Abstract
Skin macrophages play important roles in the response to external stimuli. Human skin equivalents (HSEs) incorporating the human monocytic cell line THP-1 were fabricated to generate immunocompetent human skin models. These HSEs were used to investigate the influence of the skin microenvironment and ultraviolet A (UVA) on macrophages. Transcriptomic analysis revealed that THP-1 cells in HSEs were enriched in extracellular matrix interaction hallmark but downregulated in DNA replication hallmark. Upon UVA exposure, immunocompetent HSEs presented epidermal distortion and increased DNA double-strand breaks (DSBs). The genes associated with oxidative stress and the inflammatory response were significantly upregulated in THP-1 cells. When the photoprotective agent mycosporine-2-glycine from cyanobacteria was applied to HSEs, the incidence of UVA-induced DSBs was significantly lower, and inflammatory and UV response hallmarks were downregulated in THP-1 cells. Taken together, these results suggest that immunocompetent HSEs can be used to investigate the responses of skin-resident macrophages to external stimuli.
Similar content being viewed by others
Introduction
Upon exposure to solar ultraviolet (UV) radiation, UVB (290–320 nm) is almost completely absorbed by the epidermis. UVB induces DNA damage in the skin, and unrepaired damaged DNA results in the apoptosis of keratinocytes, causing sunburn cells and contributing to carcinoma1,2. UVA (320–400 nm), which can penetrate deeply into the dermis, is efficient at generating reactive oxygen species (ROS), such as superoxide anions, hydrogen peroxide, and hydroxyl radicals3. The accumulation of ROS leads to oxidative stress and extracellular matrix (ECM) remodeling by matrix metalloproteinases (MMPs), including MMP1, MMP3, and MMP9, which reduce structural collagen or elastin4. Moreover, free radicals trigger biological responses through the activation of nuclear factor kappa B (NF-κB) and activator protein-1 (AP-1). This activation induces the expression of proinflammatory cytokines such as interleukin-6 (IL-6), interleukin-1β (IL-1β), cyclooxygenase-2 (COX-2), and tumor necrosis factor-α (TNF-α) and causes a DNA damage response via the expression of the senescence-associated secretory phenotype (SASP)5.
The skin has developed various defense mechanisms to cope with the effects caused by UV exposure to maintain tissue homeostasis. DNA photolesions and DNA fragments activate DNA repair mechanisms1. Fibroblasts also respond to damage by attempting to repair and synthesize new collagen. Moreover, endogenous antioxidant molecules are produced to detoxify ROS to protect against UV-induced skin damage. Antioxidant enzymes, including superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase, and glutathione transferase, help reduce ROS in the skin3,6. Although human skin explants have been used to evaluate the effectiveness of the active ingredients that protect the skin from UVA damage2, an in vitro three-dimensional (3D) human skin equivalent (HSE) and reconstructed human epidermis (RhE) have been utilized as alternative models for studying the harmful impacts of UVA and UVB radiation that lead to changes in gene expression7,8,9. DNA lesions, such as DNA double-strand breaks (DSBs) and oxidative DNA damage, are detected by the expression of the biomarkers gamma-histone 2 A family member X (γ-H2AX)10 or 8-oxo-2′-deoxyguanosine (8-OHdG)11,12. Moreover, transcriptomic analysis of UVA-exposed skin revealed the effects of pathways and functions of expressed genes involved in inflammation, the immune response, and the cellular metabolism of lipids and glucose13.
UV-exposed HSE and RhE models have been successfully used as skin models for evaluating UV protectants6,14; however, these models often lack immune components, which are crucial for accurately mimicking the in vivo skin microenvironment. Therefore, developing immunocompetent models of human skin can provide valuable insights into the influence of external stimuli on the skin microenvironment and its responses15,16. In prior studies, innate immune cells such as monocyte-derived Langerhans cells (mo-LCs), monocyte-derived dendritic cells (mo-DCs), and monocyte-derived macrophages (mo-Macs) have been utilized in the development of immunocompetent skin models15,17,18,19. The human leukemic monocyte lymphoma cell line U937 was cocultured with RhE to investigate the impact of UV exposure on skin barrier integrity20. Furthermore, HSE containing both mo-LCs and mo-DCs was used to study the effects of UV radiation, replicating their functions in skin environments21. In another study, human CD14+ monocytes from patients with systemic sclerosis were used to study skin pathology and the crosstalk between macrophages and fibroblasts in HSEs22. Among these studies, questions regarding the changes in the transcriptomes of skin-associated macrophages in response to UVA exposure in the context of HSEs have never been addressed.
Given the influence of UVA on cutaneous cells, enhancing our understanding of the cellular and molecular effects of UVA exposure on immune cells associated with human skin, including macrophages, is imperative. Classically activated macrophages (M1) promote inflammatory conditions during both acute and chronic exposure by producing proinflammatory mediators, recruiting immune cells, and degrading the ECM in the dermis through the release of MMPs and ROS23,24. In contrast, alternatively activated macrophages (M2) play essential roles in maintaining skin homeostasis by providing growth factors, clearing debris, and promoting tissue repair and regeneration25,26. Furthermore, prolonged exposure to UV has been associated with immunosuppression24. Cytokines induced by UV exposure, especially IL-10, exert immunosuppressive effects by suppressing antigen presentation and Th1 cell proliferation27. This exposure further suppresses the secretion of IL-12 by dendritic cells/macrophages28. Although previous studies have reported the responses of macrophages to UV radiation, understanding the impact of UVA radiation on macrophages in the context of an intact tissue microenvironment is crucial for developing strategies to mitigate the potentially harmful effects of excessive sun exposure.
Mycosporine-2-glycine (M2G), a natural sunscreen compound found in various species, such as cyanobacteria and micro- and macroalgae, is a mycosporine-like amino acid (MAA) that has strong potential photoprotective properties against UV light wavelengths, exhibiting an absorption maximum range between 310 and 365 nm29,30. In our previous study, M2G exhibited protective effects against H2O2-induced oxidative stress in human melanoma and normal human skin fibroblast. Moreover, M2G protected cells against DNA damage in melanoma cell lines, as analyzed by a comet assay31. Furthermore, the production of an inflammatory molecule, nitric oxide (NO), was reduced by the M2G compound in a lipopolysaccharide (LPS)-induced murine macrophage. M2G treatment also downregulated the expression of antioxidant-associated genes, including SOD1, CAT, HMOX1, and NRF2, in H2O2-stressed macrophages and inhibited the production of inflammatory mediators by repressing the NF-κB pathway32. However, the photoprotective effect of M2G against UV exposure in an in vitro skin model has not been investigated.
