Abstract
The recent worldwide outbreaks of mpox prioritize the development of a safe and effective mRNA vaccine. The contemporary mpox virus (MPXV) exhibits changing virological and epidemiological features, notably affecting populations already vulnerable to human immunodeficiency virus (HIV). Herein, we profile the immunogenicity of AR-MPXV5, a penta-component mRNA vaccine targeting five specific proteins (M1R, E8L, A29L, A35R, and B6R) from the representative contemporary MPXV clade II strain, in both naive and simian immunodeficiency virus (SIV)-infected nonhuman primates. Immunization with two doses of AR-MPXV5 to cynomolgus macaques effectively elicits antibody responses and cellular responses. Importantly, based on the challenge model with a contemporary MPXV clade II strain, AR-MPXV5 demonstrates effective efficacy in preventing skin lesions, eliminating viremia and reducing viral loads in multiple tissues after challenge in naive male animals. More importantly, AR-MPXV5 is well-tolerated in stable chronic SIV-infected rhesus monkeys, while eliciting comparable MPXV-specific humoral and cellular responses in both naive and SIV-infected monkeys. Together, these results support further clinical development of the AR-MPXV5 vaccine.
Similar content being viewed by others
Introduction
The re-emerging mpox (also known as monkeypox) is a zoonotic viral disease caused by the mpox virus (MPXV) which belongs to the Orthopoxvirus genus of Poxviridae family. The first documented human case of MPXV infection was reported in Democratic Republic of the Congo during 1970s1,2. Subsequently, MPXV infection has caused sporadic human cases and remained localized within countries across West and Central Africa3. Since 2022, mpox suddenly emerged and spread into more than 110 countries, leading to the declaration of public health emergency of international concern4,5. MPXV is divided into two distinct clades, the Congo Basin (Central African) clade (clade I) and West African clade (clade II), while the circulating strains during the 2022 mpox outbreak were classified as lineage B.1 of clade II6,7. The clinical symptoms and manifestations during the recent outbreak differ from those observed in previous endemics, as a reduced number of skin lesions were observed and the lesions were primarily localized in the genital and perianal regions8,9,10,11,12. Besides, in the ongoing outbreak, most confirmed and suspected cases have no direct travel history to endemic areas4,13, and a large proportion of infected cases have been found in men who had sexual intercourse with men8,10. Of note, a significant number of cases have been detected among individuals living with human immunodeficiency virus (HIV) infection who may experience HIV-associated immunodeficiency14,15,16,17,18, raising concerns regarding the potential risk of severe illness within this population. To date, persistent transmission continues to occur in some regions, necessitating the sustained endeavor for long-term disease management19,20.
Vaccination has been well evidenced to be the most effective strategy to stop Orthopoxvirus infection. Most Orthopoxvirus members share highly conserved protein-coding sequences, including MPXV, vaccinia virus (VACV) and variola virus (VARV), and have been experimentally demonstrated to confer cross-protection against other orthopoxviruses21,22,23,24. Currently, the U.S. Food and Drug Administration (FDA) has conditionally approved two smallpox vaccines for the prevention of mpox: JYNNEOS25, a nonreplicating vaccine, and ACAM200026, a traditional live attenuated vaccine. However, recent clinical findings have unveiled that smallpox vaccine induced diminished levels of neutralizing antibodies against MPXV27,28.
Messenger RNA (mRNA)-based vaccine has been developed as a promising approach in combating emerging infectious diseases, making it an attractive strategy in addressing the mpox epidemic. Similar with other orthopoxvirus, MPXV exists two infectious forms: the intracellular mature virion (IMV) and the extracellular enveloped virion (EEV). Experimental evidence has substantiated the necessity of combining antigens from both infectious forms to confer adequate protection against poxvirus infection29,30,31,32,33,34,35,36,37,38. Previously, several IMV and EEV surface proteins of orthorpoxvirus were employed in the vaccine design as protective antigens. L1R, D8L, and A27L of VACV (homologs of MPXV M1R, E8L, and A29L) are surface proteins of IMV that serve as targets for neutralizing antibodies and have been demonstrated to effectively induce protective antibodies23,29,39. B5R (homologs of MPXV B6R) is an EEV surface protein that encompasses epitopes targeted by neutralizing antibodies in the presence of complement40,41,42. A33R (homologs of MPXV A35R) is a target for antibody-dependent, complement-mediated cytolysis of infected cells22,42,43. The combination of these potential antigens holds great promise in conferring enhanced immunoprotection against poxvirus infection. However, the development of mRNA vaccines carrying multiple antigen-encoded mRNAs remains challenging, especially considering the cost and strict quality control requirement for large scale production.
Several MPXV mRNA vaccines have been reported in preclinical studies32,33,34,35,36,37,38. Based on the established mRNA-LNP platform44,45, we have shown that the penta-component vaccine candidate (AR-MPXV5) encoding five proteins of MPXV (M1R, E8L, A29L, A35R, and B6R) elicited protective immunity in mice33. In the current study, we sought to determine the safety, immunogenicity and protection efficacy in nonhuman primates, especially in the simian immunodeficiency virus (SIV)-infected animals.
Results
AR-MPXV5 elicits humoral and cellular immunity in cynomolgus macaques
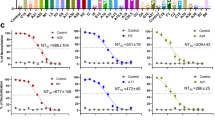
To induce a more balanced and potent immune response, our penta-component mRNA vaccine AR-MPXV5 were designed to encode five viral antigens of contemporary MPXV isolates, M1R, E8L, and A29L from IMV, and A35R and B6R from EEV, respectively (Fig. 1a). The final mRNA-LNP formulation was prepared with the well-established mRNA-LNP platform as previously described44. To investigate the immunogenicity of AR-MPXV5 in nonhuman primates, groups of cynomolgus macaques were intramuscularly immunized with 200 μg of AR-MPXV5 and boosted with the same dose 28 days later. Animals in sham group were immunized with placebo at the same procedure. Sera samples were subsequently collected on day 14, 28, and 42 post-initial immunization and subjected to antibody detection (Fig. 1b). Specifically, the total IgG antibody responses against MPXV specific antigens were determined by enzyme-linked immunosorbent assay (ELISA). The results indicated that a single dose vaccination elicited M1R, E8L, A29L, A35R, and B6R-specific IgG antibodies, and a subsequent booster immunization obviously enhanced the antibody activity (Fig. 1c). Similarly, the 50% plaque reduction neutralization test (PRNT50) demonstrated that substantial levels of neutralizing antibodies against either VACV or MPXV were detected following two immunizations (Fig. 1d, e). As expected, the IgG and neutralizing antibody titers were below the detection threshold in the sham animals.
