Abstract
Metabolic dysfunction-associated steatohepatitis (MASH) is characterized by insulin resistance and impaired hepatic metabolism, which lead to steatosis and lipotoxicity. S100A11, an alarmin upregulated in MASH, promotes steatosis in vitro, but its role in vivo remains unclear. We hypothesized that S100A11 drives MASH by upregulating hepatic lipid synthesis. Using whole-body S100a11 knockout (S100a11−/−) mice on a MASH-inducing diet, we found S100a11 deficiency reduced steatosis, inflammation, and fibrosis. Hepatotropic AAV8-mediated silencing of S100a11 confirmed these findings. Bulk RNA sequencing with Ingenuity Pathway Analysis revealed dysregulated carbohydrate and lipid metabolism in S100a11−/− livers, including downregulation of hexokinase 2 (Hk2). Since hexokinases regulate glucose flux into downstream metabolic processes, we overexpressed HK2 in S100a11−/− mice, which was sufficient to increase steatosis. Further, palmitate-induced HK2 upregulation required S100A11 in a human hepatocyte cell line. These studies identify HK2 as a downstream target of S100A11, both of which are potential therapeutic targets for MASH.
Similar content being viewed by others
Introduction
Metabolic-dysfunction-associated steatohepatitis (MASH) is the most prevalent chronic liver disease globally1. MASH is a heterogeneous disease and while its pathogenesis is multifactorial, steatosis, injury, and inflammation are central processes in the development of MASH2. It is well established that steatosis develops from the consequence of chronic positive energy balance coupled with dysfunctional adipose tissue expansion and insulin resistance. Similarly, liver injury is attributed to the direct toxicity of concurrently accumulated bioactive lipids such as saturated free fatty acids and cholesterol that can lead to lipotoxic organelle stress, such as endoplasmic reticulum stress, and hepatocyte apoptosis3. Injury is also amplified via the inflammatory activation of the innate and adaptive immune systems2. Despite significant advances, there is an incomplete understanding of signaling processes that integrate these three pivotal pathways in MASH.
Endogenous damage-associated molecular patterns or alarmins play a critical role in the chronic sterile inflammation in MASH. In this context, hepatocyte-derived danger signals play a key role in the recruitment, retention, and activation of monocyte-derived macrophages4. Several categories of endogenous alarmins have been discovered in MASH. These include high-mobility group box 1, mitochondrial DNA, heat shock proteins, cytokeratin 18, and uric acid5,6,7,8,9. Recent studies have discovered that the S100 family protein, S100A11 is upregulated in MASH and plays a role in regulating steatosis10,11,12. Yet the exact consequences of S100A11 upregulation remain incompletely understood. One of the receptors that can be activated by S100A11 is the receptor for advanced glycation end products (RAGE)13.
Our recent studies on the role of RAGE in MASH defined its upregulation on a subset of monocyte-derived macrophages13. We demonstrated that pharmacological inhibition of RAGE and myeloid cell-specific RAGE deletion led to a reduction in hepatic accumulation of monocyte-derived macrophages and subsequent inflammation13. However, our previous study did not examine potential RAGE ligands and their involvement in MASH. Continuing investigation of this endogenous alarmin-receptor signaling axis in MASH, we focused on the role of S100A11 in the liver in the current studies. We aimed to elucidate whether the RAGE ligand, S100A11, mediates MASH by employing whole-body knockout and liver-specific knockdown approaches. We demonstrate that silencing of S100A11 attenuates MASH. In the absence of S100A11, there is a reduction in hepatocyte steatosis and inflammation in the liver. Transcriptomic analysis elucidated carbohydrate and lipid metabolism as the top dysregulated pathways. Further examination of key rate-limiting enzymes demonstrated an upregulation of hexokinase 2 (HK2) in WT mice, which was attenuated in S100A11-silenced livers. Enforced expression of HK2 was sufficient to induce hepatic steatosis in S100a11−/− mouse livers. Thus, we have identified a metabolic pathway downstream of S100A11 that mediates lipogenesis and lipotoxicity.
Results
Whole body S100a11 knockout alleviates FFC diet-induced murine MASH
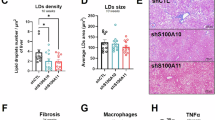
We previously identified S100A11, an alarmin, as a ceramide-dependent extracellular vesicle (EV) cargo secreted from lipotoxic hepatocytes14. Our assessment of S100A11 by immunohistochemistry demonstrated a significant increase in FFC-fed wild-type mouse livers (Supplementary Fig. 1A, B), in a hepatocellular pattern compared to CD-fed controls, at 24 weeks of dietary feeding. Correspondingly, the abundance of S100a11 mRNA was significantly increased in FFC-fed WT mouse livers compared to CD-fed mice (Supplementary Fig. 1C). We next examined S100A11 expression in human MASH livers by qPCR. Similar to the FFC-fed mouse model, which is known to have a high fidelity to human MASH15, we observed a significant upregulation in the expression of S100A11 compared to healthy controls (Supplementary Fig. 1D). To examine the role S100A11 plays in MASH pathophysiology, WT and S100A11 whole body knockout mice (S100a11−/−) were maintained on the FFC diet or CD for 24 weeks (Fig. 1A). Quantitative PCR analysis of liver tissues confirmed all the S100a11−/− mice to be knockouts, compared to the WT controls, with undetectable S100a11 transcripts (Supplementary Fig. 2A). The FFC-fed WT mice demonstrated a significant upregulation of S100a11 confirming our previous observations (Supplementary Fig. 2A). We confirmed the expression of S100A11 by immunohistochemistry in FFC-fed WT mice (Supplementary Fig. 2B). We did not detect S100A11 in S100a11−/− mice; CD-fed mice were not compared due to low basal levels observed in CD-fed WT livers (Supplementary Fig. 1A). At the start of the dietary studies, at 12 weeks of age, body mass was comparable across the groups (Supplementary Fig. 2C). Upon study completion, at 36 weeks of age, we observed a slightly lower body weight in S100a11−/− mice compared to WT on both diets (Supplementary Fig. 2D), though there was a small but significant increase in food intake in the FFC-fed S100a11−/− mice (Supplementary Fig. 2E). Food intake was similar between CD-fed WT and S100a11−/− mice. FFC-fed mice consumed fewer grams of food than CD-fed mice, consistent with the established literature that mice consume fewer grams of high-fat diets due to the higher caloric content of such diets16. Liver-to-body weight ratio demonstrated no basal changes in CD-fed mice cohorts (Fig. 1B). FFC diet significantly increased liver-to-body weight ratio, though this response was blunted in S100a11−/− mice (Fig. 1B). Plasma ALT levels were normal in CD-fed S100a11−/− and WT controls (Fig. 1C). FFC-fed WT mice had significantly higher ALT levels indicating liver injury, while FFC-fed S100a11−/− mice displayed comparatively reduced levels (Fig. 1C). No significant differences were observed in bilirubin levels (Supplementary Fig. 3A) among all cohorts. As anticipated, serum bile acid and cholesterol levels were elevated in FFC-fed mice compared to those on CD (Supplementary Fig. 3B, C). Notably, FFC-fed S100a11−/− mice exhibited significantly lower levels of serum bile acid and cholesterol levels relative to FFC-fed WT controls (Supplementary Fig. 3B, C). Histological analysis of the liver tissues using H&E stained liver sections demonstrated significant steatosis and lobular inflammation in FFC-fed WT mouse livers (Fig. 1D). FFC-fed S100a11−/− mouse livers displayed reduced steatosis and fewer inflammatory foci (Fig. 1D). The CD-fed mouse livers were histologically normal (Supplementary Fig. 3D). Steatosis and inflammatory foci were graded according to the respective components of the NAFLD activity score17, confirming reduced inflammatory foci in FFC-fed S100a11−/− mouse livers with a trend toward decreased steatosis (Fig. 1E, F). The reduction in steatosis was further assessed by lipid droplet morphometry. Total lipid droplet area and average lipid droplet size were significantly lower in FFC-fed S100a11−/− mice compared to FFC-fed WT mice (Fig. 1G, H). There was an increase in the number of lipid droplets in FFC-fed S100a11−/− mice consistent with the H&E observation of greater microvesicular steatosis (Fig. 1I).
