Abstract
We recently indicated that four-week probiotic supplementation significantly reduced depression along with microbial and neural changes in people with depression. Here we further elucidated the biological modes of action underlying the beneficial clinical effects of probiotics by focusing on immune-inflammatory processes. The analysis included a total of N = 43 participants with depression, from which N = 19 received the probiotic supplement and N = 24 received a placebo over four weeks, in addition to treatment as usual. Blood and saliva were collected at baseline, at post-intervention (week 4) and follow-up (week 8) to assess immune-inflammatory markers (IL-1β, IL-6, CRP, MIF), gut-related hormones (ghrelin, leptin), and a stress marker (cortisol). Furthermore, transcriptomic analyses were conducted to identify differentially expressed genes. Finally, we analyzed the associations between probiotic-induced clinical and immune-inflammatory changes. We observed a significant group x time interaction for the gut hormone ghrelin, indicative of an increase in the probiotics group. Additionally, the increase in ghrelin was correlated with the decrease in depressive symptoms in the probiotics group. Transcriptomic analyses identified 51 up- and 57 down-regulated genes, which were involved in functional pathways related to enhanced immune activity. We identified a probiotic-dependent upregulation of the genes ELANE, DEFA4 and OLFM4 associated to immune activation and ghrelin concentration. These results underscore the potential of probiotic supplementation to produce biological meaningful changes in immune activation in patients with depression. Further large-scale mechanistic trials are warranted to validate and extend our understanding of immune-inflammatory measures as potential biomarkers for stratification and treatment response in depression. Trial Registration: www.clinicaltrials.gov, identifier: NCT02957591.
Similar content being viewed by others
Introduction
With a global lifetime prevalence of approximately 25%, major depressive disorder (MDD), poses significant individual and societal burden [1, 2]. Antidepressants bring symptom relief, but over 50% of cases do not respond to initial treatment, with one in three patients showing resistance to antidepressants [3]. The limitations of current treatments, in light of the global impact of MDD, underscore the urgent need for alternative treatment approaches. Probiotic interventions targeting the gut microbiota show promise in relieving depressive symptoms [4]. Multiple randomized controlled trials (RCTs) demonstrated a beneficial effect of probiotic supplementation on depressive symptoms in MDD patients [5,6,7,8], including our own trial [9]. However, a significant research gap remains in understanding the specific biological mechanisms underlying the beneficial clinical effect of probiotic supplementation in MDD. Such mechanistic analyses aim to identify biological targets for patients’ stratification and developing more efficient and tailored microbial interventions.
Probiotic supplements are formulations of living microorganisms that provide a health benefit through modulation of the microbiota. They are thought to positively impact mental health, via altering the microbiota gut brain (MGB) axis [10], a set of bi-directional communication pathways between the gut and the brain, including endocrine, immune and neurotransmitters systems [11]. Dysfunctions of the MGB axis, alongside compositional and functional (e.g., metabolomic and transcriptional) changes in gut microbiota, known as dysbiosis, have been detected as important factors in the pathology and treatment of depression [12]. In patients with MDD, gut dysbiosis is associated with a disrupted gut microenvironment, harming the protective functioning of the gut epithelium which leads to intestinal barrier dysfunction [13]. The damaged intestinal barrier (commonly referred to as “leaky gut”) allows increased systemic translocation of gut metabolites, microbial cell components, or even the microbiota causing a range of negative consequences that have been implicated in the pathogenesis of depression [14,15,16].
Psychiatric disorders, including depression, exhibit a transdiagnostic pattern of gut microbial disarray, marked by a distinct pattern of depleted anti-inflammatory and enriched pro-inflammatory bacteria [17]. This pro-inflammatory microbial state is particularly noteworthy for depression, as chronic low-grade inflammation is a known pathologic feature of depression [18]. Approximately 1/3 of patients with depression have elevated immune-inflammatory markers [19], and a pro-inflammatory state is a common feature of no-responders to antidepressant medication [20, 21]. In animal models, probiotics were found to counter gut microbiota perturbation by increasing beneficial bacteria and improving overall microbial diversity, causing a reduction in circulating immune-inflammatory markers [22, 23]. But whether probiotics can produce similar reparative effects on dysbiosis and inflammatory mechanisms in patients with depression, and whether these changes can have antidepressant effects remains an area of limited exploration. A comprehensive meta-analysis on the impact of probiotic supplementation on immune-inflammatory markers in a clinical population did report reductions in a range of cytokines, including high sensitivity C-reactive protein (hs-CRP), and interleukin 6 (IL-6), while showing no effects for other markers including interleukin 1β (IL-1β) [24]. However, these implications are limited due to the diverse patient groups and their physiological states. In terms of immune-inflammatory mechanism of probiotics in depression only few RCTs assessed cytokines. Some studies found that probiotic supplementation reduces hs-CRP [8], and decreases IL-6 gene expression levels [25], although other studies reported no changes for these cytokines [26,27,28].
Hyperactivation of the hypothalamus–pituitary–adrenal (HPA) axis is a key factor linked to the pathophysiology of depression and inflammation-related gut microbiota alterations [12]. Animal models of chronic stress triggering the HPA axis, demonstrated negative effects on the gut microbiota composition and increased IL-6 levels [29]. Studies in healthy participants also indicate that psychosocial stress reduces gut microbiota diversity and beneficial microbes [30]. Probiotics may counter these effects and exert anti-inflammatory changes by modulating the gut hormone ghrelin and the peptide hormone leptin. Alterations in the gut microbiota affects ghrelin secretion and signaling [31], with specific gut bacteria affecting leptin and ghrelin levels [32, 33]. Ghrelin and leptin in turn interact with the HPA axis and influence immune-inflammatory markers. In healthy participants, exposure to a stress test increased stress ratings and serum cortisol, associated with ghrelin secretion [34], while injections of ghrelin enhanced plasma cortisol [35]. Leptin, conversely, inhibits the HPA axis. Injections of leptin in rats decreased adrenocorticotropic hormone (ACTH) levels [36], and HPA axis activity in humans reduced leptin but increased ACTH and cortisol levels [37]. Moreover, both ghrelin and leptin were found to modulate immune-inflammatory pathways linked to depression. Ghrelin injections reduced IL-6 levels in rats, an effect diminished by vagotomy, indicating that ghrelin down-regulates pro-inflammatory cytokines through vagus nerve activation [38]. Similarly, in humans, ghrelin was found to inhibit IL-6 and IL-1β [39]. In contrast, gastric leptin levels correlated positively with proinflammatory cytokines such as IL-6 and IL-1β, in patients with Helicobacter pylori infection [40]. Furthermore, leptin directly induced the secretion of IL-6 and IL-1β by human T cells [39]. In sum, changes in the gut microbiome may directly influence systemic immune-inflammatory processes, with ghrelin and leptin playing crucial roles in modulating both endocrine (HPA axis) and immunological (inflammation markers) pathways of the MGB axis.
We recently performed an RCT investigating probiotic supplementation in individuals with depression [9]. Primarily, we found that the intervention alleviated depressive symptoms (d = 0.62) and positively affected the gut microbiota composition. Here, we present additional analyses that further uncover the biological mechanisms underlying the positive impact of probiotic supplementation on depressive symptoms. Specifically, we report serum concentrations of (1) immune-inflammatory cytokines (IL-1β, IL-6, CRP, MIF), and (2) gut-related hormones (ghrelin, leptin), (3) saliva concentrations of cortisol, (4) transcriptional (gene expression) changes, and (5) subjective appetite measures.