To interrogate the crosstalk between skin and tissue-resident innate immune cells under normal or stressed conditions, skin models incorporating macrophages in HSEs are highly useful because of their ease of manipulation. In addition, skin models can be used to evaluate the potential of a novel UV protective agent against UVA damage. In this study, we utilized our previously established HSE models and employed surface plasma treatment to address these questions. THP-1, a human monocytic cell line, was incorporated into these models to generate immunocompetent full-thickness human skin, and the gene expression profiles of HSEs were characterized. Some previous reports utilized the THP-1 cell line in the context of 3D skin culture to study dermal dendritic cell activation or tissue repair processes, but the response to UV exposure has not yet been documented18,33,34. In this study, we investigated cytotoxicity responses, and gene expression changes in immunocompetent 3D skin models were assessed after UVA irradiation. Additionally, the photoprotective effects of the M2G molecule in HSEs were evaluated, and differentially expressed genes relevant to skin homeostasis after UVA irradiation were identified in resident macrophages.
Results
The THP-1 monocytic cell line exhibited mixed M1- and M2-like phenotypes in 3D HSEs
HSEs were generated through the use of a plasma-treated culture surface to increase surface hydrophilicity (Fig. 1A). To generate immunocompetent 3D HSEs incorporating immune cells, 5 × 105 THP-1 monocytic cells were cocultured with fibroblasts to generate the dermis, followed by epidermis fabrication (Fig. 1B).
The resulting HSEs, designated “HSE.THP1”, displayed a multilayered structure of differentiated epidermis overlaying the dermis and exhibited a distinct arrangement of cells underneath the epidermal-dermal junction with no significant difference in cell density compared with HSEs without monocytes, designated “HSE”, and native human skin (Fig. 2A, B). The expression levels of early (keratin 14) and intermediate (keratin 10) differentiation markers in the epidermis were comparable across all the models (Fig. 2C). Despite a lower level of expression of loricrin in terminally differentiated epidermal cells within immunocompetent HSEs, the formation of the stratum corneum (SC) signified terminal epidermis differentiation (Fig. 2C and Supplementary Fig. 1A). Moreover, the presence of tight junctions, as indicated by claudin-1 and zonula occludens-1 (ZO-1) expression, was observed (Fig. 2C). The protective barrier function, assessed by the TEER value, was mostly consistent at approximately 500 Ω•cm2, even with varying immune cell numbers in the HSEs (Supplementary Fig. 1B), confirming the stability of the fabricated HSEs, as described in our previous study35.
A Representative images of H&E-stained HSE, immunocompetent HSE (HSE.THP-1), and native human skin. The blue arrows indicate the stratum basale (SB), and the red arrows highlight the area beneath the epidermis. Scale bar: 100 μm, epidermis (E), dermis (D). B Quantification of cell numbers/mm2 in the area beneath the epidermis in the HSE and HSE.THP-1 samples. C Immunofluorescence staining of epidermal differentiation (red) and tight junction (TJ) markers (green). D Confocal microscopy images showing the expression of the dermal macrophage markers CD68 (red), CD86 (green), and CD163 (green). The black and white dotted lines indicate epidermal-dermal junctions (basement membrane). E–G Quantitation of CD68+, CD86+, and CD163+ cells/mm2 in the dermis. H The M1/M2 ratio (numbers of CD86+ cells/numbers of CD163+ cells) in the dermis. The cell nuclei were counterstained with Hoechst (blue). Photomicrograph scale bar: 50 µm; insets, 20 µm. The data represent the means ± SEMs. Statistical significance is indicated by *p < 0.05, **p < 0.01, ****p < 0.0001, and ns, not significant (n = 3 per group); unpaired t test.
To characterize the immune cells integrated into the HSE, paraffin-embedded sections were stained with monocyte/macrophage-specific markers. HSE that did not contain the THP-1 cell line showed no detectable staining for any of the other markers. THP-1 cells in the HSE.THP-1 group expressed CD68, CD86, and CD163, which are indicative of the resting-state (M0), M1, and M2 macrophage phenotypes within HSEs, thereby mirroring the characteristics observed in native human skin (Fig. 2D and Supplementary Fig. 1C). Although the quantification of immune cell densities revealed significantly greater numbers of THP-1 cells in HSE.THP-1 group, possibly as a result of the increased number of integrated THP-1 cells (Fig. 2E–G), the M1-to-M2 ratio in the dermis (CD86+/CD163+ cells) was consistent with that observed in human skin (Fig. 2H). Additionally, THP-1 cells in the HSE.THP-1 group were identified close to the epidermis, potentially influencing the cell arrangement beneath the epidermal-dermal junction, as shown by H&E staining. Based on these findings, our results indicate that THP-1 cells probably aid in the formation of new epidermal tissue. Therefore, the THP-1 cell line (5 × 105) was selected for further fabrication of immunocompetent HSEs.
The transcriptomic profiles of the integrated THP-1 cell line revealed the modulation of HSEs
In this study, THP-1 cells were stained for CD45 expression and analyzed by flow cytometry to confirm their positivity. Analysis revealed that 99.8% of the THP-1 cells were CD45 positive (Supplementary Fig. 2A, B), which allowed us to use CD45 as a marker to isolate THP-1 cells that were integrated into the HSEs from the whole-skin model (Supplementary Fig. 2C).
To characterize the phenotypes of the THP-1 cells that were incorporated into the HSEs, the transcriptomic profiles of the THP-1 cells in HSE were obtained by RNA sequencing. A total of 16,811 genes exhibited differential expression patterns between integrated macrophages (“HSE.THP1”) and 2D cultured THP-1 cell lines (“THP-1”). Among these genes, 2,145 genes were differentially expressed according to a twofold difference or greater (1760 upregulated and 385 downregulated genes (adjusted p value < 0.05) (Fig. 3A, B). As shown in the volcano plot in Fig. 3B, THP-1 cells from the HSE.THP1 group exhibited increased expression levels of genes associated with both “M1” and “M2” lineages (Supplementary Data 1). The M1-associated genes that were upregulated included CD80, CD86, IL6, and IL23A, whereas the M2-associated genes that were upregulated included ARGI, IL10, CD163, and MRC1 (Fig. 3B). Notably, CD163 exhibited the greatest increase (log2 FC = 14.74) in the integrated THP-1 cell line. Furthermore, upregulated transcripts of several key chemokines (CXCL8, CCL2, CXCL1, and CCL18), which play roles in supporting the recruitment of other immune cells, were detected. Downregulation of the expression levels of the chemokine receptors CCR2 and CXCR2 was observed in the HSE.THP-1 group, suggesting that fewer migratory and tissue-retaining behaviors of THP-1 cells were present in the HSEs.