A Schematic representation of the experimental regimen of WT and S100a11−/− mice with 24 weeks of FFC diet feeding. A total of 6 CD-WT, 7 CD- S100a11−/−, 5 FFC-WT, and 7 FFC- S100a11−/− mice were included for downstream analysis unless otherwise specified. B Relative liver mass (liver to body mass percent) of the CD and the FFC diet fed WT and S100a11−/− mice groups. C Plasma ALT levels in CD-fed and FFC-fed WT and S100a11−/− mice. D Representative H&E stained liver samples from FFC-fed WT and S100a11−/− mice. Scale bar: 50 μm. E Ten fields per H&E stained mouse liver samples were quantified for inflammatory foci at ×20 magnification. Each point represents the average value per mouse. F Steatosis was graded on H&E stained mouse liver samples per the NAFLD activity score at ×10 magnification. Each point represents the average value per mouse. G–I Lipid droplet morphometry was performed on H&E stained liver sections to calculate (G) total lipid droplet area, (H) average lipid droplet size, and (I) number of lipid droplets in CD and FFC-fed WT and S100a11−/− mouse livers (n = 3 mice per group). J Representative images of picrosirius red stained liver sections are depicted along with quantification. Percent positive area of the picrosirius red stained positive collagen fibers was quantified using polarized microscopy: Scale bar: 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Assessment of hepatic mRNA expression levels of inflammatory markers revealed a significant upregulation of pro-inflammatory genes, Ly6c and Mac2, in FFC-fed WT mice compared to CD-fed controls. Importantly, this inflammatory response was markedly attenuated in FFC-fed S100a11−/− mice (Supplementary Fig. 3E, F). Assessment of liver fibrosis using Picrosirius staining demonstrated no significant collagen fiber accumulation in CD-fed mice across the genotypes (Supplementary Fig. 3G). FFC-fed WT mouse livers displayed significant pericellular and bridging fibrosis while FFC-fed S100a11−/− mice had lower abundance of collagen fibers, verified by polarized light microscopy-based collagen quantification (Fig. 1J). mRNA analysis of fibrogenic genes in the liver demonstrated a significant increase in the expression of Col1a1 and Timp1 in FFC-fed WT mice compared to those on CD (Supplementary Fig. 3H, I). Notably, the upregulation of Col1a1 was significantly lower in FFC-fed S100a11−/− mice and Timp1 showed a similar pattern but did not reach significance. Taken together, these data demonstrate that whole-body S100a11 deletion reduces FFC-diet-induced MASH.
Liver-specific S100a11 repression alleviates FFC-diet-induced murine MASH
Due to the difference in body mass and relative liver mass in FFC-fed WT compared to S100a11−/− mice, to avoid confounding due to differences in body mass and extrahepatic effects of S100A11, we asked if hepatocyte-specific repression of S100a11 would exhibit MASH attenuation. To address this question, mice were fed CD or FFC diet for 20 weeks and then divided into four groups (Fig. 2A). Hepatotropic AAV8 viral particles encoding S100a11 shRNA or scramble shRNA were administered following 20 weeks of diet by tail vein injections. Mice were maintained on their respective diets for an additional 4 weeks. Assessment of shRNA-mediated knockdown efficacy was performed by qPCR and immunohistochemistry. We observed that FFC-shA11 (AAV8-shA11 injected FFC) mice had significantly lower S100a11 transcripts compared to FFC-scr (AAV8-scramble injected FFC) mice controls (Fig. 2B), and lower expression by immunohistochemistry (Supplementary Fig. 4A). In contrast to the whole-body knockout, both groups of FFC-fed mice gained comparable weight on the diet (Supplementary Fig. 4B). However, the relative liver mass was lower in FFC-shA11 group compared to FFC-scr group with no change in CD-fed mouse groups (Supplementary Fig. 4C). Correspondingly, there was a reduction in ALT elevation, inflammation and hepatic steatosis (Fig. 2C–F) in FFC-shA11 mice compared to FFC-scr mice while repression of S100a11 had no effect on CD fed mice (Supplementary Fig. 3D). Lipid droplet morphometry confirmed a reduction in total lipid droplet area, average droplet size, and an increase in the number of lipid droplets in the FFC-shA11 mouse livers (Fig. 2G–I) as observed in the whole-body knockout mice. Picrosirius red stained liver sections indicated no notable collagen fiber accumulation in the CD-fed mice (Supplementary Fig. 3E). FFC-scr mice cohort displayed significant fibrosis which was remarkably reduced with 4-weeks of S100a11 silencing, as noted in the FFC-shA11 group (Fig. 2J). Polarized light microscopy-based collagen quantification confirmed the histological impression of a reduction in fibrosis. Thus, hepatocyte-specific silencing of S100A11 was sufficient to attenuate MASH, while maintaining comparable body mass in mice.
A Schematic representation depicting the experimental dietary regimen and AAV8 administration timeline for the liver specific S100a11 knockdown study. Mice were treated with either sh-scramble (scr) or sh-S100A11 (shA11) virus. A total of 7 CD-scr, 7 CD-shA11, 6 FFC-scr, and 6 FFC-shA11 mice were included for downstream analysis unless otherwise specified. B qPCR expression of S100a11 in whole livers. One FFC-scr sample was a statistical outlier and removed from analysis. C Plasma ALT measurement. D Representative imagfes of H&E stained liver sections. Scale bar: 50 μm. E Ten fields per H&E stained mouse liver samples were quantified for inflammatory foci at ×20 magnification. Each point represents the average value per mouse. F Steatosis was graded on H&E stained mouse liver samples per the NAFLD activity score at ×10 magnification. Each point represents the average value per mouse. G–I Lipid droplet morphometry was performed on H&E stained liver sections to calculate (G) total lipid droplet area, (H) average lipid droplet size, and (I) number of lipid droplets per field in CD and FFC-fed scr and shA11 mouse livers (n = 3 mice per group). J Representative images of picrosirius red stained of the liver sections are demonstrated from FFC-scr and FFC-shA11 mouse liver. The percent area of the Sirius red-positive collagen fibers was quantified using polarized microscopy. One CD-scr value was a statistical outlier and removed from analysis. One CD-shA11 sample had inadequate amount of tissue for analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Carbohydrate and lipid metabolism are dysregulated in liver-specific S100a11 knockdown
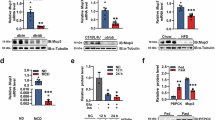
To understand the mechanism of how silencing of hepatic S100a11 impairs FFC diet-induced steatohepatitis compared to the respective controls, we employed bulk-RNA sequencing. We identified 13,379 protein coding genes wherein 2245 were significantly differentially expressed genes (DEGs) of which 1429 were downregulated and 816 were upregulated (Supplementary Fig. 5A). We observed more than five-fold reduction (Log2 FC: −2.47) of S100A11 transcripts in the FFC-shA11 livers versus FFC-scr, confirming significant S100a11 gene knockdown in this cohort (Supplementary Fig. 5A). Principal Component Analysis (PCA) confirmed significant clustering of the two independent study groups (FFC-scr and FFC-shA11) (Supplementary Fig. 5B). Ingenuity Pathways Analysis demonstrated significant dysregulation of lipid metabolism and carbohydrate metabolism in the top 10 dysregulated pathways in FFC-shA11 livers (Fig. 3A). Further interrogation of “carbohydrate metabolism” and “lipid metabolism” components, within Ingenuity Pathways Analysis, was performed by comparing the rank order list of component pathways. We noted 128 transcripts in “metabolism of carbohydrate” (p value of 6.79e−21 and activation score of 0.008) and 101 transcripts in “synthesis of fatty acids” (p value of 4.07e−20 and activation score of −2.011) with 19 transcripts that were common to both pathways (Supplementary Table 1).