Patients and Methods
This is a secondary analysis of a double-blinded RCT of probiotic supplementation in patients with depression (NCT02957591, www.clinicaltrials.gov). Clinical, microbial, and neural findings have previously been published [9, 41, 42], indicating beneficial effects of the probiotic intervention on depressive and cognitive symptoms, gut microbiota composition and fronto-limbic brain structure and function. Here we further explored probiotic effects on immune-inflammatory mechanisms.
Participants
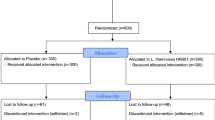
Adult inpatients (n = 60; 18–65 years of age) with a current depressive episode (F31.3-F34 according to ICD-10 criteria) were recruited at the University Psychiatry Clinics (UPK) in Basel, Switzerland between March 2017, and January 2020. All participants met the criteria for a mild depressive episode, assessed with the Hamilton Depression Rating Scale (HAMD-17 [43]) score 7 [44], and received treatment as usual (TAU) for depression (Supplementary Table 1). Excluded were patients with comorbid psychiatric disorders, with current medical conditions, with dietary restrictions and with a body mass index (BMI) > 30. Additionally, immunosuppressed and pregnant or breast-feeding patients were excluded. For more details on inclusion and exclusion criteria see [9]. All participants provided written informed consent prior to the initiation of the study, and the study was approved by the local ethics committee (Ethikkommission Nordwest- und Zentralschweiz).
Study intervention
Participants received either a placebo or a probiotic supplement in addition to TAU over four weeks. The probiotic supplement (DSFormulation; Vivomixx®; Visbiome®) consisted of eight different bacterial strains: Streptococcus thermophilus NCIMB 30438, Bifidobacterium breve NCIMB 30441, B. longum NCIMB 30435 (Re-classified as B. lactis), B. infantis NCIMB 30436 (Re-classified as B. lactis), Lactobacillus acidophilus NCIMB 30442, L. plantarum NCIMB 30437, L. paracasei NCIMB 30439, and L. delbrueckii subsp. bulgaricus NCIMB 30440 (Re-classified as L. helveticus). The daily dose consisted of two sachets containing a high dose of 900 billion colony forming units (CFU)/day that could be mixed with any cold, non-carbonated drink. In the control group, participants received a placebo containing maltose and no bacteria which was indistinguishable in color, shape, size, smell, and taste from the probiotic supplement.
Study design and procedure
Participants were randomly allocated to one of the two study arms and assessed at three time points: Week zero (baseline), week four (post-intervention) and week eight (follow-up). A standardized clinical assessment of depression (HAMD-17, Beck Depression Inventory-II (BDI-II) [45]) was conducted at all three time points. Blood samples and subjective measurements of appetite were obtained at all three time points, and saliva samples at baseline and post-intervention (see Supplementary Fig. 1). During the intervention period, all medication of the participants was registered (see Supplement) and a standardized diet containing stable amounts of fibers, starch and protein was provided. Fidelity to the protocol was assessed by the nursing personnel administering the intervention.
Blood analysis of immune-inflammatory markers and gut hormones
Blood serum concentrations of immune-inflammatory markers (IL-1β, IL-6, macrophage inhibitory factor (MIF), and CRP) and gut-related hormones (ghrelin and leptin) were obtained. Blood samples were collected at 7 am after overnight fasting according to a standardized laboratory procedure using a serum Monovette® (Sarstedt; Nümbrecht, Germany) per manufacturer’s protocol and stored at −80 °C up until further analysis. Analysis of immune-inflammatory markers and gut-related hormones was performed by an external laboratory (Labor Rothen AG, Basel, Switzerland). Quantitative CRP was determined in the laboratory using the CRP Latex reagent system on Beckman Coulter AU Analyzers, while the other immune-inflammatory markers and gut-related hormones were measured by the U-Plex® Metabolic Group 1 (Human) Multiplex Assays by Meso Scale Discovery®. Ghrelin was assessed as total ghrelin, containing both forms acylated (active) and desacylated (inactive) ghrelin.
Saliva cortisol analysis
The saliva concentration of the stress-hormone cortisol was obtained. Saliva samples were drawn at 9 pm before going to bed, and at the following morning at 7 am immediately upon awakening (S1), and after 10 min (S2), 20 min (S3), and 30 min (S4). A blue cap Salivette® (Sarstedt; Nümbrecht, Germany) with synthetic swab was employed to obtain the cortisol concentrations. Saliva samples were kept frozen at −80 °C until analysis. A time-resolved fluorescence immunoassay was used to determine cortisol concentrations by the biochemical laboratory from the Department of Biological and Clinical Psychology at the University of Trier, Germany. The cortisol awakening response (CAR) of participants was computed as the area under the curve with respect to the cortisol increase (AUCi) of the morning cortisol concentrations S1 to S4 [46].
Analysis of subjective appetite ratings
Subjective measures of appetite-related sensations were assessed using a 10-point Likert Scale in the morning after overnight fasting in addition to the blood and saliva sampling. The Likert scale was anchored by two contrasting descriptors, “not at all” and “extremely” accompanied by four measures of appetite-related sensations (“hunger”, “desire to eat”, “feeling of fullness”, “satiety”).
Transcriptomic analysis
Blood samples were collected at 7 am after overnight fasting according to a standardized laboratory procedure into a PAXgene tube (Qiagen; Hilden, Germany) per manufacturer’s protocol and stored at −80 °C up until further analysis. RNA isolation (Quantification-OD measurement, Gel electrophoresis-integrity, RNA isolation PaxGene) has been conducted by Qiagen (Qiagen; Hilden, Germany). RNA sequencing and quantification was performed at the Lausanne Genomic Technology Facility. Differential gene expression analysis (DGE) and weighted gene co-expression network analysis was performed at the Swiss Institute of Bioinformatics (SIB), University of Lausanne. Detailed descriptions of all procedures are provided in the supplement.
Statistical analysis
All analyses were conducted on a modified intention-to-treat (mITT) sample excluding non-compliant participants and drop-outs. The compliance rate cut-off of > 65% resulted in the exclusion of two patients per group from the study sample (for details see [9]). All analyses and visualizations were performed in R (v4.3.1). Unless otherwise specified, the significance level was set at p < 0.05, and multiple comparison adjustments were performed, which were selected based on variance equality and assessed outcome variable. Additional analysis details are provided in the supplement.
Effect of probiotics on blood/saliva markers and appetite measures
For blood and saliva measures, a Tukey transformation [47] was performed to reach quasi-normal distributions, and outlier values (defined at ±1.5 x interquartile range) were excluded (sensitivity analyses on the findings including all samples are provided in the supplement). Linear mixed-effects models (LMM) were applied to assess the probiotic effect including the following design formula: measure ~ group x time + age + sex + BMI + (1|ID), with group as a two-levelled factor (probiotics, placebo) and time as a three-levelled factor (baseline, post-intervention, follow-up). LMM included a random effect for participant, to account for individual differences. To avoid confounding, sex, age, and body-mass-index (BMI), were additionally added as fixed effects in the model. An analysis of variance (ANOVA, type III) was computed, and for significant main effects of group, time, and group x time interactions pairwise post-hoc multiple comparisons using estimated marginal means with t-tests were performed. Multiple comparison adjustments for post-hoc contrasts of equal variances were made using the Bonferroni method.
