Abstract
The sympathetic nervous system is crucial for responding to environmental changes. This regulation is coordinated by the spinal sympathetic preganglionic neurons (SPNs), innervating both postganglionic neurons and the adrenal gland. Despite decades of research supporting the concept of selective control within this system, the neural circuit organization responsible for the output specificity remains poorly understood. Here, by combining recent single-cell transcriptome data with viral-genetic toolkits in mice, we identify two subtypes of SPNs in the lower thoracic spinal cord, defined at the molecular level, exhibiting nonoverlapping patterns of innervation: one specifically projecting to the celiac/superior mesenteric ganglia, and the other targeting the adrenal grand. Chemogenetic manipulations on these distinct SPN subtypes revealed selective impacts on the motility of the gastrointestinal tracts or glucose metabolism mediated by the adrenal gland, respectively. This molecularly delineated parallel labeled-line organization in sympathetic outflows presents a potential avenue for selectively manipulating organ functions.
Similar content being viewed by others
Introduction
Recent advancements in single-cell transcriptome (TC) technologies have expanded neuronal cell type classification1, raising general questions regarding whether a given TC type projects exclusively to a specific set of target regions (labeled-line model, Fig. 1a1), or whether multiple TC types within a given brain region share common output targets (divergent output model, Fig. 1a2). While recent studies have exemplified varying degrees of output specificity among TC types across multiple central brain regions2,3, this issue within the efferent pathways in the spinal cord (SC) remains unexplored, despite its significant relevance to the regulation of various organs in the body.
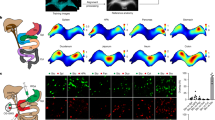
a Schematic of two extreme projection patterns for two distinct TC types. b Schematic of retrograde tracing from the CG/SMG. CA central autonomic area, IC intercalated nucleus, IML intermediolateral cell column, D dorsal, L lateral. c Representative horizontal view of the labeled CG/SMG, based on N = 3 mice. tdT tdTomato, DBH dopamine-β-hydroxylase, SMA superior mesenteric artery. Scale bar, 200 μm. d Light-sheet microscopic view of tissue-cleared SC displaying tdT+ cells (magenta). Scale bar, 500 μm. T thoracic. Number (e) and percentage (f) of tdT+ cells in the CA, IC, and IML for each SC segment. N = 3 mice. L lumbar. g Coronal sections of the T12 SC depicting tdT+ cells (magenta) and Chat (top) or nNos (bottom) immunostaining (green), with magnified images within white boxes. The arrows indicate tdT+ Chat+ cells. Scale bars, 100 μm. h Percentage of Chat+ or nNos+ cells among tdT+ cells in the CA, IC, and IML. N = 3 mice. i UMAP representation of 19 Chat+ SPN clusters (left) and a heat map showing log-normalized expression of the nNos gene (right). j Identification of enriched differentially expressed genes and expression of nNos among nNoslow clusters. The dot size reflects the population size, while the heat map shows the log-normalized expression level. k Coronal sections of the T12 SC displaying tdT+ cells (magenta) retrogradely labeled from the CG/SMG and Cartpt (top) or Chodl (bottom) mRNA expression (green), with magnified images within the white box. Arrows indicate dual-labeled cells. Scale bars, 100 μm for low-magnification images and 50 μm for magnified images. l Percentage of Cartpt+ cells (left) or Chodl+ cells (right) among tdT+ cells. N = 3 mice, except for Chodl+ data in the CA region, where N = 2 mice. m Schematic summary of the data. Data are shown as mean ± standard deviation (SD). Source data are provided as a Source Data file. For more data, see Supplementary Fig. 1 and Supplementary Movies 1 and 2.
The sympathetic nervous system is vital in maintaining homeostasis and responding to environmental changes4,5,6. This regulation is coordinated by the spinal sympathetic preganglionic neurons (SPNs), which influence various organs both through neuronal pathways via postganglionic neurons and through endocrine processes by innervating the adrenal gland. Despite decades of research supporting the concept of selective control within this system4,7,8,9,10,11,12, the neural circuit organization responsible for the specificity of sympathetic outflow remains poorly understood. To address this knowledge gap, we focus on the SPNs that target either the celiac/superior mesenteric ganglia (CG/SMG) or the adrenal medulla (AM), two crucial components of the fight-or-flight response. Upon encountering stressors, the sympathetic nervous system provides an energy supply, partly via the function of the AM13, while concurrently inhibiting the digestive functions in the gastrointestinal (GI) tract via the postganglionic neurons (postGNs) located in the CG/SMG14. The SPNs regulating these targets are located within the same lower thoracic SC4, which renders these SPNs an ideal model for studying their molecular, anatomical, and functional specificities. Notably, animals can differentially utilize these two output pathways; AM-projecting SPNs become more active in response to low plasma glucose levels9, and AM chromaffin cells are selectively activated by a brain-infused hormone10. Despite these observations supporting the presence of distinct output specificity, classical anatomical studies in rats neither reveal a definitive molecular code for SPNs nor corroborate the labeled-line organization4,9,15,16. This motivated us to utilize recent transcriptome data of SPNs17,18 in mice and viral-genetic toolkits19 to map axonal projections and manipulate the functions of SPNs targeting the CG/SMG or the AM.
Results
Cartpt+ SPNs selectively project to the CG/SMG
To unravel the molecular characteristics of SPNs projecting to the CG/SMG, we developed selective adeno-associated virus (AAV) injection procedures into the CG/SMG in mice (Supplementary Fig. 1, Supplementary Movie 1). We then injected retrogradely transducible AAVrg hSyn-Cre into the CG/SMG of Ai9 (Cre reporter tdTomato line20) mice to visualize SPNs in the SC (Fig. 1b, c). Under light-sheet microscopy after tissue clearance21, tdTomato-expressing (+) cells retrogradely labeled from the CG/SMG were predominantly found in the 8th to 13th thoracic SC (T8–T13) (Fig. 1d, Supplementary Movie 2). Histological analysis on coronal SC sections confirmed this distribution and also revealed that over 80% of tdTomato+ cells were located in the central autonomic area (CA) and intercalated nucleus (IC), contrary to the prevailing view that the majority of SPNs are located in the intermediolateral cell column (IML)22 (Fig. 1e, f). While tdTomato+ cells were nearly 100% positive for choline acetyltransferase (Chat), a functional marker of cholinergic SPNs (Fig. 1g, h), no overlap was seen between tdTomato+ cells and those labeled with neuronal nitric oxide synthase, nNos, a conventional marker for SPNs23 (Fig. 1g, h).
The detailed molecular profiles of SPNs with low or no nNos expression have not been adequately addressed in previous studies. To characterize these, we conducted a reanalysis of existing single-nucleus RNA sequencing (snRNAseq) data by ref. 17 (see Methods for details). We defined nNoslow SPN clusters as those in which 50% or fewer cells express nNos, with average expression levels (in arbitrary units) lower than zero. This led to the identification of 8 nNoslow SPN clusters (Fig. 1i, j). Histochemical analyses using selective marker genes for these nNoslow clusters revealed that tdTomato+ cells retrogradely labeled from the CG/SMG almost entirely co-localized with those expressing Cartpt (Fig. 1k, l), a dominant marker of clusters 1 and 15 with relatively weak expression in cluster 2 (Fig. 1j). We also observed that the vast majority of tdTomato+ cells expressed the Chondrolectin gene (Chodl), marking clusters 1 and 15 (Fig. 1j–l), suggesting that the tdTomato+ cells belong to SPN clusters 1 and 15. Collectively, these data indicate that SPNs retrogradely labeled from the CG/SMG form molecularly defined distinct TC subtypes marked by Cartpt (Fig. 1m).
To analyze the axonal projection targets of Cartpt+ SPNs, we introduced an AAV expressing Cre-dependent mCherry into the T8–T12 of mice that expressed Cre under the control of the Cartpt, with pan-SPN driver Chat-Cre mice24 serving as a positive control (Fig. 2a, b). Cartpt-Cre mice were generated through the CRISPR/Cas9-based method25 (Supplementary Fig. 2). At 2–3 weeks post viral injection, we confirmed that the majority of mCherry+ cells were Chat+ or Cartpt+, respectively, demonstrating high specificity (Fig. 2c). We analyzed their axons in the sections of CG/SMG, AM, and the sympathetic trunk (ST) (Fig. 2d). While Chat+ pan-SPN axons were closely co-localized with the dopamine-β-hydroxylase (DBH)+ postGNs or adrenal chromaffin cells in these structures, we detected mCherry+ axons of Cartpt+ SPNs in only the CG/SMG, with no presence in the AM or ST (Fig. 2d). These mCherry+ axons of Cartpt+ SPNs broadly projected to the entire CG/SMG (Supplementary Fig. 2c). These data confirmed our retrograde labeling result (Fig. 1) and demonstrated that Cartpt+ SPNs selectively innervate the CG/SMG without targeting the AM or ST, supporting a labeled-line organization (Fig. 1a1).
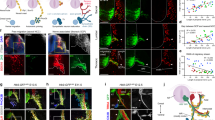
Schematics illustrating the experimental design (a) and time line (b). AM adrenal medulla, ST sympathetic trunk. c Coronal section of the SC showing mCherry (mC) expression (magenta) along with Chat, Cartpt, or Oxtr expression (green). AAV8 hSyn-DIO-mCherry was used for labeling. The right graph displays quantification for efficiency (mC+/markers+) and specificity (marker+/mC+). d mC+ axons (magenta) of Chat+, Cartpt+, or Oxtr+ SPNs to DBH+ (green) areas in the CG/SMG, AM, and 8–12th ST. The right graph shows the percentage of mC+ axons in the DBH+ regions. Magenta signals in the ST outside the DBH+ area show axons in passage. *p < 0.05, **p < 0.01, and ***p < 0.001 by one-way analysis of variance (ANOVA) followed by a two-sided Welch’s t-test with Bonferroni correction. Insets (in c, d) show magnified and channel-separated images in the boxed area. Scale bars, 100 μm for low-magnification images (c, d), and 50 μm for magnified images. The number of animals (N) is indicated in blue within the panel. Data are shown as mean ± SD. Source data are provided as a Source Data file. For more data, see Supplementary Figs. 2 and 3.
Oxtr delineates a subset of nNos+ SPNs projecting to the AM
Next, we sought SPNs targeting the AM. Based upon the literature suggesting an association between the neural hormone oxytocin (Oxt) and AM functions26,27,28, along with the documented expression of oxytocin receptor (Oxtr) mRNA in the IML of the SC29, we serendipitously found that Oxtr+ SPNs are specific projectors to the AM. First, utilizing the RNAscope single-molecule in situ hybridization method, we found that the majority of Oxtr+ cells were nNos+ in the IML of the thoracic SC (Supplementary Fig. 3a, b), consistent with the reported high-level expression of nNos in SPNs projecting to the AM15. Minimal overlap was observed between Oxtr+ and Cartpt+ cells (Supplementary Fig. 3a, b), distinguishing Oxtr+ SPNs from those projecting to the CG/SMG. Next, we repeated the anterograde axon tracing assays shown in Fig. 2 with Oxtr-Cre mice30. Post hoc histochemical analyses confirmed the mCherry was selectively expressed by Oxtr+ cells in the lower thoracic SC (Fig. 2c), and SPNs targeted by Cartpt-Cre and Oxtr-Cre were mutually exclusive (Supplementary Fig. 3c). In stark contrast to Cartpt+ SPNs, mCherry+ axons of Oxtr+ SPNs were observed in the AM, excluding the CG/SMG and ST (Fig. 2d).
