Abstract
Suppression of human megakaryocytes by dengue virus (DENV) infection significantly reduces the platelet count that eventually leads to thrombocytopenia, severe dengue and death. To understand DENV interactions with megakaryocytes, we investigated the cell cycle in leukemic human megakaryocytic in vitro cell line (MEG-01 cells). Megakaryocytes are known for complex endomitotic cell cycle leading to their polyploidy state. Our study shows that DENV uses these polyploid cells for its replication. Understanding the modulation of DENV-mediated cell cycle regulation in megakaryocytes is therefore highly important. We show that DENV2 (serotype 2) infection significantly modulates cell cycle signaling. Our protein profile microarray data showed significant upregulation of several cell cycle regulatory proteins including CDK4, CDK1, Cyclin B1 and others or downregulation of Chk1, GSK3-beta, CUL-3, and E2F-3. Quantitative real-time PCR and immunoblotting analyses further confirmed the upregulation of CDK4, CDK1, and Cyclin B1 upon DENV2 infection. Gene silencing of CDK4, CDK1 and Cyclin B1 showed significant reduction in DENV2 loads. Immunoprecipitation analysis further revealed an enhanced interaction between Cyclin B1 and CDK1 upon DENV2 infection that perhaps suggest the substantial changes noted in cell cycle regulation. Overall, our study suggests that DENV2 modulates cell cycle signaling in megakaryocytes and interferes with the critical regulatory proteins that may eventually lead to changes in endomitosis process. In conclusion, we report an important molecular insight regarding DENV2-mediated cell cycle modulation in human megakaryocytes.
Similar content being viewed by others
Introduction
Dengue viruses (DENV, existing as four serotypes; 1–4) are mosquito-borne Orthoflaviviruses that are endemic in many tropical and subtropical regions of the world1,2,3. Dengue fever (DF) is a major public health concern in many countries, especially in Southeast Asia, Pacific Islands, and the Americas4,5. It is a fast growing mosquito-borne disease around the world with an estimation of 390 million cases that are reported, annually5,6. Usually, outbreaks of DENV occurs globally with peaks of infections observed in every year7. For example, DENV2 outbreak may occur in some parts of South America and simultaneously another outbreak with DENV3 may happen in Asia. In recent years, 50–100 million symptomatic dengue infections have been annually reported with 500,000 hospitalizations and 22,000 deaths, thus suggesting dengue as a global problem. The Center for Disease Control and Prevention (CDC, USA) has closely reported the incidence of locally acquired DF in parts of Florida, Texas, and Hawaii, where the presence of Aedes mosquitoes has been continuously detected8. Very recent news from August 16, 2023, reported that DF in Broward County and Miami, FL, USA is on the rise and several cases have been reported9. Currently, there are no effective therapies/drugs, but two approved vaccines (Dengvaxia or Qdenga from Sanofi or Takeda) are available to treat dengue infections10,11,12. In some cases, reinfection with other serotypes of DENV eventually leads to the progression of (a) probable dengue, (b) dengue with warning signs, and (c) a potentially life-threatening complication called as severe dengue that is characterized by the symptoms along with signs of shock1,13,14,15. In 2016, the pan American Health Organization clinically classified these changes for symptomatic dengue infections. Severe dengue is accompanied by permeability of blood vessels and plasma leakage due to low platelet count that is termed as thrombocytopenia16,17.
Megakaryocytes are specialized bone marrow cells, primarily responsible for tailoring the biogenesis of thousands of platelets that are formed from megakaryocyte cytoplasmic extensions (called proplatelets) and released into the blood circulation18. Megakaryocytes are known for their complex endomitotic cell cycle process with partial mitosis that progress through normal prophase, pro-metaphase, metaphase, and up to anaphase A, but does not proceed or complete anaphase B, telophase, or cytokinesis. In endomitosis, megakaryocytes replicate DNA without undergoing late anaphase stage or cytokinesis, thus resulting in a high DNA content production of up to 256 N per cell19,20,21,22,23. Endoduplication process facilitates megakaryocytes to increase their DNA content and develop into large, polyploid cells with multiple nuclei that eventually leads to the platelets shedding and their release into the bloodstream19,22,24. The mechanism that regulates this endomitosis process (or typical cell cycle fate of megakaryocytes with polyploidy) is not fully understood. Several studies have reported that Cyclins and Cyclin dependent kinases (CDKs) plays crucial role(s) in this complex process22,23,25,26,27,28,29,30. Despite its importance, the knowledge of DENV-mediated modulation of cell cycle process for viral replication and propagation is critically important but largely remains unknown. Modulation of cell cycle regulatory proteins by DENV infection has been hypothesized and addressed in this current study as it may identify the important components of cell cycle that may interact with dengue and force the endomitotic megakaryocytes to undergo completion of mitosis. We hypothesize that DENV2 modulates MEG-01 cells cycle process to increase viral replication. Previously, it has been reported that modulation of polyploidy in megakaryocytes, such as inhibition of endomitosis results in reduced platelet production18. CDK1-Cyclin B1 complex, and M-phase promoting factor (MPF) plays critical role(s) in progression from G2 phase to M phase and phosphorylates various targets to promote the condensation of chromosomes and breakdown of nuclear envelope31,32,33,34,35. Recent work has suggested an involvement of CDK1 in endo-duplication process and suggested that genetic ablation of CDK1 prevents endomitosis but does not impair platelet formation, resulting in conversion from endomitosis to endocycles22. It has been noticed that changes in the expression of CDK1/Cyclin B1 leads to lack of H1 Histone kinase activity, thereby, leading to mitotic arrest in polyploid cells36. Moreover, significant reduction in CDK1 along with sustained expression of Cyclin B1 has been shown to lead alteration of CDK1/Cyclin B1 complex, thereby, resulting in mitotic arrest of polyploid cells36. Activated CDK1-Cyclin B1 enters into the nucleus, and phosphorylates plethora of substrates both in nuclear and cytoplasmic regions, thus resulting in substantial reorganization of the cell architecture upon which mitosis and cytokinesis depends34. In contrast, another study has suggested that endomitosis is promoted by downregulation of CDK1/Cyclin B1 activity23. In addition, CDK4 protein plays crucial role in regulation of G1/S phase progression along with its partner Cyclin D, where it phosphorylates Rb, thereby causing the release of the E2-F transcription factors37,38,39,40. Furthermore, CDK4 in association with other proteins facilitates the formation of mitotic spindle, which is essential for proper chromosome segregation during mitosis41,42. However, the role of CDK4 behind the cell cycle progression in endomitotic cells is not well studied as compared to CDK1 and Cyclin B1. In this study, we tested the hypothesis that DENV2 modulates the endomitotic cell cycle in human megakaryocytes by screening cell cycle regulatory proteins (in a protein profile microarray). We have performed interactome study with all the upregulated proteins to find out strong interactive proteins such as CDK4, CDK1 and Cyclin B1. We have re-validated their expression at both transcript and proteins levels and silenced these genes to find significant reduction in DENV2 loads. Our study suggests that DENV2 modulates cell cycle regulatory proteins in human megakaryocytes that eventually may lead to changes in endomitosis process.
Materials and methods
Culturing of human MEG-01 cells and dengue infection
Human megakaryocytes in vitro cell line (MEG-01 cells derived from bone marrow of a leukemic patient, catalog number CRL-2021) or human endothelial cells (EA.hy926, catalog number CRL-2922) were obtained from American Type Culture Collection (ATCC). MEG-01 or EA.hy926 cells were cultured in complete RPMI 1640 (Roswell Park Memorial Institute) or DMEM (Dulbecco’s Modified Eagle’s Medium) media (obtained from Thermo/Fisher Scientific, Inc), supplemented with 10% heat-inactivated fetal bovine serum (FBS from VWR), 1% Penicillin/Streptomycin and Glutamine mixture (SIGMA). MEG-01 and EA.hy926 cells were maintained in an incubator set at 5% CO2 and 37 °C. Dengue virus (DENV, serotype 2, New Guinea C strain) obtained from BEI resources was propagated in Aedes albopictus C6/36 cells (an in vitro cell line) maintained in an incubator set at 5% CO2 and 28 °C and as previously described43,44. C6/36 cells (obtained from ATCC, catalog number CRL 1660) were grown in EMEM (Eagle’s Minimum Essential Medium) complete media supplemented with 10% heat inactivated FBS, 1% Penicillin, Streptomycin and Glutamine solution and Amphotericin B mixture (SIGMA) and maintained according to the ATCC’s guideline(s). After 14 days post DENV2 infection, C6/36 culture supernatants were harvested by centrifugation at 1200 rpm (to pellet cell debris), and clear supernatants were aliquoted and titrated as laboratory virus stocks (with titers of 1.05 × 1e8 pfu/ml) and stored at -80 °C freezer. Laboratory virus titers were determined by end-point viral dilution assay as shown in methods and as described before44,45. To validate the modulation of cell-cycle regulatory molecules (for gene expression and immunoblotting analysis), we plated 5 × 1e5 MEG-01/EA.hy926 cells (per well of a 12 well plate) and infected with DENV2 (at multiplication of infection MOI 5, and as six replicates with additional 6 wells kept as uninfected controls). MEG-01/EA.hy926 cells were collected at day 3 (for microarray analysis or validation) or at days 1, 3 and 5 post DENV2 infection in RLT/RIPA lysis buffers for either RNA or protein extractions, respectively.