A Ingenuity Pathway Analysis of the bulk-RNA sequencing datasets demonstrate FFC-shA11 mice to have dysregulated lipid and carbohydrate metabolism. The list of pathways annotated within these two metabolic axes is enlisted as the other two sub-figures. Numbers in the box denote the number of transcripts involved in the pathway. Red asterisks (*) indicate the highlighted pathways investigated further. B FFC-shA11 mouse livers have significantly reduced Fragments per Kilobase of transcript per Million mapped reads (FPKM) compared to the FFC-scr controls (n = 3 mice per group). C qPCR assessment of critical genes important for lipid and carbohydrate metabolism. A total of 3 CD-scr, 3 CD-shA11, 5 FFC-scr, and 5 FFC-shA11 mouse livers were evaluated. D Western blot analysis of HK2 protein in CD (n = 2 per group) and FFC (n = 5 per group) fed mouse livers. Quantification is plotted to the right. Alpha-tubulin was included as a loading control. HK2 band intensity is expressed relative to corresponding alpha-tubulin. *P < 0.05, **P < 0.01, ***P < 0.001.
Of the 19 identified transcripts, we considered HK2 further, as glucose fixation by hexokinase-mediated phosphorylation is the crucial step in carbohydrate metabolism and links carbohydrate to lipid metabolism18. We observed that the expression of Hk2 was reduced in the shA11 livers in the RNA-seq data (Fig. 2B). We confirmed this by qPCR, which showed the abundance of Hk2 transcripts was low and unchanged in CD-fed mouse livers, whereas expression was significantly decreased in FFC-shA11 mouse livers compared to FFC-scr livers (Fig. 3C). Similar to the mouse data, HK2 expression was low in normal human livers (only detected in 2 of 6 normal control samples tested) and upregulated in MASH livers, though not uniformly so (detected in 4 of 9 samples tested) (Supplementary Fig. 5C). The mRNA expression of glucokinase or hexokinase 4 (Hk4), the predominant form in hepatocytes, was upregulated with FFC diet in control mice whereas hexokinase 3 (Hk3) did not significantly change (Supplementary Fig. 5D). There were no significant differences in expression between FFC-scr and -shA11 livers. We did not detect hexokinase 1 (Hk1) and hexokinase domain-containing 1 (Hkdc1) transcripts in the liver (data not shown). Among other candidate genes, we observed a similar direction in Fasn, which encodes fatty acid synthase, and peroxisome proliferator-activated receptor alpha (Pparα), which were also attenuated in FFC-shA11 (Fig. 3C). The mRNA expression of phosphoenolpyruvate carboxykinase 1 (Pck1), diacylglycerol O-acyltransferase 2 (Dgat2), and acyl-CoA oxidase 1 (Acox1) demonstrated no significant differences in FFC-shA11 mouse livers with respect to the control groups (Fig. 3C).
Next, we asked if the upregulation of Hk2 transcripts correlated with an increase in its protein expression. Western blot of HK2 in mouse livers demonstrated low basal expression in CD-fed liver (Fig. 3D). We tested this with increasing protein lysate concentrations (5 μg, 15 μg, 30 μg, and 50 μg) from CD-fed WT livers, which confirmed expression of HK2 protein in all samples tested rather than non-specific detection (Supplementary Fig. 5E). Some variability was observed among the CD-fed samples; one mouse exhibited detectable HK2 expression even at 5 μg protein input, suggesting individual variation in basal HK2 levels (Supplementary Fig. 5E). FFC-diet feeding significantly increased HK2 protein levels and liver-specific S100a11 repression lowered FFC-diet induced HK2 protein upregulation (Fig. 3D). In vitro treatment of a murine hepatocyte cell line (IMH) with palmitate, a lipotoxic stressor significantly increased the expression of HK2 protein levels in palmitate-stimulated cells compared to the controls (Supplementary Fig. 6A). De novo lipogenesis (DNL) assay in primary mouse hepatocytes demonstrated an increase in lipid synthesis in palmitate-stimulated WT hepatocytes. (Supplementary Fig. 6B). In contrast, hepatocytes isolated from S100a11−/− mice did not demonstrate an increase in DNL following PA treatment (Supplementary Fig. 6C). To confirm these findings and to determine whether S100A11 regulates HK2 expression differentially in response to carbohydrate or lipid metabolism, we treated WT and S100A11−/− Hu1545 human hepatocyte cell line for 8 h with either vehicle or 400 µM palmitate under standard or high glucose conditions. Western blot analysis demonstrated that HK2 expression was upregulated in response to palmitate treatment in WT cells and not in response to glucose, suggesting a glucose-independent effect of palmitate on HK2 induction (Supplementary Fig. 6D). In S100A11−/− cells, we did not observe an increase in HK2 expression following PA treatment. Rather, the levels of HK2 were lower than standard glucose conditions. We next measured accumulation of lipid droplets in WT and S100A11−/− cells similarly treated with palmitate under standard or high glucose conditions (Supplementary Fig. 6E, F). We found that palmitate was able to increase lipid content, whereas high glucose did not change lipid area. Furthermore, high glucose did not modify the palmitate response. We also observed that palmitate-induced steatosis was attenuated in S100A11−/− cells. Altogether, our observations suggest that S100A11 mediates lipotoxicity-induced expression of HK2, which may drive de novo lipogenesis.