To characterize the TC type of Oxtr+ SPNs, we extracted representative marker genes for each one or few nNoshigh SPN clusters (Supplementary Fig. 3d). Despite their low expression, snRNAseq data suggested the predominant presence of Oxtr in cluster 4 within the nNoshigh SPNs, which was characterized by the expression of the Palladin (Palld) gene encoding a cytoskeletal-associated protein as a marker (Supplementary Fig. 3d, e). We then detected Oxtr and Palld expression in the IML of the T8–T13 segments and found that the vast majority of Oxtr+ cells also expressed Palld, and vice versa (Supplementary Fig. 3f, g). Taken together, Oxtr+ SPNs provide a second example of the labeled-line organization (Fig. 1a1): a specific TC type of SPN projects specific output.
Next, we explored the functional aspects of the target specificity by injecting an AAV expressing Cre-dependent Gq-mCherry31 into the T8–T12 of Chat-Cre, Cartpt-Cre, or Oxtr-Cre mice (Fig. 3a, b). Within the SC, Gq-mCherry expression was specifically observed in the marker+ SPNs (Fig. 3c). Histochemical analyses of c-Fos, an immediate early gene product serving as a proxy for cellular activation, revealed that the majority of mCherry + cells turned c-Fos+ following the administration of clozapine-N-oxide (CNO), confirming the system’s capability for specific activation of molecularly defined SPN types (Fig. 3d). Activation of Chat+ SPNs led to increased c-Fos expression in both the CG/SMG and AM (Fig. 3e). By contrast, activation of Cartpt+ SPNs induced c-Fos selectively in the CG/SMG but not in the AM, whereas activation of Oxtr+ SPNs led to increased c-Fos expression in the AM but not in the CG/SMG (Fig. 3e). Of note, the timing of this c-Fos analysis relative to the CNO administration was determined through serial time course analysis (Supplementary Fig. 4a). Negative controls, omitting Gq expression, exhibited no c-Fos induction (Fig. 3e, right graphs, and Supplementary Fig. 4b). Collectively, these findings demonstrate that Cartpt+ and Oxtr+ SPNs can selectively activate the CG/SMG and AM, respectively, via their output target specificity.
Schematics illustrating the experimental design (a) and time line (b). c Coronal section of the SC showing mCherry (mC) expression (magenta) along with Chat, Cartpt, or Oxtr expression (green). AAV8 hSyn-DIO-Gq-mCherry was used for labeling. The right graph displays quantification for efficiency (mC+/markers+) and specificity (marker+/mC+). d Coronal section of the SC showing c-Fos (green) and mC (magenta) at 2 h after CNO administration. The right graph shows the percentage of c-Fos+ cells among mC+ cells in CNO-treated Gq and mC control mice. e c-Fos expression (green) in the whole CG/SMG (top) and AM sections (bottom). Magenta denotes mC+ axons. The right graph shows the number of c-Fos+ cells (CG/SMG) and the percentage of c-Fos+ per DAPI+ cells (AM) in CNO-treated Gq and mC control mice. ***p < 0.001 by a two-sided Welch’s t-test (d, e). Insets in panels (c–e) show magnified and channel-separated images in the boxed area. Scale bars, 100 μm for low-magnification images (c, d), 200 μm for low-magnification images (e), and 50 μm for magnified images. The number of animals (N) is indicated in blue within the panel. Data are shown as mean ± SD. Source data are provided as a Source Data file. For more data, see Supplementary Figs. 3 and 4.
Activation of Cartpt+ SPNs decreases intestinal motility
The presence of the molecularly defined output specificity of SPNs allows the selective manipulation of sympathetic outflows. As a proof-of-concept, we next investigated whether the chemogenetic activation of Cartpt+ SPNs (Fig. 3) influences the gastrointestinal (GI) tract, one of the major targets of postGNs in the CG/SMG32 (Fig. 4a, b). Red dye (carmine red) was orally administered, and the latency to the excretion of red-colored stools was measured as the GI transition time (GITT). While CNO administration alone resulted in a slight increase in GITT, as observed in the mCherry control experiments, activation of Chat+ SPNs substantially increased GITT by ~100 min, consistent with the known negative influence of sympathetic output on the motility of the GI tract (Fig. 4c)32. Notably, activation of Cartpt+ SPNs led to an increase in GITT by ~120 min, whereas activation of Oxtr+ SPNs did not affect GITT (Fig. 4c). We also analyzed GITT under celiac ganglionectomy conditions (Fig. 4d). Despite the activation of Cartpt+ SPNs, we observed no increase in GITT under this condition, confirming that the increase in GITT induced by the activation of Cartpt+ SPNs is mediated through the CG/SMG (Fig. 4e). As feeding and gut contents can impact GITT33, we also confirmed that the activation of Cartpt+ SPNs increased GITT even without feeding (Fig. 4f).
Schematics illustrating the experimental design (a) and time line (b). GITT, gastrointestinal transit time. The right image displays a fecal pellet stained red through carmine red administration. c GITT in Chat-Cre, Cartpt-Cre, or Oxtr-Cre mice injected with AAV8-hSyn-DIO-Gq-mCherry or AAV8-hSyn-DIO-mCherry (for control), upon administration of saline (white bar) or CNO (colored bar). The results from the same mice are connected with a line. Statistical comparisons were made using two-way ANOVA with repeated measures; (Chat-Cre) solution effect, p < 0.01, AAV effect, p < 0.01, interaction effect, ns. ***p < 0.001 by a post hoc two-sided Welch’s t-test. (Cartpt-Cre) solution effect, p < 0.01, AAV effect, p < 0.01, interaction effect, p < 0.01. ***p < 0.001 by a post hoc two-sided Welch’s t-test. (Oxtr-Cre) interaction effect, ns. d Schematics showing the Gq-mediated activation of Cartpt+ SPNs under the celiac ganglionectomy (CGX) condition. e GITT in CNO-treated Cartpt-Cre mice injected with AAV8-hSyn-DIO-Gq-mCherry under intact (left) and CGX (right) conditions. Data of intact conditions are the same as panel (c). f GITT measured without food in Cartpt-Cre mice injected with AAV8-hSyn-DIO-Gq-mCherry. g Schematic showing selective activation of SPNs projecting to the CG/SMG by injecting AAV-hSyn-DIO-Gq-mCherry into the T8–12 SC, to which Cre is retrogradely introduced from the CG/SMG via AAVrg-hSyn-Cre in wild-type mice. h GITT measured based on the panel (e) scheme upon administration of saline (white) or CNO (purple). **p < 0.01, ***p < 0.001 by two-sided Welch’s t-test (e, f, h). The number of animals (N) is indicated in blue within the panel. Data are shown as mean ± SD. Source data are provided as a Source Data file. For more data, see Supplementary Fig. 5.
In the thoracic SC, Cartpt is expressed by not only SPNs but also cholinergic interneurons17,34 (Supplementary Fig. 5). To exclude the possibility that the observed increase in GITT by the activation of Cartpt+ spinal cells is caused by cholinergic interneurons, we restricted targeted Gq-mCherry to spinal neurons retrogradely transduced with Cre from the axons in the CG/SMG (Fig. 4g). We confirmed that exclusive activation of SPNs projecting to the CG/SMG significantly increased GITT (Fig. 4h). Collectively, these data demonstrate that activation of Cartpt+ SPNs predominantly suppresses the motility of the GI tract via postGNs in the CG/SMG, indicating that a specific SPN type corresponds to the regulation of the GI tract; however, we do not exclude the possibility that Cartpt+ SPNs influence other organs (see the limitations of our study in the Discussion section).
Oxtr+ SPNs regulate glucose metabolism
We next sought to investigate the function of Oxtr+ SPNs. A prompt surge in blood glucose levels (BGLs) is a characteristic fight-or-flight response, providing energy supply to address impending danger or stressors13. Therefore, we examined BGLs upon the activation of specific SPNs. The chemogenetic activation of Chat+ SPNs using Gq-mCherry (Fig. 3) led to a significant increase in BGLs in male mice, whereas no such effect was observed in female mice (Fig. 5a, b). Although the precise reasons for this pronounced sex-specific response remain unknown, we observed that CNO administration induced comparable levels of c-Fos expression in the AM of both sexes (Supplementary Fig. 4c), suggesting that the observed sex differences in BGLs occur downstream of chromaffin cell activation in the AM. This observation aligns with previous findings supporting sex differences in the sensitivity to sympathetic activation35 and greater insulin sensitivity in the liver and adipose tissue of females compared with males36,37. Consequently, female mice appear to have a superior ability to maintain normal BGLs, which may account for the less pronounced changes in BGLs observed in females than in males. Notably, a similar male-specific increase in BGLs was observed upon activating Oxtr+ SPNs, but not Cartpt+ SPNs (Fig. 5a, b). Negative control experiments expressing mCherry alone had no impact on BGLs (Supplementary Fig. 6a, b). To assess whether Oxtr+ SPNs influence glucose metabolism through the AM, we examined their activation in male mice under adrenalectomy conditions (Fig. 5c). No increase in BGLs was observed following CNO administration, confirming that the rise in BGLs induced by the activation of Oxtr+ SPNs is mediated through the adrenal gland (Fig. 5d, e). These data demonstrate that the activation of Oxtr+ SPNs induces a sex-dependent increase in BGLs through the function of the AM. Of note, while the precise mechanism by which Oxtr+ SPN-to-AM pathways elevate BGLs remains elusive, it is unlikely that the suppression of insulin secretion is involved, as glucose-induced insulin release was unaffected by chemogenetic activation of Oxtr+ SPNs (Supplementary Fig. 7a–d).
a Change in blood glucose levels (ΔBGL) in Chat-Cre, Cartpt-Cre, or Oxtr-Cre mice injected with AAV8-hSyn-DIO-Gq-mCherry into the T8–T12 spinal segments, upon administration of saline (gray) or CNO (black). Individual data points are depicted with dots, and the average is denoted by a line. Top graphs are for males; bottom graphs are for females. +1 denotes that one data point is an outlier. Statistical comparisons were made using two-way ANOVA. Only the Chat: Gq, male and Oxtr: Gq, male groups exhibited significance; solution effect, p < 0.05; time course effect, p < 0.01; interaction effect, p < 0.05. #, p < 0.05, ##, p < 0.01 by a post hoc two-sided Welch’s t-test with Bonferroni correction showing a significant difference between the saline and CNO groups. b Area under the curve (AUC) measurements for 0 to 140 min. Two-way ANOVA with repeated measures; (Chat-Cre) sex effect, p < 0.05, solution effect, p < 0.01, interaction effect, p < 0.01. *p < 0.05 by a post hoc two-sided Welch’s t-test. (Cartpt-Cre) interaction effect, ns. (Oxtr-Cre) sex effect, p < 0.05, solution effect, ns, interaction effect, p < 0.01. ***p < 0.001 by a post hoc two-sided Welch’s t-test. c Schematics illustrating the Gq-mediated activation of Oxtr + SPNs in male mice under adrenalectomy. d ΔBGL under adrenalectomy, upon administration of saline (gray) or CNO (black). e AUC measurements for 0 to 140 min. ns, non-significant by a two-sided Welch’s t-test. The number of animals (N) is indicated in blue within the panel. Data are shown as mean ± SD. Source data are provided as a Source Data file. For more data, see Supplementary Figs. 6 and 7.
To ascertain whether the chemogenetic activation of Oxtr+ SPNs influences endocrinological cardiac modulations, we monitored heart rates (HRs) using a telemetry system (Supplementary Fig. 6c–g). Given that the SPNs regulating cardiac functions are located in the upper thoracic segments38, the Gq-mediated activation of Oxtr+ SPNs in the T8–T12 segments in this experiment should not impact them directly. While the HR in saline-injected control animals remained unaffected, a significant increase in HR was observed following CNO administration (Supplementary Fig. 6d, e). Negative control experiments expressing mCherry alone had no discernible impact on HR (Supplementary Fig. 6f, g). Collectively, these data suggest that manipulating the activity of Oxtr+ SPNs in the lower thoracic SC can influence metabolic and cardiac functions. Although these effects may be driven by adrenaline secretion from the AM, the effect size of our manipulation was insufficient to achieve statistical significance (Supplementary Fig. 7e; see the limitations in the Discussion section).
Next, we examined the potential relationship between Oxt produced in the hypothalamus and Oxtr+ SPNs in the lower thoracic SC. Oxt is a nonapeptide hormone produced by Oxt neurons in the paraventricular hypothalamus (PVH) that regulates energy metabolism39, alongside reproductive and social functions40. Mice lacking Oxt or the Oxtr gene exhibit obesity linked to reduced energy expenditure41,42. PVH Oxt neurons are activated during hypoglycemia, send axonal projections to the SC, and influence glucose tolerance43. Based on these preceding studies, we hypothesized that PVH Oxt neurons may contribute to the regulation of BGLs via the function of Oxtr+ SPNs in the lower thoracic SC. To test this hypothesis, we aimed to activate chemogenetically PVH Oxt neurons while viral-genetically ablating Oxtr+ SPNs. We introduced Oxt promoter (OXTp) driving Gq-myc44, or mCherry as a control, specifically to PVH Oxt neurons in Oxtr-Cre male mice that received an AAV injection expressing Cre-dependent active caspase (taCasp3-TEV)45, or saline as a control, into the bilateral T8–T12 SC segments (Fig. 6a, b). The targeting specificity of OXTp-Gq was validated in a previous study44. Over the subsequent 1-month post viral injection, we observed Myc-tag expression, an inference of Gq expression, in the PVH Oxt neurons (Fig. 6c). Activation of PVH Oxt neurons increased c-Fos expression in the IML of the T8–T12 segments (Fig. 6d, e). In the mCherry control group, we also observed direct axonal projections of PVH Oxt neurons in the proximity of Oxtr+ SPN cell bodies in the IML, confirming the direct projection (Fig. 6f). In the taCasp3-injected group, the number of Oxtr+ SPNs detected by RNAscope in the T8–T12 segments was significantly reduced by one-third (Fig. 6g, h). In the saline-injected control group, where the Oxtr+ SPNs remained intact, a significant increase in BGLs was observed after the chemogenetic activation of PVH Oxt neurons (Fig. 6i, j). By contrast, in animals with ablated Oxtr+ SPNs in the T8–T12 segments, no elevation in BGLs occurred after CNO administration. In the negative control group expressing only mCherry in PVH Oxt neurons, CNO administration did not impact BGLs. These data support the notion that the positive impact of Oxt on peripheral glucose metabolism is indeed mediated by Oxtr+ SPNs in the lower thoracic SC.
Schematics illustrating the experimental design (a) and time line (b). PVH, paraventricular hypothalamus. OXTp, oxytocin promoter. c Representative coronal section of the posterior PVH showing the expression of Myc (magenta) fused with Gq and Oxt (green) following the injection of AAV5-OXTp-Gq-Myc, based on N = 5 mice. Arrows indicate Myc+ Oxt+ double-positive cells. In the posterior PVH, parvocellular PVH Oxt neurons projecting the SC present71. d Coronal SC sections showing the expression of Chat (magenta) and c-Fos (green) in the IML following CNO administration in Gq-Myc or mCherry (mC) control conditions. e Number of c-Fos+ cells in five sections of the T8–12 IML. **p < 0.01 by two-sided Welch’s t-test. f Representative T8 spinal coronal section showing the expression of mC (magenta) and Oxtr (green) in OXTp-mCherry condition, based on N = 5 mice. These data demonstrate direct axonal projections from PVH Oxt neurons to the IML of lower thoracic SC. g Coronal spinal segment (T9) of indicated conditions showing Oxtr mRNA expression (magenta) by RNAscope and DAPI staining (blue). Scale bars, 100 μm (panels c, d, f, g). h Number of Oxtr+ cells in the control saline-injected group (white) and the ablation group (green) across 10 sections from the T8–12 SC. ***p < 0.001 by a two-sided Welch’s t-test. i ΔBGL in the Oxt neuron-activated group (left), Oxt neuron-activated and Oxtr+ SPN-ablated group (middle), and control group (right) upon administration of saline (gray) or CNO (black). j AUC measurements following CNO administration. Two-way ANOVA with repeated measures; solution effect, ns, AAV effect, ns, interaction effect, p < 0.05. *p < 0.05 by a post hoc Wilcoxon’s signed-rank test. k Schematic summary of the present work, showing that molecularly defined distinct TC types of SPNs regulate different downstream targets. The number of animals (N) is indicated in blue within the panel. Data are shown as mean ± SD. Source data are provided as a Source Data file.
Oxtr+ SPNs are preferentially activated during glucoprivation
Thus far, our data have demonstrated distinct functions of molecularly delineated parallel sympathetic outflows: Cartpt+ SPNs selectively innervate the CG/SMG, suppressing GI motility, whereas Oxtr+ SPNs specifically project to the AM, regulating glucose metabolism (Fig. 6k). Nevertheless, the conditions under which these neurons are activated to execute these distinct functions remain unknown. As a first step to address this issue, we sought to determine whether inducing artificial glucoprivation via 2-deoxyglucose (2-DG) injection43,46 differentially activates Cartpt+ and Oxtr+ SPNs in the lower thoracic SC (Fig. 7a). 2-DG is phosphorylated to 2-deoxyglucose-6-phosphate, which is metabolically inert, and thus inhibits glucose utilization, triggering glucoprivation46.
a Schematic representation of the experimental design to induce glucoprivation. b Representative SC sections showing c-Fos (green) and Chat (gray) after saline (left) or 2-DG (right) injection. Arrows indicate dual-labeled cells. c The number of c-Fos+ cells within Chat+ cells in the T8–10 and T11–13 segments under saline or 2-DG-treated conditions. N = 6 mice. ***p < 0.001 by a two-sided Welch’s t-test. d Representative SC sections showing c-Fos (green), Oxtr (magenta), and Chat (blue) (left), or c-Fos (green) and Cartpt (magenta) (right) after 2-DG injection. Arrows indicate triple-labeled cells. e Fraction of c-Fos+ cells within Oxtr+ cells among Chat+ cells in the 2-DG-treated condition. N = 6 mice. f Fraction of Oxtr+ or Cartpt+ cells within c-Fos+ cells in the 2-DG-treated condition. N = 6 mice. **p < 0.01 by a two-sided Welch’s t-test. Insets in panels (b) and (d) show magnified and channel-separated images in the boxed area. Scale bars, 100 μm for low-magnification images and 50 μm for magnified images (b, d). Data are shown as mean ± SD. Source data are provided as a Source Data file.
We found that 2-DG administration significantly increased c-Fos mRNA expression in the T8–13 segments of the SC (Fig. 7b, c). RNAscope analysis revealed that ~80% of Oxtr+ SPNs in the lower thoracic SC expressed c-Fos+ (Fig. 7d, e). Although we were unable to assess the fraction of Cartpt+ SPNs expressing c-Fos+ precisely because of the presence of Cartpt+ cholinergic interneurons (Supplementary Fig. 5), we noted that Cartpt+ neurons constituted only ∼20% of c-Fos+ cells in the T8–10 segments and nearly 0% in the T11–13 segments (Fig. 7d, f). By contrast, Oxtr+ neurons comprised ∼60% of c-Fos+ cells in the lower thoracic SC (Fig. 7d, f). Given that Cartpt+ SPNs are more numerous than Oxtr+ SPNs (Supplementary Fig. 3h), these data indicate the preferential activation of Oxtr+ over Cartpt+ SPNs in response to 2-DG-induced glucoprivation.
Discussion
The present study established a viral-genetic approach to studying mouse SPNs in terms of their anatomical and functional organizations. The findings demonstrate the existence of molecularly delineated parallel sympathetic outflows that regulate the CG/SMG and AM (Fig. 6k). Here, we discuss the biological insights provided by this study along with its limitations.
While early twentieth-century physiologists accentuated the homogeneous nature of sympathetic outflows47, a large body of research subsequently supported the notion of selective yet coordinated control within this system4,7,8. For instance, classical electrophysiological studies have suggested that reflex patterns exhibited by SPNs in response to stimulation from various afferents possess unique characteristics corresponding to distinct functional pathways8,11,12. Under experimental conditions, animals have demonstrated the ability to favor the activation of certain selective sympathetic outflows9,10. Sympathetic output specificity has also been suggested in recent research regarding the somatotopic nature of acupuncture48, the selective sympathetic impacts on the GI tract mediated by vagal sensing of gut microbiota32 or gut mechanosensation49, and the top-down regulation of immune responses within delimited regions of the body50. However, the precise cellular underpinnings within SPNs for these distinctive sympathetic outflows remain largely unknown.
While classical studies have suggested the nonuniform expression of various neurochemical markers among SPNs in various species4,9,15,16,51, our data enhance the resolution of molecular and anatomical profiles, thereby establishing a clearer association between molecularly defined SPN types and their output specificity through three key advancements. First, we utilized recently published snRNAseq data of cholinergic SPN populations in mice17. This facilitated the identification of molecular markers that more accurately characterize the TC types of SPNs (Fig. 1 and Supplementary Fig. 3). Second, through a viral-genetic approach, we achieved anterograde axon mapping of genetically defined SPN types exclusively from a target SC area (Fig. 2). This not only supplements information obtained via classical retrograde approaches, but also resolves ambiguities associated with the cytotoxicity of trans-neural tracer viruses52, the specificity of antibodies, and difficulties in injecting tracers into a specific peripheral organ. Third, we employed a chemogenetic approach31 to manipulate specific SPN types directly for functional assays, a capability unavailable in classical studies. Collectively, the present study opens an avenue for functionally classifying mouse SPNs. Moreover, we demonstrated that these molecularly defined output pathways can be selectively activated in response to metabolic stress (Fig. 7), refining previous studies9,10 by incorporating the resolution of TC types. To extend this concept, future research will examine activation patterns across all distinct SPN types under various stress conditions. We propose that our approach, combining transcriptome data, mouse genetics, and viral-genetic tools, has broad applicability to other SPN types throughout the SC.
Given that our identified markers (Cartpt and Oxtr) of specific SPNs represent a neural peptide and a receptor, it is intriguing to ask whether these molecular markers hold functional relevance. Despite the well-established functions of CART (encoded by Cartpt) in suppressing appetite in the arcuate nucleus of the hypothalamus53, the role of this peptide in the SC or the receptor mechanisms in the downstream CG/SMG remains unknown and thus constitutes an important focus for future studies. In particular, it is interesting to investigate the potential involvement of Cartpt+ SPNs and CART peptides in regulating the GI tract during constipation induced by the depletion of gut microbiota32 and diarrhea induced by the loss-of-mechanosensation of gut contents49. Our data, illustrating that Oxtr+ SPNs underlie Oxt-induced increase in BGLs (Fig. 6), lend support to the idea that OXTR directly senses Oxt released from the PVH Oxt neurons and activates Oxtr+ SPNs. Future studies utilizing conditional knockout of the Oxtr gene54 could address this model. In addition, considering that neuronal ablation may lead to unintended compensation and feedback mechanisms, it is important to explore more accurate loss-of-function effects. Of note, the observed hyperglycemic effect, induced by activating pan-PVH Oxt neurons, may be underestimated because Oxt secreted from the posterior pituitary into the general circulation may exert a negative influence on BGLs by activating insulin secretion55. To delineate precisely the PVH-spinal pathway, mediated by parvocellular rather than magnocellular PVH Oxt neurons56, targeted genetic manipulations of parvocellular PVH Oxt neurons should be explored in future studies. Given the broad involvement of Oxt in regulating energy expenditure28,39,57, an investigation into whether manipulating Oxtr+ SPNs could impact the progression of obesity represents a crucial step in establishing novel therapeutic targets for metabolic syndrome. In a broader context, selectively manipulating organ functions via specific SPN types may lay the basis for future therapeutic strategies addressing organ dysfunction associated with severe infectious disorders, traumatic SC lesions, and aging.
This study has several limitations. First, we did not examine the cellular logic governing postGNs in the CG/SMG, which regulate various visceral organs in addition to the GI tract. While classical studies suggested spatially restricted patterns of postGNs targeting specific organs58,59, predicting which postGNs are innervated by Cartpt+ SPNs remains challenging because of the broadly distributed axonal arbors of Cartpt+ SPNs within the CG/SMG (Supplementary Fig. 2c). A recent study further explored the molecular diversity of postGNs, suggesting that one subtype in the CG/SMG projects to upper digestive and secretory organs, thereby promoting digestion and glucagon secretion, whereas another subtype innervates the stomach and lower intestines, thereby suppressing GITT60. Another seminal study illustrated that Cartpt+ viscerofugal neurons in the small intestine project to the CG/SMG, rising BGLs presumably via postGNs targeting the liver and pancreas, without affecting GITT61. This result contrasts with our findings, where chemogenetic activation of Cartpt+ SPNs specifically reduced GITT (Fig. 4) without affecting BGLs (Fig. 5). An intriguing interpretation of these results involves labeled-line connections from Cartpt+ intestinal neurons to the liver/pancreas and Cartpt+ SPNs to the GI tract via distinct postGNs in the CG/SMG; however, we do not exclude the possibility that Cartpt+ SPNs influence multiple organs. Future studies should elucidate which molecularly defined and functionally distinct postGNs are innervated by Cartpt+ SPNs.
Second, the downstream mechanisms by which the activation of Oxtr+ SPNs regulates glucose metabolism remain unclear. While activation of the AM is presumed to increase plasma adrenaline, we observed only a slight, statistically non-significant rise in adrenaline levels following Oxtr+ SPN activation (Supplementary Fig. 7e–g). This marginal result aligns with recent findings: more severe hyperglycemia (>300 mg/dL, compared with ∼100 mg/dL observed in the present study) triggered by brain stem noradrenaline+ neurons was accompanied by only a twofold increase in plasma adrenaline62. The relatively small effect size in our study may be due to localized manipulations within the T8–T12 segments, whereas the AM-projecting SPNs are more widely distributed4,46. Additionally, SPN types other than the Oxtr+ cluster may also project to the AM. Downstream of the AM, adrenaline can elevate BGLs by suppressing insulin secretion, stimulating hepatic glucose production, promoting lipolysis, and reducing glucose utilization46. Of these pathways, our data suggest that insulin secretion remained unaffected by Oxtr+ SPN activation (Supplementary Fig. 7a–d). Future studies should elucidate further the role of adrenaline and its downstream targets in Oxtr+ SPN-mediated regulation of glucose metabolism.
Lastly, our study did not examine the upstream neural circuits organized for specific types of SPNs, both within the SC and emanating from the higher brain regions63. In the case of skeletal motor neurons, the application of viral-genetic mapping and manipulation techniques19 has unveiled complex local and long-range circuits regulating antagonistic muscles across the SC64,65. A similar approach could shed light on how multiple organ functions are selectively yet coordinately regulated.
Methods
Animals
Animals were maintained at the animal facility of the RIKEN Center for Biosystems Dynamics Research (BDR) under a 12-h light/12-h dark cycle with ad libitum access to food and water unless otherwise mentioned. Wild-type C57BL/6N mice were purchased from Japan SLC (Shizuoka, Japan). Ai9 (known as B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, Jax #007909) was originally purchased from the Jackson Laboratory by T. Imai, Chat-iresCre (also known as B6;129S6-Chattm2(cre)Lowl/J, Jax #006410) was a kind gift from B. Lowell, and Oxtr-Cre (B6;Cg-Oxtr<em2(1xPA/T2A/icre)Yinn>, RIKEN BRC11687, also known as B6; C3-Oxtrem2(icre)Yinn/J, Jax #037578) mice were provided by Y.U. Inoue. DBH-CreERT2 mice and Cartpt-Cre mice were originally generated in this study (as described in “Generation of knock-in mice”). All animal experiments were approved by the Institutional Animal Care and Use Committee of the RIKEN Kobe Branch.
We pooled data obtained from both male and female mice for retrograde tracing (Fig. 1), histochemical analyses (Fig. 1), axonal projections (Fig. 2), quantitative analysis of c-Fos (Figs. 3 and 7), and measurement of intestinal motility (Fig. 4). However, data on BGLs (Fig. 5) were presented separately because of the identification of a significant sex-dependent difference. Furthermore, experiments involving the measurement of HR (Supplementary Fig. 6), the ablation of Oxtr+ SPNs (Fig. 6), and the measurements of insulin and adrenaline (Supplementary Fig. 7) were exclusively conducted using male mice.
Generation of knock-in mice
DBH-CreERT2 (accession No. CDB0138E) and Cartpt-Cre (accession No. CDB0267E) lines (listed in https://large.riken.jp/distribution/mutant-list.html) were generated by CRISPR/Cas9-mediated knock-in in zygotes, as previously described25. For DBH-CreERT2, the ires-CreERT2 cassette (CreERT2 was derived from plasmid #46388, Addgene) was inserted just after the coding end of exon 12 using the homologous recombination-based method, and a mixture of crRNA (CRISPR RNA) 1 (5′-AGA AUA GCU UCU CAC AAG GUg uuu uag agc uau gcu guu uug) (50 ng/μl), tracrRNA (trans-activating crRNA) (100 ng/μl), donor vector (10 ng/μl), and Cas9 protein (100 ng/μl) was used for microinjection. For Cartpt-Cre, the ires2-Cre cassette (ires2 was derived from plasmid #159600, Addgene) was inserted just after the coding end of exon 3 using the microhomology-mediated end joining (MMEJ)-based method, and a mixture of crRNA 2 (AGA GGG AAU AUG GGA ACC Gag uuu uag agc uau gcu guu uug) (50 ng/μl), crRNA 3 (5′-GCA UCG UAC GCG UAC GUG UUg uuu uag agc uau gcu guu uug-3′) (50 ng/μl), tracrRNA (trans-activating crRNA) (200 ng/μl), donor vector (10 ng/μl), and Cas9 protein (100 ng/μl) was used. The guide RNA (gRNA) sites were designed by using CRISPRdirect66 to target downstream of the stop codon. crRNA and tracrRNA (5′-AAA CAG CAU AGC AAG UUA AAA UAA GGC UAG UCC GUU AUC AAC UUG AAA AAG UGG CAC CGA GUC GGU GCU) were purchased from FASMAC (Atsugi, Japan).
For DBH-CreERT2 mice, 38 F0 founder mice were obtained from 230 transferred zygotes, 32 of which were Cre-positive as identified by PCR. The line was established by one male, which harbors a targeted sequence identified by PCR and sequencing. PCR was performed using the following primers, as shown in Supplementary Fig. 1:
-
DBH-WT-F 5′-GGAAGAGGTGTGGAGTGTTCAGCATGGGAG
-
ERT2-F 5′-CACTGCGGGCTCTACTTCATCGCATTCC
-
DBH-WT-R 5′-GAGCTGTCCCTATGTGGAGTCAGCAGTGTG
For the sequence analysis, the PCR products from using the primer DBH-WT-F and DBH-3′junc-R 5′-TCTGAAGAATAGCTTCTCACAAgagctcag were subcloned into the pCR Blunt II TOPO vector (Zero Blunt TOPO PCR Cloning Kit, Thermo Fisher Scientific) and sequenced using M13-Foward and M13-Reverse primers.
For Cartpt-Cre mice, 37 F0 founder mice were obtained from 218 transferred zygotes, 13 of which were Cre-positive as identified by PCR and sequencing. The line was established by one male, which harbors a targeted sequence identified by PCR and sequencing. PCR was performed using the following primers, as shown in Supplementary Fig. 2:
-
Cartpt-F 5′-CGGATCTGACTGCTTCGACCTGAGC
-
Cre-F 5′-GTCGAGCGATGGATTTCCGTCTCTGG
-
Cartpt-R 5′-GAAGCAACAGGGAAAGAGCCCATCCG
For the sequence analysis, the PCR products from using the primer Cartpt-F and Cartpt-3′junk-R 5′-GTAAGCTGAGGTGAAGCCAGACATgtcgac or Cartpt-5′ junk-F 5′-AGAGGGAATATGGGAACgaattcgc and Cartpt-R were subcloned into the pCR Blunt II TOPO vector (Zero Blunt TOPO PCR Cloning Kit, Thermo Fisher Scientific) and sequenced using M13-Foward and M13-Reverse primers.
The germline transmission of these mouse lines was confirmed by genotyping of the F1 mice. Of note, consistent expression patterns with the endogenous genes were confirmed by the expression of the Cre reporter fluorescent protein for DBH-CreERT2 (Supplementary Fig. 1) and the histochemical analysis for Cartpt-Cre (Fig. 2). For DBH-CreERT2 mice, tamoxifen (Sigma, #10540-29-1) was dissolved in corn oil at 20 mg/ml by shaking overnight at 37 °C and stored at 4 °C for injections. A dosage of 75 mg/kg of tamoxifen (from a 20 mg/ml solution) was administered intraperitoneally to induce the activation of CreERT2.
Virus
The following AAV vectors were obtained from Addgene:
-
AAV2retro hSyn.Cre.WPRE.hGH (1.8 × 1013 gp/ml) (Addgene #105553)
-
AAV8 hSyn-DIO-hM3D(Gq)-mCherry (2.1 × 1013 gp/ml) (Addgene #44361)
-
AAV8 hSyn-DIO-mCherry (2.3 × 1013 gp/ml) (Addgene #50459)
The following AAV vector was obtained from the UNC viral core:
-
AAV1 EF1a-FLEx-taCasp3-TEVp (5.8 × 1012 gp/ml)
The following AAV vector was constructed by K. Inada44 and generated by the UNC vector core:
-
AAV5 OXTp-hM3Dq-Myc (2.4 × 1013 gp/ml)
The following AAV vector was constructed by K. Inada44 and generated by the Viral Vector Core, Gunma University Initiative for Advanced Research:
-
AAV9 OXTp-mCherry (2.9 × 1013 gp/ml)
Of note, the targeting specificity of AAV5 OXTp-hM3Dq-Myc and AAV9 OXTp-mCherry was validated in the previous study44.
Retrograde tracing from the CG/SMG
Ai9 mice (aged 3–4 weeks) underwent anesthesia through the intraperitoneal injection of a combination of 65 mg/kg ketamine (Daiichi-Sankyo) and 13 mg/kg xylazine (Sigma-Aldrich). For the CG/SMG (Fig. 1), an incision was made along the midline, and saline-soaked gauze was placed on the abdomen. To expose the CG/SMG, adjacent organs such as the stomach and intestines were carefully relocated to the moist gauze on the side of the mouse. Consistent moistening with saline was maintained during the surgical procedure to prevent dehydration. The CG/SMG, positioned around the descending aorta and left renal artery, surrounding the superior mesenteric artery, was identified.
A glass pipette prepared for a microinjector (#UMP3, WPI) was utilized to administer 500 or 100 nl of AAVrg hSyn-Cre-WPRE-hGH (Addgene #105553) with 0.1% Fast Green (#F7252, Sigma-Aldrich) into the CG/SMG of Ai9 mice, respectively, at an infusion rate of 50 nl per minute. Successful injection was confirmed by observing the ganglia staining with Fast Green. Following the injection, the abdominal cavity was rinsed with saline to prevent AAV dispersion, and saline introduction into the abdominal cavity post-surgery aimed to prevent adhesion of the abdominal organs. The incisions were then sutured. Following the surgery, an antipyretic analgesic, ketoprofen (5 mg/kg, Kissei Pharmaceutical) was administered intramuscularly, and an antibiotic, gentamicin (5 mg/kg, Takada Pharmaceutical), was administered intraperitoneally. Approximately 2 weeks after the surgical procedure, the mice were anesthetized with isoflurane, euthanized, and subsequently perfused with phosphate-buffered saline (PBS), followed by a 4% paraformaldehyde (PFA) solution in PBS. The SC was post-fixed with a 4% PFA solution in PBS.
For the tissue clearing of the CG/SMG (Supplementary Movie 1) and SC (Fig. 1d and Supplementary Movie 2), the CUBIC method21 was implemented. Specifically, after perfused fixation, the specimen underwent overnight fixation in a 4% PFA solution in PBS. Subsequently, the specimen was washed with PBS for at least 2 h, repeated twice, at 25 °C. The fixed tissues were then immersed in 5 ml of half-diluted CUBIC-L, which is a mixture of 10% (wt/wt) N-butyl diethanolamine and 10% (wt/wt) Triton X-100 in deionized water, with gentle shaking for at least 6 h at 37 °C. The solution was then replaced with 5 ml of fresh CUBIC-L, and gentle shaking at 37 °C was continued for an additional 1–2 days. To terminate the delipidation reaction, the samples were washed with 5 ml PBS, again with gentle shaking, at 25 °C for at least 2 h. This washing step was repeated more than three times. Subsequently, the samples were immersed in 5 ml of half-diluted CUBIC-R+, which is a mixture of 45% (wt/wt) antipyrine, 30% (wt/wt) nicotinamide, and 0.5% (vol/vol) N-butyl diethanolamine in deionized water, with gentle shaking at 25 °C for at least 6 h. The sample was further immersed in 5 ml of CUBIC-R+ placed in a 5 ml tube and gently shaken at 25 °C overnight.
The cleared CG/SMG or SC samples were carefully placed into glass capillaries and then positioned in the Zeiss Lightsheet Z.1 Light Sheet Fluorescence Microscope. Z-stack images were obtained using a 5× objective lens (numerical aperture [NA]: 0.1). Subsequently, the captured images were adjusted post hoc using Imaris 9.8 software, and three-dimensional reconstructions were saved as MP4 video files (Supplementary Movies 1 and 2).
For the examination of marker gene expression in retrogradely labeled cells, AAVrg hSyn-Cre-WPRE-hGH was administered into the CG/SMG of Ai9 mice. At ~2 weeks post injection, perfused fixation was carried out. The percentage of specific gene expression was calculated based on 25–70 tdTomato+ cells in the CA, IC, and IML of SC sections. Detained information on the procedures can be found in the “Histochemistry” section.
Histochemistry
For immunostaining, after conducting perfused fixation of the mice using a 4% PFA solution in PBS, the tissues were subjected to an additional overnight post-fixation in 4% PFA in PBS at 4 °C. The specimens were cryoprotected with a 30% sucrose solution in PBS at 4 °C for 24 h and then embedded in the OCT compound (#4583; Tissue-Tek). Next, 30-μm coronal sections were obtained using a cryostat (model #CM1860; Leica) and placed on MAS-coated glass slides (Matsunami). The specimens were subjected to a triple wash with PBS and treated with a 5% normal donkey serum (NDS; Southern Biotech, #0030-01) solution in PBST (PBS with 0.1% Triton X) for 30 min at 25 °C for blocking. The following primary antibodies were used in this study: goat anti-mCherry (at a dilution of 1:250; #AB0040-200, SICGEN), rabbit anti-DBH (at a dilution of 1:1000; #ab209487, Abcam), goat anti-Chat (at a dilution of 1:100; #AB144P, Sigma), rabbit anti-nNos (at a dilution of 1:250; #61-7000, Thermo Fisher Scientific), goat anti-CART (at a dilution of 1:200; #AF163, R&D Systems), rabbit anti-c-Fos (at a dilution of 1:1000; #2250S, Cell Signaling), mouse anti-Myc (at a dilution of 1:500; #sc-40, Santa Cruz), and rabbit anti-Oxt (at a dilution of 1:500, #20068, Immunostar). Before staining using anti-DBH, antigen retrieval was conducted in Tris-Cl EDTA pH9.0. These primary antibodies were diluted in 1% NDS in PBST and applied to the sections overnight at 4 °C. These sections were then washed three times with PBS and treated with the following secondary antibodies, diluted in 1% NDS in PBST and containing 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), for 1 h at 25 °C: donkey anti-rabbit AF488 (at a dilution of 1:200; #A32790, Thermo Fisher Scientific), donkey anti-goat AF555 (at a dilution of 1:200; #A32816, Thermo Fisher Scientific), donkey anti-mouse AF555 (at a dilution of 1:200; #A32773, Thermo Fisher Scientific), and donkey anti-rabbit AF488 (at a dilution of 1:200; #A32790, Thermo Fisher Scientific). The sections were washed three times with PBS and mounted with cover glass using Fluoromount (#K024; Diagnostic BioSystems).
In situ hybridization (ISH) was conducted as described previously67. To generate cRNA probes, DNA templates were amplified by PCR from SC cDNA (#MD-23; Genostaff). A T3 RNA polymerase recognition site (5′-AATTAACCCTCACTAAAGGG) was added to the 3′ end of the reverse primers. The primer sets and sequences of the probe targets were as follows:
-
Cartpt−1: 5′-ggacatctactctgccgtgg; 5′-tccgggttgtgatgtcatct
-
Cartpt−2: 5′-gccctggacatctactctgc; 5′-tccgggttgtgatgtcatct
-
Chodl: 5′-aagggaaggcagctgcttag; 5′-aactgggagctgcttccatc
-
Pitx2: 5′- tggaccaaccttacggaagc; 5′-aaacatttgtgggcgtacgc
DNA templates (500–1000 ng) that were amplified by PCR were then subjected to in vitro transcription with either DIG- (#11277073910) or Flu- (#11685619910) RNA labeling mix and T3 RNA polymerase (cat#11031163001) according to the manufacturer’s instructions (Roche). In the case of Cartpt, a combination of two probes was utilized to enhance the signal-to-noise ratio.
For single-color ISH combined with anti-mCherry staining, following the hybridization and washing steps, sections were incubated with a solution containing horseradish peroxidase (HRP)-conjugated anti-Dig (at a dilution of 1:500; #11207733910, Roche Applied Science) and goat anti-mCherry (at a dilution of 1:250; #AB0040-200, SICGEN) antibodies overnight. The signals were amplified by a 1:70 TSA-plus Cyanine 3 (#NEL744001KT; AKOYA Bioscience) for 25 min, followed by washing. mCherry-positive cells were then visualized using donkey anti-goat AF555 (at a dilution of 1:200; #32816, Invitrogen). PBS containing 50 ng/ml of DAPI (#8417, Sigma-Aldrich) was used for counter-nuclear staining.
For dual-color ISH, HRP-conjugated anti-flu antibody (at a dilution of 1:250; #NEF710001EA, AKOYA Bioscience) was used to detect Flu-labeled RNA probes by a 1:70 TSA-plus biotin (#NEL749A001KT; AKOYA Bioscience), which was then visualized with streptavidin-AF488 (at a dilution of 1:250; Life Technologies). After inactivation of HRP with a 2% sodium azide solution in PBS for 15 min at 25 °C, HRP-conjugated anti-Dig (at a dilution of 1:500; #11207733910, Roche Applied Science) and a 1:70 TSA-plus Cyanine 3 were used to detect Dig-labeled cRNA probes. The sections were then mounted with a cover glass using Fluoromount (#K024; Diagnostic BioSystems).