a Schematic diagram of the multicomponent MPXV mRNA vaccine design (created with BioRender.com). Created in BioRender. Tong, L. (2024) https://BioRender.com/k15r044. b Immunization and challenge study schedule for cynomolgus macaques. Created in BioRender. Tong, L. (2024) https://BioRender.com/k15r044. n = 3 independent biological replicates (Sham), n = 4 independent biological replicates (AR-MPXV5). c MPXV antigen-specific IgG antibody responses determined by ELISA. Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The exact P values are: P = 0.0085 (M1R, AR-MPXV5 day 14 vs AR-MPXV5 day 42), P = 0.0091 (M1R, AR-MPXV5 day 28 vs AR-MPXV5 day 42), P = 0.0053 (M1R, Sham day 42 vs AR-MPXV5 day 42); P < 0.0001 (E8L, AR-MPXV5 day 14 vs AR-MPXV5 day 42, AR-MPXV5 day 28 vs AR-MPXV5 day 42, Sham day 42 vs AR-MPXV5 day 42); P = 0.0245 (A29L, AR-MPXV5 day 14 vs AR-MPXV5 day 42), P = 0.0195 (A29L, AR-MPXV5 day 28 vs AR-MPXV5 day 42), P = 0.0097 (A29L, Sham day 42 vs AR-MPXV5 day 42); P = 0.0002 (A35R, AR-MPXV5 day 14 vs AR-MPXV5 day 42), P = 0.0003 (A35R, AR-MPXV5 day 28 vs AR-MPXV5 day 42), P = 0.0001 (A35R, Sham day 42 vs AR-MPXV5 day 42); P = 0.0059 (B6R, AR-MPXV5 day 14 vs AR-MPXV5 day 42), P = 0.0061 (B6R, AR-MPXV5 day 28 vs AR-MPXV5 day 42), P = 0.0038 (B6R, Sham day 42 vs AR-MPXV5 day 42). d, e Neutralizing antibody responses against VACV (d) and MPXV (e) 42 days after initial immunization. Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-tailed unpaired t test. *P < 0.05. P = 0.0339 (d), P = 0.0168 (e). Source data are provided as a Source Data file.
Next, the antigen-specific cellular immune response in vaccinated macaques was assessed by flow cytometry 14 days after the second immunization (Fig. 2). The stimulation of peripheral blood mononuclear cells (PBMCs) with corresponding antigen peptide pools resulted in an obvious increase of M1R-, E8L-, A29L-, A35R-, and B6R-specific CD4+ T cells producing interleukin-2 (IL-2) in AR-MPXV5-immunized animals compared to the sham group (Fig. 2). Meanwhile, no significant differences were observed in the proportion of interferon γ (IFN-γ) or interleukin-4 (IL-4) secreting CD4+ T cells between the vaccine- and sham- immunized macaques (Fig. 2). Our results demonstrated the multicomponent mRNA vaccine activates an antigen-specific CD4+ T cell response. In addition, no significant elevation in the frequency of M1R-, E8L-, A29L-, A35R-, and B6R-specific CD8+ T cells producing IFN-γ or IL-2 among the vaccinated macaques were observed (Supplementary Fig. 1), indicating the vaccine candidate predominantly stimulates CD4+ T cells in nonhuman primates. Together, these results demonstrated that AR-MPXV5 effectively elicits humoral and cellular immunity against poxvirus.
MPXV antigen-specific CD4+ cells secreting IFN-γ, IL-2, and IL-4 were determined by flow cytometry 42 days post-initial immunization. n = 3 independent biological replicates (Sham), n = 4 independent biological replicates (AR-MPXV5). Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. *P < 0.05, **P < 0.01. The exact P values are: P = 0.0013 (M1R, IL-2), P = 0.0195 (E8L, IL-2) and P = 0.0037 (B6R, IL-2). Source data are provided as a Source Data file.
AR-MPXV5 protects non-human primates from MPXV infection
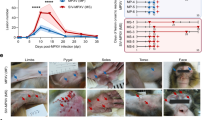
To further determine the protective efficacy of AR-MPXV5 in nonhuman primates, the infection model of MPXV was established with a contemporary circulating strain, MPXV-B.1-China-C-Tan-CQ0146,47. Groups of cynomolgus macaques immunized with two doses of mRNA vaccine or placebo were challenged with 107 TCID50 of MPXV by the intravenous route (i.v.). All challenged macaques were then monitored for signs of disease, and skin lesion as well as viral shedding were recorded at 1, 4, 7, and 10 days post-inoculation (dpi) (Fig. 1b). No significant change was observed in body weight and body temperature between AR-MPXV5 and sham groups (Supplementary Fig. 2a, b). In the sham group, skin lesions were observed in all three infected animals, encompassing the regions of the head, face, arms, chest, abdomen and legs (Fig. 3a). The lesions progressed from a vesicular or pustular rash and ultimately developed into scabs, and lesion counts appeared at 7 dpi and peaked at 10 dpi in the sham group animals (Fig. 3b). In contrast, among the four animals immunized with AR-MPXV5, two didn’t develop lesions after infection throughout the observation period; one animal developed a single small lesion and the other presented 4 small lesions at 7 dpi; all lesions faded away in the two AR-MPXV5-immunized animals at 10 dpi (Fig. 3b). The animals were then euthanized for histopathological analyses. In the sham group, the epidermis exhibited an increased thickness, accompanied by spinous layer hypertrophy and vacuolar degeneration of spinous cells. Cell necrosis was observed in both the epidermis and superficial dermis. Besides, disorganization of collagen fibers within the dermal layer can be observed, accompanied by infiltration of numerous granulocytes, lymphocytes, and macrophages (Fig. 3c). In contrast, the epidermis of the AR-MPXV5-vaccinated animals exhibited a normal and intact morphology, and collagen fibers arranged in a regular pattern within the dermal layer. Simultaneously, substantial viral proteins were detected in the damaged skin samples obtained from animals belonging to the sham group, whereas no viral protein expression was observed in immunized animals (Supplementary Fig. 3). These results clearly demonstrated that AR-MPXV5 is efficacious in delaying and alleviating skin lesions as well as pathological damage caused by MPXV infection in cynomolgus macaques.
a Photographs of skin lesions from three animals in the sham group at 10 dpi. b Skin lesions were counted at 0, 4, 7, and 10 dpi. n = 3 independent biological replicates (Sham), n = 4 independent biological replicates (AR-MPXV5). Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. ****P < 0.0001. c H&E staining of skin samples from MPXV-infected macaques. Yellow arrows: epidermis exhibited an increased thickness accompanied by spinous layer hypertrophy and vacuolar degeneration of spinous cells. Black arrows: cell necrosis in the epidermis and superficial dermis. Green arrows: disorganization of collagen fibers within the dermal layer. Red arrows: infiltration of numerous granulocytes, lymphocytes, and macrophages in dermal layer. Source data are provided as a Source Data file.
Viremia has been well documented as a hallmark for MPXV infection, and the disappearance of viremia is recognized as a correlate for protection38,48,49,50,51,52,53,54. Upon challenge, all animals in the sham group developed rapid and sustained viremia within 10 dpi, with a peak titer of 4.17 × 106 copies/mL at 7 dpi. In contrast, a minimal level of viral DNA was detected in one of the vaccinated animals at 1 dpi and subsequently disappeared in the following days (Fig. 4a). Meanwhile, MPXV genomic DNA was detected in the urine samples at 10 dpi from the sham group animals, but viral DNA was absent in all vaccinated animals (Fig. 4b). The presence of viral shedding in nasal and throat swabs was detected at 4 dpi and 7 dpi in the sham group, persisting until 10 dpi, respectively; while no viral DNA was detected in nasal and throat swabs from the vaccinated animals (Fig. 4c, d). Our results clearly demonstrated that AR-MPXV5 not only eliminates viremia but also abolished viral shedding in MPXV-infected non-human primates.
a–d Viral DNA loads in plasma (a), urine (b), nasal swab (c) and throat swab (d) at 0, 1, 4, 7, and 10 dpi. n = 3 independent biological replicates (Sham), n = 4 independent biological replicates (AR-MPXV5). Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. *P < 0.05, ***P < 0.001. The exact P values are: P = 0.0106 (a), P = 0.0004 (b), P = 0.0005 (c) and P = 0.0107 (d). e Viral DNA copies in tissues after MPXV challenge at 10 dpi. f Viral loads in tissues after MPXV challenge at 10 dpi by using plaque forming assay. Source data are provided as a Source Data file.