All the samples were analyzed in triplicate. A Heatmap of the DEGs showing the relative expression levels between integrated THP-1 cells in 3D HSE (HSE.THP1) or THP-1 cells in 2D culture (THP-1) (|log2 FC|> 2 and adjusted p value < 0.05). B Volcano plot showing all mapped DEGs with at least twofold enrichment in the HSE.THP-1 population. The red dots indicate DEGs, with the left and right sides representing downregulated and upregulated genes, respectively. The green dots indicate DEGs whose expression levels did not significantly change. The x axis represents the log2-fold change in normalized gene expression, and the y axis represents the log10 adjusted p value < 0.05. C The top 10 significantly enriched GO terms associated with the HSE.THP1 population were generated. The GO terms are ordered according to the gene ratios. The size of the dots represents the number of DEGs associated with the GO terms, and the color represents the value of the adjusted p value (p.adjust). BP biological process, CC cellular component, MF molecular function. D Bar plots showing the number of DEGs in each of the high-level significant KEGG enrichment pathways of the HSE.THP1 population. The KEGG terms are ordered according to the value of p.adjust, represented by color. E Enrichment plots of up- or downregulated genes related to gene sets identified by GSEA based on the KEGG database. Nominal p value < 0.001.
Differentially Expressed Genes (DEGs) (2145 genes) in our data set were categorized into three distinct Gene Ontology (GO) categories (Fig. 3C and Supplementary Data 2). A variety of biological processes related to the regulation of skin development and skin homeostasis were generally increased in THP-1 cells in HSEs. For example, DEGs encoding proteins linked to “blood vessel development” (GO:0001568), including VEGFA, FGF1, SERPINE1, and MMP9, were upregulated. In the CC category, terms related to structural integrity, such as “extracellular matrix” (GO:0031012), “collagen-containing extracellular matrix” (GO:0062023), and “cell‒cell junction” (GO:0005911), were enriched (adjusted p value all < 1.3 × 10−10). The most upregulated genes were members of the collagen family, such as COL1A1, COL1A2, COL3A1, COL6A3, and COL4A2, whereas matrix metalloproteinase-encoding genes (i.e., MMP1, MMP2, MMP9, MMP12, and MMP19), which regulate tissue remodeling, wound healing, and cellular migration, were also highly expressed. Moreover, DEGs associated with molecular functions related to ECM structural constituents and binding proteins, such as laminin (LAMB2, LAMA4, LAMC2, and LAMB1), glycosaminoglycan (DCN and LUM), and integrin (SPP1, TGFBI, ITGAM, and ITGB2), were intermediate to highly expressed in the HSE.THP1 population.
To corroborate the enriched GO terms and investigate the associated molecular pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and gene set enrichment analysis (GSEA) based on the KEGG database were performed (Fig. 3D, E). The results revealed the top 10 significantly enriched KEGG pathways, including “Cytokine–cytokine receptor interaction” (hsa04060), “Complement and coagulation cascades” (hsa04610), and “ECM‒receptor interaction” (hsa04512) (adjusted p value all < 2.8 × 10−6). The downregulated genes were linked to “DNA replication” and “Cell cycle” (FDR-correlated p values all <0.005). Overall, these findings, encompassing phenotypic and genotypic changes that involve cytokine and chemokine activity and the immune responses of monocyte-derived macrophages, provide insights into how the THP-1 monocytic cell line adapts to HSEs and adapts to the phenotypes of skin-resident innate immune cells.
Morphological changes, DNA damage, and responses of HSEs and THP-1 cells upon UVA exposure
Impact of UVA dose and incubation time on HSE morphology
To determine the optimal UVA dose, we next assessed the impact of UVA exposure on our reconstructed HSEs in a range from low to high UVA doses based on previous studies (Supplementary Fig. 3). Low physiological doses (nonextreme UV radiation) in the range of 2 to ranging from 2–13 J/cm2 8,9 or medium doses ranging from 20–25 J/cm2 7,36 that irradiate HSEs generate toxic signals and impact the morphology of reconstructed human skin. When relatively low doses of 10 and 20 J/cm2 were used, low percentages of γ-H2AX-positive nuclei were detected after UVA irradiation of the HSEs (Supplementary Fig. 3A, B). The application of medium doses (in the range of 30–40 J/cm2) has previously been shown to be cytotoxic to keratinocytes and fibroblasts in reconstructed human skin13,37. To evaluate the morphological changes and cytotoxicity caused by higher doses of UVA, doses of 40 and 80 J/cm2 were applied to HSEs, and the changes in morphology and DNA damage were evaluated at 6 and 24 h postexposure. Notably, increased epidermal detachment, reduced late differentiation markers, and compromised barrier function were observed at 24 h post-UVA (Supplementary Fig. 3C–E). There were cells that clearly stained positive for γ-H2AX, particularly at 80 J/cm2 (Supplementary Fig. 3F, G). Furthermore, the expression levels of genes associated with DSBs (H2AX) and the oxidative stress response (OGG1) were significantly elevated at 80 J/cm2 at 24 h postexposure (Supplementary Fig. 3H). Therefore, a UVA dose of 80 J/cm2 and a 24-h incubation after UVA exposure were chosen for subsequent experiments.