Association between probiotics’ effect on blood/saliva markers and depression
Partial correlation analyses were conducted for both treatment groups, to explore associations between significant probiotic-induced changes on blood/saliva markers and depressive symptoms (HAMD-17). Age, sex, and BMI were included as covariates. Fischer’s z test was applied to compare correlations between treatment groups. Cook’s Distance, with a cutoff of >4/N, was used for bivariate outlier detection (sensitivity analyses on the findings including all samples are provided in the supplement).
Effect of probiotics on gene expression and functional enrichment analysis
For gene expression measures, a negative binomial generalized linear model with the following design formula: expression ~ group + group x participant + group x time, was applied. Differentially expressed genes (DEGs) were identified using Wald’s test p < 0.05 and |fold-change| > 1.5. REACTOME gene set enrichment analysis (GSEA) was performed on the entire lists of expressed genes pre-ranked by signed p-value as determined by Wald’s test. Benjamini-Hochberg corrections were applied to functional enrichment p values to correct for multiple comparisons in high-dimensional data.
Association between probiotics’ effect on biological/ clinical measures and DEGs
To evaluate the link between transcriptional changes and the effect of probiotics on biological and clinical measures, LMMs were applied including gene expression as fixed effect. The LMM was built and analyzed analog to the description of LMM analysis in the previous section, with the addition of a three-way interaction group x time x gene expression. Only DEGs (log transformed and normalized gene counts) significantly different between the groups over time were analyzed. The same analysis was extended to gene modules from the WGCNA by replacing individual gene counts with “module eigengene” (ME) as fixed effects.
Results
Participant characteristics
The final study sample included 43 participants (mITT; see [9] for details). Blood samples of N = 40 (93%), saliva samples of N = 38 (88%), transcriptome data of N = 35 (81%), and appetite measures of N = 43 (100%) were available (Supplementary Figure 1). Baseline characteristics of all participants are presented in Table 1. Group comparisons showed no significant differences in demographic characteristics, medication, and clinical measures at baseline except for HAMD-17 scores, which showed a higher score for the probiotics group (W = 311, p < 0.05). Blood, saliva, and appetite measures showed no differences between the study groups at baseline except for MIF, which showed a lower concentration for the probiotics group (W = 250, p < 0.05).
Blood analysis
Immune-inflammatory markers
IL-1β and IL-6 levels did not show a significant main effect of group or time, nor a significant group x time interaction (Supplementary Table 2, Fig. 2A, D). MIF levels did not show a main effect of group, but a significant main effect of time (F(2, 96) = 11.67, p < 0.001), and a significant group x time interaction (F(2, 96) = 4.66, p < 0.05) was observed (Supplementary Table 2, Fig. 1C). Post-hoc tests demonstrated a significant increase in MIF levels from baseline to follow-up (pBonferroni < 0.01) and from post-intervention to follow-up (pBonferroni < 0.001) in the probiotics group, but not from baseline to post-intervention. No change was observed in the placebo group at any time point. CRP levels showed a main effect of group (F(1, 34.75) = 9.57, p < 0.01), but no main effect of time or group x time interaction was observed (Supplementary Table 2, Fig. 2B). Post-hoc tests demonstrated higher CRP levels across all time points in the probiotics compared to the placebo group (pBonferroni < 0.01).
Concentration levels in A–F are presented as boxplots of Median [IQR], association in G and H are presented as Pearson’s correlation (solid line) and partial correlation (dotted line) controlled for (age, sex, BMI). In A–F significant group × time interactions are depicted (*p ≤ 0.05). A Trajectory of IL-1β blood concentration levels throughout the study. B Trajectory of CRP concentration levels throughout the study. C Trajectory of MIF concentration levels throughout the study, indicative of a significant increase from post-intervention to follow-up in the probiotics group. D Trajectory of IL-6 concentration levels throughout the study. E Trajectory of leptin concentration levels throughout the study. F Trajectory of ghrelin concentration levels throughout the study, indicative of a significant increase from baseline to post-intervention in the probiotics group. G Association between changes in MIF concentration levels and HAMD-17 scores from post-intervention to follow-up for both study group. Placebo group (Pearson’s correlation: r = 0.27, p = 0.4; partial correlation: r = 0.23, p = 0.55). Probiotics group (Pearson’s correlation: r = −0.06, p = 0.86; partial correlation: r = 0.17, p = 0.64). H Association between changes in ghrelin concentration levels and HAMD-17 scores from baseline to post-intervention for both study group. Placebo group (Pearson’s correlation: r = 0.3, p = 0.22; partial correlation: r = 0.35, p = 0.20). Probiotics group (correlation: r = −0.58, p = 0.03; partial correlation: r = −0.63, p = 0.04).
Differentially expressed genes (DEGs) were defined as |fold-change| > 1.5 and p value < 0.05. A Heatmap of differential gene expression (DGE) from baseline to post-intervention assessment in the probiotics compared to placebo groups. Genes and participants are clustered based on Euclidean distance using complete linkage method. Participants are additionally split according to their treatment groups. Genes are additionally split according to their increasing (UP) or decreasing (DOWN) expression. B DEGs in the probiotics compared to placebo group between the timepoints baseline to post-intervention. C DEGs in the probiotics compared to placebo group between the timepoints post-intervention to follow-up. D Overlap of UP and DOWN DEGs in the probiotics compared to placebo groups between the study periods baseline to post-intervention (PIvsBL) and post-intervention to follow-up (FUvsPI).
Association between probiotics’ effect on MIF and depression symptoms
The partial correlation between MIF and HAMD-17 change-scores (xFollow-Up − xPost-Intervention) did not demonstrate associations in the probiotics group (r = 0.17, p = 0.64) nor in the placebo group (r = 0.23, p = 0.55) (Fig. 1G). The obtained correlations did not significantly differ between the groups (z = 0.14, p = 0.89).
Gut-related hormones
Ghrelin levels did not show a main effect of group or time, but a significant group x time interaction (F(2, 64.69) = 4.36, p < 0.05) was observed (Supplementary Table 3, Fig. 1F). Post-hoc tests demonstrated a significant increase of ghrelin levels from baseline to post-intervention (pBonferroni < 0.05) in the probiotics group, which was no longer observable at follow-up. No change was observed in the placebo group at any time point.
Leptin levels showed a significant main effect of time (F(2, 58.77) = 4.18, p < 0.05), but no main effect of group and no group x time interaction was observed (Supplementary Table 3, Fig. 1E). Post-hoc tests demonstrated an increase of leptin levels from baseline to follow-up (pBonferroni < 0.05) across both groups.
Association between probiotics’ effect on ghrelin and depression symptoms
The partial correlation between ghrelin and HAMD-17 change-scores (xPost-intervention − xBaseline) demonstrated a significant negative correlation in the probiotics group (r = −0.63, p < 0.05) but not in the placebo group (r = 0.35, p = 0.2) (Fig. 1H). The obtained correlations did significantly differ between the groups (z = 2.76, p < 0.01). That is the higher the increase in ghrelin levels from baseline to post-intervention the stronger the decrease in HAMD-17 for the probiotics but not the placebo group.
Saliva cortisol
Evening cortisol concentrations (9 pm), waking cortisol concentrations (7 am) and the CAR did not show a significant main effect of group or time, nor a significant group x time interaction (Supplementary Table 4, Supplementary Fig. 2).