To further determine the protective efficacy in various tissues of MPXV-infected macaques, animals were euthanized at 10 dpi. MPXV DNA was detected in all 11 tissues obtained from sham-immunized macaques, including the heart, liver, spleen, lung, kidney, skin, lymph node, muscle, ileum, colon and testis (Fig. 4e). The highest viral DNA load was detected in the skin, with a titer of ~ 2.44 × 108 copies/μg. Meanwhile, substantial viral DNAs were determined in spleen, testis and lymph nodes. In contrast, an obvious reduction in viral loads was detected in all tissues of vaccinated animals, with MPXV genomic DNA remaining below the detection threshold in heart, liver, and lung. Additionally, a remarkable decrease with a fold change ranging from 32 to 1.4 × 106 was observed in other tissues (Fig. 4e). Also, viral load detection in tissues from infected animals was conducted by using plaque forming assay, indicating that significant levels of infectious virus particles were detected in the skin and spleen of the sham group, whereas no infectious virus was detected in the vaccinated group (Fig. 4f). Besides, the reduction of germ cells within the seminiferous tubules and loss of spermatogenic epithelial cells were observed in the histopathological analyses in a monkey from the sham group (Supplementary Fig. 4a), and the necrosis was also found in the epithelium and lymphoid tissue of tonsil in the same macaque (Supplementary Fig. 4b).
To elucidate the tissue-specific responses following MPXV infection, transcriptomic profiles of skin and spleen from the sham group were analyzed using RNA-seq and compared with those from the vaccinated group (Supplementary Fig. 5). In skin samples, a total of 1966 genes were upregulated and 1090 genes were downregulated in sham group post-infection compared to vaccinated animals (Supplementary Fig. 5a). Gene Ontology (GO) analysis revealed that the upregulated genes were significantly associated with immune responses and defense responses (Supplementary Fig. 5b). Similar transcriptomic upregulation of genes involved in immune response was detected in spleens from sham group animals compared with vaccinated animals (Supplementary Fig. 5c, d). Together, these results suggested that compared to AR-MPXV immunized animals, MPXV infection leads to a heighted immune response across multiple tissues in sham-treated macaques.
In addition, the cytokine storm has been reported in human mpox cases, and is likely to be associated with disease exacerbation and unfavorable clinical outcomes55,56. In our study, MPXV challenge led to a substantial production of cytokines and chemokines in the sham-immunized macaques (Fig. 5a and Supplementary Tables 1 and 2). Notably, the upregulation of sera cytokines were detected at 1 dpi, peaked at 4 or 7 dpi, and faded at 10 dpi in all sham animals (Fig. 5a and Supplementary Tables 1 and 2), suggesting a strong proinflammatory response in sham-vaccinated animals. Of all upregulated cytokines, IFN-γ, SDF-1α and VEGF-A showed a remarkable increase of 365.8-, 46.5-, and 24.9-fold, respectively (Fig. 5b). Strikingly, all AR-MPXV5-vaccinated macaques showed a milder cytokine production profile upon MPXV challenge, and the levels of all selected cytokines were significantly lower than that from the sham group animals (Fig. 5a, b). These results clearly demonstrated that AR-MPXV5 vaccination has hindered the upregulation of cytokines caused by MPXV challenge in nonhuman primates.
a Cytokine and chemokine expressions were determined by Luminex and presented as fold changes compared to samples collected before challenge. b Concentrations of IFN-γ, SDF-1α and VEGF-A in plasma samples pre- and post-challenge. n = 3 independent biological replicates (Sham), n = 4 independent biological replicates (AR-MPXV5). Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. *P < 0.05, ***P < 0.001, ****P < 0.0001. The exact P values are: P < 0.0001 (IFN-γ, day 4), P = 0.0003 (SDF-1α, day 7), P = 0.0003 (VEGF-A, day 7) and P = 0.0323 (VEGF-A, day 10). c Neutralizing antibody titers against VACV in serum samples 10 days pre- and post-challenge. Source data are provided as a Source Data file.
More importantly, the neutralizing antibody titers against VACV were determined and compared pre- and post-challenge. A significantly elevated neutralizing antibody response was detected in the sham-immunized macaques at 10 dpi, while all animals immunized with AR-MPXV5 exhibited no significant increase in neutralizing antibody titers after challenge (Fig. 5c).
AR-MPXV5 induces protective immune responses in SIV-infected rhesus monkeys
To further investigate whether AR-MPXV5 induces a protective immune response in immunodeficient individuals, groups of naive or stable chronic SIV-infected rhesus monkeys were intramuscularly immunized with 200 μg of AR-MPXV5 and boosted with the same dose 28 days later (Fig. 6a). CD4+ T cell counts of naive or SIV-infected monkeys were analyzed before immunization. The result indicated an obvious reduction of CD4+ T cells in chronic SIV-infected monkeys compared with their uninfected counterparts (Fig. 6b), and the plasma SIV loads were detected prior to immunization, with ~8.36 × 104 copies/mL of viral RNAs detected in the SIV-infected monkeys (Fig. 6c). The IgG antibodies against the EEV surface protein B6R and the IMV surface protein M1R were quantified following immunization, indicating that high levels of IgG antibodies were elicited 42 days post-initial immunization, which remained detectable even 165 days post-immunization (Fig. 6d, e). Next, the neutralizing antibodies against VACV were determined after vaccination. The administration of two doses of AR-MPXV5 effectively induced neutralizing antibody responses in both of the naive and SIV-infected monkeys, with the GMTs for neutralizing antibodies against VACV reaching ~1: 1498 and ~1: 582 on day 42 after the prime immunization, respectively. The neutralizing antibody response persisted for a minimum duration of five months (Fig. 6f).
a Schematic diagram of the experimental design. Created in BioRender. Yang, C. (2024) https://BioRender.com/z08t075; Yang, C. (2024) https://BioRender.com/m48n481. n = 3 independent biological replicates. b CD4+ T cell counts in naive and SIV-infected monkeys prior to vaccination. c Plasma SIV loads in naive and SIV-infected monkeys prior to vaccination. d, e IgG antibodies against the IMV surface protein M1R (d) and EEV surface protein B6R (e) after immunization with two-doses of AR-MPXV5. Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. **P < 0.01, ns not significant. P = 0.0068 (M1R, Naive day 42 vs SIV-infected day 42). f Neutralizing antibody titers against VACV in naive and SIV-infected monkeys. Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. *P < 0.05, ns not significant. P = 0.0255 (Naive day 42 vs SIV-infected day 42). g, h MPXV antigen-specific CD4 + T cells in naive (g) and SIV-infected monkeys (h) pre- and post-immunization were determined by flow cytometry. Data are shown as the mean ± SEM. Statistical differences were analyzed by using two-way ANOVA. *P < 0.05, **P < 0.001, ****P < 0.0001. (h) P = 0.0068 (M1R, IL-2), P < 0.0001 (E8L, IL-2) and P = 0.0357 (A35R, IL-2). Source data are provided as a Source Data file.
