Abstract
The mechanisms responsible for prostate cancer metastasis are incompletely understood at both the cellular and molecular levels. In this regard, chemokines are a family of small, cytokine-like proteins that induce motility of neoplastic cells, leukocytes and cancer cells. The current study evaluates the molecular mechanisms of CXCL12 and CXCR4 in prostate cancer cell migration and invasion. We report that functional CXCR4 is significantly expressed by prostate cancer cell lines, LNCaP and PC3, when compared with normal prostatic epithelial cells (PrEC). As measured using motility and invasion chamber assays, prostate cancer cells migrated and invaded through extracellular matrix components in response to CXCL12, at rates that corresponded to CXCR4 expression. Anti-CXCR4 antibodies (Abs) significantly impaired the migration and invasive potential of PC3 and LNCaP cells. CXCL12 induction also enhanced collagenase-1 (metalloproteinase-1 (MMP-1)) expression by LNCaP and PC3 cells. Collagenase-3 (MMP-13) was expressed by prostate cancer cells, but it was not expressed by PrEC cells or modulated by CXCL12. CXCL12 increased MMP-2 expression by LNCaP and PC3; however, MMP-9 expression was elevated only in PC3 cells after CXCL12–CXCR4 ligation. PC3 cells also expressed high levels of stromelysin-1 (MMP-3) after CXCL12 stimulation. CXCL12 also significantly increased stromelysin-2 (MMP-10) expression by LNCaP cells. Stromelysin-3 (MMP-11) was expressed by LNCaP cells, but not by PC3 or PrEC cells and CXCL12 induced PC3 MMP-11 expression. Membrane type-1 MMP (MMP-14) was not expressed by PrEC or LNCaP cells, but CXCL12 significantly enhanced MMP-14 expression by PC3 cells. These studies reveal important cellular and molecular mechanisms of CXCR4/CXCL12-mediated prostate cancer cell migration and invasion.
Similar content being viewed by others
Main
Despite the obvious importance of metastasis, this process remains incompletely understood at both the cellular and molecular levels.1 Many factors have been implicated in the process of metastasis, but the precise mechanisms for the directional migration of malignant cells into different organs is unknown.2, 3, 4 In this regard, chemokines are a super family of small, cytokine-like proteins that induce—through G-protein-coupled receptor and cytoskeletal rearrangement—the adhesion of neoplastic cells or leukocytes to endothelial cells as well as the directional migration of cancer cells.5, 6
The ability of neoplastic cells to penetrate the basement membrane and then invade the interstitial stroma to initiate the metastatic process is largely mediated by proteolysis. Many proteinases are capable of degrading extracellular matrix (ECM) components, but matrix metalloproteinases (MMPs) appear to be particularly important for matrix degradation.6 These proteolytic enzymes are involved in connective tissue remodeling as well as in embryonic growth, ovulation, wound healing and menstruation.7, 8 Moreover, abnormal production of these proteinases is implicated in a number of pathological conditions.9
It has been shown that CXCL12–CXCR4 interactions may play a role in the metastasis of prostate cancer to bone.10 However, these interactions alone do not explain the metastasis pattern of prostate cancer or the potential for neoplastic prostate cells to migrate and invade other tissues. We have tested the hypothesis that prostate carcinomas use CXCR4-dependent mechanisms for migration, invasion and MMP expression. To test this hypothesis, we have elucidated the levels of CXCR4 mRNA and surface protein expression as well as CXCL12-CXCR4-induced migration, matrix invasion, and MMP expression by normal PrEC and prostate cancer cell lines. These studies suggest that the expression of CXCR4 and its interaction with CXCL12 may aid in facilitating the migration, invasion and MMP expression by prostate tumor cells.
Materials and methods
Cell Lines and Cell Culture
PC3 and LNCaP cell lines were obtained from the ATCC (Manassas, VA, USA). Normal prostate epithelial cells (PrEC) were obtained from Clonetics-Biowhittaker (Walkersville, MD, USA) and cultured in PrEMB medium (Clonetics-Biowhittaker). PC3 cells were cultured at 37°C with 5% CO2 in Ham's F12 K medium with 2 mM L-glutamine and adjusted to contain 1.5 g/l sodium bicarbonate (ATCC) with 10% fetal bovine serum (FBS) (Sigma, St Louis, MO, USA). After five passages in Ham's F12 K media, PC3 cells were switched to RPMI-1640 at 37°C and 5% CO2 with 10% FBS. LNCaP cells were cultured in RPMI-1640 with 10% FBS. Before migration and invasion studies as well as CXCR4 and MMP expression analyses were conducted, the cells were cultured for 24 h in RPMI-1640 supplemented with 2% charcoal-striped FBS. Similarly, cells were cultured for 6, 12, 18, 24, and 48 h in 2% charcoal-striped FBS RPMI-1640 during migration and invasion as well as mRNA and protein expression studies. Optimal mRNA and protein expression was measured after 12 and 24 h of culture, respectively.
RNA Isolation and Gene Expression Analysis
Human mRNA sequences for CXCR4, MMP-1, MMP-3, MMP-2, MMP-9, MMP-10, MMP-11, MMP-13, MT1-MMP (MMP-14) and 18S rRNA were obtained from NIH-NCBI gene bank database Accession numbers NM003467, NM002421, NM002422, NM004530, NM004994, NM002425, NM005940, NM002427, NM004995 and X00686.1, respectively. These sequences were then used to design primers for RT-PCR analysis, which generated amplicons 188, 83, 155, 95, 79, 94, 107, 176, 172 and 149 base pairs in size for CXCR4, MMP-1, MMP-3, MMP-2, MMP-9, MMP-10, MMP-11, MMP-13, MMP-14 and 18S rRNA, respectively. Primers were designed using the Primer 3 software program from Whitehead Institute at the Massachusetts Institute of Technology (Boston, MA, USA). Thermodynamic analysis of the primers was conducted using the following computer programs: Primer Premier™ (Integrated DNA Technologies, Coralville, IA, USA) and MIT Primer III (Boston, MA, USA). The resulting primer sets were also compared against the entire human genome to confirm specificity and ensure that the primers flanked mRNA splicing regions.