Protein microarray, imaging, scoring and data analysis
To investigate, if DENV2-mediates modulation of cell cycle regulatory proteins in human MEG-01 cells, we obtained cell cycle antibody array kit, a high-throughput ELISA based assay kit (catalog number ACC058, 2 array slides were provided per package for two independent samples) and an antibody array assay kit (catalog number KAS02, for 2 reactions, 64 antibodies; and 6 replicates per antibody) from Full Moon BioSystems (Sunnyvale, CA, USA). Each microarray slide had 6 spots coated with one antibody that will detect one protein (as 6 replicates for detection). The cell cycle antibody array features 60 highly specific proteins in cell cycle pathway and 62 well-characterized (by the Company) antibodies (6 replicates for each antibody) to quantitate/semi-quantitate protein expression profiles for human sampling (cell/tissue lysates, serum, cell culture supernatants). For more details, please see the company’s website at https://www.fullmoonbio.com/product/cell-cycle-antibody-array/. Following the instructions and Company’s protocol, we plated 3.5 × 1e5 MEG-01 cells in 6 well plates. After overnight incubation in complete media, cells were either kept as uninfected controls or infected with DENV2 (MOI of 5, at 72 h post infection, p.i.). Protein cell lysates were prepared from both uninfected or DENV2-infected groups in extraction buffer with addition of lysis beads (both extraction buffer and beads were provided in kit). After centrifugation, beads were removed, and clear supernatants were processed for purification using gel matrix columns (provided in kit). Purified protein samples were exchanged with labelling buffer (kit). Total proteins were estimated by BCA measurements (Pierce™ BCA Protein Assay Kit, Thermo/Fisher Scientific, Waltham, MA, USA). After determining the protein amounts, samples were biotinylated as per the Company’s instructions. Both labeled samples (UI/DENV2-infected) were added on to the pre-blocked antibody coated slides, respectively, followed by incubation on an orbital shaker for 2 h at 60 rpm. After incubation, Cy3-Streptavidin containing detection mixtures were added on to the slides and incubated for 20 min on a shaker at 60 rpm. All steps of the protein microarray were performed at room temperature. After several rounds of washes, both slides were shipped to the Company for image acquisition using GenePix Microarray scanner (Molecular Devices, San Jose, CA). Figure 1 shows a schematic representation on how protein microarray assay was performed. Array scanned images were analyzed using GenePix Pro7 software (Molecular Devices, San Jose, CA) installed in our laboratory computer. Briefly, array blocks were first created on GenePix Pro7 software, followed by alignment of spots. After gridding for each dot/well on the microarray, total intensity of each spot was obtained and subtracted with the empty spots. The microarray slides contain the spots for positive, empty, and negative in each block as controls. On each array slides, positives were spotted on each side of the block. Supplementary Fig. 1 shows the layout of antibodies (spotted as 6 replicates for each antibody) on the slides for each cell cycle regulatory protein (shown in column wise). Antibodies spotted for each protein are also shown according to the blocks (1–4). Spot intensities were considered to score the expression levels of each protein. Scoring was performed by arranging all proteins in order of highly upregulated or downregulated pattern and were evaluated based on the comparison between uninfected control or DENV2-infected groups as fold changes of total intensity values.
Schematic representation of protein microarray and data analysis. MEG-01 cell lysates collected from uninfected or DENV2-infected (MOI of 5, at 72 h p.i.) groups were mixed with lysis beads in extraction buffer. Following centrifugation step, beads were removed, and clear supernatants were collected and purified. Protein lysates were estimated for amounts by BCA assay and by using the Cytation 7 reader. Purified protein samples were biotinylated and added onto pre-blocked antibody-coated microarray slides (with 6 replicates for each antibody, and 64 spots in total). After coupling reaction, Cy3-tagged streptavidin was added onto the microarray slide to perform the hybridization reaction. After several rounds of washing, images were acquired using a GenePix microarray scanner. The scanned images were analyzed using GenePix Pro software from Full Moon Bio-Systems. The schematics is created in Bio Render.com.
End-point viral dilution assay
We performed the viral dilution assay as described44,45 to determine the tissue culture infectious dose (TCID50) as virus titer. Briefly, MEG-01 cells were seeded (at densities of 1 × 1e4 cells/well of a 96 well plate) in 225 µl of RPMI complete medium. After overnight incubation, MEG-01 cells were infected with DENV2 (laboratory viral stocks with known titer of 1.05 × 1e8 pfu/ml, which is serially diluted from 10–1 to 10–5) and incubated for additional 3 days. For each dilution group, at least 6 independent replicates were included in addition to the uninfected group (that is incubated with both primary and secondary antibodies) or DENV2-infected control group (treated with only secondary antibody) that served as internal controls. Images from dilutions of 10–2 to 10–5 are shown. MEG-01 cells were fixed with acetone-PBS mixture (in 3:1 ratio, for 20 min at − 20 °C) and plates were air dried, washed with 1 × PBS and blocked with 5% FBS and PBS-solution with 0.05% Sodium Azide for 15 min at RT. DENV2-Envelope (E)-protein was detected by incubation (overnight at 4 °C) with primary antibody (catalog number GTX127277, obtained from GeneTex, Inc), followed by three washes with 1 × PBS. Samples were incubated with Alexa-594 labeled rabbit secondary antibody (obtained from Thermo Scientific, Inc) for 1 h, at RT, followed by washes (3X) with 1 × PBS. After secondary antibody incubation, MEG-01 cells were counter-stained with DAPI (0.5 µg/ml for 3 min, obtained from Thermo Scientific, Inc). Plates were analyzed using Cytation7 imaging system (BioTek/Agilent). MEG-01 cells were observed for fluorescence or presence of infection in comparison to the uninfected control group. Representative images from dilutions of 10–2–10–5 (at day 3 p.i.) are shown. Images are obtained at 20 × magnification. Scale bar (200 µm) is shown on each representative image from the respective groups.
RNA extraction, cDNA synthesis and quantitative real-time PCR analysis
Total RNA isolations from MEG-01 cells or EA.hy926 endothelial cells were performed using Aurum Total RNA Mini kit (Bio-Rad) and following the manufacturer’s instructions. Briefly, 5 × 1e5 cells/well were seeded in 12 well plates followed by infection with DENV2 (MOI of 5, for 3 days p.i. or for days 1, 3 and 5 p.i.) and as five independent replicates. After infection, samples were collected in RNA lysis buffer and processed for RNA extractions. DNaseI was added on to the extraction columns during the processing of total RNA extractions. Total RNA from both groups were converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). The PCR cycle for cDNA synthesis included priming for 5 min at 25 °C, reverse transcription reaction for 20 min at 46 °C, inactivation of reverse transcription reaction at 95 °C for 1 min, and end of the cycle held at 4 °C. Quantitative-real-Time PCR (QRT-PCR) was performed using either CFX-96 or CFX-Opus system and iQ-SYBR Green Supermix (Bio-Rad). Briefly, cDNA is used as templates to determine the expression of CDK4, CDK1, Cyclin B1, other cell cycle molecules (shown in independent study with days 1, 3 and 5 p.i.) and human beta actin, respectively. To determine DENV2 viral loads, capsid transcript levels were analyzed. Following oligonucleotide sequences used to amplify the respective genes are from published studies43,44,46,47,48,49,50,51,52,53,54. For convenience, all published oligonucleotides used in this study are shown in Supplementary Table. To generate the data using initial quantity of amplified gene, a standard curve was generated using 1 to 10 ng of known quantities of respective genes by doing tenfold serial dilution, and as described previously44. To normalize templates, human beta actin or total RNA was quantified or value of equal amount of total RNA of respective genes were used, and as described previously43,45. Following protocol was used for QRT-PCR reaction: first denaturation at 95 °C for 3 min, followed by 40 cycles of steps including denaturation at 95 °C for 10 s, annealing at 58 °C for 10 s and extension at 72 °C for 30 s. Melting curve temperature starts from 65 to 95 °C with 0.5 °C increase at every 5 s intervals.