HK2 upregulation is required and sufficient to drive steatosis
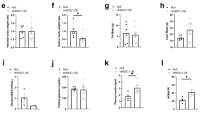
Having noted a significant upregulation of HK2 in FFC-fed livers and repression of HK2 in FFC-shA11 livers, we asked if a gain-of-function approach would help understand the relationship between S100A11 and HK2. First, we confirmed that HK2 expression was lower in FFC-fed S100a11−/− livers compared to WT (Supplementary Fig. 7A). Next, we injected 1 × 1011 genome copies of AAV8-Hk2 viral particles (Supplementary Fig. 7B) via tail vein into 12-week-old WT and S100a11−/− male mice. The mice were maintained on CD for 4 weeks after virus administration (Fig. 4A). During tissue collection, the livers of Hk2-overexpressing CD-fed mice appeared pale and enlarged compared to normal livers of scramble controls. In keeping with these morphological observations, the relative liver mass was increased significantly in the Hk2 overexpression group compared to scramble controls in both WT and S100a11−/− mice (Fig. 4B). Western blotting of liver tissues confirmed overexpression of exogenous HK2 in all the AAV8-Hk2 injected mice compared to the scramble controls (Fig. 4C). Interestingly, we also noted increased abundance of DNL-regulating master enzymes: total acetyl-CoA carboxylase (ACC) and ATP citrate lyase (ACLY) in Hk2-overexpressing WT and S100a11−/− mouse livers compared to the scramble mice (Fig. 4C). Steatosis was observed in H&E-stained liver sections from WT and S100a11−/− Hk2 overexpressing mice (Fig. 4D, E). Quantification of steatosis and of the BODIPY signal demonstrated comparable steatosis in WT and S100a11−/− Hk2 overexpressing mice (Fig. 4F, G). Lastly, biochemical measurement of liver triglyceride content confirmed that HK2 overexpression was sufficient to drive steatosis in WT and S100a11−/− mouse livers (Fig. 4H). Altogether, these observations demonstrate that even under basal (chow diet) conditions, the overexpression of HK2 is sufficient to drive steatosis in WT and S100a11−/− mouse livers.
A Schematic representation depicting the dietary regimen and AAV8-scramble (AAV8-scr) or AAV8-Hk2 administration timeline for the liver-specific Hk2 overexpression study in WT and S100a11−/− mice. A total of 4 mice per group were included for downstream analysis unless otherwise specified. B Relative liver mass normalized to total body weight. C Western blot analysis of HK2, ACLY, total ACC and α-tubulin in liver lysates. D Representative photomicrographs of H&E stained liver sections. Scale bar: 50 μm. E H&E stained liver sections were graded for steatosis per the NAFLD activity score per mouse at ×10 magnification. Each point represents the average value per mouse. F Representative photomicrographs of BODIPY stained liver sections (n = 4 mice per group). Scale bar: 20 μm. G BODIPY stained lipid droplet area was quantified using ImageJ software. Each point represents the average value per mouse. H Liver triglycerides were quantified using a biochemical assay and normalized to liver tissue weight. *P < 0.05, **P < 0.01, ***P < 0.001.
In complementary experiments, we employed BNBZ, a pharmacological inhibitor of HK2. Palmitate-treated Hu1545 cells demonstrated significant lipid-droplet accumulation (Fig. 5A, B). In contrast, in cells treated with palmitate in the presence of BNBZ there was no increase in lipid droplet accumulation. Thus, HK2 mediates palmitate-induced steatosis in hepatocytes.
A Representative BODIPY stained images from Hu1545 human hepatocyte cell line treated with palmitate (PA) with or without benitrobenrazide (BNBZ). The experiment was replicated 3 times. Vehicle control (Veh) included equivalent amounts of isopropanol and DMSO. Scale bar: 20 µm. B Quantification of BODIPY staining was performed in Image J. Each dot represents a separate experiment averaged over five fields. Approximately 16–36 cells were counted per field. Data were calculated as the mean intensity divided by the number of cells per field. **P < 0.01, ***P < 0.001, ****P < 0.0001.
S100A11 and HK2 are upregulated in hepatocytes
Though bulk liver mRNA and protein expression mostly represent hepatocytes, given the significantly greater hepatocellular mass than other cell types in the liver, we asked whether S100A11 and HK2 may be upregulated in other liver cell types. We used bone marrow-derived macrophages (BMDM) and the human hepatic stellate cell line LX-2 to address this question. Neither S100a11 nor Hk2 was upregulated in BMDM or LX2 cells (Supplementary Fig. 8A–D). We have previously demonstrated an increase in RAGE (Ager) positive recruited macrophages in MASH13. We did not detect Ager in hepatocytes (data not shown). As Ager is also expressed in hepatic stellate cells, we examined its expression in this cell type; Ager was not upregulated in palmitate-treated LX-2 cells (Supplementary Fig. 8E). Altogether, our data implicate a hepatocellular role for palmitate-induced upregulation of S100A11 and consequently HK2 in promoting MASH.
Discussion
S100A11 is an alarmin upregulated in MASH. Previous studies have implicated S100A11 in hepatic steatosis; however, the mechanism by which hepatic S100A11 mediates MASH or drives steatosis remains incompletely defined. Herein, we demonstrate that (i) S100a11−/− mice demonstrate attenuated steatohepatitis with a reduction in liver steatosis, inflammation, and injury; (ii) Liver specific knockdown of S100a11 is sufficient to mitigate FFC-diet induced steatohepatitis; (iii) De novo lipid synthesis and carbohydrate metabolism are top dysregulated pathways following S100a11 silencing; (iv) Hk2 is significantly upregulated in the liver in FFC-fed WT mice and attenuated in S100a11 silenced mice; (v) Exogenous expression of Hk2 is sufficient to drive hepatic steatosis in the absence of S100a11 and under basal nutrition conditions; (vi) Palmitate upregulates HK2 expression in an S100A11-dependent manner; and (vii) S100A11 and HK2 transcripts are upregulated in human MASH liver samples.
S100A11 belongs to the family of S100 proteins. While homeostatically expressed at a low level, it is upregulated in many liver diseases in humans and model organisms, including MASH9,10,11,19. Consistent with the literature, we observed an increase in S100A11 in human MASH liver samples and in a murine model of MASH generated by feeding mice a diet high in fat, fructose, and cholesterol. The dietary components of the FFC diet and subsequent liver phenotype are validated to be recapitulative of a human fast food diet and MASH, respectively15,20. To understand the role of S100A11 further in MASH pathophysiology, we employed whole body knockout mice (S100a11−/−) and a liver-targeted approach using a hepatotropic viral vector21. Hepatic steatosis, injury, and inflammation were attenuated with both silencing approaches. FFC-diet fed S100a11−/− mice gained less weight than WT controls. These findings suggest a role for S100A11 in the liver and in extrahepatic tissues under nutritional stress. Given the critical role of adipose tissue dysfunction in MASH, the role of S100A11 in adipose tissue should be examined in future studies employing littermate controls, which are beyond the scope of the current work. In the current study, C57BL/6 mice from Jackson Laboratories were used as controls for the S100a11−/− mice, as previously done by others22. Regardless of the contributions from extrahepatic tissues like adipose tissue, in the current study, we isolated the role of S100A11 by adopting a liver-specific knockdown approach. By employing an AAV8-shRNA, which is known to transduce hepatocytes, we obtained a significant reduction in the expression of S100A11 in the liver21. Body mass remained comparable in the shRNA and scramble control groups. In keeping with the whole-body knockout mice, silencing S100a11 in the liver was sufficient to lead to MASH attenuation supporting a liver-specific role for this alarmin in promoting steatohepatitis.