Transcriptomic impact on THP-1 cells in the HSE after UVA exposure
To explore the genome-wide impact of UVA radiation on THP-1 cells in HSEs, RNA sequencing was performed on the THP-1 cell line, and the results were compared between UVA-exposed (HSE.THP1.UV) and nonexposed (HSE.THP1) conditions. In addition, the impact of the UV protective agent M2G on the response of THP-1 cells under UVA exposure (HSE.THP1.UV.M2G) was investigated. Principal component analysis (PCA) revealed that the biological triplicates clustered well together according to treatment (Fig. 4A). First, the THP-1 cell lines in the HSEs with and without UV exposure were compared. UVA exposure resulted in distinct changes in the expression of 13,459 genes in the THP-1 cell line in HSEs. Among 509 DEGs, 177 genes were upregulated, and 332 genes were downregulated in THP-1 cells from the HSE.THP1.UV condition, with the criteria of |log2 FC| > 1 and adjusted p value < 0.05 (Fig. 4B). Genes associated with skin wound regeneration, including genes encoding serine protease inhibitors (TFPI2 and SERPINB2) and genes associated with blood vessel and skin development (i.e., NPR3, DLL1, DLL4, and PTHLH), were upregulated. Upon UVA exposure, the THP-1 cell line presented increased expression of ECM-degrading enzymes (MMP1 and MMP10) and increased leukocyte chemotaxis (C5AR2 and SERPINE1) (Fig. 4C and Supplementary Data 3).
A PCA plot based on the normalized count data from each sample. B Heatmap of the DEGs is shown with |log2 FC| >1 and adjusted p value < 0.05. C Volcano plot showing all mapped genes of the sorted THP-1 cell line. Red dots indicate DEGs with up- or downregulated genes (|log2 FC|> 1, adjusted p value < 0.05). D Dot plots of the significantly enriched GO terms are shown according to the gene ratio. The size of the dots represents the number of DEGs, and the color represents the value of the adjusted p value (p.adjust). BP biological process, CC cellular component, MF molecular function. E Bar plot of the KEGG terms ordered according to the value of p.adjust. The red and blue bars indicate the number of transcripts upregulated and downregulated in the HSE.THP1.UV group. F Enrichment plots of upregulated and downregulated genes identified by GSEA based on the hallmark database. Nominal p value < 0.001. The leading-edge genes correlated with the response to UVA exposure and contributed to the enrichment scores (ESs).
The responses of the THP-1 cell line 24 h post-UVA exposure were elucidated through GO analysis (adjusted p values all < 0.05; Fig. 4D and Supplementary Data 4), with a focus on the roles and responses of macrophages to external stimuli. The expression levels of Nrf2 target genes, including those encoding antioxidants (NQO1), a redox signaling complex (TXNIP), and transcription and growth factors for homeostasis (i.e., SRC, FOSL1, AREG, and CYP1B1), were upregulated by UVA exposure. This tendency is consistent with the GO terms, including “response to oxidative stress”, “response to reactive oxygen species”, and “response to hydrogen peroxide” (Supplementary Fig. 4). Among the DEGs related to the GO term “phagocytosis”, transcripts of MERTK, TYRO3, ITGAV, and ITGB3 encoding TAM receptors (Tyro3, Axl, and MerTK) were upregulated in the HSE.THP1.UV condition, possibly to help clear apoptotic cells. Transcripts of proinflammatory and key mediators of inflammatory signaling (i.e., CSF2, LPL, IL1R1, and IL18R1) were upregulated in the GO term “positive regulation of cytokine production” (Supplementary Fig. 4).
KEGG enrichment pathway analysis revealed modulated pathways involved in “p53 signaling pathway” (hsa04115), “nucleotide metabolism” (hsa01232), and “DNA replication” (hsa03030) (adjusted p value all < 0.05; Fig. 4E). The GSEA results from the hallmark database (FDR < 0.05, nominal p value < 0.001) were analyzed (Fig. 4F). The “inflammatory response” and “UV response up” hallmarks were both upregulated, whereas the hallmarks “E2F targets” and “G2M checkpoint” (FDR < 0.001) (Fig. 4F) were downregulated. These genes encode proteins involved in DNA replication, nucleotide metabolism, and cell proliferation, such as HELLS, MCM7, MCM2, CDK1, TK1, E2F1, E2F2, CDCA3, and EZH2 (Supplementary Fig. 5). Taken together, the THP-1 cell line in the HSEs presented differential gene expression after UVA exposure, with notable changes in pathways related to inflammation, phagocytosis, and UV responses, emphasizing the intricate role of macrophages in the response to UVA-induced oxidative stress and tissue injury.
Effect of M2G on the UVA-induced stress response of the THP-1 cell line in HSEs
The cytotoxicity of M2G on individual cells in HSEs was tested by varying the M2G concentration, and cell viability was evaluated by the WST-8 assay. The optimal concentration of M2G that did not reduce cell viability to less than 80% was determined to be 20 μM (Supplementary Fig. 6). This dose was used in the subsequent experiments.
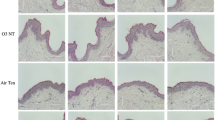
M2G was applied to the epidermis surface of the immunocompetent skin model HSE.THP1 for 2 h, followed by UVA irradiation at 80 J/cm2. M2G was subsequently removed from the HSEs before a 24 h incubation period (Fig. 5A). As shown in Fig. 5B (i), epidermal disruption was observed after UVA irradiation, where the epidermis was detached from the dermal layer. γ-H2AX-positive cells were observed in the epidermis (purple dots) (Fig. 5B, ii) and dermis. More importantly, DNA damage was significantly decreased in the HSE.THP1 population when M2G was applied, as shown by the percentages of γ-H2AX-positive nuclei (Fig. 5C). These results strongly indicate that M2G has photoprotective activity against UVA when it is applied to HSEs.
A Schematic diagram illustrating the experimental procedure used to test the effect of M2G on UVA irradiation in immunocompetent HSEs. Created with BioRender.com. B Representative images of H&E staining of HSE.THP-1 cells; non-exposure, UVA exposure, UVA exposure with or without M2G (i) and detection of DNA double-strand breaks (DSBs) by γ-H2AX IF staining (magenta) (ii). The cell nuclei were visualized by Hoechst staining (blue). The white dotted lines indicate epidermal‒dermal junctions. Scale bar: 100 µm, epidermis (E), dermis (D). C Quantitation of γ-H2AX+ cells in both the epidermal and dermal layers from (B). The data from each layer are presented as the means ± SEMs. Statistical significance is indicated by *p < 0.05, **p < 0.01, and ns, not significant, compared with the non-UVA exposure group (n = 3 per group). Two-way ANOVA followed by Holm‒Šídák’s multiple comparison test was used.
