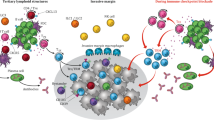
Abstract
Esophageal squamous cell carcinoma (ESCC) is a highly aggressive malignancy with a dismal prognosis. Hitherto, little has been known regarding the clinical implications of tertiary lymphoid structures (TLS) and its biological mechanisms of antitumor effect on treatment-naïve ESCC. We herein identified the presence of TLS as an independent factor for favorable survival. By characterizing the immune infiltration and genomic profiles based on transcriptomic datasets, we found TLS abundant in enriched B cells with IRF4 as a signature gene. Increased expression of IRF4 and its positive correlation with STING in activating tumor-infiltrating B cells were also investigated using a single-cell RNA sequencing dataset. CD40 as a co-regulator of IRF4 and TLS formation, in vitro experiments were conducted to further demonstrate the competitive binding relationships between CD40 and STING with TRAF2 in promoting IRF4 expression and B cell activation via the non-canonical NF-kB signaling pathway, in which CD40 reduced STING ubiquitination while promoting its phosphorylation. Our data provided deeper insights into the potential role of activated B cells and TLS in ESCC, with implications for the development of biomarkers and therapeutic targets.

Similar content being viewed by others
Introduction
Esophageal squamous cell carcinoma (ESCC) takes up an overwhelming majority of esophageal cancer (EC) in China, which is characterized by early lymphatic metastasis and poor long-term survival [1]. Recently, nivolumab [CheckMate-577] has been approved for the adjuvant treatment of patients with EC who have residual pathology after neoadjuvant chemoradiotherapy (nCRT) and complete surgical resection, which demonstrated the tremendous potential of immunotherapy for EC [2]. However, the lack of efficacy in most patients receiving PD-1/PD-L1 inhibitors [3], coupled with substantial expenses, highlighted the necessity for advancing predictive biomarkers and refining treatment strategies.
Earlier evidence has demonstrated that tertiary lymphoid structures (TLS) act as a foundation for adaptive immunity against tumors [4, 5], which could decelerate disease progression and prognosticate favorable survival in EC [6], melanoma [7], non-small cell lung cancer [8], and other malignancies. However, the specific molecular mechanisms underlying the formation process and antitumor responses of TLS remain poorly understood. Previous studies in ESCC revealed enriched B cell gene signatures and high B cell receptor clonality within TLS, affirming the presence of activated B cell populations with antitumor effects [9]. Activated B cells may recruit more lymphocytes and form TLS by releasing chemokines like IL-17, CXCL13, and CCL21 [10].
Interferon regulatory factor 4 (IRF4) has been confirmed to play a crucial role in B cell development at various stages and in adaptive immune responses, which is a member of the transcription factor family [11]. Studies have indicated that IRF4 deficiency might impair B cell proliferation and survival following BCR activation, thereby inhibiting B cell responses [12]. In contrast to other IRF proteins, IRF4 expression is triggered by multiple mitogenic signals, such as antigen receptors, LPS, and CD40 pathways, which stimulate NF-κB activity and subsequently initiate IRF4 promoter activation [13]. CD40, a transmembrane protein of type I, is present on various cell surfaces, such as B cells, monocytes, dendritic cells, endothelial cells, and epithelial cells [14]. It plays a crucial role in initiating and advancing both cellular and humoral adaptive immune responses, including the development of memory B cells and the formation of germinal centers [15]. It has been reported that CD40 can promote the formation of TLS [16]. Meanwhile, a previous study suggested that B cells could be directly activated by stimulator of interferon genes (STING) signaling triggers to promote antibody responses [17]. STING has also been confirmed to promote TLS formation in malignancies [18]. The foregoing evidence indicated the importance of CD40 and STING in B cell activation. Additionally, CD40 and STING were found to engage with TRAFs, triggering the activation of the non-canonical NF-κB signaling pathway [19, 20], but the mutual relationship between CD40 and STING in regulating IRF4 expression has been unknown.
In this study, we investigated the clinical implications and immune profiles of TLS in treatment-naïve ESCC, and found that CD40 competitively bound TRAF2 with STING to promote the IRF4-mediated B cell activation via the non-canonical NF-κB signaling pathway.
Methods
Patients and tissue samples
Patients who underwent esophagectomy for newly diagnosed ESCC at the Second Affiliated Hospital of Soochow University from January 2009 to December 2014 were retrospectively reviewed. Clinical and pathological data were extracted from electronic health records, including sex, age, smoking status, tumor site, surgical approach, vascular and perineural invasion, and TNM staging. Inclusion criteria comprised: (1) patients completed R0 resection, (2) patients had no prior systemic treatment, and (3) patients who were pathologically diagnosed with primary TxNxM0 ESCC based on the 8th TNM classification. We also excluded patients who suffered from autoimmune disease or concurrent multiple primary tumors. The collected tissues were immersed in RNAstore reagent (CoWin Biosciences, Taizhou, China) for RNA sequencing or fixed in 10% formalin for tissue sections. The study was approved by the Review Committee of the Ethics Institute of the Second Affiliated Hospital of Soochow University (JD-HG-2025-056).