We adopted a bulk RNA-sequencing followed by a candidate gene approach and identified HK2 to be significantly upregulated in FFC-fed WT mice, and the upregulation was attenuated following S100A11 silencing. We confirmed that HK2 transcripts were upregulated in human MASH liver samples, though not uniformly. Evaluating HK2 transcripts in larger MASH transcriptional datasets will be necessary to conclude that there is a correlation with disease severity. In the RNA-seq data, synthesis of fatty acids and metabolism of carbohydrates were the top dysregulated pathways. These data are interesting as they suggest that HK2 may be the driver of steatosis in FFC-fed WT mice by providing substrate for de novo lipogenesis. HK2 is the predominant form of hexokinase in adipose tissue, muscle and cancer cells, whereas HK4 is the predominant form in hepatocytes18. We found that HK2 and HK4 expression were both transcriptionally upregulated in FFC-fed mice, though only HK2 was significantly blunted in hepatocyte-specific S100A11 knockdown mice. While HK2 is predominantly expressed in muscle and heart tissues, it is reported to be regulated by several factors, including hormonal signals such as insulin and glucagon, as well as metabolic changes like fluctuations in glucose availability. These data suggest that HK2 is responsive to altered metabolic environments and its upregulation in the liver, which we report here, may be an adaptive response, albeit maladaptive, given the attenuation of steatohepatitis in low HK2 abundance (S100A11 silenced) conditions. Altogether, our data demonstrate that HK2 is upregulated in MASH and is sufficient to induce hepatic steatosis.
S100A11 does not independently function as a transcription factor. A previous study has identified S100A11, which, when overexpressed, promoted steatosis via upregulation and acetylation of the transcription factor FOXO1. ChREBP and SREBP1c play a significant role in regulating insulin resistance-driven de novo lipogenesis in hepatocytes23. ChREBP influences glucose metabolic reprogramming and transcriptionally upregulates lipogenic genes24, with a recent study identifying HK2 as a transcriptional target of ChREBP25. SREBP1c, on the other hand, regulates insulin-dependent lipogenic gene expression26 but Rome et al. demonstrate that SREBP1c also transcriptionally regulates HK2 expression in muscle cells27. Thus, the role of ChREBP and SREBP1c in regulating HK2 in MASH livers presents an intriguing area for future research. Also, ChREBP is reported to regulate the alarmin S100A6 in hepatocytes in MASH livers28. Increasing evidence of epigenetic mechanisms regulating hepatic steatosis in MASH suggests that epigenetic mechanisms may be plausible in S100A11-dependent HK2 upregulation. In support of this, Zeng et al. demonstrated S100A11 facilitated histone modification at the transketolase promoter through interaction with the SMYD3 protein. In this study, knockdown of S100A11 inhibited the pentose phosphate pathway by disrupting the mRNA synthesis of transketolase29.
Our Hk2 overexpression studies in S100a11−/− mice asked if HK2 would be sufficient to drive steatosis. In the absence of hypernutrition and without exogenous glucose or fructose in the diet, in CD-fed mice, HK2 overexpression was sufficient to drive hepatic steatosis in both WT and S100a11−/− mice. We chose to conduct the studies with CD as the FFC diet induces robust steatosis. We wanted to be able to discern the effect of HK2 overexpression on steatosis without confounding from the effects of the diet, thus our choice of CD, which does not induce significant steatosis. HK2 is distinct in substrate preference than ketohexokinase which phosphorylates fructose to fructose-1-phosphate30. Inhibition of ketohexokinase has been shown to improve hepatic steatosis and inflammation in Phase 2 human trials31,32. These studies and our novel observations support considering HK2 inhibition as a potential therapeutic strategy in MASH. In this manuscript, we describe a metabolic role for S100A11 in mediating MASH. In previously published work13, we have demonstrated that RAGE-positive recruited macrophages play a significant role in promoting inflammation in the liver in MASH. The current observations, together with our earlier data, support a model in which S100A11 may mediate MASH via pleiotropic effects, including HK2-driven lipogenesis in hepatocytes and the recruitment of RAGE-positive pro-inflammatory macrophages into the liver.
Methods
Human liver samples
Biobanked frozen human liver samples from living controls and MASH patients were obtained from Mayo Clinic Biobanks. All patients provided written informed consent and the study was approved by the Mayo Clinic IRB. The diagnosis of MASH or normal liver histology was confirmed by an expert pathologist.
Mouse studies
All animal use was approved by the Institutional Animal Care and Use Committee at the Mayo Clinic and conducted in compliance with public health regulations regarding the humane treatment of laboratory animals. Six-week-old male C56BL/6 J mice were obtained from Jackson Laboratory and acclimated for 6 weeks. During this period, the mice were randomly assigned to one of four groups with diet randomization based on cage, with all receiving a standard rodent chow diet (CD). At 12 weeks of age, the control group continued on the CD, while the experimental groups were switched to a Western diet, which consisted of 40% of calories from fat, including 34% sucrose, 20% milk fat, and 0.2% cholesterol (AIN-76A Western Diet, D12079B, TestDiet) for 24 weeks, as previously described by us14. Additionally, the experimental groups received drinking water supplemented with high fructose corn syrup (23.1 g/L fructose and 18.9 g/L glucose). This diet is referred to as the fat, fructose, and cholesterol (FFC) diet. Mice were housed in standard pathogen-free conditions, with 2–4 mice per cage, and had unrestricted access to food and water, and followed a 12-hour day-night circadian cycle. Food intake was measured in a separate cohort with one cage per condition and 4 mice per cage every week from start till week 19 of feeding by subtracting the weight of the final remaining food from the initial weight of food placed in the cage, divided by the number of mice per cage. Upon completion of the stud,y the mice were humanely euthanized by CO2 inhalation followed by cervical dislocation. Blood was collected via cardiac puncture and plasma was isolated from whole blood. Plasma was stored at -80 °C for later analysis. The liver was removed, weighed, and portions were preserved for further analysis, including RNA and protein extraction, histological analyses and cryosectioning.
Global S100A11 knockout mice
S100a11-knockout (KO) mice were a kind gift from Dr. Felix Yarovinsky and have been previously described15. Briefly, S100a11−/− mice were generated by targeting exons 2 and 3 of the S100a11 gene with two sgRNAs using CRISPR-Cas9 technology at the Mouse Genome Editing (MGE) Resource of the University of Rochester Medical Center. S100a11−/− mice were generated on a C57BL/6 background, and as published previously, age-matched C57BL/6 mice from Jackson Laboratory were employed as controls.
Hepatocyte-specific S100A11 knockdown model
AAV8 backbone with the highest known hepatotropic efficacy16,17 was chosen to create U6 promoter-driven shRNA constructs. The AAV8-GFP-U6-m-S100A11-shRNA and AAV8-GFP-U6-scramble-shRNA constructs, expressing mouse S100a11-specific shRNA and scramble shRNA, respectively, were prepared and packaged by Vector Biolabs. The S100a11 shRNA sequence (5’-CCGGGCCATGATTCTTTCATCCAAA-CTCGAG-TTTGGATGAAAGAATCATGGC-TTTTTG-3’) was validated by the company, based on ~90% knockdown of S100a11 mRNA in Hepa 1-6 cells. 12-week-old C57BL/6 wild-type male mice were placed on the CD/FFC-diet regimen for 24 weeks. At the start of the 20th week on diet, mice were randomly assigned to receive either of the AAV8-shRNAs at a dose of 2 ×1011 genome copies per mouse via tail vein injection. Mice were continued on their respective diet for another 4 weeks and then euthanized.
Hepatocyte-specific HK2 overexpression mouse model
We employed the AAV8 vector with a thyroid binding globulin promoter (AAV8-TBG) to express mouse HK2 in a hepatocyte-specific manner18. The pAAV8-TBG-mHK2 viral vector, encoding HK2 and a control with a scrambled construct (scramble), were commercially prepared and packaged (VectorBuilder Inc.). 10-week-old C57BL/6 male mice received the viral particles at a dosage of 1 × 1011 genome copies per mouse via tail vein injection, following which mice were maintained on a chow diet for an additional 4 weeks.
Histologic analyses and Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) liver tissues were sectioned into 5 µm slices and stained with hematoxylin and eosin (H&E) following standard protocols at the Mayo Clinic Histology Core. The H&E-stained slides were evaluated for steatosis and inflammatory foci. For S100A11 immunohistochemistry, FFPE liver sections were dewaxed with xylene and rehydrated through graded ethanol concentrations (100%, 90%, 80%, 70%, and 50%). The slides were then treated with 10 mM sodium citrate buffer containing 0.05% Tween (pH 6.0) for 20 minutes at 95°C for epitope retrieval, followed by cooling to room temperature. To block endogenous peroxidase activity, the slides were incubated in 3% hydrogen peroxide for 10 minutes. Non-specific immunoglobulin binding was blocked using the Goat-IgG ABC Kit (Vector Laboratories). Additional blocking of avidin and biotin was carried out for 15 minutes each using the Avidin/Biotin Blocking Kit (Vector Laboratories). The primary antibody for S100A11 (1:150 dilution) was applied and incubated overnight at 4°C in a humidified chamber. After washing, slides were incubated for 30 minutes each with biotinylated secondary antibody and ABC reagent from the ABC Kit, following the manufacturer’s instructions. The signal was developed using the DAB Peroxidase Substrate Kit (Vector Laboratories) and counterstained with Gill’s Hematoxylin (Sigma-Aldrich) for 40 seconds. The slides were then dehydrated through graded ethanols and mounted with Organo/Limonene Mount (Sigma-Aldrich). Images were captured using a Zeiss Axio at a 20X magnification, and the percentage of S100A11-positive area was quantified using ImageJ (NIH). For fibrosis assessment, the liver sections were dewaxed, rehydrated as mentioned above and stained with Picrosirius red (0.1% Sirius red (Sigma-Aldrich), and 0.1% fast green). After dehydration through graded ethanols, the slides were mounted with Organo/Limonene Mount (Sigma-Aldrich). Images were captured using a Nikon microscope with a DS-U3 camera (Nikon) at a 20X objective lens, and fibrosis quantification was performed using NIS-elements AR (Nikon) and expressed as the percentage of the total picrosirius-stained collagen-positive area in each liver section.
Quantitative lipid droplet morphometry
We utilized the LiverQuant image analysis pipeline, originally developed by Farzi et al. 19 to quantify hepatic steatosis by detecting fat globules in liver tissue sections. The LiverQuant algorithm was applied to .tiff images of histological liver samples using the detect_fat_globules() function, which segments lipid droplets based on white space detection within hematoxylin and eosin (H&E) stained slides. The detection thresholds were customized to identify droplets between 2 µm and 300 µm in diameter, with spatial calibration set a 0.17 µm per pixel. The algorithm outputs a binary mask of detected fat regions for each image, which was then processed to extract quantitative metrics such as fat area proportion, number of droplets, and average globule size in square microns. Five 20x fields were analyzed per each mouse in the groups. Following segmentation, the original and marked-up images were post-processed in Python using OpenCV and scikit-image. The mask was overlaid on the original image in red, cropped to the bounding box of detected fat globules, and saved as a .tiff file. A matching unmarked cropped image was also saved for direct comparison. The entire image area was analyzed without manual region selection, ensuring objectivity and reproducibility. Summary statistics for each image—including fat proportion area, total fat area, fat globule count, and mean globule size—were compiled into a CSV file for downstream analysis. This methodology enables robust and scalable quantification of hepatic steatosis in histological datasets, leveraging LiverQuant’s validated image analysis framework.
Quantitative real-time PCR
15–20 mg of liver tissue was homogenized in 1 mL of TRIzol using a hand-held homogenizer. After adding 200 µL of chloroform and centrifuging, the aqueous phase was transferred to new Eppendorf tubes. An equal volume of ethanol was then added, and the mixture was applied to an RNA isolation column (Direct-zol RNA MiniPrep, Zymo Research). RNA was extracted following the manufacturer’s protocol, which included an in-column DNase digestion step. The quantity and quality of the isolated RNA were evaluated using a NanoDrop ND1000 (Thermofisher Scientific). Reverse transcription was carried out using the iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR (qPCR) was performed on the Quantistudio 6 flex (Thermofisher Scientific), using the PowerSYBR Green PCR Master Mix (Applied Biosystems, Life Technologies LTD). Primers used in this study are listed in Supplementary Table 2. Relative fold change was calculated as described previously20. 18S was used as a loading control.
Western blotting
For protein biochemistry, cells or tissue were lysed in RIPA buffer containing protease inhibitors. Protein concentration was estimated using the BCA assay (ThermoFisher Scientific). 30-50 µg of protein lysate per sample was resolved on a 4-15% gradient SDS gel (Bio-Rad). After electrophoresis proteins were transferred to a 0.22 µm PVDF membrane. The membrane was blocked with 5% non-fat dry milk to prevent non-specific binding, then incubated with a primary antibody overnight at 4°C. Following three, 10 min each, washes in TBST, a horse radish peroxidase-conjugated secondary antibody was applied at room temperature. The membrane was washed again, and protein bands were detected using a chemiluminescent substrate. Antibodies used for the study are provided in Supplementary Table 3. Uncropped blots are available in Supplementary Fig. 9.
Biochemical assays
Isolation of platelet-poor plasma was done by first centrifuging 500-800 μL of citrated blood for 20 minutes at 4000 x g. The supernatant was transferred to new Eppendorf tubes and spun down at 12,000 x g for 2 minutes. The clear supernatant was then aliquoted to new tubes for analysis or storage. Approximately 100 μL of plasma was used for analysis of ALT levels using VetScan VS2 (Abaxis) a commercial veterinary chemistry analyzer.
Triglyceride assay
Liver homogenates were prepared from approximately 20 mg of liver tissue. The tissue was homogenized in 300 µL of ice-cold PBS for 30 s on ice using a tissue tearer. 100 µL aliquot of the homogenate was transferred to a new tube and mixed with 100 µL of 1% sodium deoxycholate solution (Sigma-Aldrich). The mixture was vortexed for 30 seconds and incubated at 37°C for 5 minutes. 5 µL of the solubilized liver homogenate was mixed with 150 µL of Infinity triglyceride TG reagent (Fisher Scientific) in a 96-well plate and the mixture was incubated for 15 minutes at 37°C. Absorbance was measured at 500 nm, and the TG content was extrapolated using a standard curve. Data is expressed as μg of TG per mg of liver.