Enriched oxidative phosphorylation pathway in THP-1 cell line incorporated in the M2G-treated HSEs after UVA exposure
To evaluate the global impact of M2G on the response of the THP-1 cell line to UVA irradiation in HSEs, RNA sequencing and GSEA were conducted. Oxidative phosphorylation (OXPHOS) was notably enriched in the THP-1 cell line from the M2G-treated HSEs (HSE.THP1.UV.M2G), with an FDR-corrected p value < 0.1 and NES = 1.57 (Fig. 6A). OXPHOS is a key metabolic pathway that is utilized mainly by M2 macrophages. Furthermore, the inflammatory response and UV response hallmark genes were both downregulated in the THP-1 cell line from M2G-treated HSEs (Fig. 6B and Supplementary Fig. 7). These findings suggest that the utilization of M2G in a reconstructed human skin model is effective in preventing UVA exposure through the skin layers. As a result, the use of M2G on HSEs effectively protects HSEs from UVA-induced stress and the subsequent inflammatory response and reduces the number of inflammatory phenotypes in skin-associated macrophages that maintain M2-like metabolic flux. These data highlight the potential use of M2G as a photoprotective agent that can mitigate the effects of UVA exposure on the skin and resident macrophages.
A Enrichment plot of the “hallmark_oxidative phosphorylation” gene set identified by GSEA that was enriched in the treatment group (HSE.THP1.UV.M2G) compared with the control group (HSE.THP1.UV). The heatmap on the right shows the top 30 ranks of the genes associated with OXPHOS. Signal intensities are illustrated by gradient shades of red (increased) and blue (decreased). B Genes encoding the proteins involved in “UV_response_up” and “Inflammatory response” were downregulated in the treatment group. n = 3 per group.
Discussion
The responses of dermal innate immune cells to external stimuli, especially UVA radiation, are important for understanding the ability of skin biology to coordinate responses to stress. Our study aimed to establish a human 3D skin model containing macrophages that mimic normal human skin. Based on our previous report on HSE fabrication using plasma surface treatment35, we incorporated the human monocytic cell line THP-1 into HSEs and subjected the immunocompetent HSEs to UVA exposure and treatment with a UV protective agent.
Dermal macrophages are typically replenished by adult monocytes38. An elegant study in a mouse model by Tamoutounour et al. revealed that Ly-6Chi monocytes extravasated into the skin dermis and yielded tissue-resident macrophages39,40. Furthermore, evidence has shown that circulatory monocytes can extravasate into nonlymphoid tissues and adopt tissue-resident macrophages that specialize in tissue-specific functions from the local microenvironment, as reflected in the enhancer landscapes41. The use of unmanipulated monocytes (CD14+ monocytes) for HSE fabrication likely mimics this process in the skin. Considering the current view of the origins and functional specialization of dermal macrophages, we used unmanipulated THP-1 cells as representative monocytes for HSE fabrication. We believe that our model mimicked the recruitment of circulatory monocytes to the dermal layer and adopted dermal macrophage phenotypes, indicating the potential to differentiate into tissue-resident macrophages. Our transcriptomic data revealed downregulation of the cell cycle and DNA replication hallmarks of THP-1 cells from HSEs and increased expression of genes encoding chemokine receptors, reflecting this transition, as shown in Fig. 3E.
As an immortalized human monocytic cell line, THP-1 cells offer ease of manipulation and consistency in the experimental set up, reducing variability compared with primary cells, which can vary from donor to donor. Moreover, THP-1 cells represent a valuable tool for investigating monocyte and macrophage biology42,43 and for studying an in vitro model involving the differentiation of monocyte-derived macrophages or monocyte-derived dendritic cells33,44,45. However, the use of THP-1 cells to represent primary human blood monocytes has limitations. THP-1 cells may not fully replicate the complex behaviors and accurately reflect the in vivo conditions exhibited by primary blood monocytes, such as the response to LPS42. For future research, incorporating primary human CD14+ monocytes isolated from human peripheral blood into HSEs is recommended to enhance the model’s physiological relevance.
In a simplistic view, macrophages can be divided into M1 (proinflammatory) and M2 (anti-inflammatory) types based on expression of markers, including CD68, CD86, and CD16346. Our transcriptomic results revealed mixed M1/M2 phenotypes of THP-1 cells from HSEs (Fig. 3B and Supplementary Data 1). M2-associated markers (CD163, MRC1 or CD206, and IL1R2), as well as anti-inflammatory cytokines (IL10), are expressed in THP-1 cells in HSEs, emphasizing the tendency toward the “M2” phenotype47. DEGs of integrated macrophages encoding proteins associated with blood vessel development and the regulation of tissue remodeling, including degradation enzymes (i.e., MMP1, MMP9, and MMP19) and their inhibitors (TIMP1 and TIMP4), were detected. Our findings further demonstrated the significantly elevated expression of a critical component of the complement system (C3, C1QA, C1QB, and C1QC) in the HSE.THP1 population. Similar trends were reported in previous studies, in which human monocyte-derived macrophages48,49 and dermal macrophages in mouse models50,51 were shown to be increased in skin expansion during skin wound healing.
The exposure of the skin to UV radiation triggers ROS formation, resulting in DNA DSB lesions in keratinocytes and fibroblasts13. The skin has mechanisms to repair UV-induced damage, and the immune system, including macrophages, plays a role in these repair processes. In UVA-exposed HSEs without macrophages, the expression levels of genes encoding 8-oxoguanine glycosylase (OGG1) and H2AX significantly increased, indicating the involvement of DNA repair-related signaling cascades. Moreover, keratinocytes and fibroblasts have antioxidant defense mechanisms that can be activated by increasing the expression of the Nrf2 pathway and its target genes. A previous study reported that the induction of Nrf2 target genes (HMOX1, TXNRD1, and NQO1) resulted in an immediate DNA damage response within 2 to 6 h after UVA exposure13. THP-1 cells from the HSE.THP1.UV group did not show increases in NRF2 or HMOX1 expression, but NQO1 was upregulated (Fig. 4C), indicating that the antioxidant mechanism also operates in tissue-resident immune cells. More importantly, we demonstrated that THP-1 cells from UVA-exposed HSEs exhibit a proinflammatory phenotype (M1), as indicated by the elevation of the transcription factor hypoxia-inducible factor 1 (HIF1A) (Supplementary Fig. 5). Furthermore, SPHK1 was significantly upregulated in the HSE.THP-1.UV condition, suggesting the activation by inflammatory stimuli following an increase in extracellular S1P52. The downstream pathway of SphK/S1P signaling is associated with the production of proinflammatory cytokines (i.e., IL-1β and IL-6) through the activation of nuclear transcription factors (such as NF-κB)53,54. Thus, dermal macrophages may respond to UVA exposure by activating the NF-κB signaling pathway, leading to an inflammatory response, including inflammasome activation. GSEA revealed the downregulation of E2F targets and G2M checkpoint hallmarks in THP-1 cells from UVA-exposed HSEs. The Ef2 family of transcription factors is reported to regulate UV-induced apoptosis and keratinocyte proliferation55. Unexpectedly, when the transcriptomes of THP-1 cells from UVA-exposed HSEs were examined, the expression of genes associated with epidermal development and integrity was not detected, whereas slight upregulation was detected in THP-1 cells from the HSE.THP-1 group. In general, innate immune cells do not express such genes, but it has been reported that human circulating monocytes have the potential to transdifferentiate into keratinocyte-like cells and regulate dermal fibroblast wound healing function56. Thus, it is possible that in our model of HSEs, a subset of THP-1 cells was conditioned to transdifferentiate to become keratinocyte-like cells, and UVA exposure suppressed this event. Further detailed analysis at the single-cell level may be needed to address this puzzle.