Immunohistochemistry staining and evaluation
Three consecutive 4-µm formalin-fixed and paraffin-embedded tumor sections were prepared for CD3 and CD20 labeling. Immunohistochemistry (IHC) was conducted as follows. Primarily, the tissues underwent xylene-based deparaffinization and alcohol gradient rehydration. Peroxidase activity was inhibited using 3% H2O2 for 15 min, followed by 10% goat serum blocking for 60 min. Slides were washed with 0.01 M PBS for 3 min, then treated with primary antibodies [CD3 and CD20 (Abcam, MA, USA)] at 37 °C for 1 h. Secondary antibody incubation lasted 30 min, followed by 5-min DAB (DAKO, Glostrup, Denmark) staining. Nuclear visualization was achieved through hematoxylin counterstaining, with subsequent ethanol dehydration. Sections were mounted with neutral balsam for imaging and evaluation.
Two senior pathologists (Li F. and Zhang Y.), unaware of the clinicopathological information and clinical outcomes of the patients, independently evaluated the tissue samples. CD3 and CD20 expression was quantified by assessing staining intensity and positive cell counts across five high-power fields in triplicate tumor sections. Discrepancies were resolved through consensus-based discussions. TLS identification followed the established criteria [21], with TLS+ indicating at least one detectable TLS while TLS- representing their complete absence.
Bioinformatic and statistical analyses
Transcriptomic data from 220 ESCC cases, comprising 179 treatment-naive patients from the Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.gov/geo) (GSE53625) and 41 patients from our center, were analyzed to delineate TLS-associated immune infiltration and identify differentially expressed genes (DEGs). Computational analyses were executed using R 4.1.2 (http://www.r-project.org), with immune cell meta-gene signatures for seven immune cell types were obtained from the TISIDB database (http://cis.hku.hk/TISIDB/index.php). TLS quantification employed a 12-gene signature [22], while hierarchical clustering via hclust() stratified patients into high/low IRF4 or TLS subgroups. Immune infiltration was annotated through single-sample gene set enrichment analysis (ssGSEA) implemented in GSVA (Gene Set Variation Analysis), visualized using the package pheatmap. Normalized data processed with Limma (linear models for microarray data) enabled DEG identification between TLS subgroups. Prognostic genes were determined through univariate Cox regression, with associations between TLS levels and the survival-related DEGs assessed via Student’s t-test, considering p < 0.05 as statistically significant.
Single-cell transcriptomic data were obtained from the GSE160269 repository. Subsequent processing utilized Seurat (v4.2.1) for analytical workflows. Initial quality assessment involved filtering to retain optimal cellular profiles. Since the samples were processed independently and single-cell sequencing data often contain high-dimensional variables that can introduce batch effects, we applied the “Harmony” R package and the RunUMAP function in Seurat to reduce dimensionality and mitigate batch effects.
Cell culture and reagents
TE-1 (RRID: CVCL_1759) and Eca-109 (RRID: CVCL_6898) esophageal carcinoma cell lines were purchased from Boster (Wuhan, Hubei, China). Cellular maintenance was conducted using RPMI 1640 (Gibco, NY, USA) enriched with 10% fetal bovine serum (FBS) (Sigma, MO, USA) and 1% penicillin/streptomycin/Amphotericin B solution (Beyotime Biotechnology, Shanghai, China). Cells were incubated at 37 °C under 5% CO2 conditions with controlled humidity. All human cell lines have been authenticated using STR profiling within the last 3 years. All experiments were performed with mycoplasma-free cells.
To generate ESCC-conditioned medium (TE-1-CM and Eca-109-CM), cellular cultures were established in previously described growth conditions. Upon reaching 70% confluence in 10 cm culture plates, the medium was replaced. Following 24-h incubation, the solution was centrifuged at 800 rpm for 5 min. The resultant supernatant was filtered through 0.45-μm membranes to eliminate particulate matter and cellular fragments, then preserved at 4 °C for subsequent applications.
Ubiquitination inhibitors (PYR-41, B1292, 10 μM) and phosphorylation inhibitors (phosphatase inhibitor cocktail 1, K1012, 1 ×) were purchased from Apexbio (TX, USA). Anti-phospho-STING was purchased from CST (50907, Cell Signaling Technology, MA, USA). Anti-phospho-IKK (AF3013), anti-IKK (AF6014), anti-p65 (AF5006), and anti-p52 (AF6373) were purchased from Affinity (Affinity Biosciences, Changzhou, China).
B cell isolation and culture
Briefly, B220-positive lymphocytes (positive sorting) were purified from healthy donor peripheral blood through magnetic-activated cell sorter (MACS) separation column (Miltenyi Biotec, Auburn, CA). The cells were cultivated in the ImmuCultmi Human B Cell Expansion Kit (StemCell Technologies, Vancouver, CA). Pre-B cell cultures were subcultured triweekly into fresh medium. Experimental procedures exclusively utilized cells within five passages.