To detect the expression of Oxtr, Palld, Cartpt, and c-Fos in Fig. 7 and Supplementary Fig. 3, we used the RNAscope Multiplex Fluorescent Reagent Kit v2 (#323100; ACD) following the manufacturer’s instructions. For single-color detection of Oxtr, we utilized specific probes for Oxtr (Mm-Oxtr #412171, ACD). To conduct dual-color imaging of Oxtr and Palld, we used C1-probe for Palld (Mm-Palld #822611, ACD) and C2-probe for Oxtr (Mm-Oxtr-C2 #412171-C2, ACD). For dual-color imaging of Oxtr and c-Fos, we combined probes for Oxtr (Mm-Oxtr #412171, ACD) and c-Fos (Mm-Fos-C2, #316921-C2, ACD). For triple-color imaging of Cartpt, c-Fos, and anti-Chat staining, we combined probes for Cartpt (Mm-Cartpt, #432001, ACD) and c-Fos (Mm-Fos-C2, #316921-C2, ACD), followed by anti-Chat staining. The sections were air-dried and then incubated with DEPC-treated PBS for 5 min. Subsequently, they were heated in a dry oven at 60 °C for 30 min. The sections were fixed with a 4% PFA solution in PBS for 15 min at 4 °C, followed by dehydration in 100% ethanol for 5 min and an additional 5 min of air drying. All sections were pretreated with hydrogen peroxide for 10 min at 25 °C, followed by washing with Milli-Q for 5–10 min; this step was repeated twice. Target retrieval was carried out by placing the slides in a steamer at a temperature exceeding 99 °C for 15 min, followed by a quick wash with Milli-Q for 15 s. The sections were dehydrated in 100% ethanol for 3 min and heated in a dry oven at 60 °C for 5 min. Subsequently, the sections received treatment with protease reagent (protease III) for 30 min at 40 °C, followed by another wash with Milli-Q. Sections were then incubated in the RNAscope probes for 2 h at 40 °C. Following this incubation, the slides were washed with a wash buffer. The sections were then incubated in three amplification reagents at 40 °C: AMP1 and AMP2 for 30 min each, and AMP3 for 15 min. This was followed by horseradish peroxidase-C1 (HPR-C1) amplification at 40 °C for 15 min.
For a single-color RNAscope for Oxtr combined with anti-mCherry immunostaining, sections were subsequently incubated in TSA-plus Cyanine 3 (#NEL744001KT, AKOYA Bioscience) at a 1/1500 concentration for 30 min at 40 °C. In the final step of the process, sections were subjected to HRP blocking for 15 min at 40 °C. After the final wash, the slides were prepared for IHC to detect mCherry. Following a blocking step with 5% NDS, sections were incubated overnight with goat anti-mCherry antibody (at a dilution of 1:250; #AB0040-200, SICGEN) in 1% NDS at 4 °C. After washing three times, the sections were incubated for 1 h with donkey anti-goat AF555 (at a dilution of 1:200; #32816, Thermo Fisher Scientific) and DAPI in 1% NDS. Subsequently, the sections were washed with PBS three times and mounted with a cover glass using Fluoromount (#K024; Diagnostic BioSystems).
For a dual-color RNAscope combined with anti-Chat immunostaining, sections following HPR-C1 treatment were incubated with TSA-plus Cyanine 3 (#NEL744001KT; AKOYA Bioscience) at a 1/1500 concentration for 30 min at 40 °C, followed by HRP blocking for 15 min at 40 °C. After washing, sections were included with horseradish peroxidase-C2 (HPR-C2) at 40 °C for 15 min. Subsequently, sections were incubated in TSA-plus biotin (NEL749A001KT; AKOYA Bioscience) at a 1/1500 concentration for 30 min at 40 °C. Following HRP blocking for 15 min at 40 °C and washing, sections were subjected to a blocking step with 5% NDS and then incubated overnight at 4 °C with goat anti-Chat (at a dilution of 1:250; #AB144P, Sigma) and Streptavidin-AF488 (at a dilution of 1:250; #S32354, Thermo Fisher Scientific) in 1% NDS. After washing three times, the sections were incubated for 1 h with donkey anti-Goat AF647 (at a dilution of 1:250; #A-21447, Thermo Fisher Scientific), and DAPI in 1% NDS. Subsequently, the sections were washed with PBS three times and mounted with a cover glass using Fluoromount (#K024; Diagnostic BioSystems).
To count the c-Fos+ cells in the whole mount preparation of CG/SMG (Fig. 3), 3 days before perfused fixation, a 300-μl solution containing 4 mg/ml of Fluorogold (#526-94003; Fluorochrome, LLC) in saline was administered via intraperitoneal injection to visualize the abdominal ganglia. The CG/SMG was dissected and subsequently fixed in a 4% PFA solution in PBS overnight. The specimen was trimmed under a fluorescence microscope using ultraviolet light to visualize Fluorogold. It was then subjected to triple washes with PBS and permeabilization using a solution comprising 0.5% Triton X-100, 0.05% Tween-20, and 4 μg/ml of heparin (referred to as PTxwH), followed by incubation overnight at 25 °C. Subsequently, primary antibodies, rabbit anti-c-Fos (at a 1:1000 dilution; #2250S, Cell Signaling) and goat anti-mCherry (at a 1:250 dilution, #AB0040-200, SICGEN), were introduced into the whole CG/SMG immersed in PTxwH and incubated at 4 °C for 3 days. Following the triple wash steps with PTxwH, the specimen was stained using donkey anti-rabbit AF488 (at a 1:200 dilution, #A32790, Invitrogen) and donkey anti-goat AF555 (at a 1:200 dilution, #A32816, Invitrogen) at 4 °C for 3 days. Triple washes with PTxwH followed this staining process. The CG/SMG specimen was then placed on a glass-bottom dish (#3960-035; IWAKI) and covered with Fluoromount (#K024; Diagnostic BioSystems).
The sections were imaged using an Olympus BX53 microscope equipped with a 10× objective lens (NA: 0.4). For confocal imaging of whole mount preparations, an inverted Zeiss LSM780 confocal microscope with a Plan-Apochromat 10× objective lens (NA: 0.45) was used. Z-stack images were acquired and subsequently processed into maximum projection images. The acquired images were processed using Fiji software. For the analysis of RNAscope data, we defined positive cells as those exhibiting five or more RNAscope dots, as the presence of a small number of dots could be observed even in sections derived from knockout mice54.
Reanalysis of snRNAseq data
The snRNAseq data sourced from spinal cord autonomic and skeletal motor neurons (accession No. GSE161621)17 were subjected to reanalysis in R (version 4.3.1) and RStudio (Server 2023.06.1 Build 524; supported by the RIKEN BDR DNA Analysis Facility at the Laboratory for Developmental Genome System) using Seurat (version 5.0.1)68. Specifically, the analysis focused on Chat+ clusters, excluding B cell leukemia/lymphoma 6 (Bcl6)+ motor neurons and (Pax2)+ cholinergic interneurons, narrowing the investigation to cholinergic SPNs. The standard Seurat data analysis workflow encompassed quality control, where genes were retained if at least one unique molecular identifier was detected in a minimum of three cells, followed by normalization. Subsequently, SCTransform scaling was employed, and the top 3000 variable features were identified and utilized as input for principal component analysis. The initial clustering was performed using the top 50 principal components (PCs) at a resolution of 0.05 to distinguish between neuronal and non-neuronal cell populations. Non-neuronal cells were excluded based on the expression of Solute carrier family 7 member 10 (Slc7a10) and Myelin oligodendrocyte glycoprotein (Mog). The subsetUmapClust function was then applied to perform re-clustering. Furthermore, cholinergic SPNs were segregated based on the expression of Chat, Bcl6, and Pax2 using the top 35 PCs at a resolution of 0.1. In Fig. 1, SPNs were clustered using the top 50 PCs at a resolution of 0.5. We defined nNoslow clusters as those in which 50% or fewer cells expressed nNos, with average log-normalized expression levels (in arbitrary units) lower than zero. The remaining SPN clusters were categorized as nNoshigh.
Viral injections into the SC or PVH
For viral injections into the lower thoracic SC (Figs. 2–6), Chat-Cre, Cartpt-Cre, or Oxtr-Cre mice (aged 4–5 weeks) underwent anesthesia through the intraperitoneal injection of a combination of 65 mg/kg ketamine (Daiichi-Sankyo) and 13 mg/kg xylazine (Sigma-Aldrich). An incision around the thoracic region of the SC was made. To expose the SC, the paraspinal muscles and dorsal vertebral elements were carefully excised using fine forceps. A glass pipette prepared for a microinjector (#UMP3; WPI) was utilized to administer 100 nl of the AAV vector with 0.1% Fast Green (#F7252, Sigma-Aldrich) at 10 distinct sites (infusion rate: 60–70 nl/min) bilaterally in the T8–T12 segments. In Figs. 2–6 experiments, we targeted T8–12, because SPNs in T13 and lumber region are known to project to sympathetic ganglia located lower the CG/SMG, such as inferior mesenteric ganglia22.
For the chemogenetic activation of PVH Oxt neurons (Fig. 6), Oxtr-Cre male mice (aged 6–8 weeks) underwent anesthesia as mentioned above, and were secured to stereotactic equipment (Narishige). To target the AAV into the PVH, stereotactic coordinates were determined based on the Allen Mouse Brain Atlas69, and the following coordinates were used (measured in mm from the Bregma for anteroposterior [AP] and mediolateral [ML], and from the brain surface for dorsoventral [DV]): AP: –0.8, ML: 0.2, and DV: 4.5. The volume of AAV serotype 5 pAAV OXTp-hM3Dq-Myc injected was 200 nl, administered at a rate of 50 nl per minute. Following the surgical procedures, an antipyretic analgesic, ketoprofen (5 mg/kg, Kissei Pharmaceutical), was administered intramuscularly, and an antibiotic, gentamicin (5 mg/kg, Takada Pharmaceutical), was administered intraperitoneally.
Measurement of axons and c-Fos+ cells
For the measurement of axons (Fig. 2), 30-μm sections of the sympathetic ganglia and adrenal glands were prepared and immunostained with anti-DBH and anti-mCherry as described in the “Histochemistry” section. Images of these sections were acquired using an Olympus BX53 microscope equipped with a 10× objective lens (NA: 0.4). The automatic counting of mCherry-positive axons within DBH-positive cells was carried out using Fiji software. Binarized images of DBH and mCherry were generated, and the area of mCherry-positive axons was normalized to the DBH-positive area within the image. The average counts from three sections were calculated for individual Chat-Cre, Cartpt-Cre, or Oxtr-Cre mice.
To quantify the number of c-Fos+ cells in the sections of the adrenal gland and SC (Figs. 3 and 7), immunostaining or RNAscope staining was carried out as described in the “Histochemistry” section. For the adrenal gland sections, the counting of c-Fos- and DAPI-positive cells was carried out automatically using Fiji software. The noise was removed through the “Remove Outliers” function, and the count of both c-Fos- and DAPI-positive cells was determined using the “Find Maxima” function. The average number of c-Fos-positive cells across three sections was divided by the count of DAPI-positive cells in the corresponding area. This quotient was used as the value for individual mice. For the SC sections, the quantification of c-Fos-positive cells was carried out manually. Within the T8–T12 segments, the number of c-Fos-positive cells among mCherry-positive cells was counted for five sections (one section per segment,) and the counts were averaged for individual mice. In Fig. 7, the number of c-Fos+, Oxtr+, Chat+, and Cartpt+ cells in three sections from T8–10 and three sections from T11–13 for each mouse were manually counted, and individual averages were determined.
The identification and counting of c-Fos+ cells in the whole mount CG/SMG (Fig. 3) were performed automatically using Fiji software. After removing noise through the “Remove Outliers” function, the total number of Fos+ cells was determined using the “Find Maxima” function. Of note, data were not normalized to area or volume. Each data point represents the number of c-Fos+ cells per CG/SMG.