Our group and others have investigated the effects of MAA compounds on the responses elicited by UV radiation. MAAs have potential as photoprotective agents by absorbing UV rays and preventing their harmful effects31,32,57. This study applied M2G to immunocompetent HSEs to investigate its effects on UVA-induced skin damage. Our results revealed that M2G-treated immunocompetent HSEs reduced the expression of the DNA DSB marker γ-H2AX (Fig. 5B, C), suggesting that M2G effectively mitigates the effects of UVA irradiation on HSEs. This result confirmed the findings of a previous study on the effects of M2G in 2D monocultures of keratinocytes and immune cells31,32. Glycolysis and OXPHOS are important metabolic pathways through which macrophages produce energy during homeostasis and immune responses to external stimuli58. Interestingly, our findings demonstrated that dermal macrophages enriched the transcripts associated with OXPHOS activity upon M2G treatment under UVA exposure (Fig. 6A). The results indicated that THP-1 cells utilized OXPHOS and remained “M2”-like even in the presence of UVA when M2G was applied. This finding is consistent with a study reported by Yu et al., who investigated the roles of glycolysis and OXPHOS in macrophage polarization in response to different stimuli. They reported that M2 macrophages increased OXPHOS activity during polarization, whereas a glycolytic metabolism signal was required to differentiate the M1 phenotype59.
The limitations of this study include the lack of comparisons between the responses of THP-1 cells in HSEs and macrophages from human skin and the use of the THP-1 cell line instead of primary human monocytes. To compare the response of THP-1 cells in HSE and human skin macrophages directly, the isolation of human dermal macrophages is needed for analysis. The limited number of macrophages that can be obtained from the skin may be an obstacle for such studies. In any case, these limitations of our study may require further investigation using the platforms and insights gained from our current study.
Our study reveals the dynamic behavior of macrophages in reconstructed HSEs, which exhibit “M1” and “M2” phenotypes that are crucial for tissue regeneration and responses to external cues. In response to UVA exposure, dermal macrophages display complex gene expression patterns associated with oxidative stress, phagocytosis, and inflammatory responses, underscoring their integral role in response to external threats. Additionally, the application of the photoprotectant M2G reduces DNA damage and promotes oxidative phosphorylation in macrophages, which is indicative of enhanced M2 polarization. These comprehensive insights illuminate the intricate interplay among macrophages, the skin microenvironment, and environmental stresses, paving the way for advancements in skin and UV protection strategies. Further studies on how immunocompetent HSEs respond differently to HSEs without innate immune cells are now possible using the platform provided by this study. In addition, the impact of UVB radiation on skin macrophages can also be studied using this immunocompetent platform.
Methods
Cell culture and human tissue samples
Primary normal human epidermal keratinocytes (NHEKs, FC-0007, Kurabo, Japan) were maintained in EpiLifeTM medium with 60 µM calcium (Gibco Laboratories, USA) supplemented with a HuMedia-KG2 proliferation additive set (Kurabo) and were used at passage 3. Primary normal human dermal fibroblasts (NHDF, CC2509, Lonza Biosciences, USA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with L-Gln and sodium pyruvate (Nacalai Tesque Industries, Japan), 5% (v/v) fetal bovine serum (FBS, Gibco) and 1% (v/v) antibiotic‒antimycotic mixed stock solution (Nacalai Tesque) and were used until passage 7. THP-1, a human monocytic cell line, was obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB0112, Japan) and maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Nacalai Tesque) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco) and 1% (v/v) antibiotic (Nacalai Tesque). The cells were incubated at 37 °C with humidified 5% (v/v) CO2. Human breast skin tissues were ethically collected from female donors after breast mastectomies at the Kyoto Prefectural University of Medicine Hospital. The study adhered to the principles of the Declaration of Helsinki and received approval from the Human Ethics Committee (Approval number: ERB-C-1317-1). Informed consent was obtained from all the subjects before sample collection. All ethical regulations relevant to human research participants were followed.
Human 3D skin equivalent and immunocompetent model construction
HSEs were generated as previously described35. Briefly, NHDFs (1 × 106 cells) in DMEM were mixed with fibrinogen (13.3 mg/mL final concentration) and thrombin (3.3 U/mL final concentration) (all from Sigma‒Aldrich, USA) and seeded immediately into 24-well inserts (Corning, USA) treated with plasma ions generated from a plasma generator device (YHS-R, SAKIGAKE-Semiconductor Co., Kyoto, Japan) for 2 mins. For the fabrication of immunocompetent HSEs, NHDFs (1 × 106 cells) and THP-1 cells (0.5 × 106 cells) were cocultured in fibrin hydrogels (Fig. 1A) to generate the dermal layer. After gelation, the dermal layer was submerged in DMEM for 24 h. NHEKs (1 × 106 cells) in a 1:1 v/v ratio of DMEM and EpiLife medium were seeded on the dermal layer coated with 0.04 mg/mL collagen type IV (Sigma‒Aldrich), submerged in the mixed medium and incubated for 24 h. Epidermal differentiation was initiated by replacing the medium with 2.5 µg/mL L-ascorbic acid (Fujifilm Wako, Japan), followed by air‒liquid interface cultivation for 7 days.