Besides, the antigen-specific cellular response was assessed 14 days after the second immunization by flow cytometry (Fig. 6g, h). An obvious increase of M1R-, E8L-, A35R- and B6R-specific CD4+ T cells producing IL-2 was detected in both of the naive and SIV-infected groups compared to pre-immunization levels. The animals also exhibited an elevation of M1R-, E8L-, and A35R-specific CD4+ T cells producing IFN-γ (Fig. 6g, h and Supplementary Fig. 6). The duration of antigen-specific CD4+ T cell responses were subsequently evaluated 165 days post-immunization in SIV-infected monkeys. The results indicated that M1R-, E8L-, A35R-, and B6R-specific CD4+ T cell responses maintained for more than five months (Supplementary Fig. 7). Together, our results demonstrate that MPXV mRNA vaccine AR-MPXV5 effectively elicits both humoral and cellular immune responses in stable chronic SIV-infected nonhuman primates.
AR-MPXV5 is well tolerated in naive and SIV-infected nonhuman primates
In our experiment, the administration of two doses of AR-MPXV5 was totally tolerated in cynomolgus macaques and rhesus monkeys. All naive and SIV-infected animals did not exhibit any significant clinical events after vaccination. The body weight changes as well as plasma viremia of SIV-infected animals were monitored for 42 days following the initial immunization (Supplementary Fig. 8a, b). Only a transient and minimal weight loss was observed during the administration (Supplementary Fig. 8a). No obvious increase in SIV RNA loads was detected throughout the immunization process, indicating the absence of SIV activation following vaccination (Supplementary Fig. 8b). Meanwhile, there was no significant decline in CD4+ T cell counts after immunization, while a transient increase was observed following the second immunization (Supplementary Fig. 8c). In addition, hematological parameters were recorded and analyzed in naive and SIV-infected monkeys after immunization (Supplementary Tables 3 and 4 and Supplementary Fig. 9). The white blood cell (WBC) count, neutrophil (Neu) count, and monocyte (Mon) count exhibited a transient elevation on day 1 after the first and second immunization, and return to normal ranges on day 3 (Supplementary Fig. 9). Meanwhile, a slight and transient decrease in lymphocyte (Lym) count was observed on day 1 post-immunization and subsequently returned to normal on day 3. The red blood cell (RBC) count and hemoglobin (HGB) remained within the normal range throughout the vaccination process without any significant alterations (Supplementary Fig. 9). No significant differences were observed in the alteration of hematological parameters between the naive and SIV-infected groups. Our results demonstrated that AR-MPXV5 was safe and well-tolerated in naive and SIV-infected nonhuman primates.
Discussion
A safe and effective vaccine is indispensable for the prevention and control of MPXV infection and transmission. In the present study, the immune profile and protective efficacy of a penta-component mRNA vaccine (AR-MPXV5) were well elucidated in a MPXV-infected nonhuman primate model, which have simulated and restated the natural course and clinical manifestations of MPXV infection49,50,51,52,54,57,58. Two administrations of AR-MPXV5 elicited high levels of antigen-specific IgG antibodies as well as neutralizing antibodies against VACV and MPXV, exhibiting a superior or comparable protective antibody response compared to that induced by attenuated vaccine51,54,57,59, subunit vaccine60, virus-like replicon particles (VRP)61, DNA vaccine22,29,30 as well as other mRNA vaccines38,62 in nonhuman primates as recently reported. Besides, our results revealed that AR-MPXV5 induces an elevation of MPXV antigen specific CD4+ T cells in cynomolgus macaques, suggesting the potent immunogenicity of the multiple antigens in combination. In our study, no significant elevation in the frequency of M1R-, E8L-, A29L-, A35R-, and B6R-specific CD8+ T cells producing IFN-γ or IL-2 among the vaccinated macaques were detected. Notably, our previous and current studies have demonstrated that the vaccine candidate concurrently activates both antigen-specific CD4+ and CD8+ T cells in mouse models33, but it predominantly stimulates CD4+ T cells in nonhuman primates, indicating the differential immune response elicited by mRNA vaccines across various animal species. Additional preclinical and clinical studies can provide further support to elucidate the vaccine-induced immune profiles.
The MPXV responsible for the global outbreak in 2022 belongs to clades IIb of West African clade6. Recently, another MPXV mRNA vaccine (BNT166) encoding four surface proteins (A35, B6, M1, and H3) was assessed in a cynomolgus macaques model challenged with a lethal clade I MPXV strain38. Interestingly, BNT166 vaccination failed to eliminate the viremia caused by MPXV in cynomolgus macaques38. In our current study, a nonhuman primate model infected with a circulating MPXV clade II strain was established to evaluate the protective efficacy of AR-MPXV5. This macaque model represents a non-lethal infection model characterized by substantial viremia and obvious skin lesions. AR-MPXV5 effectively prevents the development of skin lesions and absolutely eliminates viremia, suggesting that the inclusion of additional antigen components may potentially confer enhanced protection against poxvirus infection. In addition, the sham-immunized animals exhibited substantial viral DNA loads in multiple tissues, whereas a significant reduction in viral loads was observed in AR-MPXV5-immunized animals. Notably, high levels of viral DNA were detected in the testis from animals in the sham group, while an obviously decreased viral load was observed in the testis from vaccinated macaques, suggesting the potential efficacy of AR-MPXV5 in mitigating sexual transmission63. Previously, the elevation of multiple sera cytokine and chemokine expressions have been reported in MPXV-infected human or nonhuman primates, indicating a strong proinflammatory response associated with MPXV infection53,55,56,64. In our study, the administration of AR-MPXV5 significantly attenuated the cytokine storm following infection. Besides, certain orthopoxvirus proteins are capable of subverting the host immune response and modulating cytokine expression during orthopoxvirus infection56,65,66. For instance, VACV K3L functions as an inhibitor of the antiviral protein kinase PKR, facilitating evasion of innate immune responses65. VACV complement control protein (VCP/C3L) effectively inhibits the activation of the complement system65,67,68. Orthopoxviral Bcl-2-like proteins have been reported to inhibit proapoptotic signaling and the transcription of inflammatory factors56,65,66. Further investigation is still required to gain a comprehensive understanding of the regulatory role of these viral proteins in cytokine expression and their involvement in the host immune responses.