Measurements of gastrointestinal motility and blood glucose levels
Neural activation experiments targeting hM3D (Gq) to the desired cell types (Figs. 3–6) were conducted at least 2 weeks after the AAV injection. To induce activation, a dosage of 2 mg/kg of CNO (#4936, Tocris) was injected into the mice via intraperitoneal administration. Saline was injected into the same mice for the negative control. The order of CNO or Saline administration was randomized.
To measure GITT, the mice received an oral gavage of a 6% carmine red solution that was dissolved in 0.5% methylcellulose, and prepared with sterile saline. GITT was determined by measuring the time between the oral gavage and the excretion of a carmine-containing fecal pellet32. All mice had ad libitum access to food and water during the measurement period. Under the “no food condition” (Fig. 4f), food was removed just after the administration of CNO or saline. These experiments were performed during the light phase.
For celiac ganglionectomy (CGX, Fig. 4d), the CG/SMG was exposed and carefully dissected with fine forceps. Following the surgery, saline was introduced into the abdominal cavity to prevent adhesion of the abdominal organs, and the incisions were sutured. Following the surgery, ketoprofen (5 mg/kg, Kissei Pharmaceutical) was administered intramuscularly, and gentamicin (5 mg/kg, Takada Pharmaceutical) intraperitoneally. At 1 week following the surgery, GITT was measured as mentioned above.
To prevent a potential stress-induced increase in BGLs, the mice underwent a 2-day habituation period before the assays. Each mouse was individually housed, and a daily 10-min habituation was conducted to replicate the process of blood collection from the tail tip. All mice had ad libitum access to food and water throughout the habituation and measurement periods. To minimize the impact of stress further, the tail tip was gently incised at 2 h before the measurement, followed by another 10-min habituation session. Blood samples were obtained from the tail tip, and BGLs were assessed using a blood glucose meter for laboratory animals (cat# SUGL-001; ForaCare, Tokyo) at intervals of 0, 20, 40, 60, 100, and 140 min after the intraperitoneal administration of 2 mg/kg CNO or saline. In Fig. 5, the baseline BGL at time 0 min was used to calculate ΔBGL values. Of note, the baseline was not significantly different between sexes or among those injected with different AAVs (mg/dL): mean ± standard deviation, 153.70 ± 15.62 (Chat: Gq, male), 161.83 ± 19.62 (Chat: Gq, female), 143.14 ± 16.39 (Chat: mC, male), 141.83 ± 10.92 (Chat: mC, female), 143.30 ± 16.02 (Cartpt: Gq, male), 149.07 ± 24.07 (Cartpt: Gq, female), 139.50 ± 22.98 (Cartpt: mC, male), 146.88 ± 17.55 (Cartpt: mC, female), 167.43 ± 16.30 (Oxtr: Gq, male), 156.21 ± 25.48 (Oxtr: Gq, female), 162.83 ± 21.98 (Oxtr: mC, male), and 149.17 ± 22.24 (Oxtr: mC, female). In Fig. 5a, the following data points are denoted as +1 symbols in the graph as outliers: Chat: Gq, male, 100 min; 221 mg/dL, 140 min; 226 mg/dL, Oxtr: Gq, female, 40 min; –78 mg/dL, 100 min; –71 mg/dL. In Fig. 6, baseline glucose levels at time 0 min did not differ: 172.13 ± 14.89 mg/dL for Oxt: Gq, Oxtr: Saline, 165.75 ± 17.68 mg/dL for Oxt: Gq, Oxtr: taCasp, and 150.40 ± 13.14 mg/dL for control.
To perform adrenalectomy (Fig. 5c–e), Oxtr-Cre male mice (aged 5–7 weeks) were anesthetized using a combination of 65 mg/kg ketamine (Daiichi-Sankyo) and 13 mg/kg xylazine (Sigma-Aldrich) via intraperitoneal injection. An incision was made along the midline, and gauze soaked in saline was placed on the abdomen. To expose the adrenal gland, adjacent organs such as the stomach and intestines were carefully relocated to the moist gauze on the side of the mouse. Throughout the surgical procedure, these organs were consistently moistened with saline to prevent dehydration. Using fine small scissors, the bilateral adrenal glands were carefully excised from the kidneys. After the surgery, ketoprofen (5 mg/kg, Kissei Pharmaceutical) was administered intramuscularly, and gentamicin (5 mg/kg, Takada Pharmaceutical) intraperitoneally. Considering that the adrenal glands secrete aldosterone to maintain the salt balance, the drinking water was supplemented with a 1% w/v sodium chloride solution70. At 1 week following the surgery, BGLs were measured as mentioned above. The baseline was 182.10 ± 14.20 mg/dL.
Telemetry system
To monitor the HR in mice, as shown in Supplementary Fig. 6c–g, we conducted surgical implantations of telemetry devices (G2-HR E-Mitter; Starr Life Sciences Corp., Oakmont, PA, USA) following a recovery period of at least 1 month after the AAV injection. For the implantation procedure, mice underwent intraperitoneal injection of 65 mg/kg ketamine and 13 mg/kg xylazine for anesthesia. The telemeter was placed in the abdominal cavity, with the negative lead sutured subcutaneously to the right subclavian greater pectoralis muscle and the positive lead to the chest wall on the left side of the sternum, anterior to the last rib. Following the surgery, gentamicin (5 mg/kg, Takada Pharmaceutical) was promptly administered. The mice were then maintained in their home cages for 1 week to facilitate recovery. During this recovery period, the mice were habituated to intraperitoneal injection for at least 3 days. On the recording day, saline was administered at zeitgeber Time 2 (ZT2, 10:00) and CNO at ZT6 (14:00). The signals transmitted by the telemetry device were received and recorded at 1-min intervals using an ER-4000 receiver and VitalView software (Starr Life Sciences Corp.). The average HR value was calculated every 10 min. As shown in Supplementary Fig. 6d, f, the mean baseline HR during the period from –30 min to 0 min was used to calculate the ΔHR. Of note, the baseline was not significantly different between saline and CNO or those injected with different AAVs (rpm): 551.32 ± 71.70 (Oxtr: Gq, saline), 544.29 ± 73.05 (Oxtr: Gq, CNO), 565.97 ± 81.14 (Oxtr: mC, saline), and 594.83 ± 56.03 (Oxtr: mC, CNO).
Glucose-stimulated insulin secretion test
We followed the published protocol for the glucose-stimulated insulin secretion test43 (Supplementary Fig. 7a–d). Specifically, three days before the experiment, AAV-injected Oxtr-Cre male mice (AAV8-hSyn-DIO-Gq-mCherry or AAV8-hSyn-DIO-mCherry) were housed individually and habituated to i.p. injections. Food was removed for 16 h before glucose stimulation, and the mice were transferred to clean cages the day before the test. One hour before glucose stimulation, blood samples were collected from the facial vein using an animal lancet (#18310300, MEDIpoint). Additional blood samples were obtained 10 min after the i.p. injection of glucose solution (2 g/kg body weight, #16805-35, Nacalai), together with either 0.9% saline or CNO (2 mg/kg body weight, #4936, Tocris). Of note, we selected the 10-min time point following CNO administration based on literature validation of Gq-mediated activation of PVH Oxt neurons within this timeframe43. Prolonged incubation after CNO application may trigger various feedback mechanisms in response to the elevated BGLs resulting from the activation of Oxtr+ SPNs. The samples were centrifuged to isolate plasma, which was stored at −80 °C. Plasma insulin levels were measured using the Mouse Ultrasensitive Insulin ELISA kit (#80-INSMSU-01, ALPCO) according to the manufacturer’s instructions.
Glucoprivation
We followed the published protocol for the glucoprivation experiments43 (Fig. 7). Three days before the experiments, 6–8 week-old C57BL/6N mice were housed individually and habituated to i.p. injections. On the day of the experiment, the mice were transferred to clean cages and food was removed for 2 h. The mice were then injected i.p. with either 2-DG (400 mg/kg in 0.9% saline) (#HY-13966, MCE) or 0.9% saline. Two hours post-injection, the mice were anesthetized with isoflurane, and subsequently perfused with PBS, followed by a 4% PFA. The spinal cord was removed and postfixed in 4% PFA at 4 °C overnight, prior to histochemistry processing.
Measurement of plasma adrenaline levels
Blood samples were collected from the inferior vena cava (IVC) 1 h after the administration of 2 mg/kg CNO (Supplementary Fig. 7e–g). This timepoint was selected based on the peak BGLs observed after the activation of Chat+ SPNs or Oxtr+ SPNs (Fig. 5a). Plasma was separated from the blood and stored at −80 °C. Adrenaline concentrations were measured using an Epinephrine/Norepinephrine ELISA kit (#KA1877, Abnova) according to the manufacturer’s instructions.
Statistical analysis
The statistical details of each experiment, including the statistical tests used, and the exact value of the number of animals are described in each figure legend. P values < 0.05 were considered to indicate statistical significance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available in the main paper and supplementary materials. All materials, including the DBH-CreERT2 mice and Cartpt-Cre mice, are available through request to the corresponding authors. Source data are provided with this paper.
Code availability
This paper does not report the original code. All R scripts used for the reanalysis of snRNAseq data are provided with this paper.
References
Tasic, B. Single cell transcriptomics in neuroscience: cell classification and beyond. Curr. Opin. Neurobiol. 50, 242–249 (2018).
Lui, J. H. et al. Differential encoding in prefrontal cortex projection neuron classes across cognitive tasks. Cell 184, 489–506. e426 (2021).
Zhou, J. et al. Brain-wide correspondence of neuronal epigenomics and distant projections. Nature 624, 355–365 (2023).
Deuchars, S. A. & Lall, V. K. Sympathetic preganglionic neurons: properties and inputs. Compr. Physiol. 5, 829–869 (2015).
Ernsberger, U. & Rohrer, H. Sympathetic tales: subdivisons of the autonomic nervous system and the impact of developmental studies. Neural Dev. 13, 20 (2018).
Langley, J. N. On the reaction of cells and of nerve-endings to certain poisons, chiefly as regards the reaction of striated muscle to nicotine and to curari. J. Physiol. 33, 374–413 (1905).
Janig, W. & McLachlan, E. M. Characteristics of function-specific pathways in the sympathetic nervous system. Trends Neurosci. 15, 475–481 (1992).
Janig, W. & Habler, H. J. Specificity in the organization of the autonomic nervous system: a basis for precise neural regulation of homeostatic and protective body functions. Prog. Brain Res. 122, 351–367 (2000).
Parker, L. M., Kumar, N. N., Lonergan, T. & Goodchild, A. K. Neurochemical codes of sympathetic preganglionic neurons activated by glucoprivation. J. Comp. Neurol. 521, 2703–2718 (2013).
Yamaguchi-Shima, N. et al. Adrenal adrenaline- and noradrenaline-containing cells and celiac sympathetic ganglia are differentially controlled by centrally administered corticotropin-releasing factor and arginine-vasopressin in rats. Eur. J. Pharm. 564, 94–102 (2007).
