 (2019).
Brown, M. B. & Forsythe, A. B. Robust tests for the equality of variances. J. Am. Stat. Assoc. 69, 364–367 (1974).
Muggeo, V. M. R. Estimating regression models with unknown break-points. Stat. Med. 22, 3055–3071 (2003).
Muggeo, V. M. R. Testing with a nuisance parameter present only under the alternative: a score-based approach with application to segmented modelling. J. Stat. Comput. Simul. 86, 3059–3067 (2016).
Muggeo, V. M. R. Interval estimation for the breakpoint in segmented regression: a smoothed score-based approach. Aust. N. Z. J. Stat. 59, 311–322 (2017).
Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).
Rivals, I., Personnaz, L., Taing, L. & Potier, M. C. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23, 401–407 (2007).
Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Keller, H. H., Goy, R. & Kane, S. L. Validity and reliability of SCREEN II (Seniors In The Community: Risk Evaluation for eating and nutrition, Version II). Eur. J. Clin. Nutr. 59, 1149–1157 (2005).
Washburn, R. A., McAuley, E., Katula, J., Mihalko, S. L. & Boileau, R. A. The Physical Activity Scale for the Elderly (PASE): evidence for validity. J. Clin. Epidemiol. 52, 643–651 (1999).
Kessler, R. C. et al. Screening for serious mental illness in the general population. Arch. Gen. Psychiatry 60, 184–189 (2003).
Ying, K., Paulson, S., Eames, A. et al. A unified framework for systematic curation and evaluation of aging biomarkers. Nat. Aging 5, 2323–2339 (2025).
Moqri, M. et al. Validation of biomarkers of aging. Nat. Med. 30, 360–372 (2024).
Livshits, G. et al. Multi-OMICS analyses of frailty and chronic widespread musculoskeletal pain suggest involvement of shared neurological pathways. Pain 159, 2565–2572 (2018).
Huang, J. et al. Serum metabolomic profiling of all-cause mortality: a prospective analysis in the alpha-tocopherol, beta-carotene cancer prevention (ATBC) study cohort. Am. J. Epidemiol. 187, 1721–1732 (2018).
Halaschek-Wiener, J. et al. Reduced telomere length variation in healthy oldest old. Mech. Ageing Dev. 129, 638–641 (2008).
Maas, M. N., Hintzen, J. C. J., Porzberg, M. R. B. & Mecinović, J. Trimethyllysine: from carnitine biosynthesis to epigenetics. Int. J. Mol. Sci. 21, 1–33 (2020).
Vekic, J., Silva-Nunes, J. & Rizzo, M. Glucose metabolism disorders: challenges and opportunities for diagnosis and treatment. Metabolites 12, 1–4 (2022).
Bjornestad, E. O. et al. Trimethyllysine predicts all-cause and cardiovascular mortality in community-dwelling adults and patients with coronary heart disease. Eur. Heart J. Open 1, 1–10 (2021).
Li, X. S. et al. Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JCI Insight 3, 1–18 (2018).
Abela, L. et al. Plasma metabolomics reveals a diagnostic metabolic fingerprint for mitochondrial aconitase (ACO2) deficiency. PLoS ONE 12, 1–15 (2017).
Trabado, S. et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE 12, 1–17 (2017).
Johnson, L. C. et al. Amino acid and lipid associated plasma metabolomic patterns are related to healthspan indicators with ageing. Clin. Sci. 132, 1765–1777 (2018).
Cohen, A. A. et al. Statistical distance as a measure of physiological dysregulation is largely robust to variation in its biomarker composition. PLoS One 10, 1–25 (2015).
Das, S. K. et al. Metabolomic architecture of obesity implicates metabolonic lactone sulfate in cardiometabolic disease. Mol. Metab. 54, 1–10 (2021).
Vishnyakova, O. et al. Data for: Metabolomic sweet spot clock predicts mortality and age-related diseases in the Canadian Longitudinal Study on Aging. Zenodo. https://doi.org/10.5281/ZENODO.17594106.
Acknowledgements
This research is funded by the Canadian Institutes of Health Research (grant # PAD 179760) to A Brooks-Wilson and LT Elliott. LT Elliott’s research is supported by a Michael Smith Health Research BC Scholar Award. This research was made possible using the data collected by the Canadian Longitudinal Study on Aging (CLSA). Funding for the Canadian Longitudinal Study on Aging (CLSA) is provided by the Government of Canada through the Canadian Institutes of Health Research (CIHR) under grant reference: LSA 94473 and the Canada Foundation for Innovation, as well as the following provinces: Newfoundland, Nova Scotia, Quebec, Ontario, Manitoba, Alberta, and British Columbia. This research has been conducted using the CLSA Baseline Comprehensive Dataset version 7.0, Follow-up 1 Comprehensive version 4.0, Follow-up 2 Comprehensive version 1.0, Genomic data version 3.0, Epigenetics version 1.1, Metabolomics version 2.0 under Application Number 2206033. The CLSA is led by Drs. Parminder Raina, Christina Wolfson, and Susan Kirkland. The AB SCREEN™ II assessment tool is owned by Dr. Heather Keller. Use of the AB SCREEN™ II assessment tool was made under license from the University of Guelph. The opinions expressed in this manuscript are the authors’ own and do not reflect the views of the Canadian Longitudinal Study on Aging.
Author information
Authors and Affiliations
Contributions
L.T.E. and A.B.W. developed and directed the project. O.V. developed and performed the modeling pipeline and wrote the original draft. S.L. processed and aliquoted study samples and curated data. J.M., X.S., and K.R. provided critical feedback on the study and reviewed/edited the manuscript. O.V. and L.T.E. prepared the manuscript with input from all authors, and all authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Sithara Vivek, Chiara Herzog, and Valentin Vetter for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vishnyakova, O., Min, J., Leach, S. et al. Metabolomic sweet spot clock predicts mortality and age-related diseases in the Canadian Longitudinal Study on Aging. Commun Med (2026). https://doi.org/10.1038/s43856-026-01375-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01375-2