Considering the crucial role of CD4+ T cells in antibody production and their necessity for effective protection against poxvirus infection, there are concerns that people living with HIV may be at a heightened risk of severe MPXV infection, particularly those with low CD4+ T cell counts18,69,70,71. Recent studies revealed that severe MPXV infections were observed in HIV-infected individuals, especially those with advanced HIV infection or acquired immunodeficiency syndrome (AIDS) characterized by significantly diminished CD4+ T cell counts16,71,72,73,74,75. In our study, two immunizations of the multicomponent mRNA vaccine successfully induced an effective and comparable neutralizing antibody response in both of the naive and stable chronic SIV-infected monkeys. In addition, a Th1-biased, antigen-specific CD4+ T cell response was also detected in SIV-infected monkeys. Previously, several studies have shown the establishment of long-term immunity subsequent to smallpox vaccination76. In accordance with these findings, our current study also assessed the persistence of both antibody response and antigen-specific T cell responses for a minimum duration of five months. Overall, our results suggested that AR-MPXV5 was viable for eliciting a protective immune response in immunodeficiency virus infected as well as immunocompetent individuals.
Nevertheless, there are several issues to be elucidated in further studies. For instance, conducting an MPXV challenge study in SIV-infected nonhuman primates would further provide a comprehensive understanding into the protection profile of the vaccine candidate. Additionally, in August 2024, WHO has determined the upsurge in mpox cases caused by clade I MPXV infection in the Democratic Republic of the Congo (DRC) and multiple African countries constitutes a public health emergency of international concern, indicating an ongoing threat to global public health. Further studies are needed to ascertain whether the emergence of a novel MPXV clade is associated with crucial mutations that affect its transmissibility, pathogenicity as well as vaccine effectiveness.
Methods
Cells and viruses
BS-C-1 (ATCC, CCL-26) cells were cultured in Minimum Essential Medium (MEM, Procell, CM-0039), and the Vero E6 (ATCC, CRL-1586) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, C11995500BT) at 37 °C with 5% CO2. The culture mediums contained 10% fetal bovine serum (FBS, Gibco, 30044333) and 1% penicillin-streptomycin (PS, Gibco, 15140122). VACV Western Reserve (WR) strain was propagated and titrated in BS-C-1 cells before use. Cells were collected in phosphate buffered saline (PBS, Solarbio, P1020) after 48 h, and lysed by repeated freezing and thawing. Cell fragments were removed by centrifugation and virus stocks were obtained. Virus titers were determined by standard plaque assay on BS-C-1 cells. Low-passage stocks of MPXV (MPXV-B.1-China-C-Tan-CQ01) was originally isolated from a German imported case of Chinese nationality, which belongs to B.1 branch of the West African linage47. MPXV was cultivated and titrated on Vero E6 cells47, which were performed in the BSL-3 containment at the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences. For MPXV cultivation, MPXV was seeded onto monolayer Vero E6 cells with 80–90% confluence and cultured for 48 h at 37 °C with 5% CO2. The method for virus collection is the same as VACV. MPXV titration was performed on Vero E6 cells. The plaque forming units (PFUs) per mL were calculated on the basis of the number of plaques multiplied by the dilution factor.
AR-MPXV5 formulation
Lipid-nanoparticle (LNP) formulations were prepared in the same way as previously described44. Briefly, we dissolved lipids in ethanol containing an ionizable lipid, 1, 2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol and PEG-lipid (with molar ratios of 50: 10: 38.5: 1.5). The lipid mixture was then mixed with 20 mM citrate buffer (pH 4.0) containing mRNA through a T-mixer in a ratio of 1:2. Formulations were permeated into a tenfold volume of PBS with a 100 KD tangential flow filtration membrane, and filtered with a 0.22 μm filter to obtain the final LNP formulations. LNP formulations were stored at 2–8 °C. All formulations were tested for particle size, distribution, RNA concentration and encapsulation.
Vaccination and challenge experiments
All animal procedures were reviewed and approved by Institutional Animal Care and Use Committee of Academy of Military Medical Sciences (Approval Number. IACUC-DWZX-2022-055) and the Institutional Animal Care and Use Committee (IACUC) of the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences (Approval No. XJ23004). Animal experiments with infectious MPXV were conducted in animal biosafety level 3 (ABSL-3) facilities in Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences.
MPXV-seronegative cynomolgus macaques (male, 3–4 years old) were intramuscularly immunized with 200 μg of AR-MPXV5 (n = 4) or empty LNPs (n = 3) and boosted with an equal dose 28 days later. Empty LNPs were utilized as sham treatment. Plasma samples were collected prior to immunization and 14, 28, and 42 days post-initial immunization and subjected to IgG and neutralizing antibody detection. Whole blood samples were collected and peripheral blood mononuclear cells (PBMCs) were separated 42 and 165 days after initial immunization. On day 52 after initial immunization, AR-MPXV5 or sham-immunized cynomolgus macaques were intravenously challenged with 107 TCID50 of MPXV (MPXV-B.1-China-C-Tan-CQ01). Weight changes and body temperature were monitored for 10 days after inoculation. Skin lesions were observed and counted at 0, 4, 7, and 10 dpi. MPXV DNA loads in plasma, urine, nasal swab and throat swab were determined at 0, 1, 4, 7, and 10 dpi. Neutralizing antibody titers of infected macaques were determined at 10 dpi. All infected animals were euthanized at 10 dpi for tissue harvest and virological analyses. Infectious viral particles of MPXV in tissues from infected animals were detected using standard plaque assay on Vero E6 cells.
Naive and chronic SIV-infected rhesus monkeys (1 male and 5 females, 4–9 years old) were intramuscularly immunized with 200 μg of AR-MPXV5 (n = 3) and boosted with an equal dose 28 days later. Animals were immunized 27, 43, and 49 months after SIV infection. CD4+ T cell counts were determined by flow cytometry at 60 days prior to immunization and at 0, 7, 14, 28, 35, and 42 days post-initial immunization. Viral RNA loads in plasma of SIV-infected monkeys were determined at 60 days prior to immunization and at 0, 14, and 42 days post-initial immunization. The body weight changes of SIV-infected monkeys were monitored for 42 days post-initial immunization. Plasma samples were collected 42 and 165 days after initial immunization and subsequently subjected to antibody detection. Whole blood samples were collected at 0, 1, 3, 5, 7, and 14 days after the prime and boost vaccination for routine blood analyses by using hematologic analyzer (Mindray). PBMCs were separated 14 days after the boost immunization.
Enzyme-linked immunosorbent assay (ELISA)
The IgG antibody titers in serum samples were determined by ELISA. Serum samples were heated at 56 °C for 30 min before use. MPXV antigens diluted with 1× coating buffer (Solarbio, C1055) were coated on ELISA plates at 4 °C overnight, respectively. Serum samples were twofold-diluted and incubated in blocked ELISA plates for 2 h at 37 °C. Plates were washed with PBS-T (Gene-Protein Link, P05B02) for three times and then incubated with goat anti-monkey IgG-HRP at 37 °C for 1 h. ELISA plates were washed three times with PBS-T after incubation. Soluble TMB (Cwbio, CW0050S) was added to plates and incubated for 15 min at room temperature, followed by subsequent termination through the addition of stop solution (Solarbio, C1058). Synergy H1 hybrid multimode microplate reader (BioTek) was used to read the absorbance. The endpoint titers were defined as the highest reciprocal serum dilution that yielded an absorbance exceeding background values by more than twofold.
Plaque reduction neutralization test (PRNT)
Fifty percentage plaque reduction neutralization test (PRNT50) was used to detect neutralizing antibody titers in serum samples. The detection of neutralizing antibodies against VACV was performed using BS-C-1 cells, while the evaluation of neutralizing antibodies against MPXV was conducted using Vero E6 cells. BS-C-1 or Vero E6 cells were seeded in 12-well plates and incubated at 37 °C and 5% CO2. Serum samples were heated at 56 °C for 30 min before use. The threefold diluted sera were mixed with an equal volume of VACV WR strain or MPXV (MPXV-B.1-China-C-Tan-CQ01), each containing approximately 250 PFU of virus, at 37 °C for 90 min. The mixture was added to cells and incubated at 37 °C for 90 min. Then the mixture was removed, and cells were covered by 0.5% methylcellulose in Dulbecco’s minimal essential medium (DMEM, Gibco, 11965092) with 2.5% FBS. All FBS were inactivated before use. Cells were incubated at 37 °C and 5% CO2 for 2 days. The PRNT50 titers were calculated by the method of Spearman–Karber.
Flow cytometry analyses
Flow cytometry analyses was used to evaluate MPXV antigen-specific T cell responses in nonhuman primates. Briefly, a total of 106 PBMCs were co-stimulated with 1 µg/mL anti-human CD28 antibody (Biolegend, 302933), CD49d antibody (Biolegend, 304339) and the overlapping antigen peptide pools (1.5 µg/mL of each peptide, GeneScript) of M1R, E8L, A29L, A35R, and B6R at 37 °C and 5% CO2 for 1 h. PBMCs were incubated with Protein Transport Inhibitor (1:500, BD Biosciences, 554724, 555029) at 37 °C and 5% CO2 for 8 h. Cells were washed with PBS containing 1% FBS, blocked with anti-CD16/CD32 antibody (1:200, Miltenyi Biotec, 130-059-901) and then incubated with anti-human CD3 antibody (1:200, Biolegend, 562877), CD4 antibody (1:200, BD Biosciences, 550628), and CD8 antibody (1:200, Biolegend, 557834) at 4 °C for 30 min in the dark. Cells were fixed, washed by PBS containing 1% FBS and incubated with fluorescently conjugated antibodies to interferon-γ (1:200, IFN-γ, Biolegend, 559326), interleukin-2 (1:200, IL-2, Biolegend, 500325), and interleukin-4 (1:200, IL-4, Biolegend, 500811) for 30 min at 4 °C in the dark. PBMCs were resuspended with PBS containing 1% FBS, and analyzed by using the FACSVerse Flow Cytometer (BD Biosciences). Data are analyzed with Flow Jo 10.4 software.
Quantification of viral RNA and DNA loads
MPXV genomic DNA was determined by real-time quantitative PCR (qPCR). Total DNA was extracted by using DNeasy Blood & Tissue Kit (QIAGEN). The following primer-probe set was used: F3L.forward (5′-CATCTATTATAGCATCAGCATCAGA-3′), F3L.reverse (5′-GATACTCCTCCTCGTTGGTCTAC-3′), and F3L.probe (5′-TGTAGGCCGTGTATCAGCATCCATT-3′)48. qPCR was conducted in a LightCycler® 480 Instrument (Roche Diagnostics Ltd). SIV RNA loads in plasma were determined by qPCR assay77. The following primer-probe set was used: gag91 forward (5′-GCAGAGGAGGAAATTACCCAGTAC-3′), gag91 reverse (5′-CAATTTTACCCAGGCATTTAATGTT-3′), and pSHIVgag91-1 (5′-ACCTGCCATTAAGCCCGA-3′). qPCR was carried out on an ABI 7500 Real-time PCR system (Applied Biosystems) using the same protocol and primers as previously78.
RNA-sequencing and analysis
Total RNAs were extracted from the skin and spleen of euthanized animals using Trizol reagent. The construction of the RNA library and high-throughput sequencing were conducted by Beijing Annoroad Gene Technology Company following the manufacturer’s recommendations. DESeq2 v1.6.3 was used for differential gene expression analysis. Genes exhibiting a pvalue < 0.05 and an absolute log2 (fold change) value of ≥1 were identified as differentially expressed genes (DEGs). The Gene Ontology (GO) enrichment analysis of DEGs was conducted by the clusterProfiler R package. Volcano maps were generated by using GraphPad Prism 10 software.
Histopathological analysis
For histopathology, skin, testis and tonsil from infected macaques were fixed in 4% neutral-buffered formaldehyde, embedded in paraffin, sectioned, and stained with H&E. Images were captured using Olympus BX51 microscope equipped with a DP72 camera.
Immunohistochemistry
Skin samples obtained from euthanized animals were fixed in 4% paraformaldehyde for 48 h, subsequently embedded in paraffin, and sectioned. The paraffin tissue sections were deparaffinized in xylene, rehydrated and incubated with 3% hydrogen peroxide at room temperature to inactivate endogenous peroxidases. Antigen retrieval was performed by subjecting the samples to citrate buffer at 95 °C for 30 min. The slides were blocked with BSA at room temperature for 1 h and then incubated with MPXV A29 antibody (1:500, GeneTex, GTX638888) overnight at 4 °C. After three washes, the sections were incubated with anti-rabbit IgG conjugated to horseradish peroxidase (1:200, Servicebio, GB23303) at 37 °C for 50 min. The sections were counterstained with hematoxylin to facilitate microscopic observation.
Cytokine and chemokines analysis
Cytokine and chemokine expressions in plasma samples pre- and post-inoculation were measured using a Cytokine/Chemokine/Growth Factor 37-Plex NHP ProcartaPlex Panel (Thermo Fisher Scientific) according to the manufacturer’s instructions. The data were collected on Luminex 200 and analyzed by Luminex PONENT (Thermo Fisher Scientific).
Statistical analysis
Data were analyzed using GraphPad Prism 10 (GraphPad Software). The values shown in the graphs are presented as the mean ± SEM. Statistical differences between groups were analyzed using two-tailed unpaired t tests for single factor analysis or two-way ANOVA statistical tests for double factor analysis. P values are denoted as follows: ns, not significant, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. Exact P values are provided in figure legends. All experiments were conducted with at least 3 independent replicates. The investigators were not blinded to allocation during the process of data collection and analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available with the article, Supplementary Information or Source Data file. The raw RNA-seq data used in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) datasets under accession number GSE281290. Source data are provided with this paper.
References
Lum, F. M. et al. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat. Rev. Immunol. 22, 597–613 (2022).
Cho, C. T. & Wenner, H. A. Monkeypox virus. Bacteriol. Rev. 37, 1–18 (1973).
Elsayed, S., Bondy, L. & Hanage, W. P. Monkeypox virus infections in humans. Clin. Microbiol. Rev. 35, e0009222 (2022).
WHO. Multi-country monkeypox outbreak in non-endemic countries. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON385 (2022).
Wenham, C. & Eccleston-Turner, M. Monkeypox as a PHEIC: implications for global health governance. Lancet 400, 2169–2171 (2022).
Isidro, J. et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 28, 1569–1572 (2022).
Lane, H. C. & Fauci, A. S. Monkeypox - past as prologue. N. Engl. J. Med. 387, 749–750 (2022).