Total RNA was isolated from untreated and CXCL12-treated prostate cancer and normal prostatic epithelial cells using Tri-reagent™ (Molecular Research Center, Cincinnati, OH, USA), according to the manufacturer's protocols. Potential genomic DNA contamination was removed from these samples by treatment for 15 min at 37°C with RNase-free DNase (Invitrogen, San Diego, CA). RNA was precipitated and resuspended in RNA Secure™ (Ambion, Austin, TX, USA). cDNA was generated by reverse transcribing 1.5 μg of total RNA using Taqman™ reverse transcription reagent (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's protocols, and amplified with specific cDNA primers, using SYBR® Green PCR master mix reagents (Applied Biosystems). The levels of copies (>10) of mRNA relative to 18S rRNA copies of these targets were evaluated by RT-PCR analysis using the BioRad Icycler and software (Hercules, CA, USA).
Flow Cytometry Analysis of CXCR4 Surface Expression
PE-conjugated mouse anti-human CXCR4 (clone 112509) antibody was purchased from R&D Systems (Minneapolis, MN, USA). PE-conjugated mouse IgG2a monoclonal immunoglobulin isotype control was purchased from Pharmingen (San Diego, CA, USA). Prostate cancer cells and normal prostate epithelial cells were washed 3 times in PBS (supplemented with 0.5% BSA) and treated with 1.0 μg of Fc Block™ (Pharmingen) per 105 cell for 15 min at room temperature. Fc-blocked prostate cancer and normal epithelial cells were stained with 1.0 μg of PE-conjugated mouse anti-human CXCR4 or PE-conjugated mouse IgG2a isotype control antibodies per 105 cells at 4°C for 40 min. Subsequently, the cells were washed with 1.0 ml FACS buffer (1% BSA in PBS) to remove unbound antibodies. Labeled cells were fixed in 500 μl of 2% paraformaldehyde solution, and 105 cells were analyzed by flow cytometry using a FACScan flow cytometer and CellQuest™ software (BD-PharMingen).
Migration and Invasion Assays
CXCL12 was obtained from PeproTech (Rocky Hill, NJ, USA). Unlabeled mouse anti-human CXCR4 antibody and control antibody (IgG2a) was purchased from R&D Systems. Migration and invasion studies were performed using BD BioCoat™ migration or Matrigel™ invasion chambers, respectively (Becton Dickinson Labware, Bedford, MA, USA). RPMI-1640 culture media supplemented with 2% charcoal-striped FBS was added to the interior of the bottom chamber as well as to the top chamber of the inserts and allowed to hydrate for 2 h at 37°C with 5% CO2. CXCL12 or albumin (negative control) at 100 ng/ml was added to the bottom chamber. Next, 104 cells were added to the top chamber of the inserts. To block CXCL12–CXCR4 interactions, 1.0 μg/ml of mouse anti-human CXCR4 (clone 44708.111) monoclonal antibody or control antibody (R&D Systems) was added to the top chamber of Matrigel™ or control inserts and incubated overnight at 37°C and 5% CO2. After incubation, the cells on the bottom surface of the inserts were fixed with 100% methanol for 2 min, stained for 2 min in 1% toluidine blue (Sigma, St Louis, MO, USA) supplemented with 1% borax (Sigma), and then rinsed twice with deionized water (dH2O). Cells were counted by microscopic examination using a × 40 magnification.
Active MMP Protein Detection
Briefly, 105 prostate cancer (LNCaP and PC3) and PrEC cells were seeded in 24-well plates and conditioned media were collected from CXCL12 (100 ng/ml) treated or untreated cells. Active collagenases (MMP-1 and MMP-13), gelatinase A and B (MMP-2 and MMP-9) and stromelysin 2 (MMP-10) levels were measured by ELISA (R&D systems, Minneapolis, MN, USA), according to the manufacturer's protocols. Active MMP-14 was measured by ELISA according to the manufacturer's protocol (Amersham Bioscience Corp, Piscateaway, NJ, USA). Similarly, an ELISA method for active stromelysin-3 (MMP-11) detection was developed to quantify active MMP-11 expression in conditioned media. Briefly, 96-well Falcon ELISA plates (Fisher Scientific, Pittsburg, PA, USA) were coated with 100 μl of 5 μg/ml rabbit anti-human (hinge region) MMP-11 antibody (Triple Point Biologicals, Forest Grove, OR, USA) overnight at 4°C and blocked with 200 μl of 2% BSA (Sigma) in PBS for 2 h at room temperature. Serial dilutions of experimental samples and purified human MMP-11 (Triple Point Biologicals), as standard, were added and incubated overnight at 4°C. Plates were washed 4 × with PBS containing 0.05% Tween-20 (PBS-T). Mouse anti-human (active domain) MMP-11 antibody (Calbiochem, La Jolla, CA, USA) diluted in PBS (1:3000) was added in each well and incubated at room temperature for 2 h. After washing 4 × with PBS-T, 100 μl goat anti-mouse IgG-horse raddish peroxidase (HRP, 1:3000, Southern Biotechnology Associates, Birmingham, AL, USA) was added and incubated at room temperature for 1 h. After three washes, tetramethylbenzidine (TMB) substrate (eBioscience, San Diego, CA, USA) was added, allowed to react, and stopped with 50 μl of 2% H2SO4, and optical density was read at 450 nm.
