Abstract
Allergic asthma is an inflammatory condition characterized by the release of pro-inflammatory cytokines and the expansion of mucus-producing cells. This research aimed to evaluate the impact of Kaempferol 3-O-gentiobioside (K3G), extracted from Sauropus spatulifolius Beille leaves, on inflammatory cytokine secretion and mucus overproduction in IL-13-stimulated airway epithelial cells (16HBE cells) and ovalbumin (OVA)-induced allergic asthma mouse models. Studies have found that K3G significantly reduces the release of pro-inflammatory cytokines (IgE, TNF-α, histamine, IL-1β, IL-6, and IL-8) induced by IL-13. It also mitigating the expression of the mucin5AC (MUC5AC). Additionally, K3G also downregulated the expression of NLRP3, TLR4, p-IκBα, and p-P65 proteins. In an OVA-induced mouse asthma model, K3G treatment reduced the secretion of pro-inflammatory cytokines, and inhibited the increase in mucus-secreting cells in a dose-dependent manner. Furthermore, exposure to IL-13 and OVA increased the expression of NOTCH signaling receptors in both 16HBE cells and lung tissues of the mice. K3G treatment effectively targeted NOTCH1 and inhibits activation of the NOTCH pathway. In conclusion, K3G alleviates asthma-related airway alterations by suppressing inflammatory cytokines and excessive mucus secretion via the NOTCH signaling pathway. These results indicate the therapeutic promise of K3G in treating allergic asthma.
Similar content being viewed by others
Introduction
Allergic asthma impacts approximately 300 million individuals worldwide, with 5–10% of children affected, often manifesting at an early age1,2. This condition poses a significant global health challenge and imposes a substantial socioeconomic burden3,4. Allergic asthma is an atopic airway disease characterized by excessive mucus secretion, which is an important factor in asthmatic airway obstruction and mucus plug formation5,6. Studies suggest that in allergic individuals, exposure to allergens activates immune cells, such as pathogenic memory Th2 cells, and stimulates the release of cytokines like interleukin 4 (IL-4), IL-5, and IL-13, which promote increased airway mucus production7,8. Airway inflammation is a hallmark of asthma, with chronic inflammatory infiltrates often observed in affected patients9,10. In addition, activated inflammatory cells secrete cytokines, leading to excessive airway mucus secretion11, which is a significant pathological feature of asthma12 and has a major impact on its morbidity and mortality13,14. Therefore, there is an urgent need to develop asthma medications that combine anti-inflammatory effects with the ability to reduce excessive mucus production.
Recent research has emphasized the critical role of the NOTCH signaling pathway in regulating mucus secretion by modulating the differentiation of mucus-secreting cells in the airways15,16. The NOTCH pathway consists of four receptors (NOTCH1–4) and five ligands, including delta-like ligands (DLL) 1, 2, 3, and 4, as well as jagged (JAG) 1 and 217. Activation of the active domain of this pathway has been shown to influence the differentiation of basal cells in the airway epithelium, promoting their transformation into secretory cells in both humans18 and mouse19,20 airways. Moreover, various respiratory disorders have revealed the NOTCH pathway’s modulatory impact on airway inflammation. For instance, inhibition of the NOTCH pathway has been demonstrated to alleviate airway inflammation by reducing cytokine release in asthma21,22, suppressing Th2 cytokine levels, and mitigating eosinophil infiltration in allergic rhinitis23,24. IL-13, a prototypical Th2 cytokine, plays a central role in regulating various allergic airway diseases25. It is commonly used to induce the pathological changes observed in asthma, such as mucus cell metaplasia26, subepithelial fibrosis27, smooth muscle hypertrophy28, and eosinophil recruitment29. Therefore, drugs capable of modulating the NOTCH pathway are expected to emerge as one of the effective strategies for treating allergic asthma.
Toll-like receptor (TLR) 4 is central role in pathogen recognition and the initiation of immune responses30. Upon activation, TLR4 triggers downstream signaling through NF-κB, leading to the release of inflammatory cytokines31. Additionally, NF-κB activation enhances the expression of NLRP332. Studies have shown that activation of the TLR4/NF-κB/NLRP3 pathway is implicated in asthma, and its inhibition can alleviate asthma symptoms33. Thus, Modulating this pathway has shown promise as a therapeutic approach for allergic asthma.
Kaempferol 3-O-gentiobioside (K3G) is an important flavonoid compound derived from the leaves of the Chinese herb Sauropus spatulifolius Beille34, which has anti-inflammatory35 and antioxidant36 effects. In vitro experiments have demonstrated that K3G exerts anti-inflammatory effects by inhibiting inflammatory mediators such as histamine, tumor necrosis factor α (TNF-α), and IL-1β37. Additionally, K3G has been shown to protect the lungs from damage induced by respiratory viruses by inhibiting viral replication38,39. However, the impact of K3G on mucus overproduction and airway inflammation in asthma has not been thoroughly explored. Therefore, this study aimed to investigate the effects of K3G on both the 16HBE cell model and the asthma mouse model, as well as to explore its potential mechanism regulated by the NOTCH pathway.
Result
K3G does not influence the viability of 16HBE cells
K3G is a flavonoid glycoside characterized by a kaempferol aglycone core (three aromatic rings with hydroxyl groups at C-3, C-5, and C-7) linked to a gentiobiosyl disaccharide moiety at the C-3 position. This unique glycosylation pattern enhances its solubility and bioavailability, which may contribute to its anti-inflammatory effects observed in subsequent experiments (Fig. 1A). The results of the CCK-8 assay demonstrated that K3G exhibited dose- and time-dependent cytotoxicity in 16HBE cells. At 24 h, treatment with 50 µM K3G did not show significant cytotoxicity, while 100 µM K3G markedly reduced cell survival (Fig. 1B). Higher K3G concentrations reduced cell survival at 48–72 h (Fig. 1C-D). Therefore, subsequent experiments used 5, 25, and 50 µM K3G treatment for 24 h. Using Griess reagent and flow cytometry, it was found that K3G inhibited nitrite (Fig. 1E) and ROS (Figs. 1F, G) levels in a dose-dependent manner.
Effect of K3G on 16HBE Cell Viability. (A) Chemical structure of K3G (Kaempferol 3-O-gentiobioside) highlighting key functional groups: hydroxyl (-OH) at positions C-5, C-7, and C-4’, ketone (C = O) at C-4, and the β-1,6-linked gentiobiose disaccharide. (B–D) 16HBE cells were treated with various concentrations of K3G (0, 5, 25, 50, and 100 µM) for 24, 48, and 72 h. Cell viability was assessed using the CCK-8 assay. (E) Cells were pretreated with K3G (5, 25, and 50 µM) for 1 h, followed by stimulation with IL-13 at the indicated concentrations for an additional 24 h. The effect of IL-13 on nitrite production was determined using the Griess assay. (F) 16HBE cells were pretreated with K3G (5, 25, and 50 µM) for 1 h and then treated with IL-13 for 6 h; ROS levels were detected by flow cytometry. (G) Displays statistical data on ROS levels. ###P < 0.001 compared to control group (Ctrl); ***P < 0.001 compared to the IL-13-treated group.
K3G inhibited inflammatory cytokine secretion and the expression of MUC5AC in IL-13-stimulated 16HBE cells
Elevated levels of pro-inflammatory cytokines contribute to airway mucus hypersecretion in asthma patients9,10,11. To investigate the effect of K3G on IL-13-induced inflammation in airway epithelial cells, 16HBE cells were stimulated with IL-13 (10 µg/mL) for 24 h, followed by treatment with 5, 25, and 50 µM K3G. ELISA results revealed that IL-13 stimulation significantly increased the levels of IgE, TNF-α, histamine, IL-1β, IL-6, and IL-8 (Fig. 2A-F). K3G treatment, especially at 50 µM, notably reduced the levels of these cytokines. These findings indicate that K3G suppresses the release of inflammatory cytokines in IL-13-stimulated 16HBE cells.
K3G Reduces Inflammatory Cytokine Release in IL-13-Activated 16HBE Cells. (A) The levels of IgE, (B) TNF-α, (C) histamine, (D) IL-1β, (E) IL-6, and (F) IL-8 in IL-13-activated 16HBE cells treated with K3G (5, 25, 50 µM) were quantified using ELISA. ##P < 0.01, ###P < 0.001 compared to Ctrl; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the IL-13-treated group.
MUC5AC expression is elevated in the airways of asthmatic patients, stimulating mucus secretion40. To assess the effect of K3G on MUC5AC levels, both protein and mRNA expressions were measured in an allergic asthma model. The results demonstrated that IL-13 stimulation significantly upregulated MUC5AC expression, while K3G treatment reduced MUC5AC levels in a dose-dependent manner (Figs. 3A, B). MUC5AC fluorescence intensity was higher in the IL-13 group compared to the control, but significantly decreased after K3G treatment (Fig. 3C, D). These findings suggest that K3G may inhibit mucus secretion in IL-13-stimulated 16HBE cells.
K3G Inhibits Mucus Production in IL-13-Activated 16HBE Cells. (A) The mRNA levels and (B) protein concentrationsof MUC5AC were determined via qRT-PCR and ELISA, respectively. (C) Quantitative analysis of MUC5AC immunofluorescence. #P < 0.05, ##P < 0.01, and ###P < 0.001 compared to Ctrl; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the IL-13-treated group. (D) Fluorescence staining was utilized to observe the distribution of MUC5AC in 16HBE cells treated with IL-13 alone, or in combination with K3G (5, 25, or 50 µM). MUC5AC is indicated by red fluorescence. Scale bar: 50 μm.
K3G inhibits IL-13-Induced NOTCH signaling in 16HBE cells and targets NOTCH1
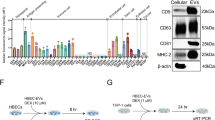
The NOTCH pathway remains persistently activated and plays a crucial role in regulating airway mucus hypersecretion15,16 and inflammation responses21,22 in asthma. qRT-PCR and Western blotting results showed that IL-13 stimulation increased the mRNA and protein levels of NOTCH1, NOTCH2, NOTCH3, and DLL4 in 16HBE cells. K3G treatment significantly downregulated the expression of these NOTCH pathway components in a dose-dependent manner (Fig. 4A-I). These results suggest that K3G suppresses the activation of the NOTCH signaling pathway in IL-13-treated 16HBE cells.
K3G Inhibits IL-13-Triggered Activation of the NOTCH Pathway in 16HBE Cells. (A–E) The protein levels of NOTCH1-3 and DLL4 were analyzed and quantified via Western blot. (F–I) The mRNA expressions of NOTCH1-3 and DLL4 were evaluated by qRT-PCR. #P < 0.05, ##P < 0.01, and ###P <0.001 compared to the Ctrl group; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the IL-13-treated group. The K3G 50 group is the group that received a high dose of K3G (50 µM) and was not treated with IL-13. (J) Molecular docking analysis explored the binding properties of K3G to NOTCH1. (K) Biacore X100 was employed to identify the kinetic and affinity parameters of the K3G-NOTCH1 interaction.
Molecular docking analysis revealed that K3G formed hydrogen bonds and hydrophobic interactions with key residues of NOTCH1, including TYR-319, PRO-2230, and VAL-1178 (Fig. 4J). The predicted binding energy was − 8.2 kcal/mol, indicating that K3G has a moderate affinity for NOTCH1. This interaction was further validated by surface plasmon resonance (SPR) analysis on the Biacore platform, which estimated the equilibrium dissociation constant (KD) of K3G binding to NOTCH1 to be approximately 57 µM (Fig. 4K).
Effect of K3G on the TLR4/NF-κB/ NLRP3 signaling pathway
Overexpression of TLR4 activates the NF-κB signaling pathway. This promotes the release of proinflammatory cytokines and activates the NLRP3 inflammasome. As a result, a cascade of inflammatory responses is initiated41. Western blot results showed that K3G at 25 and 50 µM significantly decreased the expression levels of NLRP3, TLR4, p-IκBα, and p-P65 in IL-13-stimulated 16HBE cells (Fig. 5A–E), indicating that K3G can suppress the TLR4/NF-κB/NLRP3 inflammatory pathway.
Effects of K3G on the TLR4/NF-κB/NLRP3 Signaling Pathway. (A–E) The expression levels of TLR4, NLRP3, p-IκBα and p-P65were analyzed by immunoblotting. (F) Quantitative analysis of NF-κB/p-65 immunofluorescence. (G) Immunofluorescence staining images of NF-κB/P65 in 16HBE cells under different experimental conditions. NF-κB/P65 is shown in red, and Hoechst 33,342 in blue. Images represent 5 experiments. ##P < 0.01, ###P < 0.001 compared to the Ctrl group; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the IL-13-treated group. Scale bar: 100 μm.
To further assess the effect of K3G on NF-κB/P65 protein expression, IF experiments revealed that IL-13-treated 16HBE cells displayed substantial activation signals of NF-κB/P65 compared to the control group(Fig. 5F, G), indicating that IL-13 triggers NF-κB activation within the ceslls. Notably, treatment with 50 µM K3G effectively inhibited the IL-13-induced activation of NF-κB.
K3G attenuates airway inflammation in OVA-challenged mice
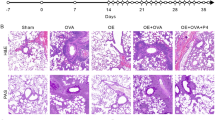
To assess the in vivo effects of K3G, different concentrations were administered to a mouse model of allergic asthma induced by OVA. The severity of inflammatory cell infiltration in lung tissue was evaluated42, demonstrating significant peribronchial and perivascular infiltration in OVA-exposed mice. Treatment with K3G reduced this infiltration and alleviated hyperplasia of mucus-secreting cell. Furthermore, K3G exhibited a dose-dependent attenuation in lung histopathological changes (Fig. 6A). In summary, K3G effectively mitigated inflammatory cell infiltration and tissue damage in the lungs.
Lung histopathological Changes in OVA-Induced Mice with or without K3G Treatment. (A) Mice were exposed to OVA alone, or to OVA combined with K3G at low- (L, 10 mg/kg), medium- (M, 20 mg/kg), or high-dose (H, 40 mg/kg) for 7 days. Lung tissues were collected 24 h post-final OVA exposure, sectioned, and stained with H&E, then observed at ×200 magnification. Scale bar: 10 μm. Inflammatory infiltration in lung tissue was quantitatively scored. The lung concentrations of pro-inflammatory cytokines, including (B) IgE, (C) TNF-α, (D) histamine, (E) IL-1β, (F) IL-6, and (G) IL-8, were measured by ELISA across all mouse groups. ##P < 0.01, ###P < 0.001 compared to the Ctrl group; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the OVA-treated group.
To further explore the the impact of K3G on inflammatory cytokine release in allergic asthma, we measured the levels of IgE, TNF-α, histamine, IL-1β, IL-6, and IL-8 (Fig. 6B-G) in lung tissues. Consistent with our in vitro findings, the levels of these pro-inflammatory cytokines were significantly elevated in OVA-challenged mice. However, treatment with K3G led to a reduction in allergic reactions and a suppression of these cytokine levels. Moreover, higher doses of K3G corresponded with a more significant decrease in inflammatory cytokine concentrations in the lung tissue. These results suggest that K3G has the potential to attenuate airway inflammation in asthma.
K3G attenuates mucus hypersecretion in OVA-challenged mice
We observed that OVA stimulation leads to excessive mucus secretion in mice, as evidenced by significant mucin cell hyperplasia. The inhibitory effects of K3G on excessive mucus production were evident in OVA-induced asthmatic mice (Fig. 7A, C). Additionally, OVA challenge increased the distribution of the mucus secretory factor MUC5AC in the airways (Fig. 7B). K3G treatment effectively suppressed the expression of MUC5AC mRNA (Fig. 7D) and protein levels (Fig. 7E).
K3G Reduces Mucus Overproduction and NOTCH Pathway Activation in OVA-Induced Mice. (A) Representative tissue sections of lungs stained with PAS from each group (n = 6). Scale bar: 10 μm. (B) Immunofluorescence staining displays the distribution of MUC5AC in mouse lung sections, indicated by green fluorescence. Scale bar: 100 μm. (C) PAS staining was quantitatively assessed. (D) MUC5AC mRNA levels in lung sections were determined by qRT-PCR. (E) MUC5AC protein concentrations were measured by ELISA. (F-J) Protein levels of NOTCH1-3 and DLL4 were analyzed and quantified through Western blotting. #P < 0.05, ##P < 0.01, and ###P < 0.001 compared to Ctrl; *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the OVA-treated group.
Furthermore, OVA stimulation activated the NOTCH signaling pathway, as indicated by the significant upregulation of NOTCH1-3, and DLL4 in the lungs. Treatment with K3G at all doses significantly reduced expression of NOTCH-related proteins in a dose-dependent manner (Fig. 7F-J), suggesting that K3G inhibits NOTCH pathway activation in OVA-challenged mice.
Discussion
The global incidence of allergic asthma is rising, particularly in urban areas43. Currently, the treatment of allergic asthma primarily consists of long-term control medications, such as inhaled corticosteroids (ICS), combinations of ICS and long-acting β2 receptor agonists (LABA), leukotriene modifiers, and long-acting muscarinic antagonists (LAMA). Additionally, rapid relief medications, including short-acting β2 receptor agonists (SABA), and allergen immunotherapy (AIT) are also utilized44. Additionally, biological agents have become increasingly common for patients with severe asthma45. Despite these treatments, the complex and multifactorial pathogenesis of allergic asthma makes effective management difficult. Thus, exploring treatment mechanisms and identifying more effective options remain central to current research on allergic asthma.
The airway epithelial barrier serves as the primary defense against respiratory pathologies induced by pathogens and environmental pollutants. Impairment of this barrier function represents a pathological feature in asthma development. The 16HBE cell line, derived from human bronchial epithelium, retains critical morphological and functional characteristics of native airway epithelium, including well-organized tight junctions, receptor expression profiles, cytokine production capacity, and mucus secretory activity46.When exposed to asthma triggers such as allergens, pollutants, or inflammation-related molecules, 16HBE cells release pro-inflammatory mediators, which exacerbate asthma-related inflammation. Notably, mucus secreted by respiratory epithelial cells plays a crucial role in defending the airways and clearing particles and pathogens. Studies have shown that the 16HBE cell line possesses mucus-secretory capabilities and can produce MUC5AC, mirroring bronchial epithelial cell functions47. Experimental evidence indicates that IL-13 stimulation significantly upregulates both inflammatory cytokine secretion and MUC5AC expression in this cellular model48. In this research, the IL-13-induced 16HBE cell model and the OVA-induced mouse model were employed to investigate the effects and mechanisms of K3G in asthma.
Excessive production of inflammatory cytokines characterizes airway inflammation in asthma. Allergens make airway epithelial cells release factors like IL-8, IL-6, etc., worsening inflammation36. IL-6 serves as a biomarker for asthma, while IL-8 promotes immune cell chemotaxis49,50. Previous studies showed upregulation of some factors in IL-13-stimulated cells51. In this study, the levels of inflammatory factors IL-1β, IL-6, and IL-8 in the control group increased, which may have been caused by the transient release of inflammatory mediators induced by the serum culture medium, mechanical digestion, and mechanical stimulation during cell culture and processing, leading to an increase in basal secretion. Although the baseline levels were higher in the normal group, the levels of pro-inflammatory cytokines were further elevated in IL-13-induced 16HBE cells and OVA-induced asthmatic mice. K3G treatment reduced these levels in vitro and in vivo, indicating its anti-inflammatory and therapeutic potential.
Here is an academic article about K3G and asthma. Airway inflammation leads to dysregulated mucus production. MUC5AC, a key mucin secreted by bronchial epithelial cells6. Studies reveal that normal human and mouse lungs maintain low baseline MUC5AC secretion in proximal airways, with minimal detection in distal regions. However, inflammatory cytokine stimulation triggers significant overexpression of this mucin52,53. Evidence shows that inhibiting MUC5AC can reverse mucus hypersecretion and alleviate asthma symptoms54,55. In our study, MUC5AC expression increased in both models and decreased after K3G treatment. Elevated MUC5AC levels are associated with airway inflammation caused by various irritants. We observed a synchronous increase in MUC5AC and inflammatory cytokines, suggesting that K3G’s effects on asthma may relate to its regulation of inflammatory cytokines and mucins.
TLR4 is a pattern-recognition receptor in vascular and airway epithelial cells, activates the NF-κB pathway upon stimulation, causing inflammatory factors release56. TLR4 induced IκBα phosphorylation and degradation enable NF-κB release and binding. Phosphorylated NF-κB translocates to the nucleus, promoting gene expression related to NLRP3 inflammasome57. Inhibiting the TLR4/NF-κB/NLRP3 pathway can reduce airway inflammation in obese asthma31. K3G decreases NLRP3, TLR4, p-IκBα, and p-P65 protein expression and inhibits IL-13-induced NF-κB activation, indicating its anti-inflammatory effect via this pathway.
The NOTCH signaling pathway is critical for regulating inflammation, immunity, ect58, and is a key regulator of airway inflammation in asthma21,59. Excessive mucus secretion in asthma is linked to persistent NOTCH activation60,61. NOTCH is a promising asthma treatment target. In IL-13-induced 16HBE cells and OVA-induced asthmatic mice, NOTCH1-3 and DLL4 expression increased, which was reversed by K3G, suggesting targeted inhibition of NOTCH1 for anti-inflammatory effects.
In conclusion, K3G reduces airway inflammation and mucus hypersecretion in IL-13-induced 16HBE and OVA-induced mouse models by decreasing inflammatory cytokine release and inhibiting MUC5AC secretion. Additionally, it also inhibits the TLR4/NF-κB/NLRP3 pathway and regulates NOTCH pathway components in asthma. which suggesting NOTCH1 is potential for allergic asthma treatment. However, K3G’s direct target in reducing airway inflammation via NOTCH pathway inhibition remains to be identified through gene editing or agonist-based experiments. The interaction between TLR4/NF-κB/NLRP3 and the NOTCH pathway also warrants more in-depth analysis to better understand the multi-target synergistic mechanism of K3G. Current studies on inflammatory factors mainly focus on IL-6, IL-8, and MUC5AC. However, future research should also consider the dynamic changes in Th2 cytokines such as IL-4 and IL-5, as well as other mucins like MUC5B, in order to comprehensively assess K3G’s regulatory effects on asthma heterogeneity.
Materials and methods
Materials
16HBE cells were obtained from ATCC (Manassas, VA). Female BALB/c mice were purchased from Vital River Laboratory Animal Co., Ltd. (Beijing, China). K3G was sourced from Pufeide Biological Technology Co., Ltd. (Chengdu, China). IL-13 was acquired from Peprotech (Cranbury, NJ, USA). The Cell Counting Kit-8 (CCK-8) reagent was provided by Kit-Dojindo (Tokyo, Japan), and ELISA kits were from Jiancheng Bioengineering Institute (Nanjing, China). MUC5AC antibody was from Abcam (Cambridge, UK), and TRIzol reagent was from Invitrogen (Carlsbad, CA, USA). The HiScript III RT SuperMix Kit and ChamQ Universal SYBR qPCR Master Mix were from Vazyme (Nanjing, China). The enhanced chemiluminescence kit was purchased from Millipore (Bedford, MA, USA). RIPA buffer was from Beyotime (Shanghai, China). Protease inhibitors and PVDF membranes were from Bio-Rad (Hercules, CA, USA). The RNeasy Mini Kit was from Qiagen (Valencia, CA, USA), and DMEM medium was purchased from Gibco (Grand Island, NY, USA). 10% fetal bovine serum, 1% penicillin-streptomycin, and ovalbumin (OVA) were from Sigma (St. Louis, MO, USA). The ultrasonic nebulizer (PARI BOY) was from Bavaria, Germany.
Cell culture and treatmen
The 16HBE cell line, derived from human airway epithelial tissue, was cultured in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin at 37 °C, in 5% CO2, and 95% humidity. For the experiments, cells were treated with different concentrations of K3G (purity > 98%) or IL-13 for 24 h, followed by treatment with K3G (5, 25, 50, or 100 µM) for another 24 h, with or without IL-13.
In vitro cell viability assay
The CCK-8 assay was used to assess cell viability. In this experiment, 16HBE cells were seeded in 96-well plates at a density of 5,000 cells per well and treated with K3G at concentrations of 0, 5, 25, or 50 µM for 24 h. After treatment, 100 µL of serum-free medium containing 10 µL of CCK-8 solution was added to each well and incubated for 2 h. Absorbance at 450 nm was measured using a Multiskan spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Asthma model and animal treatment
Thirty female BALB/c mice (4 weeks old, weighing 18–20 g) were housed in a pathogen-free environment at 22 ± 2 °C with free access to food and water. All animal experiments followed the ARRIVE guidelines and were approved by the Guangxi University of Chinese Medicine Institutional Welfare and Ethical Committee (No. DW20231214-278), ensuring compliance with relevant guidelines and regulations. The mice were divided into five experimental groups (n = 6/group): control (saline-sensitized), OVA, OVA + K3G 10 mg/kg (L), OVA + 20 mg/kg (M), and OVA + K3G 40 mg/kg (H). An asthma model was established based on previously published protocols62, with modifications. Briefly, Except for the control group, mice in the other groups were intraperitoneally injected with 200 µL of sensitization solution (100 µg OVA and 2 mg aluminum hydroxide) on days 0, 7, and 14. From days 21 to 35, all groups (except control) were exposed to nebulized 5% OVA using an ultrasonic nebulizer. Mice in the K3G groups were administered K3G solution orally for 7 consecutive days starting on day 28. On day 36, the mice were anesthetized with an intraperitoneal injection of sodium pentobarbital and euthanized by cervical dislocation. Bronchoalveolar lavage fluid (BALF) along with lung tissues were subsequently collected. Lungs without lavage were fixed in 4% paraformaldehyde (PFA) for histopathological and periodic acid-Schiff (PAS) staining, while the remaining lung tissue was stored at -80 °C for further analysis.
Histological analysis
Lung tissues were fixed in 4% PFA, embedded in paraffin, and sectioned at 5 μm. Sections were stained with PAS to evaluate goblet cell hyperplasia in the bronchi, as described previously63. PAS-positive cells were quantified using ImageJ software (California, USA) and expressed per 100 μm of the basement membrane. Each histological analysis was performed in triplicate.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent and purified with the RNeasy Mini Kit. cDNA synthesis was carried out using the HiScript III RT SuperMix Kit, following the manufacturer’s instructions. The resulting cDNA was utilized for quantitative real-time PCR (qRT-PCR) to measure the mRNA expression levels of mucin5AC (MUC5AC), NOTCH1, NOTCH2, NOTCH3, and DLL4. The relative mRNA levels were normalized to GAPDH levels, and differences in mRNA expression were calculated using the 2−ΔΔCt method. The primer sequences used are summarized in Table 1.
ELISA assay
After IL-13 and K3G treatment, supernatants from 16HBE cells were collected. Lung tissues were homogenized in RIPA buffer with protease inhibitors. The concentrations of histamine, IgE, TNF-α, IL-1β, IL-6, IL-8, and MUC5AC in the culture supernatants and lung tissues were measured using commercial ELISA kits.
Detection of nitrite levels
16HBE cells were pre-treated with K3G (5, 25, and 50 µM) for 1 h, followed by exposure to IL-13 (for 24 h. Nitrite levels in the supernatant were quantified using Griess reagent at OD 540 nm.
Flow cytometry assay for ROS
16HBE cells were cultivated with K3G (5, 25, and 50 µM) for 1 h, then treated with IL-13 for 6 h. After treatment, cells were stained with DCFH2-DA (1 µM) for 30 min. Fluorescence was analyzed using an LSRFortessa™ Cell Analyzer (BD, Franklin Lakes, NJ, USA) with the FITC channel. All experiments were performed in triplicate.
Western blot
Cells were lysed on ice for 30 min using RIPA buffer containing protease and phosphatase inhibitors. Protein concentration was quantified using the Thermo Protein Assay Kit, and equal amounts of protein (60 µg) were resolved by 6-12% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% nonfat dry milk in 0.1% Tween-20 TBS for 2 h at room temperature, then incubated overnight with primary antibody at 4 °C. After incubation with secondary antibody for 1 h at room temperature, protein bands were detected using enhanced chemiluminescence and imaged with the BIO-RAD GelDoc XR system (Bio-Rad).
Immunofluorescence (IF) staining
Tissue Sect. (5 μm) and airway epithelial cell climbing slices were processed for immunofluorescence (IF) staining. Sections were dewaxed in xylene, rehydrated, and antigen retrieval was performed in Tris-EDTA buffer (pH 9.0) at 95 °C for 15 min. Climbing slices were fixed with 4% PFA and permeabilized with 0.5% Triton X-100, and blocked with 5% BSA for 1 h. After overnight incubation with anti-MUC5AC primary antibody at 4 °C, slides were incubated with the secondary antibody, stained with FITC and DAPI, and fluorescence images were captured. Mean fluorescence intensity (MFI) of positive cells was quantified using ImageJ software.
16HBE cells (2 × 105) were plated in a 35 mm confocal dish (SPL, Pocheon, Korea) and incubated overnight. After a 1 h K3G (50 µM) treatment, cells were treated with IL-13 for 6 h. The cells were incubated overnight with primary antibody against NF-κB/P65 (1:200) at 4 °C, followed by 1 h incubation with Alexa Fluor 594-conjugated secondary antibody (1:200). After fixation, cells were stained with Hoechst 33,342 (1:1000) for nuclear visualization. Confocal images were captured using a Leica TCS SP5 microscope (Leica, Wetzlar, Germany) at 60× magnification with excitation at 588 nm and emission from 615 to 690 nm.
Molecular docking
Molecular docking of K3G with the 3D structure of NOTCH1 (UniProt ID P46531) was performed using Discovery Studio 4.5 and UCSF Chimera 1.7. The protein structure was regularized to identify key amino acids within the binding pocket. All K3G conformers were docked to the active site using LibDock, following energy minimization with the “prepare ligand” protocol. The binding affinity of the docked compound was evaluated based on its binding mode at the active site.
Surface plasmon resonance (SPR) analysis
SPR analysis was performed using a Biacore X100 system (Cytiva, USA) with a CM5 sensor chip. Recombinant human NOTCH1 was immobilized on the CM5 chip using an amine-coupling kit. K3G was dissolved in HBS-EP buffer (Cytiva), and different concentrations were injected into the flow system at 30 µL/min. The association phase lasted 180 s, followed by a 300-second dissociation phase. Binding kinetics were analyzed with Biacore X100 Control Software (version 2.0.2).
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 9.0 software (GraphPad, San Diego, CA, USA), with data presented as mean ± standard deviation from three independent experiments. One-way ANOVA followed by Bonferroni’s post-hoc test was used to compare differences between groups. A p-value of < 0.05 was considered significant.
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
References
Gan, Q. et al. The causal association between obstructive sleep apnea and Child-Onset asthma come to light: A Mendelian randomization study. Nat. Sci. Sleep. 16, 979–987 (2024).
Joulia, R. et al. Mast cell activation disrupts interactions between endothelial cells and pericytes during early life allergic asthma. J. Clin. Invest. 134, (2024).
Ahmad, K., Khanam, R., Kabir, E. & Jurges, H. The healthcare cost burden of asthma in children: A longitudinal Population-Based study. Value Health. 26, 1201–1209 (2023).
Yang, X. et al. Medical resource utilization and the associated costs of asthma in China: a 1-year retrospective study. BMC Pulm Med. 23, 463 (2023).
Reddel, H. K. et al. Global initiative for asthma strategy 2021: Executive summary and rationale for key changes. Respirology 27, 14–35 (2022).
Severine Letuve, C. S. Role of DNA methylation in Muc5AC hyperexpression in severe asthma. Eur. Respir J. 54, PA5404 (2019).
Peters, M. C. et al. A transcriptomic method to determine airway immune dysfunction in T2-High and T2-Low asthma. Am. J. Respir Crit. Care Med. 199, 465–477 (2019).
Chen, M. Z., Wang, S. A., Hsu, S. C., Silva, K. A. S. & Yang, F. M. Home dust mites promote MUC5AC Hyper-Expression by modulating the sNASP/TRAF6 axis in the airway epithelium. Int. J. Mol. Sci. 23, (2022).
Pinart, M. et al. Comorbidity of eczema, rhinitis, and asthma in IgE-sensitised and non-IgE-sensitised children in medall: A population-based cohort study. Lancet Respir Med. 2, 131–140 (2014).
Shaw, D. E. et al. Association between neutrophilic airway inflammation and airflow limitation in adults with asthma. Chest 132, 1871–1875 (2007).
Gandhi, N. A. et al. Targeting key proximal drivers of type 2 inflammation in disease. Nat. Rev. Drug Discov. 15, 35–50 (2016).
Nam, H. H. et al. Gekko gecko extract attenuates airway inflammation and mucus hypersecretion in a murine model of ovalbumin-induced asthma. J. Ethnopharmacol. 282, 114574 (2022).
Tschumperlin, D. J. Physical forces and airway remodeling in asthma. N Engl. J. Med. 364, 2058–2059 (2011).
Dost, A. F. M. et al. Organoids model transcriptional hallmarks of oncogenic KRAS activation in lung epithelial progenitor cells. Cell. Stem Cell. 27, 663–678e8 (2020).
Tasca, A. et al. Notch signaling induces either apoptosis or cell fate change in multiciliated cells during mucociliary tissue remodeling. Dev. Cell. 56, 525–539 (2021). e6.
Vladar, E. K. et al. Notch signaling inactivation by small molecule gamma-secretase inhibitors restores the multiciliated cell population in the airway epithelium. Am. J. Physiol. Lung Cell. Mol. Physiol. 324, L771–L782 (2023).
Zhou, B. et al. Notch signaling pathway: Architecture, disease, and therapeutics. Signal. Transduct. Target. Ther. 7, 95 (2022).
Gomi, K. et al. JAG1-Mediated Notch signaling regulates secretory cell differentiation of the human airway epithelium. Stem Cell. Rev. Rep. 12, 454–463 (2016).
Lafkas, D. et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528, 127–131 (2015).
Mori, M. et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development 142, 258–267 (2015).
Tindemans, I. et al. Notch signaling licenses allergic airway inflammation by promoting Th2 cell lymph node egress. J. Clin. Invest. 130, 3576–3591 (2020).
Yao, Y. E. et al. Regulation of gammadeltaT17 cells by Mycobacterium vaccae through interference with Notch/Jagged1 signaling pathway. Braz J. Med. Biol. Res. 53, e9551 (2020).
Shi, L., Ma, Y., Zheng, C. & Zhang, Q. The effect of blocking Notch signaling by gamma-secretase inhibitor on allergic rhinitis. Int. J. Pediatr. Otorhinolaryngol. 98, 32–38 (2017).
Zhong, Z. et al. Blocking Notch signalling reverses miR-155-mediated inflammation in allergic rhinitis. Int. Immunopharmacol. 116, 109832 (2023).
Maspero, J. et al. Type 2 inflammation in asthma and other airway diseases. ERJ Open. Res. 8, (2022).
Siddiqui, S. et al. Epithelial miR-141 regulates IL-13-induced airway mucus production. JCI Insight 6, (2021).
Bajbouj, K. et al. IL-13 Augments Histone Demethylase JMJD2B/KDM4B Expression Levels, Activity, and Nuclear Translocation in Airway Fibroblasts in Asthma. J Immunol Res 6629844 (2021). (2021).
Xiang, L. L. et al. IL-13 regulates Orai1 expression in human bronchial smooth muscle cells and airway remodeling in asthma mice model via LncRNA H19. J. Asthma Allergy. 15, 1245–1261 (2022).
Karkout, R. et al. Female-specific enhancement of eosinophil recruitment and activation in a type 2 innate inflammation model in the lung. Clin. Exp. Immunol. 216, 13–24 (2024).
Gerber-Tichet, E., Blanchet, F. P., Majzoub, K. & Kremer, E. J. Toll-like receptor 4 - a multifunctional virus recognition receptor. Trends Microbiol. 33, 34–47 (2025).
Yang, Z. et al. Involucrasin B suppresses airway inflammation in obese asthma by inhibiting the TLR4-NF-κB-NLRP3 pathway. Phytomedicine Int. J. Phytother Phytopharm. 132, 155850 (2024).
Xu, S., Wang, D., Tan, L. & Lu, J. The role of NLRP3 inflammasome in type 2 inflammation related diseases. Autoimmunity 57, 2310269 (2024).
Chen, X. L., Xiao, Q. L., Pang, Z. H., Tang, C. & Zhu, Q. Y. Molecular mechanisms of an-chuan granule for the treatment of asthma based on a network Pharmacology approach and experimental validation. Biosci. Rep. 41, BSR20204247 (2021).
Li, M. et al. Isolation and characterization of secondary metabolites from the leaves of sauropus spatulifolius Beille and their potential biological assays. Fitoterapia 156, 105100 (2022).
Qiu, Q. et al. Study on the Spectrum-Effect Correlation of Anti-Inflammatory Active Extract of Sauropus spatulifolius Beille. J Anal Methods Chem 5646546 (2022). (2022).
Wei, X. et al. The protective effects of sauropus spatulifolius on acute lung injury induced by lipopolysaccharide. J. Sci. Food Agric. 98, 4420–4426 (2018).
Moriyama, H., Iizuka, T., Nagai, M., Miyataka, H. & Satoh, T. Antiinflammatory activity of heat-treated Cassia Alata leaf extract and its flavonoid glycoside. Yakugaku Zasshi. 123, 607–611 (2003).
Hazni, H., Ahmad, N., Hitotsuyanagi, Y., Takeya, K. & Choo, C. Y. Phytochemical constituents from Cassia Alata with Inhibition against methicillin-resistant Staphylococcus aureus (MRSA). Planta Med. 74, 1802–1805 (2008).
Liao, Q. et al. An integrated method for optimized identification of effective natural inhibitors against SARS-CoV-2 3CLpro. Sci. Rep. 11, 22796 (2021).
Tajiri, T. et al. Pathophysiological relevance of sputum MUC5AC and MUC5B levels in patients with mild asthma. Allergol. Int. Off J. Jpn Soc. Allergol. 71, 193–199 (2022).
Zhang, C. et al. Qingwenzhike prescription alleviates acute lung injury induced by LPS via inhibiting TLR4/NF-kB pathway and NLRP3 inflammasome activation. Front. Pharmacol. 12, (2021).
Dong, L. et al. Hypoxic hUCMSC-derived extracellular vesicles attenuate allergic airway inflammation and airway remodeling in chronic asthma mice. Stem Cell. Res. Ther. 12, 4 (2021).
Song, M. et al. Geographical differences of risk of asthma and allergic rhinitis according to urban/rural area: A systematic review and Meta-analysis of cohort studies. J. Urban Health. 100, 478–492 (2023).
Papadopoulos, N. G., Miligkos, M. & Xepapadaki, P. A. Current perspective of allergic asthma: from mechanisms to management. in Allergic Diseases – From Basic Mechanisms To Comprehensive Management and Prevention (eds Traidl-Hoffmann, C., Zuberbier, T. & Werfel, T.) 69–93 (Springer International Publishing, Cham, doi:https://doi.org/10.1007/164_2021_483. (2022).
Bernstein, J. A., Llanos, J. P., Hunter, G., Martin, N. & Ambrose, C. S. Efficacy of biologics in patients with allergic severe asthma, overall and by blood eosinophil count: A literature review. Adv. Ther. 40, 4721–4740 (2023).
Peng, J., Zhao, F., Kang, X., Aierken, N. & Li, Q. Matricaria Chamomilla L. Ameliorates asthma by protecting OVA-induced rats and LPS-induced human bronchial epithelial cells through suppressing autophagy and apoptosis. Food Sci. Nutr. 13, e70030 (2025).
Callaghan, P. J., Ferrick, B., Rybakovsky, E., Thomas, S. & Mullin, J. M. Epithelial barrier function properties of the 16HBE14o- human bronchial epithelial cell culture model. Biosci. Rep. 40, BSR20201532 (2020).
Yu, X. et al. Wogonoside ameliorates airway inflammation and mucus hypersecretion via NF-κB/STAT6 signaling in Ovalbumin-Induced murine acute asthma. J. Agric. Food Chem. 72, 7033–7042 (2024).
Pan, R. et al. Diagnostic value of IL-6 for patients with asthma: A meta-analysis. Allergy Asthma Clin. Immunol. Off J. Can. Soc. Allergy Clin. Immunol. 19, 39 (2023).
Matsushima, K., Yang, D. & Oppenheim, J. J. Interleukin-8: an evolving chemokine. Cytokine 153, 155828 (2022).
Liang, J., Zhuang, R., Sun, X., Zhang, F. & Zou, B. Apremilast mitigates Interleukin (IL)-13-induced inflammatory response and mucin production in human nasal epithelial cells (hNECs). Bioengineered 12, 8583–8593 (2021).
Adler, K. B., Tuvim, M. J. & Dickey, B. F. Regulated mucin secretion from airway epithelial cells. Front. Endocrinol. 4, (2013).
Hill, D. B., Button, B., Rubinstein, M. & Boucher, R. C. Physiology and pathophysiology of human airway mucus. Physiol. Rev. 102, 1757–1836 (2022).
Takami, S. et al. Glucocorticoids inhibit MUC5AC production induced by transforming growth factor-alpha in human respiratory cells. Allergol. Int. 61, 451–459 (2012).
Li, T., Wang, Y., Huang, S. & Tang, H. The regulation mechanism of MUC5AC secretion in airway of obese asthma. Cell. Mol. Biol. Noisy–Gd. 68, 153–159 (2022).
Guo, S. et al. Polygala tenuifolia willd. Extract alleviates LPS-induced acute lung injury in rats via TLR4/NF-κB pathway and NLRP3 inflammasome suppression. Phytomedicine Int. J. Phytother Phytopharm. 132, 155859 (2024).
Zhang, T., Ma, C., Zhang, Z., Zhang, H. & Hu, H. NF-κB signaling in inflammation and cancer. MedComm 2, 618–653 (2021).
Liu, J., Sato, C., Cerletti, M. & Wagers, A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 92, 367–409 (2010).
Kang, J. H. et al. Gamma-secretase inhibitor reduces allergic pulmonary inflammation by modulating Th1 and Th2 responses. Am. J. Respir Crit. Care Med. 179, 875–882 (2009).
Liu, Y. et al. Role of Notch signaling pathway in Muc5ac secretion induced by atmospheric PM(2.5) in rats. Ecotoxicol. Env Saf. 229, 113052 (2022).
Reid, A. T. et al. Blocking Notch3 signaling abolishes MUC5AC production in airway epithelial cells from individuals with asthma. Am. J. Respir Cell. Mol. Biol. 62, 513–523 (2020).
Beigelman, A. et al. Azithromycin attenuates airway inflammation in a noninfectious mouse model of allergic asthma. Chest 136, 498–506 (2009).
Liou, C. J. & Huang, W. C. Dehydroepiandrosterone suppresses eosinophil infiltration and airway hyperresponsiveness via modulation of chemokines and Th2 cytokines in ovalbumin-sensitized mice. J. Clin. Immunol. 31, 656–665 (2011).
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81860867); Guangxi Natural Science Fund (2018JJA140356) and Innovation Project of Guangxi Graduate Education of GXUCM (YCBXJ2023011).
Author information
Authors and Affiliations
Contributions
J.S., W.L. and Y.W. Conceptualization, Y.W., Q.W. and W.Z. methodology, Y.Y. and D.W. resources, Y.W., Q.W. and W.Z. formal analysis, J.S. data curation, Y.W., Q.W. and W.Z. writing-original draft, J.S. and W.L. writing—review and editing, J.S. and W.L. supervision, W.L. and Y.W. funding acquisition, D.W., J.S. and W.L. project administration. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Y., Wang, Q., Zhu, W. et al. NOTCH pathway was involved in Kaempferol 3-O-gentiobioside attenuated airway inflammation and mucus hypersecretion. Sci Rep 15, 10383 (2025). https://doi.org/10.1038/s41598-025-95280-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95280-8
Keywords
This article is cited by
-
Distribution, Accumulation of Sabia parviflora Active Components, and Anti-inflammatory Activity of Flavonoids Monomer
Revista Brasileira de Farmacognosia (2025)