Immunoblotting analysis and immunoprecipitation assay
Western blotting analysis was performed by following the published protocols43,44,45,55. Briefly, 5 × 1e6 cells were seeded in 30 mm dishes for overnight, followed by infection with DENV2 (MOI of 5, for 3 days p.i.). Total protein lysates were collected in RIPA lysis buffer containing complete protease inhibitor cocktail (from Sigma, catalog number 11697498001). Protein concentrations were estimated by BCA assay using Pierce™ protein assay kit (Thermo/Fisher Scientific). Whole MEG-01 cell lysates (30 µg) were separated on to 12% SDS-PAGE gels. To visualize the total protein profiles, 2,2,2-Trichloroethanol (TCE) was incorporated into gel mixture during preparation of SDS-PAGE. After gel electrophoresis, total proteins were transferred on to nitrocellulose membranes by wet blotting for overnight at 4 °C. Total protein transferring was analyzed by staining the blot with Ponceau S solution. Blots were blocked in 5% skimmed milk or BSA (Sigma-Aldrich) and in 1X TBST buffer for overnight at 4 °C. After blocking, blots were probed with respective primary antibodies for overnight at 4 °C. Rabbit monoclonal/polyclonal antibodies for CDK4 (catalog number A16813), Phospho CDK4 (catalog number AP0593), CDK1 (catalog number A11420), and Cyclin B1 (catalog number A16038) in 1:1000/2500 dilutions were used (antibodies were obtained from Abclonal Biotechnology Company). DENV2 Envelope (E)-protein was detected with GeneTex, Rabbit pAb (catalog number GTX127277) in 1: 2000 dilution. Actin was detected with monoclonal antibody (catalog number ACTBD11B7 obtained from Santa Cruz Biotechnologies) and at 1:1000 dilution. After washing (3X) with 1 × TBST, blots were incubated with HRP-conjugated mouse or rabbit secondary antibodies, respectively, and at 1:5000 dilutions (at RT for 1 h). For validation of cell cycle proteins, densitometry analysis is performed for the immunoblots. Densitometry was performed using ImageJ to quantify the band intensities from Western blot images. Original blot images were saved as high-resolution, non-compressed TIFF files that were converted to 8-bit grayscale in ImageJ. The “Rectangular Selection” tool was used to draw a box around each band of interest, ensuring that same size box was used for all bands within a blot. The “Measure” function under the Analyze menu was used to obtain the raw integrated density (area × mean gray value) for each band. A background region (without bands) was also selected near each lane and its intensity was measured in the same way. The background intensity was subtracted from each band’s raw integrated density to obtain the corrected signal.
To determine Cyclin B1-CDK1 interaction upon DENV2 infection, we performed immunoprecipitation assay as previously described44. Briefly, 350 µg of total protein lysates from either uninfected or DENV2-infected (MOI of 5, for 3 days p.i.) groups were pre-cleared using protein A/G agarose beads (Pierce, Thermo/Fisher Scientific). After spin (at 13 k rpm), protein samples were collected (beads are discarded) and antigens were captured with 5 µg of CDK1 antibody (by overnight incubation on orbital shaker at 4 °C). CDK1 protein complexes were captured by incubation with fresh beads (for 4 h and at 4 °C and by incubation on shaker). Samples were washed (for 3 times) with 1 × cold PBS and collected in SDS-Laemmeli buffer (31.5 mM Tris–HCl, pH 6.8, 10% glycerol, 1% SDS, 0.005% Bromophenol Blue). Samples were boiled, cooled on ice and supernatants were loaded on to 12% SDS-PAGE gel (prepared as described above). Western blotting was performed (as described above) by using Cyclin B1 primary antibody (at 1:2500 dilution), followed by HRP-conjugated rabbit secondary antibody, respectively, and at 1:5000 dilutions. Western blots were developed by incubating with ECL reagents (Clarity™ Western ECL Substrate Kit). Chemidoc MP imaging system was used for documentation, and blots were analyzed with ImageLab software, and according to the manufacturer’s (BioRad) recommendations.
siRNA-mediated gene silencing
CDK4, CDK1 and Cyclin B1 gene silencing was performed by using siRNAs (obtained from Santa Cruz Biotechnologies, Inc.) and by following the manufacturer’s instructions and as described in our previous publication55. Briefly, 2 × 1e5 cells were seeded in 12 well plates, followed by transfection with respective siRNAs along with independent scramble siRNA controls. MEG-01 cells were transfected with 30 pmol of siRNA per well for silencing of CDK4, CDK1 and Cyclin B1 and as six independent replicates (for both silenced and scrambled groups), respectively. Following 24 h post transfection, cells were infected with DENV2 (MOI of 5, for 48 h p.i.). MEG-01 cells were collected for either RNA/protein extractions to determine silencing efficiency of respective genes/proteins and viral loads by QRT-PCR or immunoblotting analyses.
Immunofluorescence assay
To determine the co-localization of CDK4, CDK1 and Cyclin B1 proteins with DENV2 E-protein, immunofluorescence assays were performed as previously described44. Briefly, 2 × 1e5 MEG-01 cells were plated on to coverslips (kept in 12 well plates) and allowed to adhere for overnight. Following infection with DENV2 (MOI of 5, for 3 days p.i.), cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X-100 (diluted in 1 × PBS) and blocked (with 3% BSA in 1 × PBS). After blocking, cells were independently stained with anti-CDK4, CDK1 or Cyclin B1, respective antibodies (obtained from Abclonal Biotechnology Company) in 1:250 dilution, followed by detection with respective Alexa 488 (green) secondary antibody (Molecular Probes/ThermoScientific). Subsequently, MEG-01 cells were probed with DENV2-E-protein antibody 4G2, followed by detection with Alexa-Fluor 594 (red) secondary antibody, respectively. Cell nuclei were stained with DAPI to confirm cell viability. All phase contrast or fluorescent images were taken by bright field, DAPI, Texas red or GFP channels at 20X magnification and by using the Cytation 7 imaging system (BioTek, Agilent).
Statistical analysis
All the comparative results in our data sets were analyzed by GraphPad Prism6 software and Microsoft excel. For comparing two means unpaired, two-tail student’s t-test were performed for the entire analysis. For parametric tests, including the student’s t-test, we considered normal distribution of data. To account for potential deviations from normality and unequal variance, we also performed post hoc variance correction tests such as Welch’s correction if unequal variance is detected between groups. Error bars represent mean (+ SD) values. All P values of < 0.05 were considered for significant differences.
Results
Cell cycle regulatory proteins were selectively modulated upon DENV2 infection in human MEG-01 cells
To determine the modulation of cell cycle regulatory proteins upon DENV2 infection, a protein microarray analysis was performed. Detailed schematic of the protein microarray profiling is shown (Fig. 1). For protein microarray, human MEG-01 cell lysates infected with DENV2 (MOI of 5, at 72 h p.i.) or uninfected controls were processed as per the manufacturer’s instructions. Using GenePix microarray scanner, images were collected with same intensity and are shown (Fig. 2). The images were imported into GenePix Pro software to analyze the data. For protein identification in each block the layout of the microarray slides is shown (Supplementary Fig. 1). The array layout shows all the antibodies coated (as 6 replicates) to detect the proteins in each block (Supplementary Fig. 1). The antibodies are numbered as 1–64, whereas positive marker is indicated as P, empty wells are denoted as E and negative controls on each block are referred as N (Supplementary Fig. 1). Across all four blocks of both microarray slides (used for uninfected or DENV2-infected), all positive controls exhibited similar levels of expression (Fig. 2). The empty wells in both slides showed no expression whereas the negative control had no detectable spots (Fig. 2). We found that upon DENV2 infection the intensity of spot detection on the microarray slides was highest for these following proteins- CDK4, CDK1, Cyclin B1, CDK8, 14.3.3. Protein and Nuclear Mitotic Apparatus protein (NuMA) in comparison to the uninfected control group (Fig. 2). Some proteins also appeared as downregulated upon DENV2 infection (Fig. 2). However, many proteins were barely detected in both uninfected and DENV2-infected groups (Fig. 2). Following the manual analysis by visualization from the microarray images, we normalized the total intensity of each protein signal with the intensity observed for actin protein (Supplementary Fig. 2A–D). We noted that CDK4, CDK1, Cyclin B1 and NuMA were highly upregulated in our analysis of normalization to the intensity of actin protein from the respective slide (Supplementary Fig. 2A–D). The intensity of dots for all the proteins that had antibodies coated on the microarray slides is shown (Supplementary Fig. 2A–D). The empty well values were substracted and then normalized to actin protein intensity values obtained from the dots. The negative controls were not considered for measurement of total intensity as there was no detectable signal in those groups. Only one positive control is considered to represent the intensity of reference protein. The brightness of each protein from the microarray data (uninfected vs infected) is evaluated in these graphs and compared as up- or downregulated proteins between the uninfected or DENV2-infected groups (Supplementary Fig. 2A–D). These analyses further confirmed the expression data from microarray images (Fig. 2). Some of the other significantly upregulated proteins upon DENV2 infection were CDK8, CDK3, Cyclins C/E, CDC37, p18INK4c, p19Skp1, p130cas, and 14.3.3 Pan (Supplementary Fig. 2A,C,D). In addition to upregulated proteins, the normalization analyses revealed that Chk1, Cul-3, E2F-3 and GSK3-beta were significantly downregulated upon DENV2 infection (Supplementary Fig. 2B,D).