Statistics
The RT-PCR and ELISA data are expressed as the mean±s.e.m. and compared using a two-tailed Student's t-test or an unpaired Mann–Whitney U-test. The results were analyzed using the Statview II statistical program (Abacus Concepts, Inc., Berkeley, CA, USA) for Macintosh computers and were considered statistically significant if P-values were <0.05. When MMP levels were below the detectable limit of the ELISA, values were recorded as one-half of the lower detection limit (eg, 50 pg/ml for MMP-13) for statistical analysis. Kolmogorov–Smirnov (K–S) two-sample test using CELLQuest Software (BD-Pharmingen) for Macintosh computers was used to compute the statistical significance between PrEC and LNCaP or PC3 cell CXCR4 histograms.
Results
CXCR4 mRNA Expression by Prostate Cells
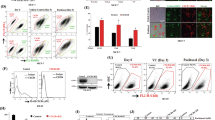
Normal prostate epithelial cells (PrEC) and prostate cancer cell lines, previously isolated from metastases to bone (PC3) or lymph nodes (LNCaP), expressed CXCR4. However, significantly higher levels of CXCR4 were observed in the malignant cell lines than in normal prostatic epithelial cells (Figure 1). LNCaP cells expressed significantly more copies of CXCR4 transcripts than did PC3 cells. Protein expression of CXCR4 by LNCaP, PC3 and PrEC cells was confirmed by flow cytometry (Figure 2). In confirmation with mRNA levels, the surface protein expression of CXCR4 was significantly higher in the cancer cell lines than normal prostatic epithelial cells. LNCaP cells also expressed significantly higher levels of cell surface CXCR4, when compared to PC3.
CXCR4 mRNA expressed by prostate cancer and normal prostatic epithelial cells. Total RNA was isolated from prostate cancer cell lines LNCaP and PC3 as well as from normal prostatic epithelial (PrEC) cells. Quantitative RT-PCR analysis of CXCR4 mRNA expression was performed in triplicate. The copies of transcripts are expressed relative to actual copies of 18S rRNA±s.e.m. Asterisk(s) indicate statistical significant differences (P<0.05) between normal and malignant cells ( ).
).
CXCR4 cell surface expression by prostate cells. LNCaP, PC3 and PrEC cells were stained with PE-conjugated isotype control or mouse anti-human CXCR4 monoclonal antibodies and quantified (in triplicate) by flow cytometry. Histograms of CXCR4 expression are shown along with the mean fluorescence intensities of stained LNCaP, PC3 and PrEC cells and significant differences between CXCR4 expression by PrEC and cancer cells. The experiments were repeated three times.
Migration and Invasion
CXCR4 expressed by the prostatic epithelial cells tested was also functional, as indicated by the ability of the LNCaP and PC3 cell lines, but not the PrEC cells to migrate toward chemotactic gradients of CXCL12 (Figure 3). Both LNCaP and PC3 cells migrated toward CXCL12. The number of LNCaP and PC3 cells that migrated in response to CXCL12 was significantly higher than for cells not exposed to CXCL12 as a chemoattractant. This CXCL12-dependent chemotaxis was neutralized by treatment with anti-CXCR4 antibody, but not by control antibody. As with migration, PrEC cells were not invasive under any conditions (Figure 4). After CXCL12 ligation, the invasive potential of PC3 and LNCaP cells significantly increased when compared with the same cells left untreated. Neutralization of CXCL12–CXCR4 interactions by anti-CXCR4 antibody treatment significantly diminished the invasiveness of LNCaP and PC3 cells but not by control antibody.
CXCR4-mediated migration of prostate cancer and normal prostatic epithelial cells. PrEC, PC3 and LNCaP cells were tested for their ability to migrate toward chemotactic gradients of 0 (□) or 100 ng/ml of CXCL12 (▧). Similarly, cells were cocultured with 1 μg/ml of monoclonal antibodies against CXCR4 (▪) or IgG2a control antibody (▨) during migration assays with CXCL12. The number of cells (±s.e.m.) that migrated to CXCL12 out of the initial 104 cells used to seed the migration chamber are shown, with asterisks ( ) indicating significant differences (P<0.05) between untreated (□) and CXCL12-induced cells (▧, ▨, ▪), while migration indexes below 1 per 104 cells are designated below detection (BD).
) indicating significant differences (P<0.05) between untreated (□) and CXCL12-induced cells (▧, ▨, ▪), while migration indexes below 1 per 104 cells are designated below detection (BD).
CXCR4-mediated invasion of prostate cancer and normal prostatic epithelial cells. PrEC, LNCaP and PC3 cells were tested for their ability to invade or translocate across a Matrigel™ matrix in response to chemotactic gradients of 0 (□) or 100 ng/ml of CXCL12 (▧). Cells were also cocultured with 1 μg/ml of monoclonal antibodies against CXCR4 (▪) or IgG2a control antibody (▨) during invasion assays using CXCL12. Asterisks ( ) indicate significant differences (P<0.05) between untreated (□) and CXCL12-induced cells (▧, ▨, ▪), while invasion indexes below 1/104 cells are designated below detection (BD).
) indicate significant differences (P<0.05) between untreated (□) and CXCL12-induced cells (▧, ▨, ▪), while invasion indexes below 1/104 cells are designated below detection (BD).