UVA irradiation and procedure
HSEs were exposed to UVA irradiation using a BlueWave® MX-250 LED lamp, RediCure™ (main peak energy emission at 365 nm) (DYMAX Corporation Torrington, CT, USA), at a single dose of 80 J/cm2. The UVA dose was calculated as follows: Dose (J/cm2) = Exposure time (secs) × Intensity (W/cm2). The UVA lamp was set upon a stable support, allowing an exposure distance of 6 cm from the sample surface. Before irradiation, the culture medium was replaced with 1× phosphate-buffered saline (PBS) solution (Nacalai Tesque) to prevent phototoxic effects. The PBS was removed, and fresh differentiated medium was added after UVA exposure. UVA‐induced damage was evaluated 24 h after UVA exposure.
Applying mycosporine-2-glycine (M2G) to HSEs
M2G was prepared as described previously32. It was diluted in Milli-Q water and was then applied for 2 h before, during, and 30 min after UVA irradiation. Afterward, the M2G was removed, and the HSEs were subsequently incubated. Harvesting and tissue histology, along with gene expression analysis, were performed 24 h after UVA exposure.
Tissue histology and immunofluorescence staining
Tissue samples were fixed in 4% paraformaldehyde phosphate buffer solution (FUJIFILM Wako) at 4 °C. The paraffin-embedded tissue prepared by Applied Medical Research Laboratory, Inc. (Osaka, Japan) was sectioned at a thickness of 5 µm. Tissue histology was performed by hematoxylin and eosin (H&E) staining. Brightfield photographs were taken with an EVOS®FL (Thermo Fisher Scientific, USA) microscope.
The sections were deparaffinized, and the FC receptors were blocked with 1% bovine serum albumin (BSA; Sigma‒Aldrich) in 1× PBS for 30 min before being stained with specific primary antibodies (Supplementary Table 1) overnight at 4 °C. The tissue sections were subsequently washed with 1× PBS, and secondary antibodies were applied to the samples for 30 min at room temperature in the dark. The nuclei were stained with Hoechst 33342 (Thermo Fisher, 1:2000) for 10 min at room temperature in the dark, and the remaining unbound dye was removed by washing. The sections were preserved with Fluoromount-GTM Mounting Medium (Invitrogen, USA). Immunostaining images were captured using a benchtop CQ1 confocal quantitative image cytometer (CellVoyager CQ1 System Highlights, Yokogawa, Japan). The mean fluorescence intensity was measured using ImageJ software (v1.53k). The image analyses were performed on at least 2 biological samples, with at least 3 randomly selected fields per sample.
Total RNA extraction and real-time quantitative polymerase chain reaction (RT‒qPCR)
Total RNA was extracted using an RNeasy mini-kit (Qiagen, Germany) according to the manufacturer’s instructions. Whole tissues were transferred to Buffer RLT (Qiagen) with 2-mercaptoethanol (Nacalai Tesque). The RNA quantity and purity were assessed using a NanoDrop 1000 spectrophotometer (Thermo Fisher). For cDNA synthesis, 1000 ng of total RNA was used with iScriptTM Reverse Transcription Supermix (Bio-Rad, CA). Comparative gene expression was analyzed using iQ™ SYBR® Green Supermix (Bio-Rad) and a StepOnePlus real-time PCR system (Applied Biosystems, USA). The mRNA levels were normalized to those of the internal control, β-actin. The primer sequences are provided in Supplementary Table 2.
Cell toxicity assay
The cytotoxic effect of M2G was assessed by WST-8 assay. NHEK, NHDF, and THP-1 cells were plated in 96-well plates (1.5 × 104, 2 × 104, and 8 × 103 cells/well, respectively) and incubated for 24 h. The cells were treated with varying M2G concentrations (0, 1, 5, 10, and 20 µM) for 24 h in 100 µL of each complete medium. After treatment, the cells were washed with 1× PBS. One hundred microliters of WST assay solution was prepared by adding 10 µL of Cell Counting Reagent SF (07553-44, Nacalai Tesque) to 100 µL of medium, and the assay solution was added to the cells for the reaction, with vacant wells used as background. The plates were subsequently incubated for 30 mins. Finally, the absorbance at 450 nm was measured using a microplate reader (Bio-Rad). The experiments were performed in triplicate, and the cell viability was calculated using the following formula:
CD45+ cell isolation, RNA extraction, and library preparation
HSEs were digested with 0.05% trypsin/0.01% ethylenediaminetetraacetic acid (EDTA, Nacalai Tesque) in 1× PBS. The cells were washed and filtered through a 100 µm VCS-100 cell strainer (AS ONE Corporation, Japan) to remove the remaining fibrin gel. Immune cells positively stained with an anti-human CD45 antibody conjugated with Alexa Fluor® 488 (BioLegend, USA) were sorted using an SH800S Cell Sorter (Sony Biotechnology Inc., CA). Harvested CD45+ cells were subjected to total RNA extraction with a PicoPureTM RNA Isolation Kit (Applied Biosystems) according to the manufacturer’s instructions. Total RNA was subjected to RNA sequencing at the Genome Information Research Center (GIRC) (Research Institute for Microbial Diseases, Osaka University, Japan).
RNA sequencing and data analysis
Sequences in FASTQ files were trimmed to remove adapter sequences and aligned to the human genome hg19 using HISAT260. The Sam and Bam files were sorted using SAMtools61. Reads were counted with HTSeq-count. The differential expression analysis (DEGs) was subsequently conducted in RStudio 2023.12.1 using the R Bioconductor package DESeq2, version 1.40.262. The analyses were conducted on triplicate samples. A combination of an adjusted p value < 0.05 and an absolute log2-fold change (|log2 FC|) value > 1 or > 2 was considered the threshold for determining the DEGs, depending on the conditions.
Functional enrichment analyses were conducted on the Gene Ontology (GO) biological process (BP), cellular compartment (CC), and molecular function (MF) categories and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The results are visualized in dot plots and bar plots. Gene set enrichment analysis (GSEA) was performed using predefined gene sets from the Molecular Signatures Database (MSigDB v2023.1.Hs)63,64 with GSEA v4.3.2 software. GSEA was conducted with the hallmarks (50 gene sets) and the KEGG results (178 gene sets) according to the statistical significance (nominal p value) of the enrichment score (ES) estimated by running 1000 gene set permutations65,66.
Statistics and reproducibility
The quantitative results of each group are presented as the means ± standard errors of the means (SEMs). Statistical analysis was performed using GraphPad Prism 10 software (GraphPad Software Inc., California, USA). A P value < 0.05 was considered to indicate a statistically significant difference. All experiments were performed in at least two biological replicates when HSEs were independently generated for each biological replicate.
Data availability
The RNA-Seq datasets generated or analyzed for this study were deposited in a public database and can be found under the GEO accession number GSE261112. Full results of the DEGs and GO enrichment analysis are provided in the Supplementary Data 1–4. Source data underlying graphs in this study are provided in Supplementary Data 5.
References
Lee, C. H., Wu, S. B., Hong, C. H., Yu, H. S. & Wei, Y. H. Molecular mechanisms of UV-induced apoptosis and its effects on skin residential cells: the implication in UV-based phototherapy. Int. J. Mol. Sci. 14, 6414–6435 (2013).
Abadie, S., Bedos, P. & Rouquette, J. A human skin model to evaluate the protective effect of compounds against UVA damage. Int. J. Cosmet. Sci. 41, 594–603 (2019).
D’Orazio, J., Jarrett, S., Amaro-Ortiz, A. & Scott, T. UV radiation and the skin. Int. J. Mol. Sci. 14, 12222–12248 (2013).
Wolfle, U. et al. Reactive molecule species and antioxidative mechanisms in normal skin and skin aging. Ski. Pharm. Physiol. 27, 316–332 (2014).
Ansary, T. M., Hossain, M. R., Kamiya, K., Komine, M. & Ohtsuki, M. Inflammatory molecules associated with ultraviolet radiation-mediated skin aging. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22083974 (2021).
Kawashima, S. et al. Protective effect of pre- and post-vitamin C treatments on UVB-irradiation-induced skin damage. Sci. Rep. 8, 16199 (2018).
Casale, C. et al. Engineering a human skin equivalent to study dermis remodelling and epidermis senescence in vitro after UVA exposure. J. Tissue Eng. Regen. Med. 12, 1658–1669 (2018).
Marionnet, C. et al. Different oxidative stress response in keratinocytes and fibroblasts of reconstructed skin exposed to non extreme daily-ultraviolet radiation. PLoS One 5, e12059 (2010).
Meloni, M., Farina, A. & de Servi, B. Molecular modifications of dermal and epidermal biomarkers following UVA exposures on reconstructed full-thickness human skin. Photochem. Photobiol. Sci. 9, 439–447 (2010).
Javeri, A., Huang, X. X., Bernerd, F., Mason, R. S. & Halliday, G. M. Human 8-oxoguanine-DNA glycosylase 1 protein and gene are expressed more abundantly in the superficial than basal layer of human epidermis. DNA Repair (Amst.) 7, 1542–1550 (2008).
Wang, K., Maayah, M., Sweasy, J. B. & Alnajjar, K. S. The role of cysteines in the structure and function of OGG1. J. Biol. Chem. 296, 100093 (2021).
Chu, Y. et al. Protective effects and mechanisms of N-phenethyl caffeamide from UVA-induced skin damage in human epidermal keratinocytes through Nrf2/HO-1 regulation. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20010164 (2019).
Marionnet, C., Pierrard, C., Golebiewski, C. & Bernerd, F. Diversity of biological effects induced by longwave UVA rays (UVA1) in reconstructed skin. PLoS One 9, e105263 (2014).
Chen, M., Liang, S., Shahid, A., Andresen, B. T. & Huang, Y. The beta-blocker carvedilol prevented ultraviolet-mediated damage of murine epidermal cells and 3D human reconstructed skin. Int. J. Mol. Sci. 21 https://doi.org/10.3390/ijms21030798 (2020).
Chau, D. Y., Johnson, C., MacNeil, S., Haycock, J. W. & Ghaemmaghami, A. M. The development of a 3D immunocompetent model of human skin. Biofabrication 5, 035011 (2013).
Vidal Yucha, S. E., Tamamoto, K. A., Nguyen, H., Cairns, D. M. & Kaplan, D. L. Human skin equivalents demonstrate need for neuro-immuno-cutaneous system. Adv. Biosyst. 3, e1800283 (2019).
Griffoni, C. et al. In vitro skin culture media influence the viability and inflammatory response of primary macrophages. Sci. Rep. 11, 7070 (2021).
Cho, Y. J., Jang, B. J., Yang, Y. J. & Shin, H. S. Transcriptional characterization of the effects of macrophages on inflammation-induced reconstructed human skin equivalents. Biotechnol. Bioprocess Eng. 28, 774–780 (2023).
Limandjaja, G. C., Waaijman, T., Roffel, S., Niessen, F. B. & Gibbs, S. Monocytes co-cultured with reconstructed keloid and normal skin models skew towards M2 macrophage phenotype. Arch. Dermatol. Res. 311, 615–627 (2019).
Ramadan, Q. & Ting, F. C. In vitro micro-physiological immune-competent model of the human skin. Lab Chip 16, 1899–1908 (2016).
Bechetoille, N. et al. Effects of solar ultraviolet radiation on engineered human skin equivalent containing both Langerhans cells and dermal dendritic cells. Tissue Eng. 13, 2667–2679 (2007).
Huang, M. et al. Self-assembled human skin equivalents model macrophage activation of cutaneous fibrogenesis in systemic sclerosis. Arthritis Rheumatol. 74, 1245–1256 (2022).
Sugaya, M. Macrophages and fibroblasts underpin skin immune responses. Exploration Immunol. 1, 226–242 (2021).
Zhuang, Y. & Lyga, J. Inflammaging in skin and other tissues–the roles of complement system and macrophage. Inflamm. Allergy Drug Targets 13, 153–161 (2014).
Yanez, D. A., Lacher, R. K., Vidyarthi, A. & Colegio, O. R. The role of macrophages in skin homeostasis. Pflug. Arch. 469, 455–463 (2017).
Minutti, C. M., Knipper, J. A., Allen, J. E. & Zaiss, D. M. Tissue-specific contribution of macrophages to wound healing. Semin. Cell Dev. Biol. 61, 3–11 (2017).
Piskin, G., Bos, J. D. & Teunissen, M. B. Neutrophils infiltrating ultraviolet B-irradiated normal human skin display high IL-10 expression. Arch. Dermatol Res. 296, 339–342 (2005).
Schmitt, D. A. & Ullrich, S. E. Exposure to ultraviolet radiation causes dendritic cells/macrophages to secrete immune-suppressive IL-12p40 homodimers. J. Immunol. 165, 3162–3167 (2000).
Kedar, L., Kashman, Y. & Oren, A. