Abstract
Childhood is a critical period for hematopoietic development and susceptibility to hematologic disease. Here we generated a multimodal single-cell atlas of healthy human bone marrow, capturing mRNA and surface protein expression in 90,710 cells, including over 20,000 hematopoietic stem and progenitor cells (HSPC) and mesenchymal stromal cells (MSC) from nine donors ranging from infancy to young adulthood (2–32 years). Young pediatric (YP) bone marrow (<10 years) was compositionally and molecularly distinct from adolescent and young adult (AYA) bone marrow (≥13 years), with hematopoietic output shifting from B cell dominance in YP bone marrow to myeloid and T cell bias in AYA bone marrow. Spatial transcriptomics of six bone marrow biopsies (0–23 years) confirmed these age-dependent changes. Two lymphoid progenitor (LyP) subsets regulated this lineage shift: CD127+ LyP cells with B cell-biased output were enriched before age 10, whereas CD127− LyP cells with combined lymphoid and myeloid features predominated thereafter. Stromal signaling showed corresponding age-dependent changes, with increased interleukin-7 production by bone marrow MSC in YP compared to AYA, indicating niche-mediated regulation of HSPC lineage potential during ontogeny. This single-cell atlas provides a comprehensive resource for understanding hematopoietic development and early-life origins of hematologic disease.
Similar content being viewed by others
Main
Hematopoiesis orchestrates the lifelong production of all differentiated blood and immune cells throughout development, adulthood and old age. Due to its close connection to virtually any organ in the human body, the hematopoietic system is a major attribute of systemic health1,2,3. Blood production by hematopoietic stem and progenitor cells (HSPC) is tightly regulated by both cell-intrinsic and extrinsic mechanisms that balance HSPC self-renewal and multilineage differentiation, maintain hematopoietic homeostasis, allow rapid and controlled responses to stress and prevent disease.
The composition and function of the hematopoietic system undergo substantial changes throughout human life. Extensive research has elucidated key features of human hematopoietic aging, including a shift from lymphoid-biased to myeloid-biased output4,5, an increase in the relative frequency of HSPC4 and a concomitant decline in their regenerative capacity6,7,8. Although differences between hematopoiesis in the developing fetus, young adults and older individuals have been studied in detail6,9,10,11, there is a relative paucity of studies defining the cellular and molecular composition of hematopoiesis during human childhood11,12,13. Comprehending pediatric hematopoiesis is crucial to understand the development of the human hematopoietic system and the origins of pediatric hematologic diseases.
A major challenge in characterizing pediatric hematopoiesis is obtaining bone marrow (BM) samples from healthy children. Additionally, several key hematopoietic and nonhematopoietic cell types, including HSPC and mesenchymal stromal cells (MSC), are rare in BM aspirates, necessitating specific enrichment strategies to capture sufficient cells for in-depth profiling14. Consequently, much of our current understanding of pediatric hematopoiesis is based on studies that may not fully capture all relevant cell types. Technologies that simultaneously measure mRNA and surface protein expression in single cells provide unprecedented opportunities to dissect the composition of hematopoiesis in health and disease15,16. Compared to flow cytometry-based approaches or unimodal single-cell RNA sequencing (scRNA-seq), these methods allow improved cell-type identification and more accurate detection of cell states17. Furthermore, as mRNA levels do not always correlate with protein expression17, the surface protein modality allows validation that specific transcripts of interest are indeed expressed and may have functional consequences.
Here, we performed multimodal profiling of BM cells from healthy individuals aged 0–32 years, revealing age-related changes in cell frequencies, states and differentiation trajectories between young pediatric (YP) donors (<10 years) and adolescent and young adult (AYA) donors (≥13 years). We identified phenotypic and transcriptional differences within the lymphoid progenitors (LyP) cell population underlying B cell-biased hematopoiesis in young children. Interaction analysis and BM cytokine measurements uncovered age-dependent changes in signaling from MSC that may differentially prime LyP toward stable or B cell-biased output. This comprehensive, multimodal pediatric BM atlas provides a valuable reference for studies on hematopoietic development and hematologic diseases, many of which may originate in childhood.
Results
Reference map shows the single-cell composition of pediatric BM
To uncover the cellular and transcriptional landscape of human pediatric BM, we established an experimental pipeline enabling single-cell analysis of rare BM cell types, while preserving information on native cell frequencies (Fig. 1a and Supplementary Fig. 1). BM samples were obtained from seven healthy pediatric donors (4 female and 3 male, age 2.1–16.7 years, median of 9.6 years), all of whom served as donors for allogeneic hematopoietic cell transplantation (Supplementary Table 1). To enable both in-depth analysis of HSPC and MSC and accurate estimation of representative cell frequencies in a cost-efficient manner, three cell fractions were isolated from each donor: a nonenriched, erythrocyte-depleted CD235a− fraction, an HSPC-enriched CD235a−CD34+ fraction and an MSC-enriched CD235a−CD45loCD90+ or CD235a−CD45loCD271+ fraction. Enriched cell fractions from each donor were pooled with the nonenriched cell fractions of a genotypically distinct donor (Supplementary Fig. 1). Donor-specific single-nucleotide variants (SNVs) enabled postsequencing assignment of cells to enriched and nonenriched fractions to determine native cell frequencies. Multiplexed libraries were processed using 10x Genomics-based simultaneous single-cell profiling of transcriptome and surface protein expression and a customized panel of 138 oligonucleotide-conjugated antibodies (Fig. 1a).
a, Schematic overview of the experimental pipeline for CITE-seq, in which BM mononuclear cells were collected from seven healthy pediatric donors aged 2.1–16.7 years (3 male and 4 female; Supplementary Table 1). CD235a−CD34+ HSPC and CD235a−CD45loCD90+ or CD235a−CD45loCD271+ MSC were enriched from each sample by flow cytometry and combined with the nonenriched fraction from a genetically distinct donor. Pooled samples were subjected to CITE-seq using 138 oligonucleotide-conjugated antibodies on the 10x Genomics platform. b, CITE-seq-based weighted nearest neighbor (WNN) uniform manifold approximation and projection (wnnUMAP) of 68,094 cells from pediatric BM aspirates (n = 7) showing 28 clusters identified and annotated based on joint transcriptomic and protein expression patterns and grouped into eight major cell populations: HSPC (8 subsets), B and plasma cells (6 subsets), T and NK cells (7 subsets), myeloid populations (3 subsets), dendritic cells (2 subsets), erythroid cells (1 subset) and stromal populations (1 subset). c, wnnUMAP depicting the relative contribution of the combined HSPC-enriched and MSC-enriched (enriched) versus the nonenriched cell fractions as in b. d, Top differentially expressed genes for all 28 clusters, grouped by major cell populations as defined in b. e, Top differentially expressed surface markers for all 28 clusters, grouped by major cell populations as defined in b. MNC, mononuclear cell; HSPC, hematopoietic stem and progenitor cell; HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte–erythroid progenitor; MkP, megakaryocyte progenitor; EryP, erythroid progenitor; MCP, mast cell progenitor; TN, naive T cell; TM, memory T cell; Treg cell, regulatory T cell; MAIT cell, mucosal-associated invariant T cell; ProgDC, dendritic cell progenitor; cDC, conventional dendritic cell; pDC, plasmacytoid dendritic cell; Ery, erythroid; DC, dendritic cell; Avg, average.
After quality control and thresholding, the pediatric dataset comprised 68,094 high-quality cells, with a median of 5,133 transcripts and 2,001 unique genes per cell for the RNA modality and 1,307 counts and 114 unique proteins per cell for the protein modality, respectively (Extended Data Fig. 1a–h). Integration across donors and measurement modalities revealed 28 high-resolution clusters. These clusters were annotated using published datasets11,12,13, canonical marker genes and differentially expressed transcripts and proteins, resulting in 8 major hematopoietic populations and 28 subpopulations: HSPC (8 subsets, including hematopoietic stem cells (HSCs), lympho–myeloid primed progenitors (LMPP), megakaryocyte–erythroid progenitors, megakaryocyte progenitors, erythroid progenitors, pro-myelocytes, mast cell progenitors and LyP), B cells and plasma cells (6 subsets), T cells and natural killer (NK) cells (7 subsets), myeloid populations (3 subsets) and dendritic cells (2 subsets; Fig. 1b and Supplementary Table 2). Erythroid and stromal cells contained no further subsets (Fig. 1b). All donors contributed to every subset (Extended Data Fig. 1i). HSPC and MSC were predominantly derived from the enriched fractions (13,225 HSPC in enriched versus 2,689 in nonenriched fractions, 4.9-fold enrichment; 541 MSC in enriched versus 29 in nonenriched fractions, 18.7-fold enrichment; Fig. 1c). All HSPC subsets expressed key HSC-defining transcripts (for example, NPM1; Fig. 1d) and surface proteins (for example, CD112 and CD34; Fig. 1e), whereas genes such as AVP (HSCs and LMPP) or MPO (pro-myelocytes) distinguished specific subsets (Fig. 1d,e and Supplementary Tables 2–6). Integration with public datasets18,19 confirmed cluster annotations (Extended Data Fig. 2). Because cell annotation in the external datasets relied on RNA expression only18,19, there was greater variability in the classification of cell types that require surface protein modality weights for accurate annotation, such as T cell subsets (Supplementary Fig. 2). This observation aligns with previous reports20, underscoring the added value of multimodal profiling for high-resolution cell-type mapping. The complete pediatric BM atlas generated in this study is available as a Seurat object and Loupe browser-compatible resource through Zenodo at https://doi.org/10.5281/zenodo.14168864 (ref. 21).
Age-related cellular and transcriptional shifts in BM
Next, we leveraged the CITE-seq reference atlas to explore developmental changes in BM composition from infancy to adolescence. Analysis of nonenriched cell fractions revealed a high abundance of B lineage cells in donors aged 2.1–9.6 years (n = 4, 2 male and 2 female; Supplementary Table 1), which occupied the majority of BM cellularity (Fig. 2a), consistent with previous observations in peripheral blood22,23. By contrast, BM composition of the donors aged 13.3–16.7 years (n = 3, 1 male and 2 female; Supplementary Table 1) exhibited a dominance of myeloid and T cells (Fig. 2a). To validate whether the observed compositional differences persisted into young adulthood, we expanded our dataset to include two adult BM donors (20.7 and 31.3 years, both female, 11,278 and 11,338 cells; Supplementary Table 1). BM composition in these two adult individuals closely resembled that of the donors aged 13.3–16.7 (Fig. 2a), allowing us to define two age categories: YP (age <10 years) and AYA (age 13–31 years; Fig. 2b). B lineage cells were significantly higher in the YP group (median 49.7% versus 13.9%), whereas T cells (median 18.6% versus 10.4%) and myeloid cells (median 51.7% versus 24.0%) were enriched in the AYA group (Fig. 2c). To validate these age-related differences, we performed spatial transcriptomics on BM trephine biopsies from an independent cohort of three YP (two male participants and one female participant, ages 0.7, 1 and 1.5 years) and three AYA donors (all male participants, ages 13.5, 17 and 23 years; Supplementary Tables 7 and 8). Analyses of cell composition in these BM biopsies revealed a higher abundance of B lineage cells in YP samples (median 34.6% in YP versus 9.1% in AYA), whereas myeloid cells were more prevalent in AYA samples (median 36.1% in YP versus 58.2% in AYA; Fig. 2d,e and Extended Data Fig. 3). Together, these findings indicate that YP BM is distinct from AYA BM and is characterized by a shift from B cell lineage bias to increased T cell and myeloid output from infancy to young adulthood.
a, Frequencies of cell subsets (as in Fig. 1b) in CITE-seq data of nonenriched BM aspirates of four YP (2.1–9.6 years, two male and two female) and five AYA (13.5–31.3 years, one male and four female) healthy donors. B lineage cells (YP BM, median 49.7%; AYA BM, median 13.9%, P = 0.016), T cells (YP BM, median 18.6%; AYA BM median 10.4%, P = 0.56) and myeloid cells (YP BM median 51.7%; AYA BM median 24.0%, P = 0.063; two-sided Wilcoxon rank-sum test). b, Principal component analysis of BM composition in YP (n = 4) and AYA (n = 5) samples as in a. Dots represent individual BM aspirates, and arrows represent vectors reflecting each cell population’s contribution to the principal components. c, Frequencies of cell subsets (as in Fig. 1b) in BM aspirates from YP (n = 4) and AYA donors (n = 5). d, Representative BM cores from Xenium-based spatial transcriptomics of BM trephine biopsies from YP (n = 3, 0.7–1.5 years, two male participants and one female participant) and AYA donors (n = 3, 13.5–23 years, all male participants). B lineage cells were overlaid to allow consistent comparisons across samples; scale bar, 300 μm; yrs, years. e, Relative abundance of BM cell subsets (as in Fig. 1b) in spatial transcriptomics data from YP (n = 3, 0.7–1.5 years, two male participants and one female participant) and AYA (n = 3, 13.5–23 years, all male participants) BM biopsies. f,h, Radar plot showing module expression of differentially expressed genes in HSCs (f), LMPP (g) and LyP (h) from YP (n = 4, 2.1–9.6 years, two male and two female participants) and AYA donors (n = 5, 13.5–31.3 years, one male and four female participants) across BM cell subsets as in Fig. 1b. Genes upregulated in YP or AYA BM cell subsets (Supplementary Table 9) were aggregated into module scores representing the average expression of age-associated transcriptional programs and assessed across all 28 cell clusters to evaluate their potential lineage-biasing effects. BaEoMaP, progenitors of basophils, eosinophils and mast cells; MkP/Mk, megakaryocyte progenitor cell or megakaryocyte; AEC/VSMC, arteriolar endothelial cell or vascular smooth muscle cells; SEC, sinusoidal endothelial cell; Nonclass., nonclassical.
YP B lineage cell bias originates from LyP cells
We next explored whether the observed age-related differences in B, T and myeloid cell frequencies were accompanied by transcriptional changes in each of these populations or their upstream progenitors. To determine whether these frequency shifts reflected a general aging program shared across hematopoietic cell types or were restricted to specific hematopoietic cell populations, we compared single-cell transcriptomes and surface marker expression of each of the 28 BM subclusters in our CITE-seq data (as defined in Fig. 1b) between YP and AYA BM. Differential expression analysis identified ~5,000 genes (Extended Data Fig. 4a and Supplementary Table 9) and ~200 surface proteins differentially expressed between YP and AYA BM populations (Extended Data Fig. 4b and Supplementary Table 10). Among the 5,000 differentially expressed genes, most were cell-type specific: 67% of genes upregulated in YP BM and 62% of genes upregulated in AYA BM were significantly altered in only a single cell population (for example, restricted to HSCs, pro-B cells or classical monocytes; Extended Data Figs. 4 and 5). By contrast, a small number of genes, such as CD59 (higher expression in AYA BM) and CD74 (higher expression in YP BM) were consistently up- or downregulated across multiple cell types (Extended Data Figs. 4 and 5). The shared genes included genes previously linked to aging in adults, such as CTSW24 or KLF6 (ref. 25; Supplementary Table 10). These observations implied that hematopoietic cells underwent predominantly cell-type-specific transcriptional changes rather than a uniform maturation program active across all populations during maturation from YP to AYA stages.
To assess whether the differentially expressed genes between YP and AYA BM imposed a lineage bias toward B cell lineage versus myeloid fates, we calculated lineage-specific module scores. These scores were generated by aggregating the differentially expressed genes between YP and AYA cell populations (Supplementary Table 9) and subsequently assigning to each individual cell in our CITE-seq data. Average module expression was then visualized across all 28 BM cell populations (as defined in Fig. 1b and Supplementary Figs. 3 and 4), focusing on populations comprising the differentiation trajectory from HSC to the B cell lineage (HSC, LMPP and LyP; Fig. 2f–h). The modules derived from the differentially expressed genes in HSC (for example, IGFBP2 and PROK2; Supplementary Table 8) and LMPP (for example, CD24; Supplementary Table 9) were uniformly distributed between B cell and myeloid populations (Fig. 2f,g and Supplementary Table 9), indicating that although early progenitors (HSC and LMPP) underwent transcriptional changes during maturation from YP to AYA, these differences did not confer a lineage differentiation bias. By contrast, the modules derived from YP-upregulated LyP genes (for example, EBF1 and CD79A; Supplementary Table 9) were enriched in B cell populations (pro-B, pre-B, naive/memory B and plasma cells) compared to other BM cell populations (Fig. 2h and Supplementary Fig. 3). Conversely, gene modules upregulated in AYA LyP versus YP LyP (for example, CD37 and SPINK2; Supplementary Table 9) were enriched in HSC, LMPP, pro-myelocytes, myelocytes, classical monocytes and nonclassical monocytes; Supplementary Fig. 3). At the single-gene level, expression of EBF1 and other B cell lineage genes, including BACH2 and CD79B, was higher in YP than in AYA samples, starting at the LyP stage and continuing through the pro-B, pre-B and naive/memory B cell stages, but not in earlier LMPP or HSC stages (Extended Data Fig. 6). Overall, these analyses indicate substantial developmental changes in BM composition and transcriptional programs between YP and AYA groups, with a prominent B cell lineage bias in HSPC emerging (and potentially regulated) at the LyP stage.
Distinct LyP subsets correlate with age-related lineage bias
Next, we further dissected the LyP population to investigate whether heterogeneity within this population could explain the differences in B cell lineage versus myeloid bias between YP and AYA BM. Subclustering of LyP cells revealed two distinct populations with unique transcriptomic and proteomic features (subclusters 1 and 2; Fig. 3a). Both subclusters exhibited hallmark features of LyP, including expression of canonical LyP transcripts (for example, IGLL1 and DNTT) and surface proteins (CD34+CD90−CD45RA+; Extended Data Fig. 7). This identity was further validated by mapping to the Deeply Integrated Human Single-Cell Omics (DISCO) external reference dataset, where both subclusters were annotated as LyP (Extended Data Fig. 8)19,26,27. Subcluster 1 showed high expression of B lineage cell-associated genes (EBF1, CD79A and VPREB1) and high surface protein expression of CD127 (interleukin-7 (IL-7) receptor; Fig. 3b,c and Supplementary Tables 11 and 12) and is referred to hereafter as B cell biased (LyP-B). Subcluster 2 showed high expression of genes associated with stemness (for example, SPINK2, ABCB1 and LRMDA), myeloid (LGALS1, CSF3R and CLEC12A) and lymphocyte lineage genes (CD37 and SPIB; Fig. 3b). Cells in this cluster also displayed increased protein expression of both myeloid (for example, CD123 and CD33) and lymphoid (for example, CD18 and CD155) markers (Fig. 3c) and are referred to as stable LyP (LyP-S). Gene Ontology term enrichment analysis supported this functional distinction, with LyP-B cells enriched for pathways related to B cell receptor signaling and lymphocyte proliferation and differentiation, whereas LyP-S cells were enriched for pathways related to myeloid leukocyte activation, mononuclear cell differentiation and B cell activation (Fig. 3d). Inference of gene regulatory networks showed increased activity of B lineage cell-associated regulons in LyP-B cells (for example, PAX5, LEF1 and TCF3)28,29, whereas LyP-S cells were enriched for regulons associated with general lymphoid development and stemness (for example, ELF4, RXRA and KLF4; Fig. 3e and Supplementary Table 13)30,31. To visualize both LyP subsets within the hematopoietic hierarchy, we created a diffusion map from our CITE-seq data, incorporating the B cell and myeloid trajectories (from HSCs to LMPP, LyP, cycling pro-B cells and pro-B cells and from HSCs to LMPP, pro-myelocytes, myelocytes and classical monocytes; Fig. 1b and Extended Data Fig. 7b). When annotated onto this trajectory, LyP-B cells localized closer to pro-B cell populations, whereas LyP-S cells positioned closer to LMPP and pro-myelocytes (Extended Data Fig. 7e), supporting the notion that although both LyP subsets arose downstream of LMPP, LyP-B cells exhibited a B cell lineage bias whereas LyP-S cells maintained broader, more multipotent potential. Although both LyP subsets were present across all individuals, LyP-B cells were significantly more abundant in YP BM, whereas LyP-S cells predominated in AYA BM (median 74.4% versus 26.9% LyP-B; Fig. 3f). Of note, in one of the two adult individuals (age 20.7), LyP-B cells were only detected in the enriched fraction (Fig. 3f), emphasizing the rarity of this subset in adults. Cell cycle analysis showed a trend toward higher proportions of cycling LyP-B cells (S + G2/M) in YP samples than in AYA samples (median 45.2% versus 24.5%; Fig. 3g), which might have contributed to their increased abundance. Moreover, the frequency of LyP-B cells in each individual was significantly correlated with the overall percentage of B lineage cells (Fig. 3h). To validate these age-associated patterns, we reanalyzed a public scRNA-seq dataset of CD34+ cells from 15 BM donors (ages 2–77 years; 8 male and 7 female donors)32 and reclassified LyP into LyP-B or LyP-S cells (Extended Data Fig. 8a). This analysis confirmed that LyP-B cells were more abundant in young donors (ages 2–12 years, median 70.1% of all LyP) than in AYA donors (ages 17–77 years, median 60.4% of all LyP; Extended Data Fig. 8b,c). Conversely, LyP-S cells were increased in AYA donors (median 29.4% in YP versus 39.6% in AYA; Extended Data Fig. 8b,c). Thus, in line with previous reports22,33, we found that two phenotypically and functionally distinct subpopulations of LyP exist in healthy BM, the abundance of which was age-dependent and associated with a systemic bias toward B lineage cells in YP BM.
a, UMAP plot of LyP-S cells (n = 690 cells) and LyP-B cells (n = 649 cells) in CITE-seq data of BM aspirates of YP and AYA donors combined (n = 9; 2.1–31.3 years, three male and six female donors). b,c, Volcano plot depicting genes (b) and surface markers (c) differentially expressed between LyP-B and LyP-S subclusters, as in a. d, Gene Ontology biological process enrichment of differentially expressed genes between LyP-B and LyP-S cells using clusterProfiler. Dot size indicates the GeneRatio, and dot color represents the adjusted P value. Enrichment significance was assessed using a hypergeometric test with Benjamini–Hochberg false discovery rate correction. e, Volcano plot of differentially active transcription factor regulons between LyP-B and LyP-S cells, inferred using single-cell regulatory network inference and clustering in Python (pySCENIC) and quantified using AUCell scores at the single-cell level. Differential regulon activity was assessed using a two-sided Wilcoxon rank-sum test as implemented in Seurat, with Bonferroni correction for multiple testing. f, Relative proportions of LyP-B and LyP-S subsets within the total LyP population (nonenriched fraction) for each individual (n = 9; Supplementary Table 1). LyP-B (YP BM, median 74.4%; AYA BM, median 26.9%; Wilcoxon rank-sum test P = 0.016). g, Stacked bar plots showing cell cycle phase assignment in LyP-B and LyP-S cells based on the combined enriched and nonenriched fractions for each individual (LyP-B cells in S + G2/M (YP, median 45.2%; AYA, median 24.5%; P = 0.11, Wilcoxon rank-sum test); LyP-S cells in S + G2/M (YP, median 35.3%; AYA, median 41.4%; P = 0.11, Wilcoxon rank-sum test)). h, Correlation plot depicting the Pearson correlation between the relative abundance of LyP-B cells within total LyP cells versus the percentage of B lineage cells (nonenriched) in each BM aspirate. Each dot represents one individual.
Age-dependent stromal signals drive LyP lineage bias
B lineage cell differentiation is regulated, in part, by cell-extrinsic signals from MSC within the BM niche34,35,36,37,38. To explore whether stromal signaling regulates LyP subsets in an age-dependent manner, we used NicheNet to infer ligand–receptor interactions that differed between LyP-B and LyP-S cells39. We prioritized expressed ligands based on their predicted regulatory influence on the differentially expressed genes between LyP-B and LyP-S cells, such as EBF1, ZCCHC7 and GLRX (LyP-B) and CD37, ATP8B4 and LGALS1 (LyP-S) and the differential expression of the receptors matching these ligands between LyP-B and LyP-S cells. The growth factor IGF2, the lymphoid-supporting cytokine IL-7 and the immunoglobulin superfamily member BTLA emerged as the top candidate ligands for LyP-B cells (Fig. 4a). IL-7 demonstrated the strongest predicted influence on B lineage cell transcription factors, including EBF1, LEF1 and PAX5 (Fig. 4a). By contrast, the pleiotropic cytokine transforming growth factor-β1 (TGFβ1) was identified as the top candidate regulator for LyP-S cells (Fig. 4a). Both ligands showed cross-regulatory potential: IL-7 affected certain LyP-S target genes (for example, LGALS1, RFLNB and MAP3K8), whereas TGFβ1 influenced certain LyP-B genes (for example, HMGB1, SOCS2 and IL7R; Fig. 4a), supporting their reciprocal regulatory relationship34,35,37. Receptor analysis of our CITE-seq data showed significantly increased mRNA expression of the IL-7 receptor α-chain (IL-7Rα, encoded by IL7R; fold change (FC): 7.01) and its co-receptor, the common γ-chain (IL2RG; FC: 1.45), on LyP-B cells. This was confirmed at the protein level via increased expression of CD127 (IL-7Rα; Fig. 3c). Conversely, LyP-S cells expressed more TGFBR1 mRNA (FC: 2.26; Fig. 4b). To identify the cellular source of these ligands in BM, we mapped IL7 and TGFB1 expression across all BM cell types in our CITE-seq data (as defined in Fig. 1b). Although TGFB1 was broadly expressed across dendritic, myeloid, NK and T cells, IL7 expression was mostly restricted to MSC (Fig. 4c). Both IL7 and TGFB1 showed age-dependent patterns: IL7 mRNA expression was higher in YP MSC than in AYA MSC, whereas TGFB1 mRNA was higher in AYA BM cells than in YP BM cells (Fig. 4d,e). To validate the age-dependent availability of IL-7, we quantified concentrations of 12 cytokines and growth factors, including IL-7, in BM plasma (defined as the cell-free fraction of BM aspirates) using a bead-based multiplex immunoassay in an independent cohort of 78 donors (‘LegendPlex cohort’, 0–20 years; median age of 8 years; 47 male and 31 female donors; Supplementary Table 14). IL-7 concentrations were significantly higher in YP than in AYA BM plasma (2.6 versus 2.0 pg ml−1; Fig. 4f). Other cytokines, such as IL-15, IL-3 or IL-34, showed no age-dependent differences (Fig. 4f). These findings aligned with prior reports identifying BM and thymic stromal cells as the primary sources of IL-7 (ref. 40) and with studies showing elevated concentrations of IL-7 in the blood plasma of young children (0–5 years) relative to AYAs (22–53 years)41.
a, NicheNet interaction analysis of differential interactions between LyP-B (n = 690 cells) and LyP-S (n = 649 cells) cells in CITE-seq data from BM aspirates of YP and AYA donors combined (n = 9; 2.1–31.3 years, three male, six female donors; Supplementary Table 1), showing the top three predicted ligands underlying the observed upregulation of LyP-B cell genes (top rows) and LyP-S cell genes (bottom rows), prioritized based on regulatory potential and differential receptor expression between the two LyP cell subsets. The genes shown represent the top 30 differentially expressed genes in LyP-B (left) or LyP-S (right) cells predicted to be regulated by these ligands. The regulatory potential of LyP-B cell ligands on LyP-S cell genes (top right quadrant) and vice versa (bottom left quadrant) is also shown. b, Dot plot showing the average expression of IL7R, IL2RG, TGFBR1 and TGFBR2 in LyP-B and LyP-S cell subsets (IL7R (LyP-B versus LyP-S; FC: 7.01, P < 0.001), IL2RG (LyP-B versus LyP-S; FC: 1.45, P = 0.002) and TGFBR1 (LyP-S versus LyP-B; FC: 2.26, P < 0.001; two-sided Wilcoxon rank-sum test with Benjamini–Hochberg false discovery rate correction)). Dot color indicates scaled receptor expression, and dot size represents the percentage of expressing cells. c–e, Dot plot showing the average mRNA expression of IL7 and TGFB1 in all BM cell types from CITE-seq data (c), in all BM cell types combined per individual (d) and in MSC only per individual (e). f, Box plots showing the concentrations of IL-7, IL-3, IL-15 and IL-34 protein measured by multiplex immunoassay (LegendPlex) in the BM of 78 individuals (age range of 0–20, median age of 8 years, 47 male and 31 female donors; Supplementary Table 13) with localized solid tumors without BM involvement. Boxes represent the median and the first and third quartiles, and whiskers indicate data points within 1.5 times the interquartile range. P values were calculated using a two-sided Wilcoxon rank-sum test.