CXCL12-Induced MMP Expression by Prostate Cancer and Normal Prostatic Epithelial Cells
The Matrigel™ system used in this study for the invasion assays consisted of laminin, collagen IV and entactin; hence, MMP-1, MMP-3, MMP-8, MMP-10, MMP-11 and/or MMP-13 may be required to efficiently digest these components. To determine if the associated increase in invasion after CXCL12–CXCR4 interaction was due to heightened MMP expression, collagenases (MMP-1 and -13), gelatinase (MMP-2 and -9), stromelysins (MMP-3, -10 and -11) and membrane type-1 MMP (MMP-14) mRNA expression were assessed by RT-qPCR and by active protein ELISA. Both untreated normal prostatic epithelial cells (PrEC) and prostate cancer cell lines (LNCaP and PC3) expressed collagenase-1 (MMP-1) mRNA and active protein. Interestingly, without CXCL12 stimulation, PC3 cells expressed significantly higher MMP-1 mRNA and active protein than did LNCaP and PrEC cells (Figure 5). CXCL12 induction resulted in higher MMP-1 mRNA and active protein expression by prostate cancer and normal prostatic epithelial cells. LNCaP and PC3 cells expressed significantly higher MMP-1 mRNA and active protein than did PrEC cells; however, these normal prostatic epithelial cells also expressed more MMP-1 mRNA and active protein after CXCL12 induction. Untreated LNCaP and PC3 cells expressed MMP-13 mRNA and active protein at significantly higher levels than did PrEC cells treated in a similar fashion; moreover, this expression was significantly higher in PC3 cells than in LNCaP cells. Remarkably, MMP-13 expression by cancer cells (LNCaP and PC3) was downregulated after CXCL12 ligation. Taken together, these data suggest that PC3 cells are predisposed to collagenase-1 (MMP-1) and collagenase-3 (MMP-13) production without CXCL12 induction, while CXCL12 ligation upregulates MMP-1 expression and downregulates MMP-13 expression by cancerous cells.
CXCL12-induced collagenase expression by normal prostatic epithelial and prostate carcinoma cells. LNCaP (▪), PC3 (▨) and PrEC (□) cells were tested for their ability to express collagenases (MMP-1 and MMP-13) mRNA and active protein. PrEC, LNCaP and PC3 cells were cultured with and without CXCL12 (100 ng/ml). RNA was isolated, and quantitative RT-PCR analysis was performed (in triplicate) for mRNA expression of collagenases (upper panel) and transcript copies are expressed relative to actual copies of 18S rRNA. Active collagenases were quantified by protein ELISA in conditioned medium (lower panel). MMP expression below the detectable limit of the RT-PCR or ELISA assay are designated as below detection (BD). Asterisk(s) indicate statistically significant differences (P<0.05) between PrEC and prostate cancer cells ( ), between untreated and treated cells (
), between untreated and treated cells ( ), or between similarly treated prostate cancer cells (★) that produced the highest levels of collagenase.
), or between similarly treated prostate cancer cells (★) that produced the highest levels of collagenase.
Gelatinases (MMP-2 and -9) were equally expressed by untreated prostatic epithelial cells and prostate cancer cell lines (Figure 6). However, CXCL12 treatment resulted in a significant increase in MMP-2 mRNA and active protein expression by LNCaP and PC3 cells but not by PrEC cells. While the untreated prostate cancer cell lines tested expressed significantly higher levels of MMP-9 mRNA and active protein than did PrEC cells, CXCL12 ligation resulted in higher MMP-9 expression by PC3 cells but not by LNCaP or PrEC cells. These results indicate that MMP-2 and -9 are differentially expressed by untreated LNCaP, PC3 and PrEC cells, while CXCL12–CXCR4 engagement induces higher mRNA and active protein levels of MMP-2 by LNCaP and PC3 cells and induced higher levels of MMP-9 mRNA and active protein by PC3 cells.
CXCL12-induced gelatinase expression by normal prostatic epithelial and prostate cancer cells. LNCaP (▪), PC3 (▨) and PrEC (□) cells were tested for their ability to express gelatinases (MMP-2 and -9). PrEC, LNCaP and PC3 cells were cultured with and without 100 ng/ml of CXCL12. Quantitative RT-PCR analysis was used to measure mRNA expression of gelatinases (upper panel) and transcript copies are expressed relative to actual copies of 18S rRNA. Active gelatinases were quantified by protein ELISA in conditioned medium (lower panel). MMP expression below the detectable limit of the RT-PCR or ELISA assays are designated as below detection (BD). Asterisk(s) indicate statistically significant differences (P<0.05) between PrEC and prostate cancer cells ( ), between untreated and treated cells (
), between untreated and treated cells ( ), or between similarly treated prostate cancer cells (★) that produced the highest levels of MMP-2 or -9.
), or between similarly treated prostate cancer cells (★) that produced the highest levels of MMP-2 or -9.
Neither of the untreated cells tested expressed substantial levels of MMP-3 (Figure 7). However, following CXCL12 induction, PC3 cells expressed significantly higher levels of MMP-3 mRNA and active protein than did the other cells tested. Untreated PC3 cells also expressed significantly higher levels of MMP-10 mRNA and active protein than did untreated LNCaP and PrEC cells, while CXCL12 induction significantly increased MMP-10 expression by LNCaP cells but not by PrEC or PC3 cells. MMP-11 mRNA and active protein was consistently expressed by untreated and CXCL12-treated LNCaP cells, but its expression by PC3 cells was dramatically increased only after CXCL12 treatment. These data suggest that CXCL12 treatment induced MMP-3 expression by PC3 cells, but not by LNCaP cells. MT-1 MMP (MMP-14) was not largely expressed by untreated PrEC, LNCaP or PC3 cells (Figure 8). However, MMP-14 expression by PC3 cells was significantly increased after CXCL12 treatment.
CXCL12-induced stromelysin expression by prostate cancer cells. PrEC (□), LNCaP (▪) and PC3 (▨) cells were tested for their ability to express stromelysin-1 (MMP-3), -2 (MMP-10) and -3 (MMP-11). PrEC, LNCaP and PC3 cells were cultured with and without 100 ng/ml of CXCL12. Quantitative RT-PCR analysis was used to measure mRNA expression of stromelysins (upper panel) and copies of transcripts are expressed relative to actual copies of 18S rRNA. Active MMP was quantified by protein ELISA of culture supernatants (lower panel). MMP expression below the detectable limit of the RT-PCR or ELISA assay are designated as below detection (BD). Asterisk(s) indicate statistically significant differences (P<0.05) between PrEC and prostate cancer cells ( ), between untreated and treated cells (
), between untreated and treated cells ( ), or between similarly treated prostate cancer cells (★) that produced the highest levels of MMP-3, -10 or -11.
), or between similarly treated prostate cancer cells (★) that produced the highest levels of MMP-3, -10 or -11.
CXCL12-induced membrane type MMP expression by prostate cancer cells. PrEC (□), LNCaP (▪) and PC3 (▨) cells were cultured with and without 100 ng/ml of CXCL12. Quantitative RT-PCR analysis was used to measure mRNA expression of MMP-14 (upper panel) and copies of transcripts are expressed relative to actual copies of 18S rRNA. Active MMP was quantified by protein ELISA of culture supernatants (lower panel). MMP expression below the detectable limit of the RT-PCR or ELISA assays are designated as below detection (BD). Asterisk(s) indicate statistically significant differences (P<0.05) between PrEC and prostate cancer cells ( ), between untreated and treated cells (
), between untreated and treated cells ( ), or between similarly treated prostate cancer cells (★) that produced the highest levels of MMP-14.
), or between similarly treated prostate cancer cells (★) that produced the highest levels of MMP-14.
Discussion
Prostate cancer is a common neoplasm and the second leading cause of cancer-related deaths in American males.11 Most of these deaths are not the result of the primary tumor growth, but are rather the result of the spread of cancer to distant organs, including bone.12 In spite of the clinical importance of metastasis, much remains to be learned about the metastatic process. Many factors have been identified that may contribute to metastasis.13 In this regard, chemokines are involved in the chemotaxis of lymphocytes and may cause cancer cells to ‘home’ to specific secondary sites to promote organ-specific metastasis.
CXCL12–CXCR4 interactions have been implicated in the pathogenesis and progression of breast cancer, pancreatic cancer, Burkett's lymphoma, non-Hodgkin's B-cell lymphoma, several types of leukemias and neuroblastomas.14, 15, 16, 17, 18, 19, 20, 21, 22 CXCR4 expression by cancer cells may also correlate with disease severity. For instance, Muller et al showed that normal breast tissues express low amounts of CXCR4, whereas breast neoplastic tissues express high levels of CXCR4. Moreover, it has been shown that CXCL12 ligation can also mediate cancer cell adhesion to endothelial cells.10
We hypothesized that directional migration and invasion of prostate cancer cells are mediated by CXCL12–CXCR4 interactions that also modulate MMP expression. To address this hypothesis, we first identified the expression of chemokine receptors by normal prostatic epithelial cells and prostate cancer cell lines, which were isolated originally from different metastatic sites (ie, lymph nodes and bone). To our knowledge, for the first time, in vitro migration and invasion studies were used to test the biological and functional significance of CXCR4 expression by prostate cancer cells. In the present study, we have shown that PrEC cells express low levels of CXCR4 mRNA and surface protein. In confirmation of this finding, Sun et al23 demonstrated that CXCR4 is expressed at higher levels by prostate cancer cells (LNCaP and PC3) as well as by prostate cancer tissue. We showed that the prostate cancer cells tested displayed significantly higher CXCR4 transcript and surface protein than did normal prostatic epithelial cells.
The extracellular matrix presents a formidable barrier to cell invasion; hence, CXCL12-mediated chemotaxis alone does not explain the metastasis of prostate cancer. MMPs have been widely studied due to their role in tumor metastasis.24, 25 Indeed, MMP-1 expression has been correlated with high mortality rates in colorectal and esophageal cancers.26, 27 Bladder tumors that invade muscle display significantly higher collagenase activity than do their noninvasive counterparts.28 Higher urinary MMP-1 expression has also been correlated with disease progression and poor survival in bladder cancer cases.29
Collagenases (MMP-1 and -13) degrade several native fibril collagens, including types I, II and III, resulting in their cleavage. In this study, we demonstrate for the first time that the expression of MMP-1 by LNCaP and PC3 cells is induced by CXCL12–CXCR4 interactions. In studies using in vitro models of rheumatoid arthritis and osteoarthritis showed that CXCL12-stimulated human chondrocytes produce MMP-1.30 The present study also shows that the CXCL12–CXCR4 interaction has an inhibitory effect on MMP-13 expression. Hence, the ability of CXCL12 to modulate MMP-1 expression may aid in migration and selective cancer dissemination.
Gelatinases (MMP-2 and -9) have been found to be associated with prostate cancer metastasis. For example, Nemeth et al31 have shown higher expression of gelatinases by prostate cancer cells when growing in bone; interestingly, CXCL12 is largely expressed by bone marrow.32 High MMP-2 and -9 levels in plasma also correlate with prostate cancer metastasis.33, 34, 35 MMP-9 obviously contributes to metastasis, since inhibition of gelatinase reduces the migration and invasion of prostate cancer cells in mice.36
CXCL12-stimulated bone marrow and peripheral blood CD34+ cells secrete MMP-2 and MMP-9.37 Similarly, this chemokine induces the expression MMP-9 by monocytes38, 39 and purified mature polyploid human megakaryocytes in a phosphatidyl-inositol 3-kinase-dependent fashion.40, 41 The CXCL12–CXCR4 axis also induces rhabdomyosarcoma cells to produce MMP-2.42 In this study, we have demonstrated that both MMP-2 and -9 are expressed at low levels by both normal prostatic epithelial cells and prostate cancer cell lines. More importantly, CXCL12–CXCR4 interactions increase MMP-2 expression by both LNCaP and PC3 cells, but not by normal prostatic epithelial cells; however, MMP-9 expression by PC3 cells was induced only after CXCL12 ligation. Hence, CXCL12 induction differentially modulates gelatinase expression by prostate cancer cell lines.
Stromelysins are expressed predominantly by normal epithelial cells, but they are also produced by carcinomas and can degrade a broad range of substrates, including type IV, V, IX and X collagens; fibronectins; laminin; elastin; gelatin and proteoglycan core proteins.43, 44, 45 MMP-3 and MMP-10 expression by head and neck carcinomas is higher than normal matched tissue.46 We show that CXCL12 induces MMP-3 upregulation by PC3 cells and MMP-10 expression by LNCaP cells. In contrast to MMP-10, LNCaP cells appear to be predisposed to MMP-11 expression, and CXCL12 further increases MMP-11 expression by both LNCaP and PC3 cells. This differential expression of stromelysins after CXCL12 induction may play a significant role in selective invasion. In addition to these soluble MMPs, membrane matrix metalloproteinases also play significant roles in matrix degradation.