De novo lipogenesis assay
Primary mouse hepatocytes were isolated according to the two-step liver collagenase perfusion method and percoll purified resulting in >97% viable primary mouse hepatocytes21. After isolation primary mouse hepatocytes were plated at a density of 120,000 cells per well in a 12-well plate and attached for 6 hours in complete medium. Next, attached cells were washed with PBS and media was switched to DMEM lipogenesis media (10% dialyzed FBS, 1 g/L glucose, without pyruvate and glutamine, 1% P/S, 0.5 μCi/mL 14C-acetate, 20 µM cold acetate, 200 µM palmitic acid in 1% low endotoxin, fatty acid-free bovine serum albumin). The cells were then incubated in a 5% CO2 incubator for 16 hours at 37°C. Media was aspirated and cells washed twice with cold PBS. Cells were lysed in 0.1 N HCl by scraping. Lipids were extracted by the addition of 750µL of 2:1 chloroform-methanol (v/v), vortexed, and incubated for 5minutes at room temperature. Samples were further diluted by adding 375 µL of water, vortexed and incubated for 5 min at room temperature, followed by centrifugation at 3000 x g at room temperature for 10 minutes. The lower phase was transferred to a new tube from which 100 µL was transferred to a scintillation vial containing 4 mL of liquid scintillation fluid and radioactivity was measured on scintillator. Counts per minute (CPM) were measured for each sample and normalized to protein concentration determined by BCA assay.
Cell culture experiments
Immortalized mouse hepatocytes (IMH)22, were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% FBS (Thermo Fisher Scientific), and 100 µg/mL Normocin (InvivoGen). Cells were maintained at 37°C in a humidified atmosphere with 5% CO₂. Palmitic acid (palmitate, Sigma-Aldrich) was dissolved in isopropyl alcohol (Sigma-Aldrich) to a stock concentration of 80 mM. Prior to treatment, the palmitic acid stock was diluted to a final concentration of 400 μM in serum-free DMEM supplemented with 1% fatty acid–free bovine serum albumin (Sigma-Aldrich). IMH cells were seeded in 6-well plates and grown to ~70–80% confluency. Cells were treated with either 400 μM palmitate or vehicle for 8 or 16 hours in serum-free DMEM containing 1% bovine serum albumin. Following treatment, cells were washed twice with cold PBS and lysed for subsequent assays. Hu1545, a human hepatocyte cell line derived by human Telomerase Reverse Transcriptase (hTERT)-mediated immortalization, was obtained from Dr. Keith Robertson23. S100A11 knockout cells were generated in the human Hu1545 cell line using Guide-it® CRISPR/Cas9 System (Takara Bio) with these single guide sequences 5’—AATTAGACAGAGTTCCTAAGCTTC and 3’—AATTGAAGCTTAGGAACTCTGTCT. Cells were transfected using Lipofectamine 3000 ® and sorted 48 h after on a BD FACS Melody® cytometer (Mayo Clinic Microscopy and Cell Analysis Core) for positive green fluorescent proteins (GFP) into single-cell wells of a 96-well plate. Positive clones were screened for mRNA and protein expression. Hu1545 wild type (WT) and S100A11 knockout (S100A11⁻/⁻) human hepatocyte cell lines were cultured in DMEM supplemented with 10% FBS, 1% normocin, and maintained at 37 °C in a humidified atmosphere with 5% CO2. Prior to treatment, cells were seeded to reach ~70–80% confluence. Cells were treated with either vehicle or 400 µM palmitate for 8 hours under two glucose concentrations: standard glucose DMEM (1 g/L, Cat # 11885-084) or high glucose DMEM (4.5 g/L, Cat # 11965-092) using commercially available cell culture media (ThermoFischer). Sodium pyruvate was added to the high-glucose DMEM to match the concentration in standard glucose DMEM. Benitrobenrazide (BNBZ, MedChemExpress), a selective pharmacological inhibitor of HK2 was obtained from a commercial manufacturer and dissolved in DMSO24. For experiments with BNBZ, cells were treated with 10 µM or 30 µM BNBZ with and without 400 µM palmitate. Prior to treating, palmitate was conjugated to bovine serum albumin at a 3:1 molar ratio and prepared freshly before use. The vehicle controls contained identical amounts of the respective vehicles (isopropanol alone or isopropanol and DMSO). Following treatment, cells were washed twice with cold PBS and lysed for assays. Bone Marrow Derived Macrophages (BMDM) were isolated from femur bones of wildtype and S100a11−/− mice as previously described by us13. Briefly, isolated and dissociated marrow was plated on 150 mm petri dishes (BD Falcon) with differentiation media changed every other day until day seven. BMDMs were removed from the plate with Accutase ® then counted and plated at two million cells per 60 mm tissue culture dish (BD Falcon) and allowed to attach for four hours. Following 2 h of culture in serum-free RPMI media, BMDM were treated with vehicle or 400µM palmitate in RPMI media containing 2.5% low-endotoxin FBS (Gemini Bio) for 8hours. BMDM were lysed in TRIzol (Invitrogen) and processed for downstream analyses. LX2 cells were cultured in DMEM (Invitrogen) with 10% FBS and anti-biotic anti-mycotic (Invitrogen). Cells were plated at 500,000 cells per well in a 6-well tissue culture treated dish (Falcon) and allowed to attach overnight. LX2 cells were treated with vehicle or 400 µM palmitate for 20 hours in DMEM media containing 2.5% FBS, 1% bovine serum albumin and later lysed in TRIzol for downstream analyses.
RNA sequencing and signaling pathway analyses
RNA was isolated as above. Total RNA concentration and quality were determined using Qubit fluorometry (ThermoFisher Scientific) and the Agilent Fragment Analyzer. The total RNA underwent further clean-up with a DNase I (Zymo Research) treatment and RNAClean XP beads (Beckman Coulter). Using the Illumina Stranded Total RNA Prep with Ribo-Zero Plus, libraries were prepared according to the manufacturer’s instructions with 100 ng of cleaned total RNA. The concentration and size distribution of the completed libraries were determined with the Agilent TapeStation’s D1000 ScreenTape and Qubit fluorometry. Libraries were sequenced at greater than 100 million read pairs per sample following Illumina’s standard protocol for the NovaSeq 6000. The NovaSeq SP flow cell was sequenced as 100 × 2 paired-end reads using the NovaSeq SP Reagent Kit v1.5 and NovaSeq Control Software v1.8.0. Base-calling was performed using Illumina’s RTA version 3.4.4. For secondary analysis, the RNAseq data were mapped to the reference genome mm10 using MAP-Rseq v.3.1.4 on RHEL8. We identified 13,379 protein-coding genes, and with a cutoff of ± 0.5 log2 fold change and adjusted p-value < 0.01, identified 2245 differentially expressed genes (DEGs). DEGs were analyzed with Ingenuity pathway analysis to uncover common regulatory pathways.
Statistical and data analyses
Data are presented as mean ± SEM in each graph. Each dot in the dot plots represents one mouse or biological replicate unless indicated otherwise. Data was assumed to fit within a normal distribution. For statistical comparison of two experimental conditions, two-tailed Student’s t test was performed in GraphPad Prism 9.5.1 for Windows (GraphPad Software). We used Ordinary one-way Analysis of Variance (ANOVA) to analyze multiple experimental groups, with Bonferroni or Šídák corrections.
For animal studies, no batch or cage effects were observed based on randomization, and all animals were housed under identical conditions. For cell culture, experimental and control samples were obtained from the same passage and treated with the same reagents. Therefore, no additional covariates were tested.
Data availability
The RNA-seq datasets generated during the current study have been deposited in the gene expression ombinus (GEO) database (GSE 304356). All other data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Younossi, Z. M. et al. The global epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among patients with type 2 diabetes. Clin. Gastroenterol. Hepatol. 22, 1999–2010.e1998 (2024).