Microarray scanned images showing highlighted proteins in uninfected and DENV2-infected groups. Microarray scanned images from uninfected (A) or DENV2-infected (MOI of 5, at 72 h p.i.) (B) groups are shown for comparison. The microarray slides had 4 blocks (blocks 1–4), and each array slide had two positive controls as duplicate and, on either side (left- and right-hand) of the slide. The empty spots and negative controls were presented on the left side of the slides. Both (A) uninfected and (B) DENV2-infected scanned images show equivalence intensity for positive marker protein signal. Negative and empty wells show no signal. Vertical boxes represent proteins with enhanced signal in DENV2-infected group in comparison to the uninfected control group.
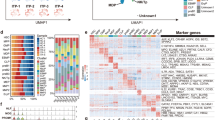
Furthermore, the heatmap dataset generated for the microarray analyses effectively visualized the differential expression patterns of the entire panel of 64 cell cycle regulatory proteins (Fig. 3A). The normalized intensity values for each gene were plotted on the Hemi2.0 (http://hemi.biocuckoo.cn:81/) platform to create a heatmap (Fig. 3A). Deeper/darker color on the heatmap revealed higher expression of those proteins. The expression patterns of these proteins were classified into distinct subcategories based on their Z scores, ranging from 0 to 1.5 (Fig. 3A). In addition, five sets of colors were used to distinguish the differences at minute level. The first bar (in dark blue) on the heatmap, without any protein name, is automatically generated by the software with the highest possible value to serve as a reference for comparing the intensity of protein expression (Fig. 3A). Minute color discrimination was observed in the positive control (from five color heatmap) but not in beta actin protein expression (Fig. 3A). Although, the positive control shows minute differences with five color heatmap, these differences are not significant, as confirmed by the statistical analysis with six replicates for each protein or the positive control (Fig. 3A). Also, by using four/three color heatmaps, there are no color discrepancies in the color intensity of positive control between the uninfected and infected groups (Supplementary Fig. 3). The discernable difference in color expression between uninfected and infected is as per the expectation in four/three colors generated heatmaps (Supplementary Fig. 3). To further analyze the distribution of significantly upregulated (Fig. 3B) or downregulated (Fig. 3C) proteins throughout the cell cycle stages upon DENV2 infection, the total intensity for each upregulated protein was normalized to the background value of the empty well. In this dataset, a total of 25 proteins exhibited altered expression levels upon DENV2 infection. Specifically, 17 proteins showed significant upregulation, while 7 proteins exhibited downregulation upon DENV2 infection (Fig. 3B and C). Among all upregulated proteins in the G1 phase, CDK4 exhibited the highest level of expression upon DENV2 infection when compared to CDK8, Cyclin E, p18INK4c, and CDK3 proteins (Fig. 3B). Likewise, 14.3.3 Pan protein showed the highest upregulation among S phase-specific induced proteins including p18INK4c, CDC-37, CDC-47, and CDC-34 (Fig. 3B). Similarly, CDK1 and Cyclin B1 proteins exhibited the highest level of upregulation when compared to other G2/S phase-specific induced proteins such as CDC-34, CDC-37, NuMA and p19Skp-1 proteins (Fig. 3B). Several other proteins showed significant downregulation upon DENV2 infection, and these included E2F-3, Chk1, GSK3-beta and CUL-3 (Fig. 3C). Based on independent intensity scoring, we found that most proteins were upregulated when compared to the number of proteins that were downregulated upon DENV2 infection (Supplementary Table 1). These data show that DENV2 infection differentially modulates the cell cycle proteins.
Cell cycle regulatory proteins are mostly upregulated upon DENV2 infection. Heatmap (A) of all modulated proteins from DENV2-infected (top row) and uninfected (bottom row) microarray slides is shown in five colors. Heatmap was generated based on the normalization to the empty well from each block. The color intensity for each color pattern is based on the scale shown at left of the heatmap that indicates lower to higher expression. Positive marker and beta actin protein expression shows no significant difference between uninfected and DENV2-infected groups. Panel (B) shows all upregulated proteins and panel (C) shows all downregulated proteins upon DENV2 infection (collected from the microarray data). Bar graphs are created by normalizing each protein intensity to total intensity of actin. All upregulated and downregulated proteins were categorized based on their cell cycle phases (G1, S, G2, and M) and their specific relative activity during cell cycle progression. The asterisk shows significance, and *, ** or *** indicates, P value from non-paired two tail student’s t-test, shown as less than 0.05, 0.01, or 0.0001, respectively. Not significant values are denoted with ns.
Independent screening of cell cycles regulatory genes further revealed differential modulation of gene expression upon DENV2 infection
To understand the importance of other cell cycle regulatory molecules that were not presented in the microarray slides, we performed an independent screen for these random molecules that also included CDK4, CDK1 and Cyclin B1. We randomly selected cell cycle genes from literature and analyzed their gene expression upon different days (1, 3 and 5, infection kinetics) post DENV2 infection. This data is considered as an independent screen and several genes analyzed here are not a part of the microarray list (as per the company’s layout- please see Supplementary Fig. 1). All published oligonucleotides used in this study are shown (Supplementary Table 2). MEG-01 cells were tested at three different time points of days 1, 3 and 5 post DENV2 (with MOI of 5) infection. QRT-PCR analysis revealed that DENV2 loads increased over the time course of infection from days 1, 3 and 5 p.i. (Fig. 4A). Also, the transcript levels of CDK1 (Fig. 4B), CDK2 (Fig. 4C), CDK4 (Fig. 4D), CDK5 (Fig. 4E), CDK6 (Fig. 4F), and CDK8 (Fig. 4G) were significantly (P < 0.05) upregulated upon DENV2 infection. All CDKs (1, 2, 5, 6 and 8) showed upregulation at all tested time points of days 1, 3 and 5 p.i. (Fig. 4B–G). However, only CDK4 showed significant (P < 0.05) upregulation at days 1 and 3 but no significant (P > 0.05) differences were noted at day 5 post DENV2 infection when compared to their respective uninfected control groups (Fig. 4D). Furthermore, we found that Cyclin A1 (Fig. 4H), Cyclin B1 (Fig. 4I), Cyclin E (Fig. 4J), and Cyclin G1 (Fig. 4K) were significantly (P < 0.05) upregulated upon DENV2 infection. Cyclins (A1, E and G1) were significantly (P < 0.05) upregulated at all tested time points of days 1, 3 and 5 p.i. (Fig. 4H, J and K). However, Cyclin B1 was upregulated at only days 1 and 3 but showed no significant (P > 0.05) differences at day 5 post DENV2 infection when compared to their respective uninfected control groups (Fig. 4I). Only Cyclin D3 (Fig. 4L) showed no significant differences. Lastly, testing of CDC2 (Fig. 4M) and CDC25A (Fig. 4N) showed upregulated transcript levels upon DENV2 infection and at all tested time points of days 1, 3 and 5 p.i., when compared to their respective uninfected control groups. These findings further supported the heatmap analyses that indicated many of the cell cycle regulatory molecules are upregulated upon DENV2 infection.
DENV2 upregulates several cells cycle regulatory gene transcripts in a time course kinetics. QRT-PCR analysis showing DENV2 capsid transcript levels (A), or CDK1 (B), CDK2 (C), CDK4 (D), CDK5 (E), CDK6 (F), CDK8, (G), Cyclin A1 (H), Cyclin B1 (I), Cyclin E (J), Cyclin G1 (K), Cyclin D3 (L), CDC2 (M) or CDC25A (N) gene expression at days 1, 3 and 5 post DENV2-infection. White bars represent uninfected group and black bars denotes DENV2-infected group. QRT-PCR data is shown as independent analysis for several cell cycle genes either present or absent in the microarray analysis. DENV2-loads and mRNA expression levels of cell cycle regulatory genes were normalized to human beta-actin mRNA levels. The asterisk shows significance, and *, or ** indicates, P value from non-paired two tail student’s t-test, shown as less than 0.05, or 0.01, respectively. Not significant values are denoted with ns.
Interactomes of cell cycle regulatory proteins upon DENV2 infection
To elucidate the influence of DENV2 infection on the modulation of cell cycle regulatory protein expression patterns across distinct phases of the cell cycle (G1, S, G2, and M), a Venn diagram (Supplementary Fig. 4A) was generated by representing binary values: 0 for upregulation, 1 for downregulation, and 2 for no significant difference in expression, corresponding to the set of 64 cell cycle phase-specific proteins (from the microarray slides), using Divenn 2.0 (https://divenn.tch.harvard.edu/v2/)56. This analysis revealed the several cell cycle proteins that were shared across the distinct phases of cell cycle. Within this context, the colors red and blue were assigned to signify upregulation and downregulation of proteins upon DENV2 infection, respectively. In Supplementary Fig. 4A, the data demonstrated that among the total proteins within the G1 phase, 70% exhibited upregulation, while in the S phase, 60% showed upregulation upon DENV2 infection. Similarly, 50% of the proteins in G2 phase and 66.6% of proteins in the M phase were observed to be upregulated upon DENV2 infection. In Supplementary Fig. 4B, only the upregulated genes were subjected to fold enrichment analysis, aimed at exploring the modulation of the top ten enriched biological processes triggered by DENV2 infection. This investigation employed a false discovery rate (FDR) threshold of 0.5, as implemented in ShinyGO v0.741 (http://bioinformatics.sdstate.edu/go74/)57. Notably, a considerable proportion of the upregulated genes exhibited functional relevance to the realm of cell cycle regulation (Supplementary Table 3, Supplementary Fig. 4B). Furthermore, examination of the top ten biological processes unveiled that the G1/S phase of the mitotic transition process was markedly overrepresented which conferred to exert a regulatory influence that accelerates the progression of cells through the G1 phase and into the S phase of the cell cycle (Supplementary Fig. 4C). Specifically, this overrepresentation was attributed to the proteins CDK4, CDK1 and Cyclin B1 (Supplementary Table 3). In pursuit of a more focused investigation, the KEGG pathway associated with cell cycle processes was selected. This selection reaffirmed the enrichment of genes related to the cell cycle, with upregulated proteins depicted in red (Supplementary Fig. 4C). Based on these findings, we proceeded to construct a protein–protein interaction network among the entirety of DENV2 upregulated proteins by employing the STRINGdb package (Supplementary Fig. 4D). Remarkably, this network displayed a robust experimental enrichment of protein–protein interactions (PPI), indicated by a p-value of 2.89e−15. Specifically, prominent within this interconnected network were CDK4, CDK1 and Cyclin B1 (Supplementary Fig. 4D). These analyses supported our experimental data in this study on understanding the roles of CDK4, CDK1, and Cyclin B1 in the context of DENV2 infection in human megakaryocytes. These bioinformatics analyses further supported us to validate the critical cell cycle regulatory protein expression and their interactions upon DENV2 infection.
Validation analysis revealed that cell cycle regulatory proteins CDK4, CDK1 and Cyclin B1 are upregulated upon DENV2 infection
Megakaryocytes become polyploid cells (N = 64/128–256) by undergoing multiple rounds of DNA replication during the S phase and escape the cytokinesis process, specifically by skipping late anaphase of the cell cycle and re-entering into the G1 phase18,22,23,24,38,58. However, the M phase-specific factors that are responsible for enabling megakaryocytes to skip anaphase remains unknown. Therefore, it is necessary to revalidate the expression of S phase specific marker, CDK4 and G2/M phase specific markers, CDK1 and Cyclin B1 upon DENV2 infection (MOI of 5, at 72 h p.i.). QRT-PCR analyses showed that DENV2-capsid transcripts were highly detected at 72 h p.i. (Fig. 5A). Tissue culture infectious dose (TCID50) determined by the end-point virus dilution assay showed that DENV2 infects MEG-01 cells with higher infectivity at the dilution of 10–2 followed by the other dilutions of 10–3, 10–4 and 10–5 (Supplementary Fig. 5A). Immunofluorescence images shown for uninfected cells treated with both primary and secondary antibodies or DENV2-infected cells stained for only secondary antibody serves as internal controls (Supplementary Fig. 5A). Also, the immunofluorescence data correlated with the percentage of DENV2 infectivity (as dilutions of 10–2, 10–3, 10–4 and 10–5) in MEG-01 cells (Supplementary Fig. 5B). Furthermore, transcript levels of CDK4 (Fig. 5B), CDK1 (Fig. 5C) and Cyclin B1 (Fig. 5D) in MEG-01 cells were significantly upregulated upon DENV2 infection in comparison to the transcript levels detected in respective uninfected control groups (Fig. 5B–D). The upregulation of these cell cycle genes (CDK4, CDK1 and Cyclin B1) is specific to DENV2 infection in MEG-01 cells. EA.hy926 endothelial cells did not show any significant differences in the gene expression of CDK4, CDK1 and Cyclin B1 upon DENV2 infection (Supplementary Fig. 6A–D). Although, DENV2 infection was detectable (Supplementary Fig. 6A), but the expression of cell cycle genes CDK4 (Supplementary Fig. 6B), CDK1 (Supplementary Fig. 6C) and Cyclin B1 (Supplementary Fig. 6D) were unaltered in comparison to their respective uninfected EA.hy926 endothelial cells. Furthermore, we determined the expression of CDK4, Phospho-CDK4, CDK1, Cyclin B1 and actin at the protein levels to validate the microarray analyses. As expected, immunoblotting analysis showed DENV2 capsid protein loads only in infected groups (Fig. 5E) and an enhanced expression of CDK4 (Fig. 5F), Phospho-CDK4 (Fig. 5G), CDK1 (Fig. 5H), and Cyclin B1 (Fig. 5I) proteins upon DENV2 infection in comparison to their respective uninfected controls. Actin protein loads were detected in both uninfected and DENV2-infected groups (Fig. 5J). Total protein profile gel images or Ponceau S-stained membrane images served as loading controls for respective blots (Fig. 5E–J). Densitometric analysis further revealed the upregulation of cell cycle proteins in DENV2-infected MEG-01 cells in comparison to their respective uninfected controls (Supplementary Fig. 7A–F). Also, full-length immunoblot images from Fig. 5 are shown for details (Supplementary Fig. 8A–F). This data further validates the results from microarray, heatmap and independent gene expression screening of the cell cycle regulatory proteins CDK4, CDK1, and Cyclin B1 upon DENV2 infection.
DENV2 infection upregulates CDK4, CDK1 and Cyclin B1 transcripts and protein levels. QRT-PCR analysis showing DENV2 capsid transcript levels (A), or CDK4 (B), CDK1 (C) and Cyclin B1 (D) gene expression in DENV2-infected (MOI of 5, at 72 h p.i.) or uninfected MEG-01 cells. DENV2-loads and mRNA expression levels of CDK4, CDK1 and Cyclin B1 genes were normalized to human beta-actin mRNA levels. Y-axis represents levels of transcript and X-axis indicates uninfected or DENV2-infected groups. P value was determined by Student’s two-tail-test is shown. Each circle or square indicates one data value and n = 6 (six independent replicates run in duplicate for QRT-PCR). Immunoblotting analyses (E–J) performed using cell lysates generated from uninfected or DENV2-infected (MOI of 5, at 72 h p.i.) MEG-01 cells are shown. Immunoblot images showing DENV2-capsid protein loads (E), or CDK4 (F), Phospho-CDK4 (G), CDK1 (H), Cyclin B1 (I) and actin (J) protein loads from uninfected or DENV2-infceted samples. Total protein profile images serve as loading controls for respective immunoblots.
Silencing of cell cycle regulatory proteins CDK4, CDK1 and Cyclin B1 reduces DENV2 loads, but dengue infection enhances CDK1 and Cyclin B1 interaction
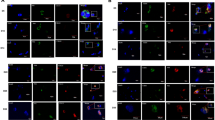
To determine whether CDK4, CDK1 and Cyclin B1 plays any role in maintaining DENV2 loads in megakaryocytes, we silenced the expression of these molecules using siRNA (Fig. 6). QRT-PCR analyses revealed significant (P < 0.5) silencing efficiencies of CDK4 (Fig. 6A), CDK1 (Fig. 6B) and Cyclin B1 (Fig. 6C) in siRNA-treated samples when compared to their respective scramble siRNA control treated groups. Immunoblotting analyses further confirmed the silencing of CDK4 (Fig. 6D), CDK1 (Fig. 6E) and Cyclin B1 (Fig. 6F) in DENV2-infected MEG-01 cells treated with respective siRNAs in comparison to the scramble siRNA-treated control groups (Fig. 6D–F). QRT-PCR analyses further revealed a significant (P < 0.5) reduction in DENV2 loads upon silencing of CDK4 (Fig. 6G), CDK1 (Fig. 6H) and Cyclin B1 (Fig. 6I) transcripts. Immunoblotting analyses demonstrated a notable reduction in DENV2 capsid protein levels upon silencing of CDK4 (Fig. 6J), CDK1 (Fig. 6K) and Cyclin B1 (Fig. 6L) proteins. Furthermore, immunoprecipitation assay revealed enhanced interaction of CDK1 and Cyclin B1 in presence of DENV2 infection (Fig. 6M). Also, full-length immunoblot images from Fig. 6 are shown for details (Supplementary Figs. 9A–D; 10A and B). A model is proposed to show the modulation in cell cycle signaling upon DENV2 infection in MEG-01 cells (Fig. 6N). To understand the importance of DENV2-mediated upregulation of CDK4, CDK1 and Cyclin B1 molecules, we performed immunofluorescence analysis. MEG-01 cells infected with DENV2 (MOI of 5, at 72 h p.i.) showed that CDK4 (Supplementary Fig. 11), CDK1 (Supplementary Fig. 12) and Cyclin B1 (Supplementary Fig. 13), co-localizes with DENV2 E-protein, thus suggesting a direct interaction of viral molecules with this cell cycle regulatory proteins. Collectively, DENV2 not only directly induces CDK4, CDK1 and Cyclin B1 proteins but also co-localizes with CDK4, CDK1 and Cyclin B1 to propagate inside the human megakaryocyte cells.
Silencing of CDK4 or CDK1 or Cyclin B1 in MEG-01 cells reduced DENV2 loads and enhanced CDK1 and Cyclin B1 interactions is enhanced upon DENV2 infection. QRT-PCR and immunoblotting analyses showing silencing of CDK4 (A, D) or CDK1 (B, E) or Cyclin B1 (C, F) in MEG-01 cells transfected with either scrambled siRNA or siRNA for respective genes (CDK4, CDK1, and Cyclin B1) for 24 h and followed by DENV2 infection (MOI of 5, at 24 h p.i.). QRT-PCR and immunoblotting analyses is shown for DENV2 capsid gene or protein loads determined upon silencing of CDK4 (D, J), or CDK1 (H, K) or Cyclin B1 (I, L) in MEG-01 cells transfected with either scrambled siRNA or siRNA for respective genes (CDK4, CDK1, and Cyclin B1) for 24 h and followed by DENV2 infection (MOI of 5, at 24 h p.i.). DENV2-loads and mRNA expression levels of CDK4, CDK1 and Cyclin B1 genes were normalized to human beta-actin mRNA levels. Y-axis represents levels of transcripts and X-axis indicates uninfected or DENV2-infected groups. P value determined by student’s t-test is shown. Scrambled siRNA-treated samples serve as negative control. (M) Immunoprecipitation assay performed with MEG-01 cell lysates from uninfected or DENV2-infected sample (MOI of 5, at 72 h p.i.) using Cyclin B1 antibody and detection with CDK1 antibody is shown for interaction between Cyclin B1 and CDK1 upon DENV2 infection. Total protein profile images serve as loading control (in panels D, E, F, J, K, L and M). (N) Schematic representation shown as model to summarize the current findings. DENV2 enters megakaryocyte cells and upregulates CDK4 protein, which eventually may enter the nucleus and facilitate cell progression from G1 to S phase. DENV2 induces the expression of CDK1 and Cyclin B1 and their interactions that may further assist in the progression of megakaryocytes through the G2 phase of the cell cycle into mitosis. Once the megakaryocytes enter the metaphase, DENV2 may reduce the expression of certain molecules, which are inhibitory to the progression of anaphase A. As a result, DENV2 modulates the cell cycle process in megakaryocytes by pushing from one phase to the next and eventually creating a favorable environment for DENV2 to selectively replicate inside megakaryocytes that undergoes polyploidy and endomitosis.
Discussion
DENV modulates human megakaryocytes and reduces the platelets count that contributes to thrombocytopenia. This medical condition perhaps leads to severe pathological symptoms such as plasma leakage, vasculature damage, failure of multiple organs and sudden death1,2,4,13,16,17,59,60,61,62. DENV has been found to decrease the number of polyploid cells in laboratory-cultured megakaryocyte cell lines, such as K562, megakaryocyte-erythrocyte progenitors, and MEG-01 cells derived from leukemia patients19,20,29,59,60. This polyploidy reduction process occurs without affecting the cell death but rather interferes with the complex cell division process known as endomitosis (partial mitosis)18,20,22,24,27,58. Polyploidy refers to cells containing more than the usual two paired sets of chromosomes, which is a characteristic feature of megakaryocytes as they mature18,20,22,24,27,58. Studies have shown that DENV infection leads to a reduction in the number of polyploid cells without inducing apoptosis but by reducing the reactive oxygen species (ROS) level22,24,58,59,60. This reduction is particularly intriguing as it points towards an intricate interference with the endomitotic cell cycle, a specialized form of cell division essential for megakaryocyte maturation22,24,58,59. Recent investigations into the molecular mechanisms underlying Zika virus (ZIKV)-induced alterations in cell cycle regulation have identified the Non-Structural protein 5 (NS5) as a key player63,64. NS5, a highly conserved protein among flaviviruses, has been found to localize to specific cellular structures such as the centrosome and spindle fibers during cell division, indicating its potential involvement in the modulation of cell cycle progression63,64. Furthermore, experiments conducted on mosquito cells (C6/36) have revealed that DENV infection can arrest the cell cycle at the G2/M phase, further emphasizing the virus’s ability to interfere with normal cellular processes14,59,60,61,65,66. There are several reports that suggested flavivirus-mediated interference of cell cycle processes67,68,69. Moreover, it has been reported that upon DENV infection, HepG2 cells exhibited significantly higher susceptibility to both infection and virus production during the G2 phase compared to other cell cycle phases66.
In our study, the protein microarray data showed that DENV2 significantly upregulates several of the cell cycle regulatory proteins such as CDK4, CDK1, Cyclin B1, CDK8, NuMA, APC-2, Cyclins E/C, p18INK4c, CDK3, 14.3.3 Pan, CDC-37, CDC-47, CDC-34, p130Cas and p19SKp-1. In addition, some other cell cycle regulatory proteins such as E2F-3, Chk1, GSK3-beta, and CUL-3 were significantly downregulated upon DENV2 infection in MEG-01 cells. Independent random analysis of other cell cycle regulatory genes (not included in the microarray analysis) further confirmed that several of the Cyclins, Cyclin-dependent kinases and others were upregulated upon a time course of DENV2-infection. Our data clearly suggested that DENV2 infection in MEG-01 cells enhances the expression of several cell cycle regulatory proteins, and this induction was very noticeable in comparison to the downregulated protein, where only few of the molecules were significantly inhibited. DENV2 induced gene expression is specific to MEG-01 cells as the key cell cycle regulatory genes (CDK4, CDK1 and Cyclin B1) remained unchanged in human endothelial cells. Furthermore, networking and gene enrichment analysis revealed that CDK4, CDK1 and Cyclin B1 proteins are the major cell cycle regulators that are responsible for the modulation of megakaryocytes by DENV2. We have subsequently confirmed the upregulation of CDK4, CDK1 and Cyclin B1 at both transcript and protein levels. Additionally, our immunofluorescence data demonstrated a colocalization of DENV2-E protein with CDK4, CDK1 and Cyclin B1 proteins. Silencing of these three key proteins- CDK4, CDK1 and Cyclin B1, significantly reduced the DENV2 loads. However, we have not explored whether silencing of these key proteins directly interferes with the viral entry into the MEG-01 cells or affects viral transmission. Our immunoprecipitation and immunoblotting analyses have further confirmed that DENV2 infection enhances the expression of CDK4, CDK1/Cyclin B1 and interactions between CDK1 and Cyclin B1, which is critical for the progression of the mitotic process and cytokinesis. Future investigations on DENV2 enhanced interaction between CDK1 and Cyclin B1 proteins, will highlight a potential mechanism by which the flaviviruses modulates cell cycle progression. We strongly believe that upregulation of CDK4 and enhanced CDK1 and Cyclin B1 interactions may interfere with the partial endomitosis process of megakaryocytes. A schematic model proposing the role of cell cycle regulatory proteins upon DENV2 infection is shown (Fig. 6N and Supplementary Fig. 14, an enlarge image of Fig. 6N). Dengue virus enters megakaryocytes and enhances the expression of CDK4 protein, which eventually may enter the nucleus and facilitate the cell progression from G1 to S phase. Furthermore, DENV2 induces the expression of CDK1 and Cyclin B1 to enhance their interactions that could result in a strong complex formation between CDK1-Cyclin B1. This complex may enter the nucleus to further assist in the progression of megakaryocytes through the G2 phase of the cell cycle into Mitosis. Once the megakaryocytes enter Metaphase, DENV2 may reduce the expression of certain molecules, which are expressed during the onset of Anaphase A, thus may eventually stabilizing the interaction between the Kinetochore of chromosomes and Microtubules. As a result, DENV likely modulates the cell cycle stages in megakaryocytes by pushing from one phase to the next. This would eventually facilitate a favorable environment for DENV2 to selectively replicate inside the newly propagated megakaryocytes.
Taken together, the observed alterations in cell cycle regulation induced by DENV infection could have a significant implication on megakaryocytes function and platelet production. DENV infection may contribute to provide a beneficial environment for flaviviral replication in human megakaryocytes by reducing the number of polyploid cells and potentially suppressing the ROS production60. Consequently, these changes will address the drastic reduction in platelet numbers. Understanding the intricate interplay between DENV and cellular processes such as cell cycle regulation is crucial for developing targeted therapeutic strategies to mitigate the impact of dengue fever on platelet production and overall disease progression. Further research aimed at elucidating the specific molecular mechanisms underlying these interactions is warranted to uncover novel therapeutic targets that may improve the clinical outcomes for dengue-infected patients.
Data availability
Any additional information or data is available from the Corresponding Author upon request. Additional data is provided as supplemental information.
Change history
02 August 2025
The original online version of this Article was revised: In the original version of this Article, the primary affiliations for authors Shovan Dutta and Supreet Khanal were incorrectly listed as their present addresses.
References
Guzman, M. G. & Harris, E. Dengue. Lancet 385, 453–465. https://doi.org/10.1016/s0140-6736(14)60572-9 (2015).
Bhatt, S. et al. The global distribution and burden of dengue. Nature 496, 504–507. https://doi.org/10.1038/nature12060 (2013).
Simmons, C. P., Farrar, J. J., van Vinh Chau, N. & Wills, B. Dengue. N. Engl. J. Med. 366, 1423–1432. https://doi.org/10.1056/NEJMra1110265 (2012).
Guzman, M. G. et al. Dengue: a continuing global threat. Nat. Rev. Microbiol. 8, S7-16. https://doi.org/10.1038/nrmicro2460 (2010).
(WHO), W. H. O. Dengue and severe dengue, https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (17 March 2023).
Carreto, C., Gutiérrez-Romero, R. & Rodríguez, T. Climate-driven mosquito-borne viral suitability index: measuring risk transmission of dengue, chikungunya and Zika in Mexico. Int. J. Health Geogr. 21, 15. https://doi.org/10.1186/s12942-022-00317-0 (2022).
Shepard, D. S. et al. Approaches to refining estimates of global burden and economics of dengue. PLoS Negl. Trop. Dis. 8, e3306. https://doi.org/10.1371/journal.pntd.0003306 (2014).
(CDC), C. f. D. C. A. P. Dengue in the US States and Territories, https://www.cdc.gov/dengue/areaswithrisk/in-the-us.html
Chen, L. H. et al. Epidemiology and burden of dengue fever in the United States: A systematic review. J. Travel Med. https://doi.org/10.1093/jtm/taad127 (2023).
Naderian, R. et al. Efficacy, immune response, and safety of dengue vaccines in adolescents: A systematic review. Rev. Med. Virol. 35, e70035. https://doi.org/10.1002/rmv.70035 (2025).
Pereira, C. A. M., Mendes, R. P. G., Silva, P. G. D., Chaves, E. J. F. & Pena, L. J. Vaccines against urban epidemic arboviruses: The state of the art. Viruses https://doi.org/10.3390/v17030382 (2025).
Saez-Llorens, X. et al. TAK-003: Development of a tetravalent dengue vaccine. Expert Rev. Vaccines https://doi.org/10.1080/14760584.2025.2490295 (2025).
Halstead, S. B. Dengue vaccine development: A 75% solution?. Lancet 380, 1535–1536. https://doi.org/10.1016/s0140-6736(12)61510-4 (2012).
Yung, C. F. et al. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, singapore. Am. J. Trop. Med. Hyg. 92, 999–1005. https://doi.org/10.4269/ajtmh.14-0628 (2015).
Rico-Hesse, R. Microevolution and virulence of dengue viruses. Adv. Virus Res. 59, 315–341. https://doi.org/10.1016/s0065-3527(03)59009-1 (2003).
Srikiatkhachorn, A. Plasma leakage in dengue haemorrhagic fever. Thromb. Haemost. 102, 1042–1049. https://doi.org/10.1160/th09-03-0208 (2009).
Rajapakse, S. Dengue shock. J. Emerg. Trauma Shock 4, 120–127. https://doi.org/10.4103/0974-2700.76835 (2011).
Patel, S. R., Hartwig, J. H. & Italiano, J. E. Jr. The biogenesis of platelets from megakaryocyte proplatelets. J. Clin. Invest. 115, 3348–3354. https://doi.org/10.1172/jci26891 (2005).
Machlus, K. R. & Italiano, J. E. Jr. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 201, 785–796. https://doi.org/10.1083/jcb.201304054 (2013).
Italiano, J. E. Jr. Unraveling mechanisms that control platelet production. Semin. Thromb. Hemost. 39, 15–24. https://doi.org/10.1055/s-0032-1331157 (2013).
Machlus, K. R., Thon, J. N. & Italiano, J. E. Jr. Interpreting the developmental dance of the megakaryocyte: A review of the cellular and molecular processes mediating platelet formation. Br. J. Haematol. 165, 227–236. https://doi.org/10.1111/bjh.12758 (2014).
Trakala, M. et al. Functional reprogramming of polyploidization in megakaryocytes. Dev. Cell 32, 155–167. https://doi.org/10.1016/j.devcel.2014.12.015 (2015).
Lee, H. O., Davidson, J. M. & Duronio, R. J. Endoreplication: Polyploidy with purpose. Genes Dev. 23, 2461–2477. https://doi.org/10.1101/gad.1829209 (2009).
Nurden, P., Poujol, C. & Nurden, A. T. The evolution of megakaryocytes to platelets. Baillieres Clin. Haematol. 10, 1–27. https://doi.org/10.1016/s0950-3536(97)80048-0 (1997).
Muntean, A. G. et al. Cyclin D-Cdk4 is regulated by GATA-1 and required for megakaryocyte growth and polyploidization. Blood 109, 5199–5207. https://doi.org/10.1182/blood-2006-11-059378 (2007).
Baccini, V. et al. Role of p21(Cip1/Waf1) in cell-cycle exit of endomitotic megakaryocytes. Blood 98, 3274–3282. https://doi.org/10.1182/blood.v98.12.3274 (2001).
Kikuchi, J. et al. Polyploidization and functional maturation are two distinct processes during megakaryocytic differentiation: Involvement of cyclin-dependent kinase inhibitor p21 in polyploidization. Blood 89, 3980–3990 (1997).
Zhang, Y., Wang, Z. & Ravid, K. The cell cycle in polyploid megakaryocytes is associated with reduced activity of cyclin B1-dependent cdc2 kinase. J. Biol. Chem. 271, 4266–4272. https://doi.org/10.1074/jbc.271.8.4266 (1996).
Wilhide, C. C., Van Dang, C., Dipersio, J., Kenedy, A. A. & Bray, P. F. Overexpression of cyclin D1 in the Dami megakaryocytic cell line causes growth arrest. Blood 86, 294–304 (1995).
Datta, N. S. & Long, M. W. Modulation of MDM2/p53 and cyclin-activating kinase during the megakaryocyte differentiation of human erythroleukemia cells. Exp. Hematol. 30, 158–165. https://doi.org/10.1016/s0301-472x(01)00780-9 (2002).
Lemonnier, T., Dupré, A. & Jessus, C. The G2-to-M transition from a phosphatase perspective: A new vision of the meiotic division. Cell Di.v 15, 9. https://doi.org/10.1186/s13008-020-00065-2 (2020).
Gavet, O. & Pines, J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 18, 533–543. https://doi.org/10.1016/j.devcel.2010.02.013 (2010).
Jones, K. T. Turning it on and off: M-phase promoting factor during meiotic maturation and fertilization. Mol. Hum. Reprod. 10, 1–5. https://doi.org/10.1093/molehr/gah009 (2004).
Gavet, O. & Pines, J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189, 247–259. https://doi.org/10.1083/jcb.200909144 (2010).
Bentley, A. M., Normand, G., Hoyt, J. & King, R. W. Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis. Mol. Biol. Cell 18, 4847–4858. https://doi.org/10.1091/mbc.e06-06-0539 (2007).
Datta, N. S. et al. Novel alterations in CDK1/cyclin B1 kinase complex formation occur during the acquisition of a polyploid DNA content. Mol. Biol. Cell 7, 209–223. https://doi.org/10.1091/mbc.7.2.209 (1996).
Topacio, B. R. et al. Cyclin D-Cdk 4,6 drives cell-cycle progression via the retinoblastoma protein’s C-terminal helix. Mol. Cell 74, 758-770.e754. https://doi.org/10.1016/j.molcel.2019.03.020 (2019).
Bertoli, C., Skotheim, J. M. & de Bruin, R. A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14, 518–528. https://doi.org/10.1038/nrm3629 (2013).
Narasimha, A. M. et al. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife https://doi.org/10.7554/eLife.02872 (2014).
Schade, A. E., Oser, M. G., Nicholson, H. E. & DeCaprio, J. A. Cyclin D-CDK4 relieves cooperative repression of proliferation and cell cycle gene expression by DREAM and RB. Oncogene 38, 4962–4976. https://doi.org/10.1038/s41388-019-0767-9 (2019).
Ding, L. et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21061960 (2020).
Lee, S. H., Rodriguez, L. R. L., Majumdar, R., De Marval, P. L. M. & Rodriguez-Puebla, M. L. CDK4 has the ability to regulate Aurora B and Cenpp expression in mouse keratinocytes. Oncol. Lett. 22, 732. https://doi.org/10.3892/ol.2021.12993 (2021).
Dutta, S. et al. Coordination of different ligands to copper(II) and cobalt(III) metal centers enhances Zika virus and dengue virus loads in both arthropod cells and human keratinocytes. Biochim. Biophys. Acta Gen. Subj. 1862, 40–50. https://doi.org/10.1016/j.bbagen.2017.10.004 (2018).
Vora, A. et al. Arthropod EVs mediate dengue virus transmission through interaction with a tetraspanin domain containing glycoprotein Tsp29Fb. Proc. Natl. Acad. Sci. U. S. A. 115, E6604–E6613. https://doi.org/10.1073/pnas.1720125115 (2018).
Zhou, W. et al. Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cells and facilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 14, e1006764. https://doi.org/10.1371/journal.ppat.1006764 (2018).
Cornell, L., Wander, S. A., Visal, T., Wagle, N. & Shapiro, G. I. MicroRNA-mediated suppression of the TGF-beta pathway confers transmissible and reversible CDK4/6 inhibitor resistance. Cell Rep. 26, 2667–2680. https://doi.org/10.1016/j.celrep.2019.02.023 (2019).
Dantas, M., Oliveira, A., Aguiar, P., Maiato, H. & Ferreira, J. G. Nuclear tension controls mitotic entry by regulating cyclin B1 nuclear translocation. J. Cell Biol. https://doi.org/10.1083/jcb.202205051 (2022).
Hu, X. et al. Phosphorylation of VP1 mediated by CDK1-cyclin B1 facilitates infectious bursal disease virus replication. J Virol 97, e0194122. https://doi.org/10.1128/jvi.01941-22 (2023).
Ji, X., Humenik, J., Yang, D. & Liebhaber, S. A. PolyC-binding proteins enhance expression of the CDK2 cell cycle regulatory protein via alternative splicing. Nucleic Acids Res. 46, 2030–2044. https://doi.org/10.1093/nar/gkx1255 (2018).
Khanal, S., Sultana, H., Catravas, J. D., Carlyon, J. A. & Neelakanta, G. Anaplasma phagocytophilum infection modulates expression of megakaryocyte cell cycle genes through phosphatidylinositol-3-kinase signaling. PLoS ONE 12, e0182898. https://doi.org/10.1371/journal.pone.0182898 (2017).
Khanal, S., Taank, V., Anderson, J. F., Sultana, H. & Neelakanta, G. Arthropod transcriptional activator protein-1 (AP-1) aids tick-rickettsial pathogen survival in the cold. Sci. Rep. 8, 11409. https://doi.org/10.1038/s41598-018-29654-6 (2018).
Maryu, G. & Yang, Q. Nuclear-cytoplasmic compartmentalization of cyclin B1-Cdk1 promotes robust timing of mitotic events. Cell Rep. 41, 111870. https://doi.org/10.1016/j.celrep.2022.111870 (2022).
Tian, R. Q. et al. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. J. Biol.Chem. 286, 25556–25563. https://doi.org/10.1074/jbc.M111.221564 (2011).
Tsikitis, M., Zhang, Z., Edelman, W., Zagzag, D. & Kalpana, G. V. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc. Natl. Acad. Sci. U. S. A. 102, 12129–12134. https://doi.org/10.1073/pnas.0505300102 (2005).
Zhou, W., Woodson, M., Sherman, M. B., Neelakanta, G. & Sultana, H. Exosomes mediate Zika virus transmission through SMPD3 neutral Sphingomyelinase in cortical neurons. Emerg. Microbes Infect. 8, 307–326. https://doi.org/10.1080/22221751.2019.1578188 (2019).
Sun, L. et al. DiVenn: An interactive and integrated web-based visualization tool for comparing gene lists. Front. Genet. 10, 421. https://doi.org/10.3389/fgene.2019.00421 (2019).
Ge, S. X., Jung, D. & Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. https://doi.org/10.1093/bioinformatics/btz931 (2020).
Mazzi, S., Lordier, L., Debili, N., Raslova, H. & Vainchenker, W. Megakaryocyte and polyploidization. Exp. Hematol. 57, 1–13. https://doi.org/10.1016/j.exphem.2017.10.001 (2018).
Banerjee, A., Tripathi, A., Duggal, S., Banerjee, A. & Vrati, S. Dengue virus infection impedes megakaryopoiesis in MEG-01 cells where the virus envelope protein interacts with the transcription factor TAL-1. Sci. Rep. 10, 19587. https://doi.org/10.1038/s41598-020-76350-5 (2020).
Kaur, J. et al. Replication of dengue virus in K562-megakaryocytes induces suppression in the accumulation of reactive oxygen species. Front. Microbiol. 12, 784070. https://doi.org/10.3389/fmicb.2021.784070 (2021).
Kaur, J. et al. Megakaryopoiesis in Dengue virus infected K562 cell promotes viral replication which inhibits endomitosis and accumulation of ROS associated with differentiation. bioRxiv https://doi.org/10.1101/2020.06.25.172544 (2020).
Simmons, C. P., Farrar, J. J., Chau, N. V. V. & Wills, B. Dengue. N. Engl. J. Med. 366, 1423–1432. https://doi.org/10.1056/NEJMra1110265 (2012).
Lebeau, G. et al. Cell cycle and mitosis progression during ZIKA virus infection: The viral non-structural protein NS5 as a master regulator of the APC/cyclosome?. Biochimie 221, 75–80. https://doi.org/10.1016/j.biochi.2024.01.016 (2024).
Liu, J. et al. Zika virus envelope protein induces G2/M cell cycle arrest and apoptosis via an intrinsic cell death signaling pathway in neuroendocrine PC12 cells. Int. J. Biol. Sci. 14, 1099–1108. https://doi.org/10.7150/ijbs.26400 (2018).
Chong, M. K. & Ng, M. L. Significance of cell cycle manipulation in the establishment of dengue virus and west Nile virus infection. Int. J. Infect. Dis. 12, e168. https://doi.org/10.1016/j.ijid.2008.05.418 (2008).
Phoolcharoen, W. & Smith, D. R. Internalization of the dengue virus is cell cycle modulated in HepG2, but not Vero cells. J. Med. Virol. 74, 434–441. https://doi.org/10.1002/jmv.20195 (2004).
Mussgay, M. Growth cycle of arboviruses in vertebrate and arthropod cells. Prog. Med. Virol. 6, 193–267 (1964).
Mussgay, M., Enzmann, P. J., Horzinek, M. C. & Weiland, E. Growth cycle of arboviruses in vertebrate and arthropod cells. Prog. Med. Virol. 19, 257–323 (1975).
Vial, T. et al. Mosquito metabolomics reveal that dengue virus replication requires phospholipid reconfiguration via the remodeling cycle. Proc. Natl. Acad. Sci. U. S. A. 117, 27627–27636. https://doi.org/10.1073/pnas.2015095117 (2020).
Acknowledgements
We sincerely acknowledge the financial support from University of Tennessee at Knoxville, College of Veterinary Medicine and Old Dominion University, College of Sciences as startup funds to H.S and G.N. We would also like to sincerely acknowledge the kind support from BEI resources. The following reagents were obtained through BEI Resources, NIAID, NIH: Dengue virus type 2-New Guinea C (NGC), NR-84; and Monoclonal Anti-Flavivirus Group Antigen, Clone D1-4G2-4-15 (produced in vitro), NR-50327. Lastly, we sincerely thank the company Full Moon Biosystems for all their support with scanning images and for their help with GenePix Pro Software instructions.
Author information
Authors and Affiliations
Contributions
S.B., and H.S. performed experiments, discussed, analyzed, and interpreted the data in several settings. G.N. helped with molecular and sequencing analysis and is involved in all data analysis and discussions. S.B, performed all molecular/biochemical/bioinformatics analysis with the help of H.S. and G.N. S.D and S.K generated QRT-PCR data shown in Fig. 5. S.B wrote the draft and H.S re-wrote and revised the entire paper. H.S. modified, revised, and finalized the paper. All authors read and revised/edited the completed manuscript. H.S. collected all required materials and reagents, conceived, designed, and coordinated the entire study, organized all the data, compiled, and supervised overall investigations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Basak, S., Dutta, S., Khanal, S. et al. Dengue virus modulates critical cell cycle regulatory proteins in human megakaryocyte cells. Sci Rep 15, 19016 (2025). https://doi.org/10.1038/s41598-025-02640-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-02640-5