MT1-MMP (MMP-14) is a membrane-associated protease that regulates cell matrix expression through proteolysis.47, 48, 49, 50 MMP-14 degrades native type I collagen, fibronectins, laminin, fibrin, gelatin and the proteoglycan core of the proteins of cartilage. Studies have shown that MMP-14 is expressed at higher levels by tumors than by normal (adjacent) tissue.46, 51 Other studies have shown an association between MMP-14 and the transformation of benign prostate epithelium to high-grade prostatic intraepithelial neoplasia52 and the increased invasive property of prostate cancer cells.53 We show that, following CXCL12 induction, PC3 cell lines significantly express MMP-14 but LNCaP cell lines do not. Most recently, it has been shown that CXCL12 also promotes melanoma cell invasion across basement membranes that involve MMP-14 interactions.54 Hence, PC3 cells' higher level of MMP-14 expression may partially explain their ability to preferentially home to and survive in bone.
Our results show that CXCL12 can increase the expression of certain MMPs. We also show that MMP-1, -9, -10, -11 and -13 are expressed at relatively high levels by untreated cancer cells compared to untreated normal prostate epithelial cells. However, these high levels will not result in invasion without a simultaneous signal for chemotaxis. A host factor (eg, CXCL12) that modulates MMP production as well as provides a signal for cancer cell movement has important implications in prostate cancer progression. In many instances, CXCL12 increased MMP-1, -2, -3, -9, -10, -11 and -14 expression and activity by cancer cells. However, CXCL12 decreased basal levels of MMP-13 by both LNCaP and PC3 cells. Certain host factors, for example, IL-1, have been shown to increase MMP-1 expression by four-fold, while increasing MMP-13 expression by >500-fold.55 This varied collagenase expression is a result of related, yet different, induction pathways. To explain, MMP-13 induction by chondrocytes requires p38 mitogen-activated protein kinase (MAPK) and c-Jun N-terminal kinase (JNK) activation as well as nuclear factor (NF)-kappaB translocation.56 In contrast, MMP-1 induction requires p38 MAPK and MAPK of the extracellular signal-regulated kinase (MEK) activation, but not JNK activation or NF-kappaB translocation.
Consequently, CXCL12 increases p38 MAPK41, 57 and MEK activity58 in melanoma and lymphocytes, respectively. While p38 MAPK and MEK activity is increased by CXCL12 stimulation of hematopoietic progenitor and lymphocytes, JNK is not activated by this chemokine.59, 60 The modulation of NF-kappaB translocation by CXCL12 is not as certain.41, 61 Taken together, the ability of CXCL12 to simultaneously increase p38 MAPK and MEK activity, while lowering and/or not affecting JNK activity or NF-kappaB translocation, would presumably lead to higher increases in MMP-1 expression compared to MMP-13, which is confirmed by our findings. While additional studies will be needed to determine the precise molecular mechanism(s) of CXCL12-mediated MMP-1 and -13 modulation, our findings may highlight the physiologic significance of this differential regulation. MMP-1 and -13 are uniquely expressed for epithelial cell regrowth (re-epithelialization) and tissue remodeling in chronic wounds, respectively.62 Perhaps carcinoma cell growth without motility (ie, epithelial cell regrowth) is supported in part by MMP-1 but not MMP-13 expression, while tumor cell motility/invasion and mechanisms, used during chronic wound repair, require selective MMP-1 and MMP-13 activity.
In summary, our current study suggests that differential expression and ligation of CXCR4 with CXCL12 could determine prostate cancer cell migration and invasion. However, there are a number of limitations inherent in any in vitro study. The milieu of host factors that are present in the microenvironment of tumor cells including, but not limited to, androgen, colony stimulating factor, fibroblast/monocyte-derived MMPs, cytokines, etc all play a complex role in prostate cancer development and progression. Hence, many more in vivo and in vitro studies will be needed to determine the precise mechanism(s) and clinical significance of CXCL12-mediated MMP modulation and prostate tumor cell invasion.
Expression of functional CXCR4 by prostate cancer cells together with selective CXCL12 expression by bone and lymph node support our hypothesis that prostate cancer cell migration and invasion are in part mediated by CXCL12. Moreover, the effect of CXCL12 on MMP expression suggests that this chemokine plays an important role in invasion via MMP modulation, which would represent an important molecular mechanism of prostate cancer progression. While additional studies will be necessary to evaluate the precise cellular and molecular mechanisms of CXCL12-mediated migration and invasion of prostate cancer cells, the current study shows that CXCL12–CXCR4 interactions might play significant roles in prostate cancer progression.
References
Fidler IJ . Critical determinants of metastasis. Sem Cancer Biol 2002;12:89–96.
Nicolson GL . Paracrine and autocrine growth mechanisms in tumor metastasis to specific sites with particular emphasis on brain and lung metastasis. Cancer Metast Rev 1993;12:325–343.
Wang JM, Deng X, Gong W, et al. Chemokine and their role in tumor growth and metastasis. J Immunol Methods 1998;220:1–17.
Yeatman TJ, Nicolson GL . Molecular basis of tumor progression: mechanisms of organ-specific tumor metastasis. Semin Surg Oncol 1993;9:256–263.
Zlotnik A, Yoshie O . Chemokines: a new classification system and their role in immunity. Immunity 2000;12:121–127.
Butcher EC, Willima M, Youngman K, et al. Lymphocyte trafficking and regional immunity. Adv Immunol 1999;72:209–253.
Nagase H, Woessner Jr JF . Matrix metalloproteinases. J Biol Chem 1999;274:21491–21494.
Hulboy DL, Rudolph LA, Matrisian M . Matrix metalloproteinases as mediators of reproductive function. Mol Hum Reprod 1997;3:27–45.
Stetler-Stevenson WG, Aznavoorian S, Liotta LA . Tumor cell interactions with the extracellular matrix during invasion and metastasis. Ann Rev Cell Biol 1993;9:541–573.
Tiachman RS, Cooper C, Keller ET, et al. Use of stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res 2002;62:1832–1837.
Pienta KJ, Esper PS . Risk factors for prostate cancer. Ann Internal Med 1993;118:793–803.
Jacobs SC . Spread of prostatic cancer to bone. Urology 1983;21:337–344.
Liotta LA, Kohn EC . The microenvironment of the tumour–host interface. Nature 2001;411:375–379.
Sehgal A, Keener C, Boynton AL, et al. CXCR-4, a chemokine receptor, is overexpressed in and required for proliferation of glioblastoma tumor cells. J Surg Oncol 1998;69:99–104.
Slanicka KM, Nissen C, Manz CY, et al. The membrane-bound isoform of stem cell factor synergizes with soluble flt3 ligand in supporting early hematopoietic cells in long-term cultures of normal and aplastic anemia bone marrow. Exp Hematol 1998;26:356–373.
Burger JA, Burger M, Kipps TJ . Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood 1999;94:3658–3667.
Mohle R, Failenschmid C, Bautz F, et al. Overexpression of the chemokine receptor CXCR4 in B cell chronic lymphocytic leukemia is associated with increased functional response to stromal cell-derived factor-1 (SDF-1). Leukemia 1999;13:1954–1959.
Gupta SK, Pillarisetti K, Lysko PG . Modulation of CXCR4 expression and SDF-1alpha functional activity during differentiation of human monocytes and macrophages. J Leuk Biol 1999;66:135–143.
Koshiba T, Hosotani R, Miyamoto Y, et al. Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: a possible role for tumor. Clin Cancer Res 2000;6:3530–3535.
Arai J, Yasukawa M, Yakushijin Y, et al. Stromal cells in lymph nodes attract B-lymphoma cells via production of stromal cell-derived factor-1. Eur J Haematol 2000;64:323–332.
Mohle R, Schittenhelm M, Failenschmid C, et al. Functional response of leukaemic blasts to stromal cell-derived factor-1 correlates with preferential expression of the chemokine receptor CXCR4 in acute myelomonocytic and lymphoblastic leukaemia. Br J Haematol 2000;110:563–572.
Geminder H, Sagi-Assif O, Goldberg L, et al. A possible role for CXCR4 and its ligand, the CXC chemokine stromal cell-derived factor-1, in the development of bone marrow metastases in neuroblastoma. J Immunol 2001;167:4747–4757.
Sun YX, Wang J, Shelburne CE, et al. Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem 2003;89:462–473.
Benaud C, Dickson RB, Thompson EW . Roles of the matrix metalloproteinases in mammary gland development and cancer. Breast Cancer Res Treat 1998;50:97–116.
Cockett MI, Murphy G, Birch ML, et al. Matrix metalloproteinases and metastatic cancer. Biochem Soc Symp 1998;63:295–313.
Murray GI, Duncan ME, O'Neil P, et al. Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat Med 1996;2:461–462.
Murray GI, Duncan ME, O'Neil P, et al. Matrix metalloproteinase-1 is associated with poor prognosis in oesophageal cancer. J Pathol 1998;185:256–261.
Wirl G, Frick J . Collagenase—a marker enzyme in human bladder cancer? Urol Res 1979;7:103–108.
Durkan GC, Nutt JE, Tajjayabun PH, et al. Prognostic significance of matrix metalloproteinase-1 and tissue inhibitor of metalloproteinase-1 in voided urine samples from patients with transitional cell carcinoma of the bladder. Clin Cancer Res 2001;7:3450–3456.
Kanbe K, Takagishi K, Chen Q . Stimulation of matrix metalloprotease 3 release from human chondrocytes by the interaction of stromal cell-derived factor 1 and CXC chemokine receptor 4. Arthritis Rheum 2002;46:130–137.
Nemeth JA, Yousif R, Herzog M, et al. Matrix metalloproteinase activity, bone matrix turnover, and tumor cell proliferation in prostate cancer bone metastasis. J Natl Cancer Inst 2002;94:17–25.
Ponomaryov T, et al. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J Clin Invest 2000;106:1331–1339.
Gohji K, et al. Serum matrix metalloproteinase-2 and its density in men with prostate cancer as a new predictor of disease extension. Int J Cancer 1998;79:96–101.
Raab G, Moses MA, Adam RM, et al. Increased incidence of matrix metalloproteinases in urine of cancer patients. J Cell Biochem 1998;69:143–153.
Moses MA, Wiederschain D, Loughlin KR, et al. Increased incidence of matrix metalloproteinases in urine of cancer patients. Cancer Res 1998;58:1395–1399.
Sehgal G, Hua J, Bernhard EJ, et al. Requirement for matrix metalloproteinase-9 (gelatinase B) expression in metastasis by murine prostate carcinoma. Am J Pathol 1998;152:591–596.
Janowska-Wieczorek A, Marquez LA, Dobrowsky A, et al. Differential MMP and TIMP production by human marrow and peripheral blood CD34(+) cells in response to chemokines. Exp Hematol 2000;28:1274–1285.
Klier CM, Nelson EL, Cohen CD, et al. Chemokine-induced secretion of gelatinase B in primary human monocytes. Biol Chem 2001;382:1405–1410.
Yu X, Collin-Osdoby P, Osdoby P . SDF-1 increases recruitment of osteoclast precursors by upregulation of matrix metalloproteinase-9 activity. Connect Tissue Res 2003;44 (Suppl 1):79–84.
Lane WJ, Dias S, Hattori K, et al. Stromal-derived factor 1-induced megakaryocyte migration and platelet production is dependent on matrix metalloproteinases. Blood 2000;96:4152–4159.