Subjective appetite ratings
The appetite sensations, hunger, satiety, fullness, and desire to eat did not show a significant main effect of group or time, nor a significant group x time interaction (Supplementary Table 5, Supplementary Fig. 3). The partial correlation between ghrelin and subjective appetite ratings change-scores (xPost-intervention − xBaseline) demonstrated for the feeling of fullness a significant positive correlation in the probiotics group (r = 0.61, p < 0.05) but not in the placebo group (r = 0.36, p = 0.31) (Supplementary Table 6). The obtained correlations did not significantly differ between the groups (z = 1.24, p = 0.21), indicating that the association between higher ghrelin levels and increased feelings of fullness observed in the probiotics group is not unique to the intervention. All additional partial correlations between changes in ghrelin and subjective appetite ratings did not demonstrate significant results for both groups (Supplementary Table 6).
Transcriptomics analysis
DGE and functional enrichment
From baseline to post-intervention, DGE analysis revealed the upregulation of 51 genes (fold change > 1.5) and downregulation of 57 genes (fold change < −1.5) (unadjusted p < 0.05) in the probiotics compared to the placebo group (Fig. 2A, B). Gene set enrichment analysis (GSEA) of REACTOME pathways demonstrated the DGE to be a coordinated upregulation of genes involved in functional pathways of immune activation. Probiotic supplementation was associated with an upregulation of biological processes of “Neutrophil degranulation”, “Antigen processing cross presentation”, “Signaling by CSF3 (G-CSF)”, “Antimicrobial peptides”, “Negative regulation of NOTCH4 signaling”, “Inactivation of CSF3 (G-CSF) signaling”, all functionally associated to immune mechanisms (Fig. 3A). Analyses of overlapping DEG and enriched pathways identified the DEG (HBB, ELANE, DEFA4, OLFM4, KRT1) in “Neutrophil degranulation”, and the DEG (ELANE, DEFA4) in “Antimicrobial peptides” (Fig. 3B, C). The GSEA of downregulated genes revealed no functional coordination, as no association to consensus REACTOME pathways was obtained. The DGE from post-intervention to follow-up demonstrated no upregulation of genes functionally associated to immune activity, indicating a transient and immediate effect of probiotic supplementation on DGE related to immune activation (Fig. 3A). Changes in multiple DEGs were significantly linked to changes in ghrelin levels in the probiotics compared to the placebo groups from baseline to post-intervention (Fig. 4A). For two DEGs (TREML1 and ELANE) the change score (xPost-intervention − xBaseline) demonstrated a significant partial correlation to the ghrelin change score (xPost-intervention − xBaseline) in the probiotics group (Fig. 4B).
A Comparison of enrichment profiles of REACTOME pathways between the baseline (BL), post-intervention (week 4; PI), and follow-up (week 8; FU) assessment. Significantly upregulated pathways between PI and BL, in probiotics compared to placebo groups were filtered for display (adjusted p ≤ 0.01 and |NES| > 2). Color of cells indicates the NES. Corresponding BH-adjusted p values are also indicated (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). B Core enriched DEGs for GSEA enriched REACTOME pathways at post-intervention vs baseline. C Trajectories of norm count of core enriched DEGs throughout the study period associated to the pathway neutrophil degranulation. Norm counts are present as M (SE), axes have been edited according to respective norm count for clearer interpretation.
A Effect of group x time x gene expression interaction on the biological/clinical measures assessed by LMM. The color scale represents -log scaled ANOVA p-values. Significant interactions are depicted (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). B Association between the change of ghrelin and significantly associated genes (*p ≤ 0.05) from baseline to post-intervention for both study group. Only DEGs with a significant association to ghrelin are presented. Associations were assessed as Pearson’s correlation (solid line) and partial correlation (dotted line) controlled for (age, sex, BMI). ELANE: Placebo group (correlation: r = −0.42, p = 0.1; partial correlation: r = −0.69, p = 0.01). Probiotics group (correlation: r = 0.32, p = 0.28; partial correlation: r = 0.7, p = 0.02). TREML1: Placebo group (correlation: r = 0.08, p = 0.75; partial correlation: r = 0.061, p = 0.83). Probiotics group (correlation: r = −0.51, p = 0.07; partial correlation: r = −0.6, p = 0.07).
Weighted gene correlation network analysis
To capture correlation pattern among genes in response to the probiotic intervention, we grouped co-expressed genes into 37 modules using a network-based approach [48]. We found that the Module 24 (M24) exhibited the highest degree of overlap to DEGs, containing 47 (8.5%) upregulated DEGs (Fig. 5B). The REACTOME over-representation analysis of module genes found M24 to be enriched for functional pathways which we already identified as upregulated in the DGE analysis (“Neutrophil degranulation” and “Antimicrobial peptides”) (Fig. 5D). This indicates that the module M24 recapitulates probiotics-specific transcriptional changes on immune activity. Interestingly, we also found that the interaction group × time × ME (“module eigengene”) of M24 was significantly associated to ghrelin (Fig. 5C, E).
A Identification of co-expressed gene modules based on a weighted gene co-expression network analysis. Colors are randomly assigned to facilitate module identification. B Percentage of probiotics-induced DEGs in modules. N next to module indicates absolute number of DEGs in modules. C Effect of group × time × ME (“module eigengene”) interaction on the biological/clinical measures assessed by LMM. The color scale represents −log scaled ANOVA p-values. Significant interactions are depicted (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). D Significantly enriched REACTOME pathways in modules associated to ghrelin. BH-adjusted p values for the enrichment test (*p ≤ 0.05). E Identification of genes with a high Module Membership (correlation to ME) and association to ghrelin (LMM assessed significant group × time × gene expression) in M24 and M33.
Discussion
We previously reported a beneficial impact of a multi-strain probiotic intervention on both depressive symptoms and gut microbiota composition in individuals with depression receiving TAU [9]. Following this, the current secondary analysis focusing on immune-inflammatory mechanisms of probiotics revealed three key outcomes: Firstly, probiotics exhibited a significant, albeit transient, increase in circulating levels of ghrelin over the intervention period. This effect was associated with the improvement in depressive symptoms during the intervention phase. Secondly, probiotics showed no immediate effect on pro-inflammatory cytokines, cortisol concentrations, and leptin. However, MIF levels showed a significant increase at follow-up in the probiotic group. Thirdly, probiotics induced changes in gene expression patterns functionally associated with the immune system. Similar to the elevation of circulating ghrelin, the transcriptional changes were only evident during the intervention period and no longer evident at follow-up.
The association between the probiotic-induced increase of ghrelin levels and decrease of depressive symptoms adds to the existing literature suggesting antidepressant effects of ghrelin [49, 50]. Multiple preclinical studies reported antidepressant-like properties of ghrelin in rodents [51,52,53,54,55]. Clinical data show a more mixed picture of ghrelin’s effect on depressive symptoms [49, 56]. There is evidence for improved depressive symptoms following ghrelin administration in a study on patients with MDD [57], while another study reported an association between higher severity of depressive symptoms and increased ghrelin concentrations in patients with depression [58]. However, other studies reported no such association in patients with depression [59,60,61]. In addition, compared to healthy individuals, in patients with depression higher [62,63,64], lower [65] and comparable [61, 66, 67] ghrelin concentrations were reported, indicating variability amongst patients with depression in terms of ghrelin concentrations. In this study, the obtained increase in ghrelin levels was due to the intervention effect of a multi-strain probiotic over four weeks, which returned to baseline concentrations at follow-up four weeks after the intervention was completed. The finding of higher ghrelin concentrations following probiotic supplementation in patients with depression is in line with preclinical results reporting higher ghrelin gene expression in mice treated with the same multi-strain probiotic [68]. Furthermore, mice treated with the probiotic were found to have increased numbers of ghrelin secreting cells in the mouse intestine, alongside higher numbers of cells shielding the gastric epithelium [68]. In addition, probiotics (Lactobacillus spp.) were reported to enhance ghrelin gene expression and ghrelin secretion in other animal studies [69, 70]. These findings stand in contrast to another animal study reporting reduced ghrelin concentrations after administering the same multi-strain probiotic we used [71]. However, none of these preclinical findings were obtained using an animal model of depression, limiting the translation to a clinical population of patients with depression. Nonetheless, in accordance with our initial finding of increased abundances of the genus Lactobacillus following the probiotic supplementations [9], most animal studies demonstrated that probiotics (Lactobacillus spp.) have the potential to enhance ghrelin gene expression and secretion.
We found no treatment effect over time of probiotic supplementation on all immune-inflammatory blood markers except for MIF, for which we obtained a significant increase at follow-up. The lack of effects at post-intervention is in line with other clinical trials in patients with depression, which did not find the proposed anti-inflammatory properties of probiotics reported in animal models [72, 73]. In accordance with our results, no changes in IL-1β and IL-6 levels, were reported in a human study administering the same multi-strain probiotic supplement we used and in a recent meta-analysis evaluating probiotics trials [4, 74]. However, while most results point towards no changes in immune-inflammatory markers, two trials using the same probiotic strains in different supplements reported decreased hs-CRP levels in patients with depression [8], and reduced IL-6 levels in patients with multiple sclerosis [75]. These mixed findings on probiotic supplement effects on immune-inflammatory markers might be due to the variability in used probiotic strains and targeted clinical populations [76]. Some probiotics may exhibit anti-inflammatory properties, while others might function as immune stimulants, thereby enhancing physiological inflammation [77]. In line with this, for the probiotic supplement used in our study, increased macrophage activation has been reported [78, 79]. This may explain our probiotic-induced increase in MIF concentrations at follow-up. Macrophages are known to release cytokines, including MIF, and activate the innate immune system [80]. MIF, in turn, acts as a regulator of the innate immune activity but has also been reported to play a crucial role in maintaining the barrier function of intestinal epithelial cells [81] and is associated with enhanced macrophage-dependent pathogen clearance [82]. Furthermore, previous research reported for probiotics of the genus Lactobacillus to stimulate MIF production [83]. The obtained increase in MIF concentration levels from post-intervention to follow-up suggests a delayed response in the immune system, where the effect of probiotics on MIF became more apparent only after the intervention ceased. In the context of depression, delayed or dysregulated immune responsivity is a common feature [84]. Our results, together with previous findings, propose that probiotics play a modulatory role in immune-inflammatory processes, activating both stimulatory and inhibitory responses depending on context and probiotic strains.
Gene expression analysis revealed transcriptional changes in the probiotic group compared to the placebo group during the intervention. Our study found 108 DEGs following a multi-strain probiotic intervention. Only one other study has examined gene expression changes after probiotic administration in patients with depression, reporting higher IL-6 expression post-intervention [85]. Other studies have observed anti-inflammatory gene expression changes with probiotic supplementation in neurodegenerative disorders [86, 87]. Our findings showed that DEGs were related to immune activation pathways rather than anti-inflammatory processes, aligning with the literature suggesting different probiotic strains have distinct biological mechanisms [88]. Specifically, neutrophil-associated immune activation was the most significant effect, consistent with preclinical research indicating that the same multi-strain probiotic activates the epithelial innate immune system [77]. This “physiologic inflammation” induced by probiotics is proposed to enhance defense against harmful gut bacteria and support the innate immune system. Using WGCNA, we identified module M24, which captured upregulated immune activity and was associated with probiotic-induced ghrelin changes over time. M24 includes DEGs such as Olfactomedin 4 (OLFM4), Elastase (ELANE), and Defensin Alpha 4 (DEFA4), which are important for gastrointestinal defense. OLFM4 and DEFA4, in particular, have been linked to host defense in the mature intestine [89], and increased expression following probiotics supplementation of the genus Lactobacillus in mice [90]. OLFM4 is part of the gastrointestinal mucosal surface and may play a role in epithelial defense [91], while DEFA4 is involved in killing Gram-negative bacteria associated with gut dysbiosis [92].
This study had several strengths and limitations that necessitate careful consideration. A key strength lies in the systematic and comprehensive examination of diverse biological parameters associated with the mechanisms of the MGB axis involved in depression. By employing a combination of transcriptomics and blood concentration analyses, our methodology facilitated an in-depth exploration of the biological underpinnings relevant to the reported antidepressant mechanisms of probiotics. Notwithstanding these strengths, several limitations should be noted. The small sample size limits the generalizability of our findings, and the analyses conducted were exploratory and not a priori powered. Consequently, for the transcriptomics results of DGE, unadjusted p-values are reported which should be interpreted with caution. Additionally, the CAR was sampled until 30 min post-awakening, while consensus guidelines suggest sampling up to 30–45 mins post-awakening [93]. Moreover, we analyzed and reported total ghrelin levels without distinguishing between the acylated and desacylated forms. Considering the contrasting roles on energy balance reported for both forms (desacylated ghrelin is proposed to be a functional inhibitor of acylated ghrelin) [94, 95], distinguishing both forms would have provided more precise insights. Especially in the context of depression, as there is no conclusive evidence yet on the link of acylated, desacylated, or total ghrelin to depression and treatment response. Lastly, despite our efforts to record and control for medication effects, we cannot definitively ascertain whether the observed effects are specific to interactions with certain antidepressants, and the inclusion criteria (HAMD-17 > 7) led to a heterogeneous sample in terms of symptom severity. Future studies with larger sample sizes are necessary to validate and potentially extend our findings to patients with moderate to severe depression. Additionally, differentiating between both forms of ghrelin is crucial in future research to better understand their respective link to probiotics and their impact on depressive symptoms.
In conclusion, the biological mechanisms of add-on multi-strain probiotic supplementation in patients with depression were linked to the gut hormone ghrelin and the upregulation of genes of immune activation. Higher ghrelin levels after probiotic supplementation were furthermore related to improved depressive symptoms, hinting at a potential link between ghrelin secretion and antidepressant mechanisms. These findings emphasize probiotics’ biological mechanism of action as promoting immune activation ultimately associated to symptom relief in patients with depression. Moving forward, our results warrant replication in large-scale mechanistic trials of probiotic supplementation to test the potential of immune-inflammatory measures as stratification and treatment response biomarkers in depression. This could pave the way for more targeted and personalized approaches in the treatment of depression.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Rotenstein LS, Ramos MA, Torre M, Bradley Segal J, Peluso MJ, Guille C, et al. Prevalence of depression, depressive symptoms, and suicidal ideation among medical students: a systematic review and meta-analysis. JAMA. 2016;316:2214.
Abbafati C, Abbas KM, Abbasi M, Abbasifard M, Abbasi-Kangevari M, Abbastabar H, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. The Lancet. 2020;396:1204–22.
Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D Report. Am J Psychiatry. 2006;163:1905–17.
Zhang Q, Chen B, Zhang J, Dong J, Ma J, Zhang Y, et al. Effect of prebiotics, probiotics, synbiotics on depression: results from a meta-analysis. BMC Psychiatry. 2023;23. https://doi.org/10.1186/S12888-023-04963-X.
Chahwan B, Kwan S, Isik A, van Hemert S, Burke C, Roberts L. Gut feelings: a randomised, triple-blind, placebo-controlled trial of probiotics for depressive symptoms. J Affect Disord. 2019;253:317–26.
Nikolova VL, Cleare AJ, Young AH, Stone JM. Acceptability, tolerability, and estimates of putative treatment effects of probiotics as adjunctive treatment in patients with depression: a randomized clinical trial. JAMA Psychiatry. 2023;80:842.
Nikolova VL, Cleare AJ, Young AH, Stone JM. Updated review and meta-analysis of probiotics for the treatment of clinical depression: adjunctive vs. stand-alone treatment. J Clin Med. 2021;10:1–14.
Akkasheh G, Kashani-Poor Z, Tajabadi-Ebrahimi M, Jafari P, Akbari H, Taghizadeh M, et al. Clinical and metabolic response to probiotic administration in patients with major depressive disorder: a randomized, double-blind, placebo-controlled trial. Nutrition. 2016;32:315–20.
Schaub AC, Schneider E, Vazquez-Castellanos JF, Schweinfurth N, Kettelhack C, Doll JPK, et al. Clinical, gut microbial and neural effects of a probiotic add-on therapy in depressed patients: a randomized controlled trial. Translational Psychiatry. 2022;12:1–10.
Dinan TG, Stanton C, Cryan JF. Psychobiotics: a novel class of psychotropic. Biol Psychiatry. 2013;74:720–6.
Dinan TG, Cryan JF. Melancholic microbes: a link between gut microbiota and depression? Neurogastroenterol Motil. 2013;25:713–9.
Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12.
Kronsten VT, Tranah TH, Pariante C, Shawcross DL. Gut-derived systemic inflammation as a driver of depression in chronic liver disease. J Hepatol. 2022;76:665–80.
Ghosh S, Whitley CS, Haribabu B, Jala VR. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. 2021;11:1463–82.
Slyepchenko A, Maes M, Jacka FN, Köhler CA, Barichello T, McIntyre RS, et al. Gut microbiota, bacterial translocation, and interactions with diet: pathophysiological links between major depressive disorder and non-communicable medical comorbidities. Psychother Psychosom. 2016;86:31–46.
Kiecolt-Glaser JK, Wilson SJ, Bailey ML, Andridge R, Peng J, Jaremka LM, et al. Marital distress, depression, and a leaky gut: Translocation of bacterial endotoxin as a pathway to inflammation. Psychoneuroendocrinology. 2018;98:52–60.
Nikolova VL, Hall MRB, Hall LJ, Cleare AJ, Stone JM, Young AH. Perturbations in gut microbiota composition in psychiatric disorders: a review and meta-analysis. JAMA Psychiatry. 2021;78:1343–54.
Osimo EF, Baxter LJ, Lewis G, Jones PB, Khandaker GM. Prevalence of low-grade inflammation in depression: a systematic review and meta-analysis of CRP levels. Psychol Med. 2019;49:1958–70.
Raison CL, Miller AH. Is Depression an inflammatory disorder? Curr Psychiatry Rep. 2011;13:467.
Strawbridge R, Arnone D, Danese A, Papadopoulos A, Herane Vives A, Cleare AJ. Inflammation and clinical response to treatment in depression: a meta-analysis. Eur Neuropsychopharmacol. 2015;25:1532–43.
Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF, et al. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry. 2013;70:31–41.
Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013;6:39–51.
Yong SJ, Tong T, Chew J, Lim WL. Antidepressive mechanisms of probiotics and their therapeutic potential. Front Neurosci. 2019;13. https://doi.org/10.3389/FNINS.2019.01361.
Milajerdi A, Mousavi SM, Sadeghi A, Salari-Moghaddam A, Parohan M, Larijani B, et al. The effect of probiotics on inflammatory biomarkers: a meta-analysis of randomized clinical trials. Eur J Nutr. 2020;59:633–49.
Reiter A, Bengesser SA, Hauschild AC, Birkl-Töglhofer AM, Fellendorf FT, Platzer M, et al. Interleukin-6 gene expression changes after a 4-week intake of a multispecies probiotic in major depressive disorder-preliminary results of the PROVIT study. Nutrients. 2020;12:1–18.
Romijn AR, Rucklidge JJ, Kuijer RG, Frampton C. A double-blind, randomized, placebo-controlled trial of Lactobacillus helveticus and Bifidobacterium longum for the symptoms of depression. Aust N Z J Psychiatry. 2017;51:810–21.
Kazemi A, Noorbala AA, Azam K, Eskandari MH, Djafarian K. Effect of probiotic and prebiotic vs placebo on psychological outcomes in patients with major depressive disorder: a randomized clinical trial. Clin Nutr. 2019;38:522–8.
Rudzki L, Ostrowska L, Pawlak D, Małus A, Pawlak K, Waszkiewicz N, et al. Probiotic Lactobacillus Plantarum 299 v decreases kynurenine concentration and improves cognitive functions in patients with major depression: a double-blind, randomized, placebo controlled study. Psychoneuroendocrinology. 2019;100:213–22.
Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun. 2011;25:397–407.
Ma L, Yan Y, Webb RJ, Li Y, Mehrabani S, Xin B, et al. Psychological stress and gut microbiota composition: a systematic review of human studies. Neuropsychobiology. 2023;82:247–62.
Schalla MA, Stengel A. Effects of microbiome changes on endocrine ghrelin signaling – a systematic review. Peptides (NY). 2020;133:170388.
Ravussin Y, Koren O, Spor A, Leduc C, Gutman R, Stombaugh J, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20:738–47.
Queipo-Ortuño MI, Seoane LM, Murri M, Pardo M, Gomez-Zumaquero JM, Cardona F, et al. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS ONE. 2013;8. https://doi.org/10.1371/JOURNAL.PONE.0065465.
Rouach V, Bloch M, Rosenberg N, Gilad S, Limor R, Stern N, et al. The acute ghrelin response to a psychological stress challenge does not predict the post-stress urge to eat. Psychoneuroendocrinology. 2007;32:693–702.
Schmid DA, Held K, Ising M, Uhr M, Weikel JC, Steiger A. Ghrelin stimulates appetite, imagination of food, GH, ACTH, and cortisol, but does not affect leptin in normal controls. Neuropsychopharmacology. 2005;30:1187–92.
Kawakami A, Okada N, Rokkaku K, Honda K, Ishibashi S, Onaka T. Leptin inhibits and ghrelin augments hypothalamic noradrenaline release after stress. Stress. 2008;11:363–9.
Rubin RT, Rhodes ME, Czambel RK. Sexual diergism of baseline plasma leptin and leptin suppression by arginine vasopressin in major depressives and matched controls. Psychiatry Res. 2002;113:255–68.
Wu R, Dong W, Zhou M, Zhang F, Marini CP, Ravikumar TS, et al. Ghrelin attenuates sepsis-induced acute lung injury and mortality in rats. Am J Respir Crit Care Med. 2012;176: 805–13. https://doi.org/10.1164/rccm200604-511OC.
Dixit VD, Schaffer EM, Pyle RS, Collins GD, Sakthivel SK, Palaniappan R, et al. Ghrelin inhibits leptin- and activation-induced proinflammatory cytokine expression by human monocytes and T cells. J Clin Investig. 2004;114:57–66.
Nishi Y, Hiejima H, Hosoda H, Kaiya H, Mori K, Fukue Y, et al. Ingested medium-chain fatty acids are directly utilized for the acyl modification of ghrelin. Endocrinology. 2005;146:2255–64.
Yamanbaeva G, Schaub AC, Schneider E, Schweinfurth N, Kettelhack C, Doll JPK, et al. Effects of a probiotic add-on treatment on fronto-limbic brain structure, function, and perfusion in depression: secondary neuroimaging findings of a randomized controlled trial. J Affect Disord. 2023;324:529–38.
Schneider E, Doll JPK, Schweinfurth N, Kettelhack C, Schaub AC, Yamanbaeva G, et al. Effect of short-term, high-dose probiotic supplementation on cognition, related brain functions and BDNF in patients with depression: a secondary analysis of a randomized controlled trial. J Psychiatry Neurosci. 2023;48:E23–E33.
Hamilton M. Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol. 1967;6:278–96.
Zimmerman M, Martinez JH, Young D, Chelminski I, Dalrymple K. Severity classification on the Hamilton Depression Rating Scale. J Affect Disord. 2013;150:384–8.
Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–71.
Pruessner JC, Kirschbaum C, Meinlschmid G, Hellhammer DH. Two formulas for computation of the area under the curve represent measures of total hormone concentration versus time-dependent change. Psychoneuroendocrinology. 2003;28:916–31.
Tukey J. Exploratory data analysis. 1977.http://theta.edu.pl/wp-content/uploads/2012/10/exploratorydataanalysis_tukey.pdf. Accessed 15 Dec 2023.
Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:1–13.
Wittekind DA, Kluge M. Ghrelin in psychiatric disorders – a review. Psychoneuroendocrinology. 2015;52:176–94.
Stone LA, Harmatz ES, Goosens KA. Ghrelin as a stress hormone: implications for psychiatric illness. Biol Psychiatry. 2020;88:531–40.
Walker AK, Rivera PD, Wang Q, Chuang JC, Tran S, Osborne-Lawrence S, et al. The P7C3 class of neuroprotective compounds exerts antidepressant efficacy in mice by increasing hippocampal neurogenesis. Mol Psychiatry. 2015;20:500–8.
Poretti MB, Rask-Andersen M, Kumar P, Rubiales de Barioglio S, Fiol de Cuneo M, Schiöth HB, et al. Ghrelin effects expression of several genes associated with depression-like behavior. Prog Neuropsychopharmacol Biol Psychiatry. 2015;56:227–34.
Spencer SJ, Xu L, Clarke MA, Lemus M, Reichenbach A, Geenen B, et al. Ghrelin regulates the hypothalamic-pituitary-adrenal axis and restricts anxiety after acute stress. Biol Psychiatry. 2012;72:457–65.
Jensen M, Ratner C, Rudenko O, Christiansen SH, Skov LJ, Hundahl C et al. Anxiolytic-Like Effects of Increased Ghrelin Receptor Signaling in the Amygdala. Int J Neuropsychopharmacol. 2016;19. https://doi.org/10.1093/IJNP/PYV123.
Lutter M, Sakata I, Osborne-Lawrence S, Rovinsky SA, Anderson JG, Jung S, et al. The orexigenic hormone ghrelin defends against depressive symptoms of chronic stress. Nat Neurosci. 2008;11:752–3.
Spencer SJ, Emmerzaal TL, Kozicz T, Andrews ZB. Ghrelin’s role in the hypothalamic-pituitary-adrenal axis stress response: implications for mood disorders. Biol Psychiatry. 2015;78:19–27.
Kluge M, Schüssler P, Dresler M, Schmidt D, Yassouridis A, Uhr M, et al. Effects of ghrelin on psychopathology, sleep and secretion of cortisol and growth hormone in patients with major depression. J Psychiatr Res. 2011;45:421–6.
Algul S, Ozcelik O. Evaluating the levels of nesfatin-1 and ghrelin hormones in patients with moderate and severe major depressive disorders. Psychiatry Investig. 2018;15:214.
Wittekind DA, Kratzsch J, Biemann R, Mergl R, Riedel-Heller S, Witte V, et al. Association between self-rating depression scores and total ghrelin and adipokine serum levels in a large population-based sample. Front Psychiatry. 2022;13. https://doi.org/10.3389/FPSYT.2022.891325.
van Andel M, van Schoor NM, Korten NC, Heijboer AC, Drent ML. Ghrelin, leptin and high-molecular-weight adiponectin in relation to depressive symptoms in older adults: results from the Longitudinal Aging Study Amsterdam. J Affect Disord. 2022;296:103–10.
Kluge M, Schüssler P, Schmid D, Uhr M, Kleyer S, Yassouridis A, et al. Ghrelin plasma levels are not altered in major depression. Neuropsychobiology. 2009;59:199–204.
Gecici O, Kuloglu M, Atmaca M, Tezcan AE, Tunckol H, Emül HM, et al. High serum leptin levels in depressive disorders with atypical features. Psychiatry Clin Neurosci. 2005;59:736–8.
Kurt E, Guler O, Serteser M, Cansel N, Ozbulut O, Altinbaş K, et al. The effects of electroconvulsive therapy on ghrelin, leptin and cholesterol levels in patients with mood disorders. Neurosci Lett. 2007;426:49–53.
Ozsoy S, Besirli A, Abdulrezzak U, Basturk M. Serum ghrelin and leptin levels in patients with depression and the effects of treatment. Psychiatry Investig. 2014;11:167.
Barim AO, Aydin S, Colak R, Dag E, Deniz O, Sahin I. Ghrelin, paraoxonase and arylesterase levels in depressive patients before and after citalopram treatment. Clin Biochem. 2009;42:1076–81.
Matsuo K, Nakano M, Nakashima M, Watanuki T, Egashira K, Matsubara T, et al. Neural correlates of plasma acylated ghrelin level in individuals with major depressive disorder. Brain Res. 2012;1473:185–92.
Schanze A, Reulbach U, Scheuchenzuber M, Gröschl M, Kornhuber J, Kraus T. Ghrelin and eating disturbances in psychiatric disorders. Neuropsychobiology. 2008;57:126–30.
Khoder G, Al-Yassir F, Al Menhali A, Saseedharan P, Sugathan S, Tomasetto C, et al. Probiotics Upregulate Trefoil Factors and Downregulate Pepsinogen in the Mouse Stomach. Int J Mol Sci. 2019;20. https://doi.org/10.3390/IJMS20163901.
Giorgia G, Elia C, Andrea P, Cinzia C, Stefania S, Ana R, et al. Effects of lactogen 13, a new probiotic preparation, on gut microbiota and endocrine signals controlling growth and appetite of Oreochromis niloticus juveniles. Microb Ecol. 2018;76:1063–74.
Saito H, Nakakita Y, Segawa S, Tsuchiya Y. Oral administration of heat-killed Lactobacillus brevis SBC8803 elevates the ratio of acyl/des-acyl ghrelin in blood and increases short-term food intake. Benef Microbes. 2019;10:671–7.
Yadav H, Lee JH, Lloyd J, Walter P, Rane SG. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J Biol Chem. 2013;288:25088–97.
Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–88.
Wall R, Ross RP, Shanahan F, O’Mahony L, Kiely B, Quigley E, et al. Impact of administered bifidobacterium on murine host fatty acid composition. Lipids. 2010;45:429–36.
Rajkumar H, Mahmood N, Kumar M, Varikuti SR, Challa HR, Myakala SP Effect of probiotic (VSL#3) and omega-3 on lipid profile, insulin sensitivity, inflammatory markers, and gut colonization in overweight adults: a randomized, controlled trial. Mediators Inflamm. 2014;2014. https://doi.org/10.1155/2014/348959.
Rahimlou M, Hosseini SA, Majdinasab N, Haghighizadeh MH, Husain D. Effects of long-term administration of Multi-Strain Probiotic on circulating levels of BDNF, NGF, IL-6 and mental health in patients with multiple sclerosis: a randomized, double-blind, placebo-controlled trial. Nutr Neurosci. 2022;25:411–22.
Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. The impact of probiotics and prebiotics on the immune system. Nat Rev Immunol. 2012;12:728–34.
Pagnini C, Saeed R, Bamias G, Arseneau KO, Pizarro TT, Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci USA. 2010;107:454–9.
Biagioli M, Carino A, Di Giorgio C, Marchianò S, Bordoni M, Roselli R, et al. Discovery of a novel multi-strains probiotic formulation with improved efficacy toward intestinal inflammation. Nutrients. 2020;12:1945.
Biagioli M, Capobianco D, Carino A, Marchianò S, Fiorucci C, Ricci P, et al. Divergent effectiveness of multispecies probiotic preparations on intestinal microbiota structure depends on metabolic properties. Nutrients. 2019;11:325.
Duque GA, Descoteaux A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front Immunol. 2014;5:117833.
Vujicic M, Saksida T, Despotovic S, Bajic SS, Lalić I, Koprivica I, et al. The role of macrophage migration inhibitory factor in the function of intestinal barrier. Sci Rep. 2018;8:1–12.
Roger T, Delaloye J, Chanson AL, Giddey M, Le Roy D, Calandra T. Macrophage migration inhibitory factor deficiency is associated with impaired killing of Gram-negative bacteria by macrophages and increased susceptibility to Klebsiella pneumoniae sepsis. J Infect Dis. 2013;207:331–9.
Jeffrey MP, MacPherson CW, Mathieu O, Tompkins TA, Green-Johnson JM. Secretome-mediated interactions with intestinal epithelial cells: a role for secretome components from Lactobacillus rhamnosus R0011 in the attenuation of Salmonella enterica serovar typhimurium secretome and TNF-α–induced proinflammatory responses. J Immunol. 2020;204:2523–34.
Tubbs JD, Ding J, Baum L, Sham PC. Immune dysregulation in depression: evidence from genome-wide association. Brain Behav Immun Health. 2020;7:100108.
Reiter A, Bengesser SA, Hauschild AC, Birkl-Töglhofer AM, Fellendorf FT, Platzer M, et al. Interleukin-6 gene expression changes after a 4-week intake of a multispecies probiotic in major depressive disorder—preliminary results of the PROVIT study. Nutrients. 2020;12:2575.
Tamtaji OR, Kouchaki E, Salami M, Aghadavod E, Akbari E, Tajabadi-Ebrahimi M, et al. The effects of probiotic supplementation on gene expression related to inflammation, insulin, and lipids in patients with multiple sclerosis: a randomized, double-blind, placebo-controlled trial. J Am Coll Nutr. 2017;36:660–5.
Borzabadi S, Oryan S, Eidi A, Asemi Z. The effects of probiotic supplementation on gene expression related to inflammation, insulin and lipid in patients with Parkinson’s disease: a randomized, double-blind, placebocontrolled trial. Arch Iran Med. 2018;21:289–95.
Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A. Mechanisms of action of probiotics. Advances in Nutrition. 2019;10:S49.
Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, et al. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Reports. 2015;4:1140–55.
Lee H, Jung KB, Kwon O, Son YS, Choi E, Yu WD, et al. Limosilactobacillus reuteri DS0384 promotes intestinal epithelial maturation via the postbiotic effect in human intestinal organoids and infant mice. Gut Microbes. 2022;14. https://doi.org/10.1080/19490976.2022.2121580.
Grover PK, Hardingham JE, Cummins AG. Stem cell marker olfactomedin 4: critical appraisal of its characteristics and role in tumorigenesis. Cancer Metastasis Rev. 2010;29:761–75.
Basingab F, Alsaiary A, Almontashri S, Alrofaidi A, Alharbi M, Azhari S, et al. Alterations in immune-related defensin alpha 4 (DEFA4) gene expression in health and disease. Int J Inflam. 2022;2022. https://doi.org/10.1155/2022/9099136.
Stalder T, Kirschbaum C, Kudielka BM, Adam EK, Pruessner JC, Wüst S, et al. Assessment of the cortisol awakening response: Expert consensus guidelines. Psychoneuroendocrinology. 2016;63:414–32.
Delhanty PJD, Neggers SJ, Van Der Lely AJ. Mechanisms in endocrinology: Ghrelin: the differences between acyl- and des-acyl ghrelin. Eur J Endocrinol. 2012;167:601–8.
Asakawa A, Inui A, Fujimiya M, Sakamaki R, Shinfuku N, Ueta Y, et al. Stomach regulates energy balance via acylated ghrelin and desacyl ghrelin. Gut. 2005;54:18–24.
Acknowledgements
The study was supported by the Gertrud Thalmann Foundation of the University Psychiatric Clinics (UPK) Basel (AS, UEL), the Kämpf-Bötschi Foundation (UEL), the research fund junior researchers from University of Basel (Appln 3MS1041, AS), the research fund of the UPK Basel (AS) and the Stiftung zur Förderung der gastroenterologischen und allgemeinen klinischen Forschung sowie der medizinischen Bildauswertung (AS). MENDES S.A., Switzerland, supplied the investigational medicinal product. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
AS had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. LM, UEL, and AS designed the study. LS, JPKD, VL, ACS, CK, LM, NS and AS contributed to the acquisition, analysis, or interpretation of data. Statistical analyses of blood/saliva concentration and appetite measures were performed by LS and VL. Transcriptomic analyses and interpretations were performed by FM and MI from the Vital-IT Competence Center in Bioinformatics, SIB Swiss Institute of Bioinformatics, University of Lausanne. Administrative, technical, or material support was provided by LM, AS, and UEL. LS and AS drafted the manuscript. All authors critically reviewed the article and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sempach, L., Doll, J.P.K., Limbach, V. et al. Examining immune-inflammatory mechanisms of probiotic supplementation in depression: secondary findings from a randomized clinical trial. Transl Psychiatry 14, 305 (2024). https://doi.org/10.1038/s41398-024-03030-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-024-03030-7
This article is cited by
-
The Role and Mechanisms of Probiotic Supplementation on Depressive Symptoms: A Narrative Review
Current Nutrition Reports (2025)
-
Probiotics Encapsulated via Biological Macromolecule for Neurological Therapy and Functional Food: A Review
Probiotics and Antimicrobial Proteins (2025)