Abstract
This pioneering genome-wide association study examined surrogate markers for insulin resistance (IR) in 147,880 Taiwanese individuals using data from the Taiwan Biobank. The study focused on two IR surrogate markers: the triglyceride to high-density lipoprotein cholesterol (TG:HDL-C) ratio and the TyG index (the product of fasting plasma glucose and triglycerides). We identified genome-wide significance loci within four gene clusters: GCKR, MLXIPL, APOA5, and APOC1, uncovering 197 genes associated with IR. Transcriptome-wide association analysis revealed significant associations between these clusters and TyG, primarily in adipose tissue. Gene ontology analysis highlighted pathways related to Alzheimer’s disease, glucose homeostasis, insulin resistance, and lipoprotein dynamics. The study identified sex-specific genes associated with TyG. Polygenic risk score analysis linked both IR markers to gout and hyperlipidemia. Our findings elucidate the complex relationships between IR surrogate markers, genetic predisposition, and disease phenotypes in the Taiwanese population, contributing valuable insights to the field of metabolic research.
Similar content being viewed by others
Introduction
Insulin resistance (IR) is a well-documented pathophysiological condition characterized by the diminished responsiveness of cells to insulin, a hormone essential for the regulation of glucose metabolism1. In individuals with IR, cellular sensitivity to insulin is markedly reduced, resulting in elevated blood glucose levels and compensatory hyperinsulinemia as the pancreas attempts to overcome this resistance2. This metabolic dysregulation is closely associated with an increased risk of several metabolic disorders, including type 2 diabetes (T2D), obesity, dyslipidemia, and cardiovascular diseases3,4. The impaired insulin action in key target tissues— especially skeletal muscle, liver, and adipose tissue—plays a central role in the pathogenesis of these conditions5. This underscores the critical importance of IR in the development and progression of metabolic disorders, necessitating targeted therapeutic strategies to mitigate its impact6.
The euglycemic-hyperinsulinemic clamp7 is considered the most reliable test for evaluating IR, but its complexity limits its use in large-scale studies. Prior research has demonstrated a robust correlation between IR scores derived from the homeostasis model assessment (HOMA) approach and IR assessed by glucose clamp techniques8. The triglyceride to high-density lipoprotein cholesterol (TG:HDL-C) ratio is an alternative IR marker that is more cost-effective and accessible in larger population studies compared to the HOMA test9,10,11,12. A recent genome-wide association study (GWAS) on TG:HDL-C in European populations discovered 114 single-nucleotide polymorphisms (SNPs) associated with IR13. However, this previous GWAS on TG:HDL-C was limited to populations of European ancestry. Expanding genetic studies to diverse populations presents a valuable opportunity to uncover insights into the complex genetic foundations of IR14. In addition, IR is associated with impaired fatty acid utilization; increased free fatty acid flow from adipose to nonadipose tissue leads to aberrant fat metabolism and worsens IR. Hence, the TyG index, calculated from fasting plasma glucose and triglycerides, has emerged as another simple surrogate marker for IR15,16. The TyG index has high sensitivity for recognizing IR among apparently healthy subjects, compared with the HOMA-IR index16. As of the time of this writing and to our current knowledge, GWASs focusing on TyG have not been undertaken.
In this study, we conducted the GWAS on IR surrogate markers, including TyG, TG:HDL-C, and the logarithmically transformed TG:HDL-C ratio (log(TG:HDL-C)), in the Taiwanese population utilizing the Taiwan Biobank. We also carried out comparisons of heritability and genetic correlations among these markers. Due to the strong correlation between these IR surrogate markers and the lack of GWASs on TyG—an emerging and robust marker of IR—our subsequent analyses were centered on TyG. Additionally, fine mapping and polygenic risk score (PRS) analysis were performed for TyG. Moreover, we explored the relationships between disease phenotypes and IR surrogate markers. Furthermore, because sex differences in IR have been suggested17, we undertook sex-stratified and sex-differentiated analyses of TyG in both female and male cohorts, along with a transcriptome-wide association study (TWAS) of TyG in the whole cohort. Finally, pathway analysis was accomplished on genes associated with TyG to reveal additional clues to the causes and consequences of IR-related phenotypes.
Results
Taiwan Biobank study cohort
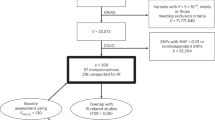
Supplementary Table 1 presents baseline characteristics of the Taiwanese population within the Taiwan Biobank, stratified by whole, female, and male cohorts. In our study utilizing the Taiwan Biobank, we analyzed a total of 136,735 individuals with measurements for TyG and 138,303 individuals for TG:HDL-C and log(TG:HDL-C) (Supplementary Table 1). The gender distribution in both samples consisted of ~35.5% male participants and 64.5% female participants.
GWASs of IR markers
We conducted GWASs on three IR surrogate markers–TyG, TG:HDL-C, and log(TG:HDL-C)–within the Taiwan Biobank sample. The GWAS test statistics were calibrated at a genome-wide level, ensuring robustness and reliability (Supplementary Fig. 1). Figure 1 presents the Manhattan plot illustrating the distribution of association p values across the genome for SNPs associated with these markers.
The Manhattan plots for the identified SNPs associated with the three IR surrogate markers, including a the TyG index, b TG:HDL-C ratio, and c log(TG:HDL-C) ratio. IR insulin resistance, TG:HDLC the triglyceride to high-density lipoprotein cholesterol ratio, TyG the product of fasting plasma glucose and triglycerides (Ln[fasting triglycerides (mg/dL) × fasting glucose (mg/dL)/2]). This GWAS analysis utilized BOLT-LMM’s mixed linear models with a two-sided chi-square test. The conventional genome-wide significance threshold of P < 5 × 10−8 was applied. Source data is provided as a Source Data file.
Supplementary Table 2 provides the number of significant SNPs identified at various significant thresholds for each marker, offering a comprehensive overview of the genetic landscape associated with IR. Furthermore, Supplementary Table 3 presents the GWAS results of the top 20 significant SNPs after clumping for each marker, highlighting the genetic variants with the strongest associations with IR.
Four gene clusters are associated with TyG
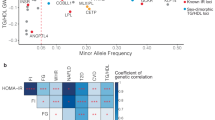
Supplementary Tables 4 and 5 provide an overview of all identified variants and the top 20 variants/genes with genome-wide significance for TyG, respectively. These variants primarily clustered within four gene regions, centered on GCKR (chromosome 2), MLXIPL (chromosome 7), APOA5 (chromosome 11), and APOC1 (chromosome 19). Further analysis identified additional significant genes within these clusters: 29 genes in the GCKR cluster, 17 in the MLXIPL cluster, 15 in the APOA5 cluster, and 6 in the APOC1 cluster (Tables 1–4 and Fig. 2).
The locus zoom plot for the a GCKR, b MLXIPL, c APOA5, and d APOC1 gene clusters concerning the TyG index in the Taiwan Biobank illustrates single nucleotide polymorphisms (SNPs) by their chromosomal positions and their association with TyG (−log10 P). The SNPs are color-coded to indicate their linkage disequilibrium with the top SNPs a rs1260326; b rs3812316; c rs651821; and d rs483082. Additionally, estimated recombination rates are depicted in cyan, derived from Asian subjects in the 1000 Genomes Project. The plot was generated using LocusZoom. This GWAS analysis utilized BOLT-LMM’s mixed linear models with a two-sided chi-square test. The conventional genome-wide significance threshold of P < 5 × 10−8 was applied. The topologically associating domain (TAD) TAD-1 spans from 25,920,000 to 27,320,000 (GRCh38) in pancreatic tissue. The TAD-2 spans from 27,680,000 to 28,520,000 (GRCh38) in pancreatic tissue. The TAD-3 spans from 73,160,000 to 74,760,000 (GRCh38) in pancreatic tissue. The TAD−4 spans from 116,760,000 to 119,360,000 (GRCh38) in pancreatic tissue. Notably, these TADs cover a significant portion of the GCKR, MLXIPL, and APOA5 gene clusters. Source data are provided as a Source Data file.
We conducted additional GWASs for TG:HDL-C and glucose levels (Supplementary Tables 6-7). Supplementary Tables 8–9 present the top variants exhibiting genome-wide significance for TG:HDL-C and glucose levels, respectively. The top 20 variants for TG:HDL-C are identical to those for TyG, with different rankings (Table 5). GCKR, a major factor in T2D risk18, is the only gene consistently associated with glucose levels, TG:HDL-C, and TyG (Table 5).
Supplementary Table 10 summarizes genome-wide significance for variants associated with insulin-related traits in previous studies. Our analysis revealed 76 genome-wide significant genes in TyG that had been previously associated with IR in GWAS studies on European and/or East Asian populations (Supplementary Table 11).
Genes for IR identified in the Taiwan Biobank
Our GWAS on TyG within the Taiwan Biobank unveiled 197 genes not previously documented in the context of IR (Supplementary Table 12). According to the NHGRI-EBI GWAS Catalog19, these genes have been associated with TG (99 genes), HDL-C (58 genes), T2D (29 genes) separately, as well as traits known to be associated with T2D risk including Alzheimer’s Disease (AD; 14 genes), fasting glucose levels (22 genes), and body mass index (49 genes).
The top 20 genes, previously unreported in the literature, exhibiting genome-wide significance for TyG (Table 6) are primarily clustered within four gene regions: GCKR, MLXIPL, APOA5, and APOC1. Supplementary Fig. 2 displays a Manhattan plot of these top 20 TyG genes, emphasizing SNPs neutral in European studies but significant in our cohort, indicating population-specific effects. Analysis of LD between top SNPs and SNPs previously unreported in the literature for TyG (Supplementary Table 13) revealed varying degrees of LD within each gene region, providing insights into potential functional relationships. For example, we observed strong LD between MLXIPL rs3812316 and TBL2 rs13246490 (r2 = 0.835), suggesting a possible shared genetic effect. In contrast, APOA1 rs12718464 and APOA5 rs651821 exhibited weak LD (r2 = 0.008), indicating potentially independent genetic influences on TyG within this region.
Conditional analyses on the top 20 genes, previously unreported in the literature, associated with TyG (Supplementary Table 14) helped discern independent genetic effects within closely located loci. For instance, APOA1 rs12718464 maintained a significant association with TyG (P = 1.1E-13) when conditioning on APOA5 rs651821, suggesting an independent effect. Conversely, conditioning on MLXIPL rs3812316 nullified the association of TBL2 rs13246490 (P = 0.32), indicating these variants likely represent the same genetic signal. These findings enhance our understanding of the complex interplay between genetic variants influencing TyG and help identify truly independent genetic associations.
Heritability and genetic correlations for IR surrogate markers
We estimated the heritability20 of TyG, TG:HDL-C, and log(TG:HDL-C) to be 15.5% (standard deviation (stdev) = 2.5%), 13.9% (stdev = 2.3%), and 17.3% (stdev = 2.5%), respectively (Supplementary Table 15). These values indicate the proportion of phenotypic variance attributed to genetic factors for each IR surrogate marker. Intercept values ranging from 1.07 to 1.09 suggest minimal biases due to confounding factors.
We also assessed genetic correlations to determine the extent to which genetic factors influencing TyG also affect TG:HDL-C and log(TG:HDL-C)20. The genetic correlations among TyG, TG:HDL-C, and log(TG:HDL-C) were notably high, ranging from 0.96 to 0.99 (Supplementary Table 15). This strong correlation underscores the close relationship among these three IR surrogate markers, highlighting their interconnectedness in the context of genetic influences on IR.
Fine mapping analysis for TyG
We employed fine mapping analysis to identify candidate causal variants linked to TyG21, consolidating overlapping loci into 11 distinct genomic regions of interest (Supplementary Table 16 and Supplementary Fig. 3). This approach enhanced analytical precision and prioritized areas for further investigation. For instance, within the chromosomal region 43378777–46429300 base pair on chromosome 19, five credible sets were identified with purity values ranging from 0.92 to 1.0 (Supplementary Table 16), suggesting a high degree of independence among the SNPs. Coverage values for these sets ranged from 0.95 to 1.0, indicating a high probability that the true causal variant for IR is represented within the credible sets. Certain SNPs exhibited PIP values exceeding 0.9, signifying a heightened probability of these variants being linked to IR (Supplementary Fig. 3). This indicates a robust likelihood that these specific genetic variants may play a significant role in IR manifestation or susceptibility, based on evidence from the Bayesian variable selection approach.
PRS analysis for TyG
We conducted PRS analysis to quantify the cumulative impact of multiple genetic variants on TyG22. The analysis used 854,050 SNPs to calculate the PRS for TyG (Supplementary Table 17 and Supplementary Fig. 4). The PRS model demonstrated substantial explanatory power, with 23.80% of the variance in TyG attributed to genetic factors. The incremental predictive value (R2–R2 Null) achieved by integrating genetic information into the PRS model was 10.57% compared to a model without genetic predictors. The R2 and R2–R2 Null values for TyG surpassed those reported in diverse phenotypes from prior investigations (e.g., triglyceride23 in Supplementary Table 17 and Supplementary Fig. 5), underscoring the robustness and effectiveness of the PRS model in elucidating the genetic underpinnings of TyG in the context of IR.
Association of disease phenotypes with IR surrogate markers
This study investigated the relationship between 27 disease phenotypes and IR surrogate markers (TyG, TG:HDL) and their PRS within the Taiwan Biobank (Table 7). Statistical significance was determined using the Bonferroni correction threshold of p = 0.00185. TyG exhibited associations with 20 different diseases, including coronary heart disease and mood disorders (P < 0.0001). TG:HDL showed associations with 14 diseases, such as gout and hyperlipidemia (P < 0.0001). PRS of TyG was linked to gout (P = 0.0016) and hyperlipidemia (P < 0.0001). PRS of TG:HDL was also associated with gout (P = 0.001) and hyperlipidemia (P < 0.0001). These findings highlight the relationships between IR surrogate markers, their PRS, and various disease phenotypes within the Taiwan Biobank.
Sex-stratified and sex-differentiated analyses of TyG
Sex-specific genetic differences have been found previously in the context of IR, and exploring sex-specific genetic effects may reveal a substantial number of important genes, previously unreported in the literature17. We conducted sex-stratified analyses, revealing 5787 significant SNPs in females (Supplementary Table 18) and 2817 SNPs in males (Supplementary Table 19).
Sex-differentiated analyses identified 915 SNPs with statistically heterogeneous effects between sexes (heterogeneity p value < 0.05) among those with genome-wide significance (Supplementary Table 20). For instance, the SNP rs7412 in APOE showed a heterogeneity p value of 2.37E-4 between sexes, with a p value of 2.29E-33 (effect size = 0.0796) for males and a p value of 4.43E-24 (effect size = 0.0494) for females. We identified 42 genes with sex-specific effects in females and 25 in males (Supplementary Table 21). Notably, female-specific genes were found in the MLXIPL cluster (FZD9 and BAZ1B) and APOA5 cluster (APOA4, APOA1, PAFAH1B2, SIDT2, TAGLN, and PCSK7). Male-specific genes such as MRPL33 (GCKR cluster), BACE1, CEP164, DSCAML1 (APOA5 cluster), and APOC1P1 (APOC1 cluster) were identified. The top loci with stronger female-specific effects were mapped to KLF14, APOA4, NID2, and TNFAIP8. Conversely, loci mapping to GMIP, TRIB1, and PBX4 showed stronger male-specific effects. Specifically, the locus rs1364422 (KLF14) demonstrated the strongest female-specific effect (heterogeneity p value = 1.51E-05). The locus rs190712692 (APOC1) showed the strongest male-specific effect (heterogeneity p value = 1.24E-05). However, some APOC1 loci also exhibited female-specific effects.
In our parent-offspring cohort, we detected parent-of-origin effects at the KLF14 locus (rs1364422), with 0 paternal and 1173 maternal transmissions of the minor allele in 1173 heterozygous offspring, indicating strong maternal inheritance (p value < 2.2E-16; Supplementary Table 22).
TWAS of TyG in the whole cohort
We used a TWAS approach24 to identify genes with expression differences associated with TyG. Supplementary Tables 23–27 present genes exhibiting significant associations with TyG in TWAS at genome-wide significance across chromosomes 2, 7, 11, 19, and in aggregate. In the GCKR gene cluster, 23 of 29 genes showed significant associations in both TWAS and GWAS, predominantly in adipose-subcutaneous, muscle-skeletal, and thyroid tissues (Table 1). The MLXIPL gene cluster revealed 9 of 17 genes with significant associations in both analyses, primarily in adipose-subcutaneous tissue (Table 2). In the APOA5 gene cluster, 13 of 15 genes demonstrated significant associations, mainly in adipose-visceral-omentum tissue (Table 3). The APOC1 gene cluster showed 4 of 6 genes with significant associations in both TWAS and GWAS (Table 4).
Pathway analysis of TyG
Pathway analysis was employed to elucidate key pathways influenced by TyG and gene-metabolic interactions25. The analysis included genome-wide significant genes, sex-specific genes, and genes from previous studies (Supplementary Tables 28–31). A summary of GO analysis results for four gene groups is presented in Table 8 and Supplementary Table 32.
Genome-wide significant genes are involved in AD pathogenesis, glucose homeostasis, insulin resistance and signaling, neurodegenerative pathways, Wnt signaling cascade, and plasma lipoprotein dynamics (Table 8). These enriched GO terms and pathways demonstrate substantial overlap with those associated with genes from previous studies (Table 8), suggesting a consistent functional profile across multiple investigations.
Female-specific genes exhibited similar enrichment to genome-wide significant genes (Table 8). The congruence between female-specific and genome-wide significant genes implies a potentially prominent role for these pathways in female-specific disease mechanisms. Conversely, male-specific genes revealed a distinct functional profile (Table 8) and lacked several pathways enriched in female-specific genes (e.g., estrogen signaling pathway). The male-specific genes showed enrichment in the tight junction pathway, which was absent in the female-specific gene set. This sexual dimorphism in pathway enrichment suggests potential differences in disease mechanisms between sexes.
Discussion
A recent European GWAS pinpointed 114 genetic loci that play a role in TG:HDL-C13. However, despite the large number of identified loci, they do not fully account for the heritability of IR, suggesting the existence of additional undiscovered genetic variants. Because the majority of GWAS studies on IR have focused predominantly on European populations, genetic variants that contribute to variations in traits across different populations are likely to be discovered by expanding genetic investigations to diverse populations14,26. Indeed, this inaugural GWAS of IR markers in the Taiwan Biobank has provided a wealth of loci, previously unreported in the literature, enhancing our understanding of the genetic underpinnings of IR-related diseases.
In our GWAS of IR markers, we identified genome-wide significant associations with loci in four distinct genomic regions: GCKR, MLXIPL, APOA5, and APOC1. SNPs with minimal impact in European studies are significant here (Supplementary Fig. 2), indicating potential unique genetic or environmental influences in Asians, warranting further investigation. The enrichment of IR-associated loci over such large regions might be attributed to the unique population structure and haplotype patterns present in the Taiwanese population. However, we found that these clusters are included within haploblocks identified in broader population studies14,27,28,29,30,31. For instance, our analysis of the East Asian population from the 1000 Genomes Project revealed peaks in recombination rates at the boundaries of the APOA5 gene cluster (Fig. 2). Furthermore, although they have not been highlighted in previous studies, similar clusters over the same genetic regions have also appeared in studies within the European, African, Korean, South Asian, and diverse populations14,27,28,29,30,31. For example, a recent study identified these four gene clusters in a diverse cohort, associating them with blood glucose, HDL-C, and TG levels (Supplementary Table 33)28. These clustered associations, therefore, may reflect low recombination rates within the regions that lead to a high probability of co-inheritance driven by strong IR-related loci, such as GCKR32,33.
Another explanation for genetic association to extend across such broad genomic regions is speculative but bears some mention here. We noted that the clustered high-association regions were mostly contained within the same or closely adjacent topologically associated domains (TADs), three-dimensional chromatin structures that enclose adjacent genes that are subject to some degree of co-regulation and co-expression in vivo (Tables 1–4 and Fig. 2). Regulatory elements within TAD structures can influence the disease-related expression of multiple neighboring genes34,35, and especially considering the TWAS results indicating the common mis-expression for these clustered genes in adipocytes (Tables 1–4), we speculate that the extended association clusters we identified could reflect the influence of noncoding variants that alter regulatory relationships and extend across the TADs. In contrast to the other three clusters, the APOC1 cluster is not enclosed within an established TAD (Fig. 2); however, the genes in this cluster are known to interact with the same long-distance enhancers36,37, suggesting that enhancer variation could contribute to a cluster-wide association with IR in this region as well.
Our sex-differentiated analyses of TyG identified 40 female-specific and 25 male-specific genes. Notably, KLF14 exhibited the most pronounced sex-specific effect in females, consistent with recent findings in European populations13. KLF14-associated SNPs have consistently shown sex-specific associations with metabolic traits, including T2D, waist-to-hip ratio, TG, HDL-C, and low-density lipoprotein cholesterol38,39,40,41, and corroborating these human studies, KLF14 deficiency in adipocytes leads to increased adiposity in female but not male mice42. The observed stronger associations in females are hypothesized to stem from the modulation of KLF14 expression rather than from hormonal influences43,44. We also identified several female-specific genes located within the APOA5 gene cluster, aligning with previous research45,46,47,48. These findings underscore sex-dependent mechanisms in lipid regulation and metabolism, exemplifying the importance of considering sex-specific effects in genetic studies of metabolic traits.
The imprinting analysis of KLF14 not only corroborates its known role in sex-specific genetic influence but also highlights the utility of large biobank datasets with familial information for dissecting complex genetic mechanisms49. Our findings suggest that the imprinting status of KLF14 could influence its biological roles, potentially affecting metabolic pathways or disease risk in a manner dependent on the sex of the parent from whom the allele is inherited44,50. Future studies could expand this approach to other genes with suspected imprinting effects, furthering our understanding of how genetic and epigenetic factors interplay in human health and disease.
Comparing gene sets from previous studies and the current genome-wide significant genes revealed substantial concordance in enriched pathways, particularly those implicated in insulin resistance/signaling, lipid metabolism, and glucose homeostasis, highlighting the critical role of these pathways in the etiology of IR. Interesting, GO analysis of the four distinct gene groups (genome-wide significant, female-specific, male-specific, and previously identified genes) also consistently demonstrated significant involvement in neurodegenerative processes; pathways associated with AD, including those related to nervous system development, are prominently represented across all groups. This shared enrichment suggests a potential mechanistic link underlying the high association between IR and neurodegenerative disorders51,52,53.
Our pathway analysis revealed some potential differences in male and female groups. For example, the GO analysis of female-specific genes revealed a distinct enrichment in pathways associated with estrogen signaling; this pathway’s prominence in female-specific genes is consistent with its proposed role in modulating disease risk or progression in women54. Conversely, the male-specific gene set exhibits a unique enrichment in the tight junction pathway, which is crucial for maintaining cellular barriers55. This pathway is absent in the female-specific gene set, indicating a potential sex-based difference in cellular mechanisms. Furthermore, insulin signaling pathway genes were more highly represented in the female-specific genes, pointing to potentially divergent metabolic regulation between sexes56,57. The identification of distinct molecular pathways in male and female-specific gene sets underscores the potential for developing sex-specific therapeutic approaches58,59. Sex-specific genetic factors and disease mechanisms interplay complexly17, suggesting that sex-tailored interventions may improve treatment efficacy60. Further research is warranted to elucidate the functional implications of these sex-specific pathway enrichments and their impact on disease risk and treatment strategies61,62.
The absence of medication data limits our understanding of drug effects on insulin resistance markers. Our sensitivity analysis, excluding self-reported diabetes mellitus (DM) cases (Supplementary Fig. 6 and Supplementary Table 34), shows minimal change in genetic associations for the TyG index, suggesting robustness despite potential DM treatment effects. Nonetheless, this does not substitute for medication records, emphasizing the need for future studies to include such data to explore medication’s influence on genetic associations with metabolic traits.
Methods
Taiwan Biobank
The study cohort comprised 147,880 Taiwanese subjects from the Taiwan Biobank63,64,65,66,67,68,69. Detailed inclusion and exclusion criteria26 are presented in Supplementary Methods. Ethical approval for the study was obtained from the Institutional Review Board of Taipei Veterans General Hospital (approval number: 2023-04-007CC#1). All participants provided informed consent in accordance with established guidelines and regulations.
Genotyping and imputation
Stringent quality control measures were applied to ensure reliable SNP analysis66,70. SNPs were excluded if they deviated from Hardy-Weinberg equilibrium (P < 1 × 10−6), exhibited a genotyping call rate below 95%, or a minor allele frequency less than 1%. From an initial set of 686,370 directly genotyped and 15,851,039 imputed SNPs, 7,604,854 SNPs remained after quality control and were used for functional prediction.
Statistical analysis
We conducted a GWAS to investigate the genetic underpinnings of three IR surrogate markers: TyG16, TG:HDLC13, and log(TG:HDLC). The TyG index was derived using the formula Ln[fasting triglycerides (mg/dL)×fasting glucose (mg/dL)/2]. The GWAS analysis employed mixed linear models in BOLT-LMM (version 2.4.1)71, adjusting for sex and the top 10 principal components (PCs) to account for potential confounding factors. Manhattan and quantile-quantile (Q-Q) plots were generated utilizing the R package ‘qqman’. The study used a genome-wide significance threshold of P < 5 × 10−8.
To identify independent genetic variants, we employed the clumping method in PLINK70, which accounts for linkage disequilibrium (LD) patterns in the genome. We performed fine mapping using susieR (v.0.12.35)21, which includes two metrics: Posterior Inclusion Probability (PIP) and Residual Sum of Squares Posterior Inclusion Probability (RSS PIP). We estimated heritability and genetic correlations using LD Score Regression (LDSC) v.1.0.120. We constructed PRS analysis using PRS-CS (v.1.1.0)22. Detailed methodologies for clumping, fine mapping, LDSC statistics, and PRS models are available in Supplementary Methods.
Sex-stratified and sex-differentiated analyses
Sex-stratified analyses used mixed linear models in BOLT-LMM (version 2.4.1)71, separately for male (n = 48,189) and female (n = 88,564) subjects, adjusting for sex and the top 10 PCs. Sex-differentiated analysis72 utilized GWAMA73 with the “-sex” option to identify sex-specific allelic effects and test for heterogeneity between sexes.
Transcriptome-wide association studies
We conducted TWAS to investigate the relationship between gene expression and TyG. Using FUSION24, we combined gene expression measurements with GWAS summary statistics. We obtained precomputed expression reference weights from GTEx(v7) for 11 human tissues, including various adipose, brain, and organ tissues. FUSION was then used to calculate TWAS P-values, identifying genes with significant associations between their expression levels and TyG susceptibility.
Pathway analysis
We constructed protein-protein interaction (PPI) networks utilizing NDEx-The Network Data Exchange74 with significant genes associated with TyG. The Human Integrated Protein–Protein Interaction Reference database75 was employed for PPI analysis within NDEx. We then conducted pathway analysis on the genes within the PPI network using ClueGO25, a Cytoscape76 plugin. ClueGO leverages various ontology source databases to identify gene ontology (GO) terms and pathway networks, elucidating the functional significance of identified genes within specific biological processes and pathways25.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The summary statistics data generated in this study are available in the Supplementary Information file. The Taiwan Biobank genetic data are subject to controlled access due to privacy policy requirements. Access can be obtained by submitting a formal request to the Taiwan Biobank at biobank@gate.sinica.edu.tw, with responses typically provided within 30 days, subject to approval. Approved users must adhere to data use agreements restricting secondary distribution and non-research use. Source data are provided with this paper.
Change history
28 April 2024
In the version of this article initially published, the information "This study was supported by the Population Health Research Center from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan (grant number NTU-112L9004)." was missing from the acknowledgement section. The original has been updated.
References
Petersen, M. C. & Shulman, G. I. Mechanisms of insulin action and insulin resistance. Physiol. Rev. 98, 2133–2223 (2018).
Thomas, D. D., Corkey, B. E., Istfan, N. W. & Apovian, C. M. Hyperinsulinemia: an early indicator of metabolic dysfunction. J. Endocr. Soc. 3, 1727–1747 (2019).
Gluvic, Z. et al. Link between metabolic syndrome and insulin resistance. Curr. Vasc. Pharmacol. 15, 30–39 (2017).
Yaribeygi, H., Farrokhi, F. R., Butler, A. E. & Sahebkar, A. Insulin resistance: review of the underlying molecular mechanisms. J. Cell. Physiol. 234, 8152–8161 (2019).
White, M. F. & Kahn, C. R. Insulin action at a molecular level–100 years of progress. Mol. Metab. 52, 101304 (2021).
Lee, S.-H., Park, S.-Y. & Choi, C. S. Insulin resistance: from mechanisms to therapeutic strategies. Diabetes Metab. J. 46, 15 (2022).
DeFronzo, R. A., Tobin, J. D. & Andres, R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am. J. Physiol. Endocrinol. Metab. 237, E214 (1979).
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Chiang, J.-K., Lai, N.-S., Chang, J.-K. & Koo, M. Predicting insulin resistance using the triglyceride-to-high-density lipoprotein cholesterol ratio in Taiwanese adults. Cardiovasc. Diabetol. 10, 1–6 (2011).
Gong, R. et al. Associations between TG/HDL ratio and insulin resistance in the US population: a cross-sectional study. Endocr. Connect. 10, 1502–1512 (2021).
Nur Zati Iwani, A. K. et al. TG: HDL-C ratio is a good marker to identify children affected by obesity with increased cardiometabolic risk and insulin resistance. Int. J. Endocrinol. 2019, 8586167 (2019).
McLaughlin, T. et al. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann. Intern. Med. 139, 802–809 (2003).
Oliveri, A. et al. Comprehensive genetic study of the insulin resistance marker TG: HDL-C in the UK Biobank. Nat. Genet. 56, 212–221 (2024).
Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019).
Guerrero-Romero, F. et al. The product of triglycerides and glucose, a simple measure of insulin sensitivity. Comparison with the euglycemic-hyperinsulinemic clamp. J. Clin. Endocrinol. Metab. 95, 3347–3351 (2010).
Simental-Mendía, L. E., Rodríguez-Morán, M. & Guerrero-Romero, F. The product of fasting glucose and triglycerides as surrogate for identifying insulin resistance in apparently healthy subjects. Metab. Syndr. Relat. Disord. 6, 299–304 (2008).
Ober, C., Loisel, D. A. & Gilad, Y. Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 9, 911–922 (2008).
Spracklen, C. N. et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 582, 240–245 (2020).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2018).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B Stat. Methodol. 82, 1273–1300 (2020).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Graham, B. E., Plotkin, B., Muglia, L., Moore, J. H. & Williams, S. M. Estimating prevalence of human traits among populations from polygenic risk scores. Hum. Genom. 15, 1–16 (2021).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Lin, E. et al. Genome-wide association study in the Taiwan Biobank identifies four novel genes for human height: NABP2, RA SA2, RNF41, and SLC39A5. Hum. Mol. Genet. 30, 2362–2369 (2021).
Lamina, C. et al. A genome-wide association meta-analysis on apolipoprotein A-IV concentrations. Hum. Mol. Genet. 25, 3635–3646 (2016).
Sinnott-Armstrong, N. et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 53, 185–194 (2021).
Koskeridis, F. et al. Pleiotropic genetic architecture and novel loci for C-reactive protein levels. Nat. Commun. 13, 6939 (2022).
Lee, S.-B. et al. Dyslipidaemia—genotype interactions with nutrient intake and cerebro-cardiovascular disease. Biomedicines 10, 1615 (2022).
Hoffmann, T. J. et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 50, 401–413 (2018).
Dupuis, J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42, 105–116 (2010).
Park, S. Association of polygenic risk scores for insulin resistance risk and their interaction with a plant-based diet, especially fruits, vitamin C, and flavonoid intake, in Asian adults. Nutrition 111, 112007 (2023).
da Costa-Nunes, J. A. & Noordermeer, D. TADs: dynamic structures to create stable regulatory functions. Curr. Opin. Struct. Biol. 81, 102622 (2023).
van Mierlo, G., Pushkarev, O., Kribelbauer, J. F. & Deplancke, B. Chromatin modules and their implication in genomic organization and gene regulation. Trends Genet. 39, 140–153 (2023).
Trusca, V. G. et al. Macrophage-specific up-regulation of apolipoprotein E gene expression by STAT1 is achieved via long range genomic interactions. J. Biol. Chem. 286, 13891–13904 (2011).
Zannis, V. I., Kan, H.-Y., Kritis, A., Zanni, E. & Kardassis, D. Transcriptional regulation of the human apolipoprotein genes. Front. Biosci. 6, d456–d504 (2001).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Pulit, S. L. et al. Meta-analysis of genome-wide association studies for body fat distribution in 694,649 individuals of European ancestry. Hum. Mol. Genet. 28, 166–174 (2019).
Shungin, D. et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Yang, Q. et al. Adipocyte-specific modulation of KLF14 expression in mice leads to sex-dependent impacts on adiposity and lipid metabolism. Diabetes 71, 677–693 (2022).
Small, K. S. et al. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nat. Genet. 50, 572–580 (2018).
Yang, Q. & Civelek, M. Transcription factor KLF14 and metabolic syndrome. Front. Cardiovasc. Med. 7, 91 (2020).
Angelov, A., Connelly, P. J., Delles, C. & Kararigas, G. Sex-biased and sex hormone-dependent regulation of apolipoprotein A1. Curr. Opin. Physiol. 33, 100654 (2023).
Bai, W. et al. Functional polymorphisms of the APOA1/C3/A4/A5-ZPR1-BUD13 gene cluster are associated with dyslipidemia in a sex-specific pattern. PeerJ 6, e6175 (2019).
Lee, G., Jeon, H. K. & Yoo, H. Y. Sex-related differences in single nucleotide polymorphisms associated with dyslipidemia in a Korean population. Lipids Health Dis. 21, 124 (2022).
Wang, Y. E. et al. SNPs in apolipoproteins contribute to sex-dependent differences in blood lipids before and after a high-fat dietary challenge in healthy US adults. BMC Nutr. 8, 95 (2022).
Hofmeister, R. J. et al. Parent-of-Origin inference for Biobanks. Nat. Commun. 13, 6668 (2022).
So-Youn 2 Richards Hannah B 7 the GIANT Consortium MCSKSHÅKGENACTGKATUS, Investigators M, Consortium D. et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 43, 561–564 (2011).
Dierssen, M. & Barone, E. Brain insulin resistance in neurodevelopmental and neurodegenerative disorders: mind the gap!. Front. Neurosci. 15, 730378 (2021).
Kshirsagar, V., Thingore, C. & Juvekar, A. Insulin resistance: a connecting link between Alzheimer’s disease and metabolic disorder. Metab. Brain Dis. 36, 67–83 (2021).
Ristow, M. Neurodegenerative disorders associated with diabetes mellitus. J. Mol. Med. 82, 510–529 (2004).
Tecalco-Cruz, A. C., López-Canovas, L. & Azuara-Liceaga, E. Estrogen signaling via estrogen receptor alpha and its implications for neurodegeneration associated with Alzheimer’s disease in aging women. Metab. Brain Dis. 38, 783–793 (2023).
Harhaj, N. S. & Antonetti, D. A. Regulation of tight junctions and loss of barrier function in pathophysiology. Int. J. Biochem. Cell Biol. 36, 1206–1237 (2004).
Shingleton, A. W. & Vea, I. M. Sex-specific regulation of development, growth and metabolism. Semin. Cell Dev. Biol. 138, 117–127 (2023).
Tramunt, B. et al. Sex differences in metabolic regulation and diabetes susceptibility. Diabetologia 63, 453–461 (2020).
Sadeghi, I., Rodríguez‐Fernández, B., Navarro, A., Gispert, J. D. & Vilor‐Tejedor, N. Understanding sex‐specific molecular mechanisms in preclinical and Alzheimer’s disease through brain proteomics and transcriptomics. Alzheimer’s Dement. 19, e076194 (2023).
Silveira, P. P., Pokhvisneva, I., Howard, D. M. & Meaney, M. J. A sex-specific genome-wide association study of depression phenotypes in UK Biobank. Mol. Psychiatry 28, 2469–2479 (2023).
Bernstein, S. R., Kelleher, C. & Khalil, R. A. Gender-based research underscores sex differences in biological processes, clinical disorders and pharmacological interventions. Biochem. Pharmacol. 215, 115737 (2023).
Garratt, M. Why do sexes differ in lifespan extension? Sex-specific pathways of aging and underlying mechanisms for dimorphic responses. Nutr. Healthy Aging 5, 247–259 (2020).
Bourquard, T. et al. Functional variants identify sex-specific genes and pathways in Alzheimer’s disease. Nat. Commun. 14, 2765 (2023).
Chen, C. H. et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet. 25, 5321–5331 (2016).
Fan, C. T., Lin, J. C. & Lee, C. H. Taiwan Biobank: a project aiming to aid Taiwan’s transition into a biomedical island. Pharmacogenomics 9, 235–246 (2008).
Hou, S.-J. et al. An association study in the Taiwan Biobank reveals RORA as a novel locus for sleep duration in the Taiwanese Population. Sleep Med. 73, 70–75 (2020).
Lin, E. et al. Association and interaction of APOA5, BUD13, CETP, LIPA, and health-related behavior with metabolic syndrome in a Taiwanese population. Sci. Rep. 6, 36830 (2016).
Lin, E. et al. Effects of circadian clock genes and health-related behavior on metabolic syndrome in a Taiwanese population: evidence from association and interaction analysis. PLoS ONE 12, e0173861 (2017).
Lin, E. et al. The ADAMTS9 gene is associated with cognitive aging in the elderly in a Taiwanese population. PLoS ONE 12, e0172440 (2017).
Lin, E. et al. The rs1277306 variant of the REST gene confers susceptibility to cognitive aging in an elderly Taiwanese population. Dement Geriatr. Cogn. Disord. 43, 119–127 (2017).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Magi, R., Lindgren, C. M. & Morris, A. P. Meta‐analysis of sex‐specific genome‐wide association studies. Genet. Epidemiol. 34, 846–853 (2010).
Mägi, R. & Morris, A. P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinforma. 11, 1–6 (2010).
Pratt, D. et al. NDEx, the network data exchange. Cell Syst. 1, 302–305 (2015).
Alanis-Lobato, G., Andrade-Navarro, M. A. & Schaefer, M. H. HIPPIE v2. 0: enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res. 45, D408–D414 (2016).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
We thank Lisa Stubbs of Pacific Northwest Research Institute for her valuable suggestions and thorough proofreading of our manuscript. This work was supported in part by grants from the National Science and Technology Council (Taiwan) (grant NSTC112-2314-B-075-013-MY3, NSTC112-2321-B-A49-021, NSTC112-2321-B-A49-013; SJT) and from the Taipei Veterans General Hospital (grant VTA112-V1-8-1, V112C-008; SJT). This study was supported by the Population Health Research Center from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan (grant number NTU-112L9004).
Author information
Authors and Affiliations
Contributions
E.L. had a role in study conception and design, analysis and interpretation of data, draft manuscript. Y.T.Y. had a role in analysis and interpretation of data. P.H.K. had a role in study conception and design. M.H.C. and A.C.Y. had a role in acquisition of data. S.J.T. had a role in study conception and design. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yi-Cheng Chang and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, E., Yan, YT., Chen, MH. et al. Gene clusters linked to insulin resistance identified in a genome-wide study of the Taiwan Biobank population. Nat Commun 16, 3525 (2025). https://doi.org/10.1038/s41467-025-58506-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58506-x