Western blotting
Cellular lysis was performed using RIPA solution (Beyotime Biotechnology, Shanghai, China) supplemented with phenylmethylsulfonyl fluoride (PMSF) (Beyotime Biotechnology, Shanghai, China) under ice-cold conditions for 30 min. Protein concentrations were determined through the BCA assay (Boster, Wuhan, China), followed by denaturation in loading buffer (Fdbio Science, Hangzhou, China). Equivalent protein quantities were resolved through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, MA, USA). Non-specific binding sites were blocked with 5% BSA solution (Beyotime Biotechnology, Shanghai, China) for 60 min at room temperature (RT). Membranes were probed with primary antibodies overnight at 4 °C, then incubated with HRP-labeled secondary antibodies for 1 h at RT. Chemiluminescent signals were visualized using an e-Blot Touch Imager (e-Blot, Shanghai, China) with western HRP detection reagent (Fdbio Science, Hangzhou, China).
Cell proliferation
B lymphocytes (2 × 103/well) were seeded in 96-well plates containing standard culture medium. Following 24-h incubation, the medium was replaced with TE-1-CM or Eca-109-CM. Cellular proliferation was monitored daily from day 1 to 5 using the CCK-8 method. The CCK-8 reagent was obtained from Dojindo Chemical Co., Japan, for proliferation analysis.
Fluorescence-activated cell sorter (FACS) analysis
Cellular suspensions were initially treated with 2% rat serum or Fc-Block (2.4G2) prior to staining. Optimal concentrations of biotinylated or fluorophore-labeled antibodies were applied for immunostaining. Anti-CD19 (APC) and anti-pre-BCR (PE) monoclonal antibodies were acquired from BD Biosciences (San Jose, CA). Flow cytometric evaluation was conducted using a FACS Calibur instrument.
Reverse transcription-polymerase chain reaction
Cellular RNA was isolated using the RNA-Quick Purification Kit (ESScience, Shanghai, China). Nucleic acid quantification was determined through the ABI-7300 analyzer (Applied Biosystems ABI, USA). cDNA synthesis and RT-PCR amplification were executed with TransScript one-step RT-PCR SuperMix (TransGen Biotech, Beijing, China). The following primer pairs were utilized [forward (F) and reverse (R) primers]:
IL10 F 5’-CGAGATGCCTTCAGCAGAGT-3’, R 5’-GGCAACCCAGGTAACCCTT
A-3’
TGFβ F 5’-GGAAATTGAGGGCTTTCGCC-3’, R 5’-CCGGTAGTGAACCCGTTG
AT-3’
CXCL13 F 5’-CCTCTCTCCAGTCCAAGGTGT-3’, R 5’-TCTTGGACAACCATTC
CCACG-3’
IL35 F 5’-CATTGCCACGTACAGGCTCG-3’, R 5’-CGTAGGGAGCCATGGAGA
AC-3’
IL-17 F 5’-CACCTTGGAATCTCCACCGC-3’, R 5’-GGATCTCTTGCTGGATGGG
G-3’
CCL21 F 5’-GCTCTGGCCTCTTACTCACC-3’ R 5’-CTCCATCACTGCCTTGGGT
C-3’
GAPDH F 5′-ATCACTGCCACCCAGAAG-3′, R 5′-TCCACGACGGACACATTG-3′.
Co-immunoprecipitation (Co-IP) and immunoblot analysis
Protein extraction was conducted following stimulation with CD40 ligand (recombinant human sCD40L, 1 µg/ml, PeproTech, NJ, USA) or STING agonist (STING agonist-1, 10 µM, Apexbio, TX, USA). Antibody-bead complexes were formed by incubating 10 µg/mL primary antibodies (anti-TRAF2, Boster, BM4677, Wuhan, China; anti-TRAF3, Proteintech, 66310-1-lg, Wuhan, China; anti-TRAF6, abcam, ab137452, MA, USA) with 20 µl Protein A/G beads (Beaver, 22202-20, Suzhou, China) for 15 minutes at room temperature. Cellular protein solutions (50 µg/mL) were subsequently incubated with these complexes for 60 minutes at 4 °C with constant agitation. Electrophoretic separation was performed using SDS-PAGE, followed by protein transfer onto PVDF membranes (Thermo Fisher Scientific, MA, USA). Immunoblotting was carried out using specific primary antibodies and HRP-conjugated secondary antibodies, with detection achieved through western HRP substrate (Fdbio Science, Hangzhou, China). The input group was the positive control, and the IgG group was the negative control.
Immunofluorescence (IF)
Cellular specimens were immobilized using 4% paraformaldehyde in phosphate-buffered saline (pH 7.3) for 15 min, then subjected to membrane permeabilization with 0.5% Triton X-100 for an equivalent duration at RT. Following one-hour blocking with 3% bovine serum albumin in PBS, specimens were incubated with primary antibodies (anti-CD40, anti-STING, and anti-TRAF2 rabbit monoclonal antibodies, Affinity Biosciences, Changzhou, China) at a 1:200 dilution for 60 min at RT. Multiplex fluorescent labeling was performed using FITC, CY3, and CY5 conjugates, with subsequent triple washing steps. Mounting was accomplished using ProLong Gold anti-fade reagent containing DAPI (Life Technologies, MD, USA). Microscopic visualization was conducted using a Zeiss LSM 780 confocal imaging system.
Ubiquitination assay
To investigate STING ubiquitination dynamics, B lymphocytes were activated using CD40 ligand (recombinant human sCD40 Ligand, 1 µg/ml, PeproTech, NJ, USA). Two days after stimulation, cells were treated with cycloheximide (CHX, protein synthesis inhibitor, 10 μM, Apexbio, TX, USA) at different times to observe the stability of STING. Proteasomal degradation was blocked through MG132 treatment (10 μM, Apexbio, TX, USA) for 4 h prior to cell harvesting and lysis. Then, 10 µg/mL of primary antibody working solution (anti-STING, abcam, ab239074, MA, USA) was added to 20 µL of Protein A/G beads (Beaver, 22202-20, Suzhou, China) for 15 min at RT. Subsequently, 50 µg/mL of cellular protein solution were added to the magnetic bead-antibody complex and permitted to precipitate for 1 h at 4 °C on a rocker platform, followed by immunoblot analysis with an anti-ubiquitin antibody (Zen-Bioscience, R26024, Chengdu, China) to detect ubiquitinated STING.
Statistical analysis
Statistical analyses were performed utilizing SPSS version 25.0 (IBM Corporation, Armonk, NY, USA). The sample size calculation was employed by the Schoenfeld method, demonstrating that a total of 112 patients need to be enrolled to meet statistical requirements. Associations between TLS and clinical parameters were assessed through chi-square testing and Spearman’s correlation analysis, with results presented as frequency distributions (n, %). Logistic regression modeling identified independent predictors of TLS presence. Survival outcomes, including recurrence-free survival (RFS) and overall survival (OS), were evaluated using Kaplan–Meier methodology, with intergroup comparisons conducted via log-rank testing. The prognostic significance of TLS for survival endpoints was determined through time-dependent Cox proportional hazards modeling. Variables demonstrating p < 0.2 in univariate analyses were incorporated into multivariate Cox regression models. Statistical significance was established at p < 0.05 for all two-tailed tests.
Result
Characterization of TLS in ESCC and its association with clinical outcomes
The clinicopathological features of 183 treatment-naive ESCC patients are summarized in Table S1. The presence of TLS within the tumor microenvironment was determined through combined hematoxylin-eosin (H&E) staining and immunohistochemical analysis (Fig. 1A). These structures were localized either within the tumor core or at the invasive front.
A Representative image of TLS+ and TLS- detected in formalin-fixed paraffin-embedded tumor sections by haematoxylin and eosin (H&E) staining and images of TLS+ detected by immunohistochemistry staining showing CD3 + T cell zones and CD20 + B cell zones (scale bar = 100 μm). Kaplan–Meier curves for RFS (B) and OS (C) in ESCC patients based on the presence of TLS. TLS tertiary lymphoid structures, ESCC esophageal squamous cell carcinoma, RFS recurrence-free survival, OS overall survival.
As demonstrated in Table S1, the formation of TLS showed no statistically significant correlations with various clinicopathological parameters, including patient age, gender, smoking status, and perineural invasion, etc. Notably, multivariable logistic regression analysis identified TNM stage as an independent predictor of the presence of TLS [odds ratio (OR) = 0.175, 95% confidence interval (95% CI): 0.058-0.52, p = 0.002, Table S2].
As shown in Table S3, the multivariate analysis showed that advanced TNM stages [hazard ratio (HR) = 3.704, 95% CI: 1.772-7.743, p < 0.001], elder age (HR = 1.618, 95% CI: 1.052-2.490, p = 0.028), and positive PD-1 expression (HR = 2.777, 95% CI: 1.756–4.393, p < 0.001) were independent risk factors for RFS, whereas presence of TLS (HR = 0.251, 95% CI: 0.144-0.437, p < 0.001) was an indicator of prolonged RFS (Fig. 1B). Furthermore, the presence of TLS was also identified as a prognosticator for better OS (HR = 0.148, 95% CI: 0.077–0.282, p < 0.001) in the multivariate analysis (Fig. 1C).
Immune infiltration and genomic profiles of TLS in ESCC patients
We analyzed the sequencing data of 179 patients from the GEO database and those of 41 patients from our institution, aiming to elucidate the immune infiltration characteristics associated with TLS in ESCC. The clinicopathological features of 41 ESCC patients are summarized in Table S4. Based on the median expression levels of TLS-associated genes, the study population was divided into two subgroups characterized by high and low-TLS infiltration. As shown in Fig. 2A, B, ESCC within the high-TLS subgroup were characterized by substantial enrichment of immunocompetent cells, particularly activated B lymphocytes and CD4 + T cell populations.
A Immune infiltration patterns of ESCC stratified by the presence of TLS in the GEO dataset GSE53625. B Immune infiltration patterns of ESCC stratified by the presence of TLS in 41 surgical specimens from the internal cohort. C Impact of the intersecting gene IRF4 on the prognosis of ESCC. D GO enrichment analysis of the intersecting genes. ESCC esophageal squamous cell carcinoma, DEGs differentially expressed genes, TLS tertiary lymphoid structures, IRF4 interferon regulatory factor 4, GO gene ontology.
To achieve the survival-related DEGs, we investigated the DEGs between the high-TLS group and the low-TLS group. Only IRF4 was obtained in the intersection of the DEGs from the two cohorts, which possibly accounted for the survival advantages regarding TLS. Specifically, Kaplan–Meier curves indicated that high-IRF4 expression in ESCC was associated with better prognosis (Fig. 2C). It was noted that IRF4 exhibited a higher expression level in the high-TLS group. GO enrichment analysis also indicated that IRF4 was significantly associated with immune response (Fig. 2D).
Identification of IRF4-related immunophenotypes and its relationship with STING
Tumor-infiltrating B cells have been identified in TLS within ESCC [23]. IRF4, as an immune response-related marker [24], tended to be expressed in activated B cells in the single-cell RNA sequencing dataset (Fig. 3A, B). We also identified a positive correlation between the expression level of IRF4 and the proportion of activated B cells (Fig. 3C). Additionally, it was suggested that the ESCC samples in the high-IRF4 group were abundant in activated B cells (Fig. 3D, E). Furthermore, we found that both STING and IRF4 were enriched in the NF-κB pathway and exhibited a positive correlation (Fig. 3F, G).
A Characterization of B cells with IRF4 expression in the single-cell sequencing dataset GSE160269. B Association between IRF4 and B cell subtypes within the TLS-enriched samples in the single-cell sequencing dataset GSE160269. C Positive correlation between IRF4 gene expression and activated B cell infiltration. D Immune infiltration patterns of ESCC stratified by IRF4 expression levels in the GEO dataset GSE53625. E Immune infiltration pattern of ESCC stratified by IRF4 expression levels in 41 surgical specimens from our internal cohort. F Enrichment of IRF4 and STING in the NF-κB pathway. G Relationship between IRF4 and STING expression levels.
CD40 promoted the expression level of IRF4 and B cell activation in ESCC
It was revealed that TE-1 CM and Eca-109 CM failed to modulate IRF4 expression in B lymphocytes (Fig. S1A, B). However, upon stimulation with CD40 agonist, there was a significant increase in IRF4 expression (Fig. 4A, B). Although TE-1 CM/Eca-109 CM failed to stimulate B cell proliferation, CD40 agonist co-stimulation in these media significantly augmented cellular proliferation, as quantified by CCK-8 assay (Figs. 4C and S1C). Our results also suggested that activation of CD40 and its downstream significantly enhanced the B cell activation and expression of TLS-related chemokines in these ESCC cells through flow cytometry and RT-qPCR (p < 0.05) (Figs. 4D, E and S1D, E).
A, B Impact of CD40 stimulation on expression of IRF4 in B cells under different ESCC-CM (n = 3). C Impact of CD40 stimulation on the inhibitory effect of B cells on tumor cell proliferation (n = 3). D Flow cytometric analysis indicated that CD40 stimulation altered the degree of B cell differentiation (APC-CD19, PE-pre-BCR). E The effect of CD40 stimulation on the production of TLS-related chemokines in B cells under TE-1 and EC109 CM (n = 3). CM conditioned medium. *p < 0.05; **p < 0.01; ***p < 0.0001.
CD40 competitively bound TRAFs with STING
Previous evidence has uncovered that CD40 possesses cytoplasmic regions capable of interacting with TRAF2, TRAF3, and TRAF6 within both classical and alternative NF-κB signaling cascades [25]. Analogously, STING operates through TRAF3 and TRAF6 recruitment to regulate NF-κB activation triggered by double-stranded DNA via distinct pathways [26]. To explore potential TRAF-mediated signaling convergence between CD40 and STING, we initially examined their respective associations with TRAF proteins. Our data demonstrated direct interactions between both CD40 and STING with multiple TRAF family members, with particularly strong binding with TRAF2 (Fig. 5A). Similarly, exogenous activation of CD40 could protect STING from degradation (Fig. 5B). Immunofluorescence results revealed that CD40 exhibited stronger binding affinity to TRAF2 compared with STING (Fig. 5C). In other word, CD40 exhibited competitive TRAF2 binding capacity against STING. Enhanced CD40-TRAF2 interaction consequently diminished STING-TRAF2 complex formation, resulting in attenuated STING proteolysis. Western blot showed that the exogenous activation of CD40, STING, or both greatly enhanced the NF-κB activation (Fig. 5D). Flow cytometry further suggested that both STING and CD40 could activate the non-canonical NF-κB pathways by recruiting TRAF2 and therefore promote B cell activation (Fig. 5E).
A Co-immunoprecipitation (co-IP) of CD40 or STING. B Co-IP using anti-TRAF2 to pull down STING and immunoblot (IB) detection of CD40 and STING molecules. C Multiplex immunofluorescence demonstrated the interactions of CD40 and STING with TRAF2 (scale bar = 100 μm). D Impact of CD40 and STING stimulation in the NF-κB pathway. E Flow cytometry revealed the degrees of B cell differentiation under different stimuli (APC-CD19, PE-pre-BCR).
CD40 interacted with STING through TRAF2 and reduced STING ubiquitination
To elucidate the molecular bases of CD40-mediated STING regulation, we examined STING ubiquitination patterns in B lymphocytes under conditions of CD40 overexpression and its absence. After adding the protein synthesis inhibitor CHX, we observed that CD40 can stabilize the expression of STING (Fig. 6A). Experimental data revealed that CD40 agonist administration significantly decreased STING ubiquitination levels while enhancing its phosphorylation status (Fig. 6B). Meanwhile, we observed that IRF4 expression, STING phosphorylation and NF-κB activation were remarkably attenuated after stimulation of CD40 agonist and phosphorylation inhibitor (Fig. 6C). Similarly, B cell activation was impeded when exogenous phosphorylation inhibitor was added, indicating that CD40 promoted compensatory phosphorylation of STING (Fig. 6D). STING maintained its ability to enhance IRF4 expression, NF-κB activation and B cell activation when we added the ubiquitination inhibitor, and lost the aforementioned effects when the phosphorylation inhibitor was administered (Fig. 6E, F).
A, B Detection of STING ubiquitination after CD40 stimulation. C Detection of IRF4 expression, STING phosphorylation, and NF-κB pathway regulation after CD40 stimulation or administration of phosphorylation inhibitors. D Flow cytometry revealed the degrees of B cell differentiation after CD40 stimulation or administration of a phosphorylation inhibitor (APC-CD19 and PE-pre-BCR). E Detection of IRF4 expression, STING phosphorylation, and NF-κB pathway regulation after STING/phosphorylation inhibitor/ubiquitination inhibitor administration. F Flow cytometry revealed the degrees of B cell differentiation after STING/phosphorylation inhibitor/ ubiquitination inhibitor administration (APC-CD19 and PE-pre-BCR).
Discussion
With the advancement of immunotherapy, a growing body of studies indicated that TLS was associated with better prognosis in malignancies [27,28,29], which supported the findings from our study. Nevertheless, the mechanisms of TLS delaying tumor progression have not been well understood. A theory supports TLS as a humoral response site against tumor antigens, which can drive adaptive immune responses in tumors [30]. In 2020, Barros et al. [9] demonstrated that ESCC frequently exhibited elevated B cell gene signatures, increased BCR clonality, and inflammatory infiltration, highlighting the crucial role of B lymphocytes in TLS formation. In our study, comprehensive immune characterization of ESCC revealed that the density of TLS correlated with increased infiltration of tumor-suppressive immune populations, particularly activated B lymphocytes, corroborating observations of Barros et al. [9]. All these evidences highlighted B cells in exerting antitumor efficacy within TLS.
To investigate the underlying biological pathways contributing to the prognostic benefits associated with TLS presence, we sought DEGs between high- and low-TLS groups as well as their prognostic implications. Interestingly, only one survival-related DEG (IRF4) was found to be responsible for the favorable prognosis of the presence of TLS. Additionally, immunological analysis demonstrated that elevated IRF4 levels were associated with enhanced recruitment of activated B lymphocytes and tumor-suppressive immune populations in ESCC.
A previous study has reported that IRF4, a transcription factor regulated by BCR and CD40 pathways, plays a critical role in plasma cell differentiation and germinal center B cell development [12]. Our study revealed that CD40 stimulation upregulated IRF4 expression and enhanced B lymphocyte activation independent of co-culture with tumor cells. Activation of CD40 on B cells, in the appropriate cytokine context, triggered immunoglobulin isotype switching and promoted B cell activation and proliferation [31]. The two major effects of CD40 activation on B cells appear to be the activation of NF-κB and the activation of protein tyrosine kinases (PTKs) [32]. These findings collectively indicated the significant potential of CD40 in promoting B cell activation and the subsequent formation of TLS.
It has been demonstrated that both CD40 and STING agonists could induce the generation of TLS and can bind to TRAF molecules, thereby activating the NF-κB pathway [33, 34]. To investigate potential crosstalk between CD40 and STING, we pulled down the TRAFs, which showed that TRAF2 played an important role in the relationship between CD40 and STING. Our experimental findings demonstrated that CD40 preferentially interacted with TRAF2, thereby inhibiting STING ubiquitination and proteasomal degradation. Furthermore, administration of CD40 agonists significantly elevated STING phosphorylation levels, substantially amplifying NF-κB pathway activation and consequent B lymphocyte stimulation. On this basis, our study made an important discovery that CD40 competitively bound TRAF2 with STING, which enhanced the expression of IRF4 and activated B cells.
There are several limitations in our study. Firstly, the retrospective data of our investigation inherently contained unavoidable selection and performance-related biases. Secondly, only treatment-naïve ESCC patients were included in our current study, which limited the significance of TLS as a robust biomarker in ESCC, especially in ESCC receiving neoadjuvant treatment. Thirdly, we did not assess the crosstalk between exogenous activation of CD40 and STING in activating the canonical NF-κB pathway. In addition, in vivo experiments were unavailable to determine whether CD40 could enhance IRF4-mediated B cell activation and TLS formation.
In summary, the presence of TLS was a promising prognosticator for improved survival in ESCC. Our data provided deeper insights into the potential role of activated B cells and TLS in ESCC, with implications for the development of biomarkers and therapeutic targets.
Data availability
All data generated or analyzed during this study are included in this published article (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
References
Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022;135:584–90.
Kelly RJ, Ajani JA, Kuzdzal J, Zander T, Van Cutsem E, Piessen G, et al. Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N Engl J Med. 2021;384:1191–203.
Schumacher TN, Thommen DS. Tertiary lymphoid structures in cancer. Science. 2022;375:eabf9419.
Yap DWT, Leone AG, Wong NZH, Zhao JJ, Tey JCS, Sundar R, et al. Effectiveness of immune checkpoint inhibitors in patients with advanced esophageal squamous cell carcinoma: a meta-analysis including low PD-L1 subgroups. JAMA Oncol. 2023;9:215–24.
Sautès-Fridman C, Petitprez F, Calderaro J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19:307–25.
Nakamura S, Ohuchida K, Hayashi M, Katayama N, Tsutsumi C, Yamada Y, et al. Tertiary lymphoid structures correlate with enhancement of antitumor immunity in esophageal squamous cell carcinoma. Br J Cancer. 2023;129:1314–26.
Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–5.
Sun X, Liu W, Sun L, Mo H, Feng Y, Wu X, et al. Maturation and abundance of tertiary lymphoid structures are associated with the efficacy of neoadjuvant chemoimmunotherapy in resectable non-small cell lung cancer. J Immunother Cancer. 2022;10:e005531.
Barros LRC, Souza-Santos PT, Pretti MAM, Vieira GF, Bragatte MAS, Mendes MFA, et al. High infiltration of B cells in tertiary lymphoid structures, TCR oligoclonality, and neoantigens are part of esophageal squamous cell carcinoma microenvironment. J Leukoc Biol. 2020;108:1307–18.
Chaurio RA, Anadon CM, Lee Costich T, Payne KK, Biswas S, Harro CM, et al. TGF-β-mediated silencing of genomic organizer SATB1 promotes Tfh cell differentiation and formation of intra-tumoral tertiary lymphoid structures. Immunity. 2022;55:115–128.e9.
Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495.
Cook SL, Sievert EP, Sciammas R. B cell-intrinsic IRF4 haploinsufficiency impairs affinity maturation. J Immunol. 2021;207:2992–3003.
Grumont RJ, Gerondakis S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor kappaB. J Exp Med. 2000;191:1281–92.
Portillo JC, Yu JS, Vos S, Bapputty R, Lopez Corcino Y, Hubal A, et al. Disruption of retinal inflammation and the development of diabetic retinopathy in mice by a CD40-derived peptide or mutation of CD40 in Müller cells. Diabetologia. 2022;65:2157–71.
Luo W, Conter L, Elsner RA, Smita S, Weisel F, Callahan D, et al. IL-21R signal reprogramming cooperates with CD40 and BCR signals to select and differentiate germinal center B cells. Sci Immunol. 2023;8:eadd1823.
van Hooren L, Vaccaro A, Ramachandran M, Vazaios K, Libard S, van de Walle T, et al. Agonistic CD40 therapy induces tertiary lymphoid structures but impairs responses to checkpoint blockade in glioma. Nat Commun. 2021;12:4127.
Walker MM, Crute BW, Cambier JC, Getahun A. B Cell-Intrinsic STING signaling triggers cell activation, synergizes with B cell receptor signals, and promotes antibody responses. J Immunol. 2018;201:2641–53.
Chelvanambi M, Fecek RJ, Taylor JL, Storkus WJ. STING agonist-based treatment promotes vascular normalization and tertiary lymphoid structure formation in the therapeutic melanoma microenvironment. J Immunother Cancer. 2021;9:e001906.
Strohm L, Ubbens H, Münzel T, Daiber A, Daub S. Role of CD40(L)-TRAF signaling in inflammation and resolution-a double-edged sword. Front Pharm. 2022;13:995061.
Wang P, Li S, Zhao Y, Zhang B, Li Y, Liu S, et al. The GRA15 protein from Toxoplasma gondii enhances host defense responses by activating the interferon stimulator STING. J Biol Chem. 2019;294:16494–508.
Li H, Wang J, Liu H, Lan T, Xu L, Wang G, et al. Existence of intratumoral tertiary lymphoid structures is associated with immune cells infiltration and predicts better prognosis in early-stage hepatocellular carcinoma. Aging. 2020;12:3451–72.
Tan Q, Huang Y, Deng K, Lu M, Wang L, Rong Z, et al. Identification immunophenotyping of lung adenocarcinomas based on the tumor microenvironment. J Cell Biochem. 2020;121:4569–79.
Wang H, Su C, Li Z, Ma C, Hong L, Li Z, et al. Evaluation of multiple immune cells and patient outcomes in esophageal squamous cell carcinoma. Front Immunol. 2023;14:1091098.
Alvisi G, Brummelman J, Puccio S, Mazza EM, Tomada EP, Losurdo A, et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J Clin Invest. 2020;130:3137–50.
van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17.
Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol. 2014;88:5328–41.
Li R, Huang X, Yang W, Wang J, Liang Y, Zhang T, et al. Tertiary lymphoid structures favor outcome in resected esophageal squamous cell carcinoma. J Pathol Clin Res. 2022;8:422–35.
Wang Q, Shen X, An R, Bai J, Dong J, Cai H, et al. Peritumoral tertiary lymphoid structure and tumor stroma percentage predict the prognosis of patients with non-metastatic colorectal cancer. Front Immunol. 2022;13:962056.
Zhang Q, Wu S. Tertiary lymphoid structures are critical for cancer prognosis and therapeutic response. Front Immunol. 2023;13:1063711.
Meylan M, Petitprez F, Becht E, Bougoüin A, Pupier G, Calvez A, et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity. 2022;55:527–541.e5.
Cheng L, Li G, Pellegry CM, Yasui F, Li F, Zurawski SM, et al. TLR9- and CD40-targeting vaccination promotes human B cell maturation and IgG induction via pDC-dependent mechanisms in humanized mice. Front Immunol. 2021;12:672143.
Ren CL, Fu SM, Geha RS. Protein tyrosine kinase activation and protein kinase C translocation are functional components of CD40 signal transduction in resting human B cells. Immunol Invest. 1994;23:437–48.
Diebold M, Sievers C, Bantug G, Sanderson N, Kappos L, Kuhle J, et al. Dimethyl fumarate influences innate and adaptive immunity in multiple sclerosis. J Autoimmun. 2018;86:39–50.
Messaoud-Nacer Y, Culerier E, Rose S, Maillet I, Rouxel N, Briault S, et al. STING agonist diABZI induces PANoptosis and DNA mediated acute respiratory distress syndrome (ARDS). Cell Death Dis. 2022;13:269.
Acknowledgements
We appreciate the technical support from Dr. Xi Zhang from the Department of Rehabilitation, Huashan Hospital, Fudan University. We also appreciate the technical support from Dr. Feng Li and Dr. Yongsheng Zhang from the Department of Pathology, the Second Affiliated Hospital of Soochow University.
Funding
Funded by National Natural Science Foundation of China (82172076); Key Scientific Program of Jiangsu Provincial Health Commission (ZD2021033); Project of Medical New Technology Assistance of the Second Affiliated Hospital of Soochow University (23ZL004 and 23ZL012); The Project of Capacity Enhancement of Institutional Clinical Trials in Suzhou (SLT2023030); The Project of Clinical Innovation and Interdisciplinary Translation of Soochow University (ML12301623); The Construction Project of High-End Clinical Medical Technology Platform and Translational Base of Soochow University (ML12301123); Discipline construction project of the Second Affiliated Hospital of Soochow University (Phase II); Gusu Health Talent Program of Suzhou (GSWS2023073, MR-32-25-016221); Project of Medical Innovation Application Research of Suzhou (SKY2022094); Pre-research Fund of the Second Affiliated Institute of Soochow University - National Nature Pre-Research Project (SDFEYGZ2222); Youth Funding of Zhongshan Hospital (grant numbers Residency 2022-007, 2023ZSQN48); Shanghai Science and Technology Innovation Action Plan Sailing Program (24YF2704200); China Postdoctoral Science Foundation (2024M750521).
Author information
Authors and Affiliations
Contributions
Y Zheng and D Chen contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Y Zheng, Y Xu, X Xu, P Song, and X Wu. The administrative and fund-raising work were provided by L Tan, Y Mao, and Y Chen. The first draft of the manuscript was written by Y Zheng, D Chen, and Y Xu, and reviewed by L Tan, Y Mao, and Y Chen. All authors polished and approved the final manuscript. All authors reviewed the results and contributed to the final manuscript. All authors approved this manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and consent to participate
All methods of the study were performed in accordance with the relevant guidelines and regulations. The study was approved by the Review Committee of the Ethics Institute of the Second Affiliated Hospital of Soochow University (JD-HG-2025-056). Informed consent was obtained from all participants. This study has been registered with the Chinese Clinical Trial Registry (registration number: ChiCTR2500101699).
Consent for publication
Informed consent for images was obtained from all individual participants included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, Y., Chen, D., Xu, Y. et al. Characterization of tertiary lymphoid structure identifies competitive binding of CD40 and STING with TRAF2 driving IRF4-mediated B cell activation in esophageal squamous cell carcinoma. Cancer Gene Ther (2025). https://doi.org/10.1038/s41417-025-00944-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-025-00944-2