Majka M, Janowska-Wieczorek A, Ratajczak J, et al. Stromal-derived factor 1 and thrombopoietin regulate distinct aspects of human megakaryopoiesis. Blood 2000;96:4142–4151.
Libura J, Drukala J, Majka M, et al. CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood 2002;100:2597–2606.
Birkedal-Hansen H, Moore WG, Bodden MK, et al. Matrix metalloproteinases: a review. Crit Rev Oral Biol Med 1993;4:197–250.
Shapiro SD . Matrix metalloproteinase degradation of extracellular matrix: biological consequences. Curr Opin Cell Biol 1998;10:602–608.
Nagase H . Activation mechanisms of matrix metalloproteinases. Biol Chem 1997;378:151–160.
Birkedal-Hansen B, Pavelic ZP, Gulckman JL, et al. MMP and TIMP gene expression in head and neck squamous cell carcinomas and adjacent tissues. Oral Dis 2000;6:376–382.
Sato H, Takino T, Okada Y, et al. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature 1994;370:61–65.
Zucker S, Drews M, Conner C, et al. Tissue inhibitor of metalloproteinase-2 (TIMP-2) binds to the catalytic domain of the cell surface receptor, membrane type 1-matrix metalloproteinase 1 (MT1-MMP). J Biol Chem 1998;273:1216–1222.
Werb Z . ECM and cell surface proteolysis: regulating cellular ecology. Cell 1997;91:439–442.
Stanton H, Gavrilovic J, Atkinson SJ, et al. The activation of ProMMP-2 (gelatinase A) by HT1080 fibrosarcoma cells is promoted by culture on a fibronectin substrate and is concomitant with an increase in processing of MT1-MMP (MMP-14) to a 45 kDa form. J Cell Sci 1998;111:2789–2798.
Nomura H, Sato H, Seiki M, et al. Expression of membrane-type matrix metalloproteinase in human gastric carcinomas. Cancer Res 1995;55:3263–3266.
Upadhyay J, Shekarriz B, Nemeth JA, et al. Membrane type 1-matrix metalloproteinase (MT1-MMP) and MMP-2 immunolocalization in human prostate: change in cellular localization associated with high-grade prostatic intraepithelial neoplasia. Clin Cancer Res 1999;5:4105–4110.
Nagakawa O, Murakami K, Yamaura T, et al. Expression of membrane-type 1 matrix metalloproteinase (MT1-MMP) on prostate cancer cell lines. Cancer Lett 2000;155:173–179.
Bartolome RA, Galvez BG, Longo N, et al. Stromal cell-derived factor-1alpha promotes melanoma cell invasion across basement membranes involving stimulation of membrane-type 1 matrix metalloproteinase and Rho GTPase activities. Cancer Res 2004;64:2534–2543.
Fuchs S, Skwara A, Bloch M, et al. Differential induction and regulation of matrix metalloproteinases in osteoarthritic tissue and fluid synovial fibroblasts. Osteoarthritis Cartilage 2004;12:409–418.
Mengshol JA, Vincenti MP, Coon CI, et al. Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor kappaB: differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum 2000;43:801–811.
Robledo MM, Bartolome RA, Longo N, et al. Expression of functional chemokine receptors CXCR3 and CXCR4 on human melanoma cells. J Biol Chem 2001;276:45098–45105.
Laakko T, Juliano RL . Adhesion regulation of stromal cell-derived factor-1 activation of ERK in lymphocytes by phosphatases. J Biol Chem 2003;278:31621–31628.
Ganju RK, Brubaker SA, Meyer J, et al. The alpha-chemokine, stromal cell-derived factor-1alpha, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J Biol Chem 1998;273:23169–23175.
Yonezawa A, Hori T, Sakaida H, et al. SDF-1 has costimulatory effects on human T cells: possible involvement of MAPK (ERK2) activation. Microbiol Immunol 2000;44:135–141.
Lee Y, Gotoh A, Kwon HJ, et al. Enhancement of intracellular signaling associated with hematopoietic progenitor cell survival in response to SDF-1/CXCL12 in synergy with other cytokines. Blood 2002;99:4307–4317.
Vaalamo M, Mattila L, Johansson N, et al. Distinct populations of stromal cells express collagenase-3 (MMP-13) and collagenase-1 (MMP-1) in chronic ulcers but not in normally healing wounds. J Invest Dermatol 1997;109:96–101.
Acknowledgements
This work benefited from many fruitful conversations with investigators at the Morehouse School of Medicine and at the Wallace Tumor Institute at the University of Alabama at Birmingham via the National Cancer Institute-sponsored ‘Comprehensive Minority Institution/Cancer Center Partnership.’ This work was supported in part by the Department of the Army's US Army Medical Research Acquisition Activity Grant DAMD17-01-1-0079 as well as the Public Health Service's National Institutes of Health Grants RR03034, GM08248 and CA92078.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, S., Singh, U., Grizzle, W. et al. CXCL12–CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest 84, 1666–1676 (2004). https://doi.org/10.1038/labinvest.3700181
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/labinvest.3700181
Keywords
This article is cited by
-
Interactions of the chemokines CXCL11 and CXCL12 in human tumor cells
BMC Cancer (2022)
-
The multifaceted roles of the chemokines CCL2 and CXCL12 in osteophilic metastatic cancers
Cancer and Metastasis Reviews (2021)
-
Light-controlled calcium signalling in prostate cancer and benign prostatic hyperplasia
Future Journal of Pharmaceutical Sciences (2020)
-
Modulation of CXC-motif chemokine receptor 7, but not 4, expression is related to migration of the human prostate cancer cell LNCaP: regulation by androgen and inflammatory stimuli
Inflammation Research (2020)
-
Transcriptome Network Analysis Identifies CXCL13-CXCR5 Signaling Modules in the Prostate Tumor Immune Microenvironment
Scientific Reports (2019)