Adult stromal cell atlases have defined various subsets of BM MSC expressing distinct HSPC-supporting factors and identified adipo-MSC and Thy-1+ MSC as the main producers of IL-7 in BM14. When mapped onto this atlas, most MSC in our CITE-seq data were annotated as adipo-MSC (Extended Data Fig. 9a,b). However, the limited number of MSC in these data (n = 588) precluded subset-specific analyses (Extended Data Fig. 9a). In the spatial transcriptomics dataset, which captured 8.4-fold more stromal cells (n = 4,950 versus 588 in CITE-seq; Fig. 5a–c), the stromal cell population in YP samples showed significantly increased frequencies of osteo-MSC (12.2% versus 1.4% of total MSC) and reduced frequencies of adipocytes (2.0% versus 14.0% of total MSC) compared to AYA samples (Fig. 5c–e). Adipo-MSC frequencies were comparable between YP and AYA samples (73.6 versus 66.1% of total MSC; Fig. 5c–e). These findings suggest that the elevated concentrations of IL-7 protein in YP BM might arise from increased per-cell production by adipo-MSC or from a more promiscuous stromal source than in AYA BM. In summary, age-dependent expression of IL7 and TGFB1, along with their respective receptors, paralleled the age-dependent dynamics of LyP subsets, supporting a model where signals from BM stromal cells cooperate with LyP-intrinsic differences to shape lineage bias during human development.
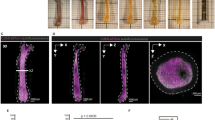
a–c, Xenium spatial transcriptomics-based UMAP of 163,325 cells from BM trephine biopsies of YP (n = 3, age 0.7–1.5 years, two male and one female donor) and AYA donors (n = 3, age 13.5–23 years, all male donors; Supplementary Table 7), showing the identification of 27 clusters, annotated and grouped into 11 major populations: HSPC (3 subsets), B and plasma cells (5 subsets), T and NK cells (3 subsets), myeloid populations (5 subsets), stromal cells (5 subsets), erythroid cells, dendritic cells, megakaryocytes, erythroid cells, sinusoidal endothelial cells and arteriolar endothelial cell or vascular smooth muscle cells, annotated per major population (a) and subcluster (b). c, UMAP of cells as in a showing adipo-MSC (n = 3,571 cells), adipocytes (n = 478 cells), fibro-MSC (n = 38 cells), fibro/osteo-MSC (n = 598 cells), osteo-MSC (n = 265 cells) and osteoclasts (n = 82 cells). Key markers for each subpopulation are available in Supplementary Table 2. d, Representative BM cores from each individual as in a showing the spatial distribution of adipo-MSC, adipocytes, fibro-MSC, fibro/osteo-MSC, osteo-MSC and osteoclasts; scale bar, 300 μm. e, Stacked bar plot showing the relative abundance of adipo-MSC, adipocytes, fibro-MSC, fibro/osteo-MSC, osteo-MSC and osteoclasts in each individual. Osteo-MSC (YP BM median 12.2% of total MSC; AYA BM median of 1.4% of total MSC, mixed-effects logistic regression with donor as random intercept, Benjamini–Hochberg-adjusted P = 3.2 × 10−16), adipocytes (YP BM median 2.0% of total MSC; AYA BM median 14.0% of total MSC; Benjamini–Hochberg-adjusted P = 2.3 × 10−36) and adipo-MSC (YP BM median 73.6% of total MSC; AYA BM median 66.1% of total MSC; Benjamini–Hochberg-adjusted P = 0.60).
Discussion
Here, we compiled a comprehensive multimodal single-cell analysis of healthy pediatric BM, including single-cell transcriptomics, cell surface proteomics and spatial transcriptomics, to map HSPC lineage fate decisions during human development from infancy to young adulthood (0–32 years). We showed that YP BM (<10 years) was biased toward B lineage differentiation, whereas AYA BM was dominated by T cell and myeloid populations. This age-dependent lineage shift was driven by two distinct LyP subsets: CD127+ LyP, transcriptionally primed toward B cell output (LyP-B) and enriched before 10 years of age, and CD127+ LyP with T cell and myeloid features, predominating from ≥13 years onward. These changes correlated with age-related differences in BM MSC signaling, characterized by increased IL-7 production in early childhood and increased TGFβ signaling in AYA, indicative of niche-derived signals as critical regulators of HSPC lineage bias across development.
We identified two transcriptionally and phenotypically distinct subpopulations of LyP cells that differed in surface marker expression, lineage differentiation programs and age-dependent prevalence. Our data are consistent with previous flow cytometry-based studies that reported similar heterogeneity within human LyP22,33,42. Up to 57% of multipotent LyP (CD34+CD38−Thy-1neg-loCD45RA+) from human adult BM generate B cell, T cell and myeloid progeny in vitro and in vivo35, reflecting residual myeloid potential. Studies in humanized mice have demonstrated that CD34+CD38loCD45RA+ITGB7+ progenitor cells can be subdivided into CD127− and CD127+ LyP subsets biased toward NK cell–innate lymphoid cell–T cell or B cell lineages, respectively22,33,42. Although CD127− LyP cell differentiation has been linked to FLT3 signaling, expansion of CD127+ LyP cells has been considered largely cell autonomous28. Our data refined this model by identifying niche-derived IL-7 as an HSPC-extrinsic regulator of CD127+ LyP cells, shaping the developmental balance between NK cell–innate lymphoid cell–T and B cell lineage output.
Our work highlighted the critical role of BM stromal signaling in driving age-dependent changes in hematopoiesis. Stromal-derived factors, including FLT3L, CXCL12 and IL-7, are established regulators of B cell lymphopoiesis36,37,38. We showed that stromal expression of IL7 progressively declined from birth to young adulthood, coinciding with a shift from B cell lineage to myeloid differentiation in LyP cells. Measurement of IL-7 protein concentrations in BM-derived plasma supported the functional relevance of our transcriptomic data. The mechanisms underlying age-related cytokine production by BM MSC remain incompletely known. Sex hormones represent one plausible contributor: both testosterone and estrogen suppress IL-7 production, and estrogen inhibits lymphopoiesis in mice, partly by inducing apoptosis of LyP43. Estrogen depletion increases BM IL-7 expression44. Additionally, pregnancy-induced elevations in estrogen and progesterone are associated with reduced IL-7 levels and B cell lymphopoiesis, effects reversible by recombinant IL-7 (ref. 45). Our dataset did not allow for the identification of the precise age at which the shift from B cell lineage to myeloid bias occurs, nor its relationship with pubertal onset in individual donors. Rather than a discrete event, this shift is likely to represent a gradual developmental process, occurring between ages 10 and 13, with interindividual variability in onset and duration. Alternatively, or in conjunction with sex hormones, developmental changes in MSC composition may contribute to the increased IL-7 production in YP BM. Adult stromal atlases have identified distinct MSC populations, with adipo-MSC and Thy-1+ MSC as the principal sources of BM IL-7 (ref. 14), alongside contributions of epithelial cells and reticular cells. In our CITE-seq data, MSC most closely resembled adipo-MSC, but numbers were insufficient for robust subset-specific analyses. Although our spatial datasets captured larger numbers of MSC, panel design precluded precise identification of IL-7-producing stromal subsets, which might differ between YP individuals and adults. Further studies are needed to define the precise stromal source of BM IL-7 across human development.
Finally, the developmental states observed within the healthy pediatric lymphoid lineage closely resembled the transcriptomic heterogeneity reported in B cell acute lymphoblastic leukemia46. Bulk leukemic samples from young children displayed stronger B lineage transcriptional signatures than those from AYA individuals, which had more multipotent transcriptional profiles46. These age-dependent leukemic states mirrored the age-dependent transcriptomic programs of LyP cells identified here. Because the cell of origin for pediatric B cell acute lymphoblastic leukemia likely emerges years before clinical diagnosis, possibly during fetal development47,48,49, these findings suggest that BM niche-mediated instruction of B cell lineage differentiation is preserved in the malignant setting. Elevated IL-7 in YP BM, which can induce a preleukemic state49,50, may therefore facilitate acquisition of additional oncogenic events. Given the increasing application of lymphoid-targeted therapies and the potential for lineage switch as a mechanism of escape, manipulating lineage-instructive signals from the BM niche may provide a relevant therapeutic strategy.
In conclusion, we provide a comprehensive reference of healthy pediatric BM spanning from birth to young adulthood (0–32 years) and have made our dataset openly available as an online resource accompanying this manuscript (https://doi.org/10.5281/zenodo.14168864)21. The differences between pediatric and adult BM underscore the importance of age-matched references when interpreting disease-associated changes. By enabling such comparisons, our pediatric single-cell map provides a foundation for future research on pediatric blood disorders. In addition, it reveals age-specific regulatory mechanisms that may underlie the timing and susceptibility of hematologic malignancies in childhood and adolescence.
Methods
Human donors
BM samples were obtained from three independent cohorts, each used for distinct analyses. All samples were obtained through the Biobank of the Princess Máxima Center for Pediatric Oncology, Utrecht, the Netherlands. Cohort demographics, including donor numbers, age ranges and sex distributions, are provided in Supplementary Tables 1, 7 and 14. For CITE-seq, BM aspirates from healthy individuals (n = 9, 2.1–31.3 years, three male and six female donors; Supplementary Table 1) were acquired. BM aspirates had been collected bilaterally from the posterior superior iliac crests as part of a stem cell donation to an affected relative. None of the healthy donors received any HSC-mobilizing treatment before donation. Written informed consent was obtained from all donors or legal guardians. Approval was granted by the Institutional Review Board of the Princess Máxima Center (PMCLAB2022.0328).
For Xenium-based spatial transcriptomics, formalin-fixed paraffin-embedded (FFPE) BM trephine biopsies were analyzed from an independent cohort of YP (n = 3, 0.7–1.5 years, two male and one female donor) and AYA donors (n = 3, 13.5–23 years, all male donors). As BM biopsies are invasive and not routinely performed in healthy individuals, samples were obtained from individuals undergoing diagnostic evaluation for suspected BM metastasis of solid tumors. All selected individuals were ultimately diagnosed with localized disease and showed no evidence of BM involvement (Supplementary Table 7). Written informed consent was obtained from all donors or legal guardians. Approval was granted by the Institutional Review Board of the Princess Máxima Center (PMCLAB2022.0351).
For BM cytokine measurements, BM plasma from cancer-free BM aspirates (n = 78, ages 0–20 years, 47 male and 31 female donors; Supplementary Table 14) was acquired. All aspirates were derived from individuals diagnosed with localized solid tumors, including low-grade embryonal rhabdomyosarcoma, Ewing sarcoma, ganglioneuroblastoma, malignant rhabdoid tumor, neuroblastoma or small cell sarcoma, without any evidence for BM involvement. Informed consent was obtained from all donors or legal guardians. Approval was granted by the Institutional Review Board of the Princess Máxima Center (PMCLAB2025.0620).
Sample preparation for CITE-seq
Residual mononuclear cells from BM aspirates, leftover after graft infusion, were isolated by Ficoll (Cytiva Life Sciences) density gradient centrifugation and cryopreserved in liquid nitrogen until further use. For CITE-seq, cryopreserved BM samples were thawed rapidly in a water bath at 37 °C. An equal volume of prewarmed thawing medium (DMEM, high glucose, pyruvate, no glutamine and 20% fetal calf serum) was added to the viably frozen cell suspension in a dropwise fashion. The cell suspension was transferred to a 50-ml Falcon tube and was further diluted (1:10) by dropwise addition of prewarmed thawing medium. Cells were centrifuged at 400g for 5 min at 4 °C. The cell pellet was resuspended in thawing medium containing DNase (100 µg ml−1; Roche) with MgCl2 (10 mM; Merck) and incubated for 30 min at 4 °C. After incubation, cells were centrifuged at 400g for 5 min at 4 °C and resuspended in cell staining buffer (CSB; Biolegend). The number of cells in the suspension was established using the Countess II cell counter (Invitrogen).
Cell sorting and multiplexing for CITE-seq
Before library preparation, each individual’s sample was enriched for HSPC and MSC, which were multiplexed with the nonenriched cell fraction of another genetically distinct individual. For this, FcR blocking reagent (Human Trustain FcX, Biolegend) was added to the cells at a 1:10 dilution and incubated on ice for 5 min. Cells were then incubated with Zombie NIR viability dye (BioLegend) on ice in the dark for 15 min. After incubation, the cell suspension was washed by adding CSB and centrifuging at 400g at 4 °C, followed by resuspension. Next, the cells were incubated on ice in the dark for 30 min with a customized mix of fluorophore-conjugated antibodies and oligonucleotide-conjugated antibodies (Supplementary Tables 15 and 16). Following incubation, the cells were washed three times with CSB and centrifuged at 400g at 4 °C after each wash. The cell suspension was then filtered using a 35-µm filter and sorted using a Sony SH800S cell sorter (SONY SH800S system software v2.1), with a uniform gating strategy for all samples (Supplementary Fig. 1). In general, we sorted 2.5 × 104 nonenriched, erythrocyte precursor-depleted cells (Zombie NIR−CD235a−) from one individual and combined with 1 × 104 HSPC (Zombie NIR−CD235a−CD45−CD34+) and up to 2 × 103 MSC (Zombie NIR−CD235a−CD45loCD34−CD271+ or Zombie NIR−CD235a−CD45loCD34−CD90+) of another genetically distinct individual. During subsequent data analysis (described below), cells were demultiplexed and assigned to their original sample based on SNVs specific for each individual. After mixing, the resulting cell suspensions were counted using Trypan Blue and a Bürker counting chamber.
Library preparation and sequencing
Approximately 40,000 cells per multiplexed sample were loaded onto a Chromium Single Cell G chip and used for library preparation using a Chromium Next GEM Single Cell 3′ Library and Gel Bead Kit v3.1 (10x Genomics) according to the manufacturer’s instructions. For each multiplexed sample, two libraries were prepared, one for the RNA and one for the antibody capture modality. Each library was sequenced using a NovaSeq 6000 (Illumina) and the following number of cycles: read 1: 28; read i7: 10; read i5: 10; read 2: 91.
Xenium experiments
In collaboration with the Department of Pathology, representative regions from each BM biopsy (n = 6 biopsies, one to three regions per biopsy; Supplementary Table 7) were identified and incorporated into a tissue microarray (TMA), comprising one to three BM cores per individual (1.5 mm in diameter each). A 5-µm TMA section was prepared for spatial transcriptomics according to the Demonstrated Protocols Xenium In Situ for FFPE Tissue Preparation Guide (CG000578, 10x Genomics) and Xenium In Situ for FFPE Tissues Deparaffinization and Decrosslinking (CG000580, 10x Genomics). The Human Multi-Tissues and Cancer Panel probe set was supplemented with probes targeting 100 custom genes (Supplementary Table 8) and Cell Segmentation Reagents. Probe hybridization, ligation and rolling circle amplification were performed by the Leiden Genome Technology Center at the Leiden University Medical Center, following the manufacturer’s protocol (CG000582 Rev E, 10x Genomics).
CITE-seq data preprocessing
CITE seq data were processed using CellRanger count with feature barcoding (version 7.1.0, 10x Genomics) using the refdata-gex-GRCh38-2020-A transcriptome and a modified Feature Reference file (Supplementary Table 14).
CITE-seq genotype demultiplexing and barcode filtering
Cells from multiplexed samples were SNV-based genotype-demultiplexed using souporcell (singularity image created 1 December 2021)51. Barcodes with less than 1,500 transcripts and/or a percentage of mitochondrial genes above 10% were removed. Also, barcodes classified as doublets or unassigned genotypes by souporcell51 and barcodes classified as doublets in over five of ten runs using scDblFinder (version 1.18.0)52 were discarded.
CITE-seq normalization, dimensional reduction, feature deconfounding and integration
Further processing and analyses were performed in R (version 4.4.0) using Seurat (version 5.1.0)20. For each individual, donor gene expression data were SCTransform normalized with SCTransform (v2, number of variable features = 3,000). Dimensional reduction was conducted with the RunPCA function from Seurat. SCT integration features were calculated using the SelectSCTIntegrationFeatures function from Seurat. Using gene lists from the SCutils package (version 1.123), the following genes were filtered out from the SCT integration features, provided they were also found as variable features: genes specific to the S or G2M cell cycle phase, donor-specific genes correlating with the S or G2M phase, male and female-specific genes, stress-related genes and ribosomal protein genes. Gene expression data were integrated across donors with canonical correlation analysis (CCA), using the IntegrateLayers function in Seurat and the filtered SCT integration features. Antibody capture data were normalized per library using DSBantibody normalization (version 1.0.3)20. For each individual donor, FindVariableFeatures and ScaleData were run, and dimensional reduction was performed with the RunPCA function from Seurat. Data were also integrated with CCA using the IntegrateLayers function in Seurat and all antibodies as integration features.
CITE-seq visualization, clustering and cell-type annotation
A WNN graph was created with the FindMultiModalNeighbors function in Seurat, using the integrated reductions of both RNA and ADT modalities. A wnnUMAP was created from this WNN graph with RunUMAP, using 30 principal components for both modalities. Clustering was performed using FindClusters from Seurat with the wsnn graph, the SLM modularity optimization algorithm, as recommended by the Weighted Nearest Neighbor Analysis vignette from Seurat, and a resolution of 0.4.
Cell-type annotation was performed by combining three complimentary annotation approaches, performed with gene expression log-normalized data. First, cell types were inferred with SingleR (version 2.6.0)53. Second, individual cells were mapped to two external reference BM datasets11,13. Mapping to the CITE-seq dataset bmcite (version 0.3.0) from SeuratData (version 0.2.2.9001) was performed as recommended by the Seurat multimodal reference mapping vignette18. The Gene Expression BM dataset in DISCO19 was downsampled to a maximum of 3,000 cells per cell type and SCTransform normalized, followed by generation of a principal component analysis and UMAP, which were in turn used for reference mapping, according to the Seurat Mapping and annotating query datasets vignette. Third, antibody capture data and RNA expression data from known marker genes were used to confirm cell-type annotations.
For in-depth analysis of the erythroid lineage, myeloid lineage and T and NK cells, the respective clusters were subsetted, followed by subset-specific SCT normalization, dimensional reduction, feature deconfounding, integration and clustering at resolutions of 0.7, 0.5 and 0.3, respectively. Cell-type annotations were then redefined as described above. The resulting Seurat object provided cell-type annotations at multiple levels of resolution, ranging from broad categories (major groups, for example, T cells) to more detailed classifications (high-resolution clusters, for example, γδT cells).
CITE-seq subclustering of LyP cells
For in-depth analysis of LyP cells, cells were subsetted and analyzed as described above, with the following deviations: for clustering, a resolution of 0.1 was used; CCA integration between donors was not performed.
CITE-seq differential gene and protein expression
To compare each major group and high-resolution cell type against all other cell types, we used the wilcoxauc function in presto (version 1.0.0) to find cell-type-specific, significantly differentially expressed genes and proteins. We used the following thresholds: p-adj < 0.01, pct_in – pct_out ≥ 0, pct_in > 20, auc > 0.5 and logFC > 0. For each cell type, the FindMarkers function (test.use = ‘wilcox’, logfc.threshold = 0, min.pct = 0.01) was used to identify genes and proteins with significant differential expression between YP and AYA cells (thresholds: p-adj < 0.01 and FC > 1.5 and pct.1 > pct.2 or p-adj < 0.01 and FC < –1.5 and pct.1 < pct.2). From these cell-type-specific young and old gene lists, we computed module scores using Seurat’s AddModuleScore function and assessed scores per cell. Module scores were visualized as mean module scores per cluster using the ggradar package (version 0.2), capping negative average scores at 0. For LyP cell subclusters, the FindMarkers function (test.use = ‘wilcox’, logfc.threshold = 0, min.pct = 0.01) was used to identify significantly differentially expressed genes and proteins between clusters 0 and 1 (thresholds: p-adj < 0.01 and FC > 2 and pct.1 > pct.2 or p-adj < 0.01 and FC < –2 and pct.1 < pct.2). For all differential gene expression analyses, male- and female-specific genes were removed. For comparisons of major groups and high-resolution clusters, confounder genes mentioned above were also removed. Gene Ontology enrichment analysis was performed using the enrichGO function from the clusterProfiler (version 4.12.0) package, using biological process terms.
Transcription factor activity analysis in CITE-seq data
Inference of transcription factor network activity was performed using pySCENIC (image version aertslab-pyscenic-0.11.2.sif)54. For this, we used a loom file with raw transcript counts of a downsampled dataset as input, along with the ‘hs_hgnc_tfs.txt’ transcription factor list, the ‘motifs-v9-nr.hgnc-m0.001-o0.0.tbl’ motifs and the ‘hg38__refseq-r80__10kb_up_and_down_tss.mc9nr.feather’ input databases. Only transcription factors that were identified in two or more of three independent runs were analyzed. Activity per cell was calculated as the mean AUCell values across runs. For LyP cell subclusters, FindMarkers (test.use = ‘wilcox’, logfc.threshold = 0, min.pct = 0.01) was used to identify significant differential regulon activity between clusters 0 and 1 (thresholds: p-adj < 0.01 and FC > 1).
Cell interaction analysis in CITE-seq data
Interaction analysis and ligand prioritization were performed using NicheNet (nichenetr package, version 2.2.0)39. Ligand activity analysis was performed on genes upregulated in LyP-B or LyP-S cells (identified using the FindMarkers function as described above), using the top n = 5,000 downstream targets for each ligand and a quantile cutoff of 0.001. Ligands were prioritized by equally weighing the following two criteria: (1) the predicted ligand activity and (2) LyP subset-specific receptor expression.
Xenium data preprocessing, filtering, normalization and dimensional reduction
Imaging and signal decoding were performed using the Xenium on-board analysis pipeline (10x Genomics, version xenium-3.1.1.0). The Xenium ranger relabel function was applied after removing the FLT3LG feature, due to lack of specificity of this custom probe. Cell segmentation was performed using a probabilistic segmentation method (ProSeg) to infer refined cell boundaries55. Regions of interest corresponding to the individual cores were selected in Xenium Explorer (version 3). For the selected cells, metadata and gene expression profile matrices were imported into scanpy (version 1.11.3)56 for downstream analyses. Quality assessment matrices were performed on the cells, including normalizing gene counts to total cell volume. Cells with a sum log1 (P value)-scaled gene count greater than 3.5 were retained for downstream analyses. An autoencoder variational model, ResolVI (scvi-tools, version 1.3.3), was applied, treating all BM cores as one batch (model parameters: n_hidden = 32, n_latent = 10, n_layers = 2, dropout_rate = 0.05)57 to obtain a low-dimensional representation with ten dimensions. The latent representation was used for constructing a UMAP.
Xenium visualization, clustering and cell-type annotation
Leiden clustering (resolution 2.4, ten iterations) was performed based on the ResolVI latent space (resolution 2.4, ten iterations). For cell-type annotation, we used reference mapping with Tangram58 against the adult BM atlas as published by Bandyopadhyay et al.14 and Zeng et al.59 and CITE-seq data from this manuscript. Cells were then subdivided into major groups: HSPC, T/NK cells, megakaryocyte lineage, B cell lineage, myeloid lineage, erythroid lineage and nonhematopoietic clusters. These subsets were reclustered at resolutions of 0.3, 0.5 and 1.0. Final annotations were assigned by integrating reference-based predictions with known marker gene expression, yielding both broad and fine-grained cell-type labels.
Comparison of LyP cell subtypes in CITE-seq data and a public dataset of CD34+ cells
A public scRNA-seq dataset of CD34+ cells isolated from BM aspirates of 15 donors (ages 2–77 years; 8 male and 7 female donors)32 was downloaded from the Gene Expression Omnibus (GEO) under accession number GSE189161. Transcripts in all cells were log normalized per sample (orig.ident) to a total of 10,000 transcripts. LyP exhibiting LyP score >0 with more than 1,500 transcripts were retained for downstream analyses. Module scores were constructed from differentially expressed genes in the LyP, LyP-B and LyP-S populations from the CITE-seq data in this manuscript, excluding sex- and cell cycle-related genes. Relative cell fractions of cells with an LyP-B score of >0 and LyP-S score ≤0 (LyP-B); cells with a LyP-S score >0 and LyP-B score ≤0 (LyP-S) and cells with LyP-B score ≤0 and LyP-S score ≤0 (double-negative LyP) were compared.
Cytokine measurements in BM plasma
BM plasma (the cell-free fraction of BM aspirates) was collected by Ficoll (Cytiva Life Sciences) density gradient centrifugation of EDTA-anticoagulated BM aspirates (n = 78 samples; Supplementary Table 14). The resulting plasma was stored at –80 °C until further analysis. IL-7 and IL-15 were measured using the LEGENDplex Human Hematopoietic Stem Cell Panel (Biolegend). Samples were processed, and cytokine levels were measured according to the manufacturer’s instructions, with the sole deviation being an extended overnight incubation during the initial incubation step to optimize cytokine detection. Cytokine concentrations were analyzed in R and visualized using the ggplot2 (version 3.5.1) and ggbeeswarm (version 0.7.2) packages. Cytokine concentrations lower than the detection limit (IL-15, <271.56 pg ml−1; IL-7, <0.317 pg ml−1) were assigned half the value of the detection limit. Outliers were identified and removed.
Statistics and reproducibility
Dimensional reduction of BM composition data was performed using principal component analysis and the prcomp function from the stats package (version 4.4.0). The relative abundance of cell types, LyP-B and LyP-S cell subsets and cycling versus noncycling LyP between YP BM and AYA BM was compared using a two-sided Wilcoxon rank-sum test. Correlations between B cell and LyP-B cell frequencies were calculated using the stat_correlation function (method = ‘pearson’) from the ggpmisc package (version 0.6.0). To allow reliable comparisons of rare cell types (cycling versus noncycling LyP and MSC subsets), the enriched and nonenriched were pooled. For all other comparisons, the nonenriched fractions were used. For comparison of spatial cell-type composition, mixed-effects logistic regression models were used (lme4 package, version 1.1-35.5) with donor as a random effect to account for multiple cores per individual; P values were adjusted for multiple testing using the Benjamini–Hochberg false discovery rate method. YP and AYA sample cytokine concentrations were compared using a Wilcoxon rank-sum test. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications60.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All generated data and the corresponding analysis scripts have been deposited in Zenodo at https://doi.org/10.5281/zenodo.14168864 (ref. 21). The publicly available data used in the study are DISCO BM atlas19 (https://www.immunesinglecell.com/atlas/bone_marrow), the Stuart et al. dataset (GEO: GSE128639)18, the Li et al. dataset (GEO: GSE189161)32 and the Bandyopadhyay et al. dataset (GEO: GSE253355)14.
Code availability
No new code was generated in this study. All code used in the analysis scripts have been deposited in Zenodo at https://doi.org/10.5281/zenodo.14168864 (ref. 21).
References
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
Jaiswal, S. & Ebert, B. L. Clonal hematopoiesis in human aging and disease. Science 366, eaan4673 (2019).
Carpenter, R. S. & Maryanovich, M. Systemic and local regulation of hematopoietic homeostasis in health and disease. Nat. Cardiovasc. Res. 3, 651–665 (2024).
Pang, W. W. et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl Acad. Sci. USA 108, 20012–20017 (2011).
Ross, J. B. et al. Depleting myeloid-biased haematopoietic stem cells rejuvenates aged immunity. Nature 628, 162–170 (2024).
de Haan, G. & Lazare, S. S. Aging of hematopoietic stem cells. Blood 131, 479–487 (2018).
Su, T.-Y. et al. Aging is associated with functional and molecular changes in distinct hematopoietic stem cell subsets. Nat. Commun. 15, 7966 (2024).
Cain, T. L., Derecka, M. & McKinney-Freeman, S. The role of the haematopoietic stem cell niche in development and ageing. Nat. Rev. Mol. Cell Biol. 26, 32–50 (2024).
Weng, C. et al. Deciphering cell states and genealogies of human haematopoiesis. Nature 627, 389–398 (2024).
Hennrich, M. L. et al. Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline. Nat. Commun. 9, 4004 (2018).
Chen, C. et al. Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood 139, 2198–2211 (2022).
Jardine, L. et al. Blood and immune development in human fetal bone marrow and Down syndrome. Nature 598, 327–331 (2021).
Hay, S. B., Ferchen, K., Chetal, K., Grimes, H. L. & Salomonis, N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp. Hematol. 68, 51–61 (2018).
Bandyopadhyay, S. et al. Mapping the cellular biogeography of human bone marrow niches using single-cell transcriptomics and proteomic imaging. Cell 187, 3120–3140 (2024).
Tang, F. et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Triana, S. et al. Single-cell proteo-genomic reference maps of the hematopoietic system enable the purification and massive profiling of precisely defined cell states. Nat. Immunol. 22, 1577–1589 (2021).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Li, M. et al. DISCO: a database of deeply integrated human single-cell omics data. Nucleic Acids Res. 50, D596–D602 (2022).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Belderbos, M. E. Scripts and data supporting the manuscript ‘Single-cell multiomic atlas of healthy pediatric bone marrow reveals age-dependent differences in lineage differentiation driven by stromal signaling’. Zenodo https://doi.org/10.5281/zenodo.18018504 (2025).
Keita, S. et al. Distinct subsets of multi-lymphoid progenitors support ontogeny-related changes in human lymphopoiesis. Cell Rep. 42, 112618 (2023).
Caldwell, C. W., Poje, E. & Helikson, M. A. B-cell precursors in normal pediatric bone marrow. Am. J. Clin. Pathol. 95, 816–823 (1991).
de Almeida Chuffa, L. G. et al. Aging whole blood transcriptome reveals candidate genes for SARS-CoV-2-related vascular and immune alterations. J. Mol. Med. 100, 285–301 (2022).
Roisman, A. et al. A FLI1–KLF6 axis regulates aging in human hematopoietic stem and progenitor cells and normalization of KLF6 levels in aged cells leads to their rejuvenation. Blood 138, 19 (2021).
Mende, N. et al. Unique molecular and functional features of extramedullary hematopoietic stem and progenitor cell reservoirs in humans. Blood 139, 3387–3401 (2022).
Kim, Y. et al. Terminal deoxynucleotidyl transferase and CD84 identify human multi-potent lymphoid progenitors. Nat. Commun. 15, 5910 (2024).
Nutt, S. L., Heavey, B., Rolink, A. G. & Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor PAX5. Nature 402, 14–20 (1999).
Nutt, S. L. & Kee, B. L. The transcriptional regulation of B cell lineage commitment. Immunity 26, 715–725 (2007).
Lacorazza, H. D. et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9, 175–187 (2006).
Menéndez-Gutiérrez, M. P. et al. Retinoid X receptor promotes hematopoietic stem cell fitness and quiescence and preserves hematopoietic homeostasis. Blood 141, 592–608 (2023).
Li, H. et al. The dynamics of hematopoiesis over the human lifespan. Nat. Methods 22, 422–434 (2025).
Alhaj Hussen, K. et al. Molecular and functional characterization of lymphoid progenitor subsets reveals a bipartite architecture of human lymphopoiesis. Immunity 47, 680–696 (2017).
Laurenti, E. et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 14, 756–763 (2013).
Doulatov, S., Notta, F., Laurenti, E. & Dick, J. E. Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136 (2012).
Abbuehl, J.-P., Tatarova, Z., Held, W. & Huelsken, J. Long-term engraftment of primary bone marrow stromal cells repairs niche damage and improves hematopoietic stem cell transplantation. Cell Stem Cell 21, 241–255 (2017).
Ding, L. & Morrison, S. J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013).
Zehentmeier, S. & Pereira, J. P. Cell circuits and niches controlling B cell development. Immunol. Rev. 289, 142–157 (2019).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Andrew, D. & Aspinall, R. Age-associated thymic atrophy is linked to a decline in IL-7 production. Exp. Gerontol. 37, 455–463 (2002).
Fry, T. J. et al. A potential role for interleukin-7 in T-cell homeostasis. Blood 97, 2983–2990 (2001).
Alhaj Hussen, K. et al. Multimodal cartography of human lymphopoiesis reveals B and T/NK/ILC lineages are subjected to differential regulation. iScience 26, 107890 (2023).
Medina, K. L. et al. Identification of very early lymphoid precursors in bone marrow and their regulation by estrogen. Nat. Immunol. 2, 718–724 (2001).
Medina, K. L. & Kincade, P. W. Pregnancy-related steroids are potential negative regulators of B lymphopoiesis. Proc. Natl Acad. Sci. USA 91, 5382–5386 (1994).
Bosco, N., Ceredig, R. & Rolink, A. Transient decrease in interleukin-7 availability arrests B lymphopoiesis during pregnancy. Eur. J. Immunol. 38, 381–390 (2008).
Iacobucci, I. et al. Single cell dissection of developmental and transcriptional heterogeneity in B-cell acute lymphoblastic leukemia. Preprint at bioRxiv https://doi.org/10.1101/2023.12.04.569954 (2023).
Hong, D. et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science 319, 336–339 (2008).
Sanjuan-Pla, A. et al. Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia. Blood 126, 2676–2685 (2015).
Greaves, M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer 18, 471–484 (2018).
Geron, I. et al. An instructive role for interleukin-7 receptor α in the development of human B-cell precursor leukemia. Nat. Commun. 13, 659 (2022).
Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17, 615–620 (2020).
Germain, P.-L., Lun, A., Meixide, C. G., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979 (2022).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020).
Jones, D. C. et al. Cell simulation as cell segmentation. Nat. Methods 22, 1331–1342 (2025).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Ergen, C. & Yosef, N. ResolVI—addressing noise and bias in spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2025.01.20.634005 (2025).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Zeng, A. G. X. et al. Single-cell transcriptional mapping reveals gentic and non-genetic determinants of aberrant differentiation in AML. Preprint at bioRxiv https://doi.org/10.1101/2023.12.26.573390 (2023).
Ye, Q. et al. Single-cell multi-omic landscape reveals anatomical-specific immune features in adult and pediatric sepsis. Nat. Immunol. 27, 150–165 (2025).
Acknowledgements
We thank the study participants and their guardians for facilitating this research. We thank P. Lijnzaad (Princess Máxima Center) and J. De Martino (Princess Máxima Center) for direct technical assistance with single-cell data generation and analysis, L. Visser (Princess Máxima Center) for assistance in making the dataset publicly available, M. Bleijs (Princess Máxima Center) for proofreading and R. Moeniralam (Princess Máxima Center) for assistance in sample selection and tissue processing for spatial transcriptomics. We thank the Leiden Genome Technology Center for their support in performing the Xenium spatial transcriptomics experiments. This project was supported financially by the European Hematology Association (Physician Scientist Grant to M.E.B.), the Dutch Research Council’s Veni Grant (VI.Veni.202.021 to M.E.B.), the Landsteiner Foundation for Blood Transfusion Research (2305F to M.E.B.), the European Research Council (101114895, to M.E.B.) and the Horizon Europe/Marie Sklodowska-Curie Action co-fund project (101081481). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.E.B. and T.M. conceptualized the study. E.S.H. led most of the wet lab experiments, with assistance from N.E., A.K.B., P.F. and B.M.t.P. K.F.M. and I.J.K. analyzed the single-cell transcriptomic and proteomic data. E.S.H. and M.S.-V. generated the spatial transcriptomics data. I.J.K., A.K.S.J., L.-T.C. and M.v.d.M. analyzed the spatial transcriptomics data. P.F. and T.C. contributed to the computational analysis. K.F.M., E.S.H. and I.J.K. interpreted the data. P.F., W.J.d.J. and T.C. contributed to data interpretation and provided scientific advice. O.H. provided intellectual input. K.F.M. and E.S.H. wrote the manuscript, with special support from I.J.K. and input from all authors. M.E.B. generated funding. T.M. and M.E.B. supervised all aspects of the study, including wet lab experiments, data analysis, interpretation and writing of the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Peter van Galen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ioana Staicu, in collaboration with the Nature Immunology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quality control plots of CITE-seq data.
a) Bar plot showing the number of bone marrow (BM) cells collected from seven healthy pediatric donors aged 2.1-16.7 years (3 males, 4 females). From each sample, HSPC (CD235a−CD34+) and mesenchymal stromal cells (CD235a−CD45loCD90+ or CD235a−CD45loCD271+) were enriched by flow cytometry and combined with the non-enriched fraction from a genetically distinct donor. Pooled samples were subjected to CITE-seq using 138 oligonucleotide-conjugated antibodies, on the 10x Genomics platform. b) Boxplot of the percentage of mitochondrial reads per cell. c) Boxplot of the number of RNA counts per cell. d) Boxplot of the number of unique RNA features per cell. e) Boxplot of the number of ADT counts per cell. f) Boxplot of the number of unique ADT features per cell. g) Violin plot showing the relative expression level of the female-specific XIST gene per donor sample used to confirm sample sex. h) Violin plot showing the relative expression level of the male-specific UTY gene per donor sample used to confirm sample sex. i) CITE-seq-based weighted nearest neighbor uniform manifold approximation and projection (wwnUMAP) of cells from pediatric BM aspirates (n = 7) and adult BM aspirates (n = 2) per individual. Twenty-eight clusters were identified and annotated based on joint transcriptomic and protein expression patterns. MSC, mesenchymal stromal cell; HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte-erythroid progenitor; MkP, megakaryocyte progenitor; EryP, erythroid progenitor; McP, mast cell progenitor; TN, naive T cell, TM: memory T cell; Treg cell: regulatory T cell; MAIT cell: mucosal-associated invariant T cell; NK cell, natural killer cell; progDC, dendritic cell progenitor; cDC, conventional DC; pDC, plasmacytoid DC, Ery, erythroid; DC, dendritic cell.
Extended Data Fig. 2 External references validate cell type annotations and multimodal clustering.
a) Tile plots showing mapping of each of the 28 cell clusters in the CITE-seq dataset (n = 9 donors, age 2.1-31.3, 3 male, 5 female) to the external dataset of Stuart et al.18. Colors depict the proportion of cells in each cluster assigned to each reference cell type. b) As in a), but for the external DISCO single-cell reference map19. HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte-erythroid progenitor; MkP, megakaryocyte progenitor; EryP, erythroid progenitor; McP, mast cell progenitor; LyP, lymphoid progenitor; TN, naive T cell; TM, memory T cell; Treg cell, regulatory T cell; MAIT, mucosal-associated invariant T cell; NK, natural killer cell; prog DC, dendritic cell progenitor; cDC, conventional DC; pDC, plasmacytoid DC; MSC, mesenchymal stromal cells; LMPP, lymphoid-primed multipotent progenitor; Prog_Mk, megakaryocyte progenitor; Prog_RBC, red blood cell progenitor; GMP, granulocyte-monocyteprogenitor; Mono, monocyte; Prog_DC, dendritic cell progenitor; MPP, multipotent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; ILC, innate lymphoid cell; CDP,common dendritic cell progenitor; cDC1, conventional DC type 1; cDC2, conventional DC type 2; pre-pDC, precursor plasmacytoid DC.
Extended Data Fig. 3 Spatial transcriptomics validates age-dependent differences in BM composition between YP and AYA.
a) Representative BM cores from each individual showing the spatial distribution of major cell populations within the biopsy sections. Scale bar = 300 μm. b) Stacked bar plots showing the relative abundance of major BM cell populations within the biopsy sections. c) as in b), but at the subset level. Abbreviations: HSPC, hematopoietic stem and progenitor cell; NK cell, natural killer cell; MSC, mesenchymal stromal cell; SEC, sinusoidal endothelial cell; AEC/VSMC, arterial endothelial cell or vascular smooth muscle cell; EryP, erythroid progenitor cell; BaEoMaP, progenitors of basophils, eosinophils and mast cells; MkP/Mk, megakaryocyte progenitor cell or megakaryocyte; pDC, plasmacytoid dendritic cell; Adipo-MSC, adipolineage MSC; Fibro-Osteo-MSC, fibro-osteolineage MSC; Fibro-MSC, fibrolineage MSC; Osteo-MSC, osteolineage MSC; YP, young pediatric; AYA, adolescents and young adults.
Extended Data Fig. 4 Differentially expressed genes and cell surface proteins between YP and AYA per cell type and their overlap.
a) CITE-seq-based stacked bar plots showing the number of differentially expressed genes between YP (age 2.1-9.6 years, n = 4 donors, 2 males, 2 females) and AYA (age 13.3-16.7, n = 3 donors, 1 male, 2 female), categorized by the number of cell types in which they are identified. b) As in a), but for cell surface proteins. c) Bar plot showing the number of differentially expressed genes upregulated in YP and AYA per cell type. d) As in c), but for cell surface proteins. Abbreviations: YP, young pediatric; DE, differentially expressed; AYA, adolescents and young adults; HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte-erythroid progenitor; MkP, megakaryocyte progenitor; EryP, erythroid progenitor; McP, mast cell progenitor; LyP, lymphoid progenitor; TN, naive T cell; TM, memory T cell; Treg cell, regulatory T cell; MAIT cell, mucosal-associated invariant T cell; NK cell, natural killer cell; progDC, dendritic cell progenitor; cDC, conventional DC; pDC, plasmacytoid DC; Ery, erythroid cell; DC, dendritic cell; MSC, mesenchymal stromal cell.
Extended Data Fig. 5 Shared and differentially expressed genes between YP and AYA cell clusters.
a) Heatmap showing the overlap of differentially expressed genes between pairs of cell types from CITE-seq data of BM aspirates of YP donors (n = 4, age 2.1-9.6 years, 2 males, 2 females). b) As in a), but for AYA donors (n = 3, age 13.3-16.7, 1 male, 2 female). Abbreviations: YP, young pediatric; DE, differentially expressed; AYA, adolescent and young adults. HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte-erythroid progenitor; MkP, megakaryocyte progenitor; EryP, erythroid progenitor; McP, mast cell progenitor; LyP, lymphoid progenitor; TN, naive T cell; TM, memory T cell; Treg cell, regulatory T cell; MAIT cell, mucosal-associated invariant T cell; NK cell, natural killer cell; progDC, dendritic cell progenitor; cDC, conventional DC; pDC, plasmacytoid DC; Ery, erythroid cell; DC, dendritic cell; MSC, mesenchymal stromal cell.
Extended Data Fig. 6 B cell lineage bias in YP BM originates at the level of LyP.
Heatmap showing the relative expression of the top 10 differentially expressed genes identified in CITE-seq data across B cell populations, HSPC and myeloid cell populations (as defined in Fig. 1b), comparing young pediatric (YP; age 2.1–9.6 years; n = 4 donors, 2 male and 2 female) and adolescent and young adult (AYA; age 13.3–16.7 years; n = 3 donors, 1 male and 2 female) samples. Abbreviations: YP, young pediatric; AYA, adolescent and young adult; HSPC, hematopoietic stem and progenitor cells; HSC, hematopoietic stem cell; LMPP, lymphoid–myeloid primed progenitor; MEP, megakaryocyte–erythroid progenitor; McP, mast cell progenitor; LyP, lymphoid progenitor.YP, young pediatric; AYA, adolescents and young adults; HSPC, hematopoeitic stem and progenitor cells; HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte-erythroid progenitor; McP, mast cell progenitor; LyP, lymphoid progenitor.
Extended Data Fig. 7 Validation of LyP subcluster annotation and differential lineage bias.
a) CITE-seq based violin plots showing the relative expression levels of DSB-normalized ADT counts for shown surface markers in LMPP, LyP-S, LyP-B and pro B cells (cycling), as identified in Fig. 1b and Fig. 3a. b) Violin plots showing the relative expression of the top10 marker genes for lymphoid progenitor cells from the DISCO single-cell reference atlas19. c) Tile plot showing the percentage of cells for each LyP subcluster mapping to each of the cell clusters in the DISCO single-cell reference atlas19. d) CITE-seq based diffusion map, showing the differentiation trajectory from HSC to B cell and myeloid lineages, visualizing all the intermediary cell types. e) As in d) highlighting the Lyp-B and LyP-S subclusters. Abbreviations: LyP, lymphoid progenitor; pDC, plasmacytoid dendritic cell; GMP, granulocyte-monocyte progenitor; CDP, common dendritic cell progenitor; pre-pDC, precursor plamacytoid DC; CMP, common myeloid progenitor; HSC, hematopoietic stem cell; LMPP, lympho-myeloid primed progenitor.
Extended Data Fig. 8 External data validate of age-dependent shifts in LyP subset abundance.
a) Dot plot showing the consistency between the LyP-transcriptional signatures in our BM reference map (n = 9 donors) and those reported by Li et al. (n = 15, ages = 2-77 years22). b) Bar plot showing the frequencies of LyP-B and LyP-S subclusters by age in the reference map of Li et al. Cells were selected based on LyP-identity in both datasets (LyP score>0) and subclassified as LyP-B (blue) or LyP-S (orange) based on subset-specific scores. Cells positive for both were classified according to the highest score; cells that did not meet either criterion were labeled unclassified (grey). c, Boxplot showing LyP-B and LyP-S frequencies in male individuals stratified by age. Abbreviations: LyP: lymphoid progenitor; Prog: progenitor; Ly: lymphoid; G: granulocyte; Mk: megakaryocyte; E: erythroid; pDC: plasmacytoid dendritic cell; G/M: granulocyte/monocyte; MPP: multipotent progenitor; Mono/DC: monocyte/dendritic cell; Baso/Mast: basophil/mast cell.
Extended Data Fig. 9 Analysis of mesenchymal stromal cell subsets in the CITE-seq dataset.
a) Tile plot showing the percentage of mesenchymal stromal cells (MSC, n = 588 cells) in CITE-seq data of BM aspirates of YP and AYA donors (n = 9; 2.1-31.3 years, 3 males 6 females), mapping to the MSC clusters in an external reference (Bandyopadhyay et al. 14). Cell numbers are shown in brackets. b) Dot plot showing the mRNA expression of IL7 in the MSC in our CITE-seq dataset, annotated using the MSC cluster labels from Bandyopadhyay et al. Abbreviations: MSC, mesenchymal stromal cell.
Supplementary information
Supplementary Information
Table of Contents and Supplementary Figs. 1–4.
Supplementary Table
Supplementary Tables 1–16.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hanemaaijer, E.S., Müskens, K.F., Kal, I.J. et al. Single-cell multiomic atlas of healthy pediatric bone marrow reveals age-dependent differences in lineage differentiation driven by stromal signaling. Nat Immunol (2026). https://doi.org/10.1038/s41590-026-02422-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41590-026-02422-9