Gilbey, M. P. & Stein, R. D. Characteristics of sympathetic preganglionic neurones in the lumbar spinal cord of the cat. J. Physiol. 432, 427–443 (1991).
Boczek-Funcke, A. et al. Classification of preganglionic neurones projecting into the cat cervical sympathetic trunk. J. Physiol. 453, 319–339 (1992).
Nonogaki, K. New insights into sympathetic regulation of glucose and fat metabolism. Diabetologia 43, 533–549 (2000).
Browning, K. N. & Travagli, R. A. Central nervous system control of gastrointestinal motility and secretion and modulation of gastrointestinal functions. Compr. Physiol. 4, 1339–1368 (2014).
Hinrichs, J. M. & Llewellyn-Smith, I. J. Variability in the occurrence of nitric oxide synthase immunoreactivity in different populations of rat sympathetic preganglionic neurons. J. Comp. Neurol. 514, 492–506 (2009).
Fenwick, N. M., Martin, C. L. & Llewellyn-Smith, I. J. Immunoreactivity for cocaine- and amphetamine-regulated transcript in rat sympathetic preganglionic neurons projecting to sympathetic ganglia and the adrenal medulla. J. Comp. Neurol. 495, 422–433 (2006).
Blum, J. A. et al. Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat. Neurosci. 24, 572–583 (2021).
Alkaslasi, M. R. et al. Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord. Nat. Commun. 12, 2471 (2021).
Luo, L., Callaway, E. M. & Svoboda, K. Genetic dissection of neural circuits: a decade of progress. Neuron 98, 256–281 (2018).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Strack, A. M., Sawyer, W. B., Marubio, L. M. & Loewy, A. D. Spinal origin of sympathetic preganglionic neurons in the rat. Brain Res. 455, 187–191 (1988).
Espinosa-Medina, I. et al. The sacral autonomic outflow is sympathetic. Science 354, 893–897 (2016).
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011).
Abe, T., Inoue, K. I., Furuta, Y. & Kiyonari, H. Pronuclear microinjection during S-phase increases the efficiency of CRISPR-Cas9-assisted knockin of large DNA donors in mouse zygotes. Cell Rep. 31, 107653 (2020).
Stanic, D. et al. Oxytocin in corticosterone-induced chronic stress model: focus on adrenal gland function. Psychoneuroendocrinology 80, 137–146 (2017).
Plecas, B., Ugresic, N., Hristic, M., Popovic, A. & Jovovic, D. The response of rat adrenal medulla to oxytocin. Arch. Int. Physiol. Biochim. 97, 303–308 (1989).
Jovanovic, P., Stefanovic, B., Spasojevic, N., Puskas, N. & Dronjak, S. Effects of oxytocin on adreno-medullary catecholamine synthesis, uptake and storage in rats exposed to chronic isolation stress. Endocr. Res. 41, 124–131 (2016).
Wrobel, L. et al. Distribution and identity of neurons expressing the oxytocin receptor in the mouse spinal cord. Neurosci. Lett. 495, 49–54 (2011).
Inoue, Y. U. et al. Targeting neurons with functional oxytocin receptors: a novel set of simple knock-in mouse lines for oxytocin receptor visualization and manipulation. eNeuro 9, ENEURO.0423-21.2022 (2022).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163–5168 (2007).
Muller, P. A. et al. Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature 583, 441–446 (2020).
Muller, M., Canfora, E. E. & Blaak, E. E. Gastrointestinal transit time, glucose homeostasis and metabolic health: modulation by dietary fibers. Nutrients 10, 275 (2018).
Eleftheriadis, P. E. et al. Peptidergic modulation of motor neuron output via CART signaling at C bouton synapses. Proc. Natl Acad. Sci. USA 120, e2300348120 (2023).
Drake, K., Gateva, E., Deutsch, J. & Cohen, W. R. Sex differences in the adrenal catecholamine response to hypoglycemia in rats. Metabolism 47, 121–124 (1998).
Goren, H. J., Kulkarni, R. N. & Kahn, C. R. Glucose homeostasis and tissue transcript content of insulin signaling intermediates in four inbred strains of mice: C57BL/6, C57BLKS/6, DBA/2, and 129X1. Endocrinology 145, 3307–3323 (2004).
Benede-Ubieto, R., Estevez-Vazquez, O., Ramadori, P., Cubero, F. J. & Nevzorova, Y. A. Guidelines and considerations for metabolic tolerance tests in mice. Diabetes Metab. Syndr. Obes. 13, 439–450 (2020).
Coote, J. H. & Chauhan, R. A. The sympathetic innervation of the heart: Important new insights. Auton. Neurosci. 199, 17–23 (2016).
Onaka, T. & Takayanagi, Y. Role of oxytocin in the control of stress and food intake. J. Neuroendocrinol. 31, e12700 (2019).
Froemke, R. C. & Young, L. J. Oxytocin, neural plasticity, and social behavior. Annu. Rev. Neurosci. 44, 359–381 (2021).
Camerino, C. Low sympathetic tone and obese phenotype in oxytocin-deficient mice. Obesity 17, 980–984 (2009).
Takayanagi, Y. et al. Oxytocin receptor-deficient mice developed late-onset obesity. NeuroReport 19, 951–955 (2008).
Papazoglou, I. et al. A distinct hypothalamus-to-beta cell circuit modulates insulin secretion. Cell Metab. 34, 285–298.e287 (2022).
Inada, K. et al. Plasticity of neural connections underlying oxytocin-mediated parental behaviors of male mice. Neuron 110, 2009–2023 e2005 (2022).
Yang, C. F. et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 153, 896–909 (2013).
Verberne, A. J., Korim, W. S., Sabetghadam, A. & Llewellyn-Smith, I. J. Adrenaline: insights into its metabolic roles in hypoglycaemia and diabetes. Br. J. Pharm. 173, 1425–1437 (2016).
Cannon W. B. The Wisdom of the Body. (Norton, New York, 1939).
Liu, S. et al. Somatotopic organization and intensity dependence in driving distinct NPY-expressing sympathetic pathways by electroacupuncture. Neuron 108, 436–450.e437 (2020).
Servin-Vences, M. R. et al. PIEZO2 in somatosensory neurons controls gastrointestinal transit. Cell 186, 3386–3399.e3315 (2023).
Koren, T. et al. Insular cortex neurons encode and retrieve specific immune responses. Cell 184, 5902–5915.e5917 (2021).
Gonsalvez, D. G., Kerman, I. A., McAllen, R. M. & Anderson, C. R. Chemical coding for cardiovascular sympathetic preganglionic neurons in rats. J. Neurosci. 30, 11781–11791 (2010).
Ugolini, G. Viruses in connectomics: viral transneuronal tracers and genetically modified recombinants as neuroscience research tools. J. Neurosci. Methods 346, 108917 (2020).
Douglass, J., McKinzie, A. A. & Couceyro, P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J. Neurosci. 15, 2471–2481 (1995).
Inada, K., Tsujimoto, K., Yoshida, M., Nishimori, K. & Miyamichi, K. Oxytocin signaling in the posterior hypothalamus prevents hyperphagic obesity in mice. eLife 11, e75718 (2022).
Mohan, S., Khan, D., Moffett, R. C., Irwin, N. & Flatt, P. R. Oxytocin is present in islets and plays a role in beta-cell function and survival. Peptides 100, 260–268 (2018).
Althammer, F. & Grinevich, V. Diversity of oxytocin neurons: beyond magno- and parvocellular cell types? J. Neuroendocrinol. https://doi.org/10.1111/jne.12549 (2017).
Li, E. et al. Control of lipolysis by a population of oxytocinergic sympathetic neurons. Nature 625, 175–180 (2024).
Trudrung, P., Furness, J. B., Pompolo, S. & Messenger, J. P. Locations and chemistries of sympathetic nerve cells that project to the gastrointestinal tract and spleen. Arch. Histol. Cytol. 57, 139–150 (1994).
Torres, H. et al. Sympathetic innervation of the mouse kidney and liver arising from prevertebral ganglia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 321, R328–R337 (2021).
Wang, T., Teng, B., Yao, D. R., Gao, W. & Oka, Y. Organ-specific sympathetic innervation defines visceral functions. Nature https://doi.org/10.1038/s41586-024-08269-0 (2024). Online ahead of print.
Muller, P. A. et al. Enteric neurons autonomously regulate blood glucose. Science 370, 314–321 (2020).
Abe, C. et al. Hepatic glycogenolysis and hypometabolism induced by chemogenetic stimulation of C1 neurons. J. Physiol. 601, 2293–2306 (2023).
Winter, C. C. et al. A transcriptomic taxonomy of mouse brain-wide spinal projecting neurons. Nature 624, 403–414 (2023).
Arber, S. Motor circuits in action: specification, connectivity, and function. Neuron 74, 975–989 (2012).
Tripodi, M., Stepien, A. E. & Arber, S. Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature 479, 61–66 (2011).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Ishii, K. K. et al. A labeled-line neural circuit for pheromone-mediated sexual behaviors in mice. Neuron 95, 123–137.e128 (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Choi, S. et al. Corticosterone inhibits GAS6 to govern hair follicle stem-cell quiescence. Nature 592, 428–432 (2021).
Li, H. et al. Single-neuron projectomes of mouse paraventricular hypothalamic nucleus oxytocin neurons reveal mutually exclusive projection patterns. Neuron 112, 1081–1099.e1087 (2024).
Acknowledgements
We thank the staff at the RIKEN BDR animal facility for the animal care and in vitro fertilization, Hideki Enomoto (Kobe University), Takeshi Imai (Kyushu University), and members of the Miyamichi Laboratory for the critical reading of the manuscript, Addgene, the University of North Carolina Vector Core, and the Viral Vector Core of Gunma University Initiative for Advanced Research for the AAV productions, and the Laboratory for Developmental Genome System for support with the snRNA-seq reanalysis. This study was supported by the RIKEN Special Postdoctoral Researchers Program, JSPS KAKENHI (21K1521), and JSPS Restart Postdoctoral (RPD) Research Fellowship to Y.H., and the JST CREST Program (JPMJCR2021), JSPS KAKENHI (20K20589 and 21H02587), grants from Uehara Memorial Foundation, G-7 Scholarship Foundation, and a RIKEN BDR QMIN project grant to K.M.
Author information
Authors and Affiliations
Contributions
Y.H. and K.M. conceived the experiments. Y.H. performed the experiments and analyzed the data, with technical support by S.Y., S.U., K.I., M.H., and S.I. M.T. conducted a reanalysis of the snRNAseq data. DBH-CreERT2 and Cartpt-Cre mice were generated by M.S., T.A., and M.H. Y.U.I. and T.I. provided the Oxtr-iCre mice. Y.H. and K.M. wrote the paper, with contributions from all coauthors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Marc Schneeberger, Henning Fenselau and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Harima, Y., Tsurutani, M., Yamada, S. et al. Parallel labeled-line organization of sympathetic outflow for selective organ regulation in mice. Nat Commun 15, 10478 (2024). https://doi.org/10.1038/s41467-024-54928-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54928-1
This article is cited by
-
Molecular and functional diversity of the autonomic nervous system
Nature Reviews Neuroscience (2025)