Thornhill, J. P. et al. Monkeypox virus infection in humans across 16 countries - April-June 2022. N. Engl. J. Med. 387, 679–691 (2022).
Tarin-Vicente, E. J. et al. Clinical presentation and virological assessment of confirmed human monkeypox virus cases in Spain: a prospective observational cohort study. Lancet 400, 661–669 (2022).
Girometti, N. et al. Demographic and clinical characteristics of confirmed human monkeypox virus cases in individuals attending a sexual health centre in London, UK: an observational analysis. Lancet Infect. Dis. 22, 1321–1328 (2022).
Patel, A. et al. Clinical features and novel presentations of human monkeypox in a central London centre during the 2022 outbreak: descriptive case series. BMJ 378, e072410 (2022).
van Ewijk, C. E. et al. Mpox outbreak in the Netherlands, 2022: public health response, characteristics of the first 1,000 cases and protection of the first-generation smallpox vaccine. Euro Surveill. 28, 2200772 (2023).
Zumla, A. et al. Monkeypox outbreaks outside endemic regions: scientific and social priorities. Lancet Infect. Dis. 22, 929–931 (2022).
Mitja, O. et al. Mpox in people with advanced HIV infection: a global case series. Lancet 401, 939–949 (2023).
Mitja, O. et al. Monkeypox. Lancet 401, 60–74 (2023).
Yinka-Ogunleye, A. et al. Outbreak of human monkeypox in Nigeria in 2017-18: a clinical and epidemiological report. Lancet Infect. Dis. 19, 872–879 (2019).
Curran, K. G. et al. HIV and sexually transmitted infections among persons with monkeypox - eight U.S. Jurisdictions, May 17-July 22, 2022. MMWR Morb. Mortal. Wkly. Rep. 71, 1141–1147 (2022).
Heath, S. L. & Bansal, A. Mpox infection in people living with HIV. AIDS 37, 701–703 (2023).
Liverpool, L. The disease will be neglected’: scientists react to the WHO ending mpox emergency. Nature 617, 662–663 (2023).
Vakaniaki, E. H. et al. Sustained human outbreak of a new MPXV clade I lineage in eastern Democratic Republic of the Congo. Nat. Med. 30, 2791–2795 (2024).
Gilchuk, I. et al. Cross-Neutralizing and Protective Human Antibody Specificities to Poxvirus Infections. Cell 167, 684–694.e689 (2016).
Hooper, J. W. et al. Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. J. Virol. 78, 4433–4443 (2004).
Fogg, C. et al. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J. Virol. 78, 10230–10237 (2004).
Wyatt, L. S., Earl, P. L., Eller, L. A. & Moss, B. Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. Proc. Natl. Acad. Sci. USA 101, 4590–4595 (2004).
FDA. FDA approves first live, non-replicating vaccine to prevent smallpox and monkeypox. https://www.fda.gov/news-events/press-announcements/fda-approves-first-live-non-replicating-vaccine-prevent-smallpox-and-monkeypox (2019).
Greenberg, R. N. & Kennedy, J. S. ACAM2000: a newly licensed cell culture-based live vaccinia smallpox vaccine. Expert Opin. Investig. Drugs 17, 555–564 (2008).
Zaeck, L. M. et al. Low levels of monkeypox virus-neutralizing antibodies after MVA-BN vaccination in healthy individuals. Nat. Med. 29, 270–278 (2023).
Tian, L. et al. Vaccinia virus tiantan strain is inefficient in eliciting cross-reactive immunity against the emerging monkeypox virus strain. Emerg. Microbes Infect. 13, 2306967 (2024).
Hooper, J. W., Custer, D. M. & Thompson, E. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology 306, 181–195 (2003).
Hirao, L. A. et al. Multivalent smallpox DNA vaccine delivered by intradermal electroporation drives protective immunity in nonhuman primates against lethal monkeypox challenge. J. Infect. Dis. 203, 95–102 (2011).
Mucker, E. M. et al. A nucleic acid-based orthopoxvirus vaccine targeting the vaccinia virus L1, A27, B5, and A33 proteins protects rabbits against lethal rabbitpox virus aerosol challenge. J. Virol. 96, e0150421 (2022).
Freyn, A. W. et al. An mpox virus mRNA-lipid nanoparticle vaccine confers protection against lethal orthopoxviral challenge. Sci. Transl. Med. 15, eadg3540 (2023).
Zhang, R. R. et al. Rational development of multicomponent mRNA vaccine candidates against mpox. Emerg. Microbes Infect. 12, 2192815 (2023).
Sang, Y. et al. Monkeypox virus quadrivalent mRNA vaccine induces immune response and protects against vaccinia virus. Signal Transduct. Target. Ther. 8, 172 (2023).
Zeng, J. et al. Mpox multi-antigen mRNA vaccine candidates by a simplified manufacturing strategy afford efficient protection against lethal orthopoxvirus challenge. Emerg. Microbes Infect. 12, 2204151 (2023).
Hou, F. et al. mRNA vaccines encoding fusion proteins of monkeypox virus antigens protect mice from vaccinia virus challenge. Nat. Commun. 14, 5925 (2023).
Fang, Z. et al. Polyvalent mRNA vaccination elicited potent immune response to monkeypox virus surface antigens. Cell Res. 33, 407–410 (2023).
Zuiani, A. et al. A multivalent mRNA monkeypox virus vaccine (BNT166) protects mice and macaques from orthopoxvirus disease. Cell 187, 1363–1373.e12 (2024).
Sakhatskyy, P., Wang, S., Chou, T. H. & Lu, S. Immunogenicity and protection efficacy of monovalent and polyvalent poxvirus vaccines that include the D8 antigen. Virology 355, 164–174 (2006).
Benhnia, M. R. et al. Heavily isotype-dependent protective activities of human antibodies against vaccinia virus extracellular virion antigen B5. J. Virol. 83, 12355–12367 (2009).
Benhnia, M. R. et al. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J Virol. 83, 1201–1215 (2009).
Cohen, M. E., Xiao, Y., Eisenberg, R. J., Cohen, G. H. & Isaacs, S. N. Antibody against extracellular vaccinia virus (EV) protects mice through complement and Fc receptors. PLoS ONE 6, e20597 (2011).
Matho, M. H. et al. Structural and functional characterization of anti-A33 antibodies reveal a potent cross-species orthopoxviruses neutralizer. PLoS Pathog. 11, e1005148 (2015).
Zhang, N. N. et al. A thermostable mRNA vaccine against COVID-19. Cell 182, 1271–1283.e1216 (2020).
Zhang, N. N. et al. Rapid development of an updated mRNA vaccine against the SARS-CoV-2 Omicron variant. Cell Res. 32, 401–403 (2022).
Zhao, H. et al. The first imported case of monkeypox in the Mainland of China - Chongqing Municipality, China, September 16, 2022. China CDC Wkly. 4, 853–854 (2022).
Huang, B. et al. Isolation and characterization of monkeypox virus from the first case of monkeypox - Chongqing Municipality, China, 2022. China CDC Wkly. 4, 1019–1024 (2022).
Aid, M. et al. Mpox infection protects against re-challenge in rhesus macaques. Cell 186, 4652–4661.e4613 (2023).
Earl, P. L. et al. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 428, 182–185 (2004).
Stittelaar, K. J. et al. Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature 439, 745–748 (2006).
Earl, P. L. et al. Rapid protection in a monkeypox model by a single injection of a replication-deficient vaccinia virus. Proc. Natl. Acad. Sci. USA 105, 10889–10894 (2008).
Goff, A. J. et al. A novel respiratory model of infection with monkeypox virus in cynomolgus macaques. J. Virol. 85, 4898–4909 (2011).
Johnson, R. F. et al. Comparative analysis of monkeypox virus infection of cynomolgus macaques by the intravenous or intrabronchial inoculation route. J. Virol. 85, 2112–2125 (2011).
Hatch, G. J. et al. Assessment of the protective effect of Imvamune and Acam2000 vaccines against aerosolized monkeypox virus in cynomolgus macaques. J. Virol. 87, 7805–7815 (2013).
Johnston, S. C. et al. Cytokine modulation correlates with severity of monkeypox disease in humans. J. Clin. Virol. 63, 42–45 (2015).
da Silva, G. B., de Carvalho Braga, G., Simoes, J. L. B., Kempka, A. P. & Bagatini, M. D. Cytokine storm in human monkeypox: a possible involvement of purinergic signaling. Cytokine 177, 156560 (2024).
Wei, H. et al. Coadministration of cidofovir and smallpox vaccine reduced vaccination side effects but interfered with vaccine-elicited immune responses and immunity to monkeypox. J. Virol. 83, 1115–1125 (2009).
Huggins, J. et al. Nonhuman primates are protected from smallpox virus or monkeypox virus challenges by the antiviral drug ST-246. Antimicrob. Agents Chemother. 53, 2620–2625 (2009).
Denzler, K. L. et al. Attenuated NYCBH vaccinia virus deleted for the E3L gene confers partial protection against lethal monkeypox virus disease in cynomolgus macaques. Vaccine 29, 9684–9690 (2011).
Buchman, G. W. et al. A protein-based smallpox vaccine protects non-human primates from a lethal monkeypox virus challenge. Vaccine 28, 6627–6636 (2010).
Hooper, J. W. et al. Molecular smallpox vaccine delivered by alphavirus replicons elicits protective immunity in mice and non-human primates. Vaccine 28, 494–511 (2009).
Mucker, E. M. et al. Comparison of protection against mpox following mRNA or modified vaccinia Ankara vaccination in nonhuman primates. Cell 187, 5540–5553 (2024).
Liu, J. et al. Retrospective detection of monkeypox virus in the testes of nonhuman primate survivors. Nat. Microbiol. 7, 1980–1986 (2022).
Tree, J. A. et al. Sequence of pathogenic events in cynomolgus macaques infected with aerosolized monkeypox virus. J. Virol. 89, 4335–4344 (2015).
Bratke, K. A., McLysaght, A. & Rothenburg, S. A survey of host range genes in poxvirus genomes. Infect. Genet. Evol. 14, 406–425 (2013).
Shchelkunov, S. N. Orthopoxvirus genes that mediate disease virulence and host tropism. Adv. Virol. 2012, 524743 (2012).
Kotwal, G. J., Isaacs, S. N., McKenzie, R., Frank, M. M. & Moss, B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science 250, 827–830 (1990).
Isaacs, S. N., Kotwal, G. J. & Moss, B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. USA 89, 628–632 (1992).
Gruner, E. et al. Mpox-specific immune responses elicited by vaccination or infection in people living with HIV. J. Infect. Dis. 230, 1110–1119 (2024).
Maan, I., Kohli, M. & Gilson, R. Mpox in people living with HIV. Curr. Opin. Infect. Dis. 37, 1–7 (2024).
Wong, M., Damon, I. K., Zucker, J., Foote, M. M. K. & El-Sadr, W. ART initiation for people living with HIV with severe mpox. Lancet 402, 1750 (2023).
Iroezindu, M. O., Crowell, T. A., Ogoina, D. & Yinka-Ogunleye, A. Human Mpox in people living with HIV: epidemiologic and clinical perspectives from Nigeria. AIDS Res. Hum. Retroviruses 39, 593–600 (2023).
Ogoina, D. et al. Clinical course and outcome of human monkeypox in Nigeria. Clin. Infect. Dis. 71, e210–e214 (2020).
Adler, H. et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect. Dis. 22, 1153–1162 (2022).
Miller, M. J. et al. Severe monkeypox in hospitalized patients - United States, August 10-October 10, 2022. MMWR Morb. Mortal. Wkly. Rep. 71, 1412–1417 (2022).
Hammarlund, E. et al. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat. Med. 11, 1005–1011 (2005).
Chong, H. et al. Monotherapy with a low-dose lipopeptide HIV fusion inhibitor maintains long-term viral suppression in rhesus macaques. PLoS Pathog. 15, e1007552 (2019).
Xue, J. et al. Efficient treatment and pre-exposure prophylaxis in rhesus macaques by an HIV fusion-inhibitory lipopeptide. Cell 185, 131–144.e118 (2022).
Acknowledgements
This work was supported by the National Key Research and Development Project of China (2021YFC2302400, 2023YFC2309000, and 2022YFC2304101), and the National Natural Science Foundation of China (82241069, 82241068). C.-F.Q. was supported by the National Science Fund for Distinguished Young Scholars (81925025), and the Innovation Fund for Medical Sciences (2019-I2M-5-049) from the Chinese Academy of Medical Sciences. J.X. was supported by the National Science Fund for Excellent Young Scholars (82222041). Q.Y. was sponsored by Beijing Nova Program (20240484525).
Author information
Authors and Affiliations
Contributions
C.F.Q., J.X., and Q.Y. designed the experiments and wrote the manuscript. Q.Y., D.Z., R.R.Z., Q.X., X.Y.H., and M.X.S. performed the majority of the experiments. Z.C., L.Z., J.R.M., N.L., J.J.Z., T.C., J.H.L., and Y.Z.H. performed the animal experiments and sample collection. X.C., H.T.L., C.Z., R.T.L., M.W., Z.J.W., J.Y., Y.F.Q., and B.Y. contributed specific experiments and data analysis. Y.B.H. and W.T. contributed the MPXV strain. All authors read and approved the contents of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.F.Q., Q.Y., R.R.Z., and Z.J.W. have filed a patent related to the vaccine reported in the study. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ye, Q., Zhang, D., Zhang, RR. et al. A penta-component mpox mRNA vaccine induces protective immunity in nonhuman primates. Nat Commun 15, 10611 (2024). https://doi.org/10.1038/s41467-024-54909-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54909-4
This article is cited by
-
Comparative analysis of Mpox clades: epidemiology, transmission dynamics, and detection strategies
BMC Infectious Diseases (2025)
-
Opposite regulation of immune genes in blood and skin highlights tissue-specific dynamics of mpox virus
Scientific Reports (2025)
-
An mRNA vaccine against monkeypox virus inhibits infection by co-activation of humoral and cellular immune responses
Nature Communications (2025)
-
Potent neutralization by antibodies targeting the MPXV A28 protein
Nature Communications (2025)