Parthasarathy, G., Revelo, X. & Malhi, H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol. Commun. 4, 478–492 (2020).
Venkatesan, N., Doskey, L. C. & Malhi, H. The role of endoplasmic reticulum in lipotoxicity during metabolic dysfunction-associated steatotic liver disease (MASLD) pathogenesis. Am. J. Pathol. 193, 1887–1899 (2023).
Lefere, S. & Tacke, F. Macrophages in obesity and non-alcoholic fatty liver disease: crosstalk with metabolism. JHEP Rep. 1, 30–43 (2019).
Zhou, J. E. et al. Hepatic macrophage targeted siRNA lipid nanoparticles treat non-alcoholic steatohepatitis. J. Control Release 343, 175–186 (2022).
Garcia-Martinez, I. et al. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J. Clin. Invest. 126, 859–864 (2016).
Nakatsu, Y. et al. The xanthine oxidase inhibitor febuxostat suppresses development of nonalcoholic steatohepatitis in a rodent model. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G42–G51 (2015).
Feldstein, A. E. et al. Cytokeratin-18 fragment levels as noninvasive biomarkers for nonalcoholic steatohepatitis: a multicenter validation study. Hepatology 50, 1072–1078 (2009).
Nie, Z., Xiao, C., Wang, Y., Li, R. & Zhao, F. Heat shock proteins (HSPs) in non-alcoholic fatty liver disease (NAFLD): from molecular mechanisms to therapeutic avenues. Biomark. Res. 12, 120 (2024).
Sobolewski, C. et al. S100A11/ANXA2 belongs to a tumour suppressor/oncogene network deregulated early with steatosis and involved in inflammation and hepatocellular carcinoma development. Gut 69, 1841–1854 (2020).
Zhang, L. et al. S100A11 Promotes liver steatosis via FOXO1-mediated autophagy and lipogenesis. Cell Mol. Gastroenterol. Hepatol. 11, 697–724 (2021).
Teng, F. et al. The S100 calcium-binding protein A11 promotes hepatic steatosis through RAGE-mediated AKT-mTOR signaling. Metabolism 117, 154725 (2021).
Parthasarathy, G. et al. Macrophage RAGE activation is proinflammatory in NASH. JCI Insight 9, e169138 (2024).
Nakao, Y. et al. A comparative proteomic analysis of extracellular vesicles associated with lipotoxicity. Front. Cell Dev. Biol. 9, 735001 (2021).
Krishnan, A. et al. A longitudinal study of whole body, tissue, and cellular physiology in a mouse model of fibrosing NASH with high fidelity to the human condition. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G666–g680 (2017).
de Moura e Dias, M. et al. Diet-induced obesity in animal models: points to consider and influence on metabolic markers. Diabetol. Metab. Syndr. 13, 32 (2021).
Brunt, E. M., Kleiner, D. E., Wilson, L. A., Belt, P. & Neuschwander-Tetri, B. A. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology 53, 810–820 (2011).
Guo, D., Meng, Y., Jiang, X. & Lu, Z. Hexokinases in cancer and other pathologies. Cell Insight 2, 100077 (2023).
Niu, L. et al. Noninvasive proteomic biomarkers for alcohol-related liver disease. Nat. Med. 28, 1277–1287 (2022).
Mauer, A. S., Hirsova, P., Maiers, J. L., Shah, V. H. & Malhi, H. Inhibition of sphingosine 1-phosphate signaling ameliorates murine nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G300–g313 (2017).
Vilà, L. et al. AAV8-mediated Sirt1 gene transfer to the liver prevents high carbohydrate diet-induced nonalcoholic fatty liver disease. Mol. Ther. Methods Clin. Dev. 1, 14039 (2014).
Safronova, A. et al. Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat. Immunol. 20, 64–72 (2019).
Linden, A. G. et al. Interplay between ChREBP and SREBP-1c coordinates postprandial glycolysis and lipogenesis in livers of mice. J. Lipid Res. 59, 475–487 (2018).
Daniel, P. V. et al. NF-κB p65 regulates hepatic lipogenesis by promoting nuclear entry of ChREBP in response to a high carbohydrate diet. J. Biol. Chem. 296, 100714 (2021).
Benichou, E. et al. The transcription factor ChREBP Orchestrates liver carcinogenesis by coordinating the PI3K/AKT signaling and cancer metabolism. Nat. Commun. 15, 1879 (2024).
Dogra, S. et al. Zinc oxide nanoparticles attenuate hepatic steatosis development in high-fat-diet fed mice through activated AMPK signaling axis. Nanomedicine 17, 210–222 (2019).
Rome, S. et al. Microarray analyses of SREBP-1a and SREBP-1c target genes identify new regulatory pathways in muscle. Physiol. Genomics 34, 327–337 (2008).
Dogra, S. et al. Liver-derived S100A6 propels β-cell dysfunction in NAFLD. Diabetes 71, 2284–2296 (2022).
Zeng, X. et al. S100A11 activates the pentose phosphate pathway to induce malignant biological behaviour of pancreatic ductal adenocarcinoma. Cell Death Dis. 13, 568 (2022).
Shepherd, E. L. et al. Ketohexokinase inhibition improves NASH by reducing fructose-induced steatosis and fibrogenesis. JHEP Rep. 3, 100217 (2021).
Kazierad, D. J. et al. Inhibition of ketohexokinase in adults with NAFLD reduces liver fat and inflammatory markers: a randomized phase 2 trial. Med 2, 800–813.e803 (2021).
Saxena, A. R. et al. A phase 2a, randomized, double-blind, placebo-controlled, three-arm, parallel-group study to assess the efficacy, safety, tolerability and pharmacodynamics of PF-06835919 in patients with non-alcoholic fatty liver disease and type 2 diabetes. Diab. Obes. Metab. 25, 992–1001 (2023).
Acknowledgements
This work is supported by NIH grant DK111378 (to H.M.), the Mayo Foundation (to H.M.), the Optical Microscopy Core and the Clinical Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567), and the Histology Core of the Mayo Clinic. P.V.D. gratefully acknowledges the American Liver Foundation Postdoctoral Research Fellowship Award 2024. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. The authors are grateful to Ms. Courtney Hoover for outstanding administrative assistance. The IMH cells were kindly provided by Dr. Randal Kaufman.
Author information
Authors and Affiliations
Contributions
H.M. conceived the project. H.M., G.P., and P.V.D. designed the experiments. G.P., P.V.D., Y.L., N.G., T.N., H.L.E., G.M., A.S.M., and D.P. conducted the experiments and interpreted the results. N.K.N. performed bioinformatics analysis of RNA-seq. H.M. provided all the reagents. G.P., P.V.D., Y.L., and H.M. wrote the manuscript. All authors contributed to manuscript editing and provided a critical review and approval of the manuscript. We affirm that authorship was offered only to individuals who made significant intellectual contributions, and no one who met the criteria for authorship was omitted. All contributors who do not meet authorship criteria are acknowledged appropriately. This research reflects a collaborative effort that values integrity, transparency, and inclusion. Contributions were evaluated solely on the basis of merit and relevance.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Daniel, P.V., Puri, G., Luo, Y. et al. Silencing of S100A11 attenuates murine metabolic dysfunction-associated steatohepatitis. npj Gut Liver 2, 32 (2025). https://doi.org/10.1038/s44355-025-00044-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44355-025-00044-w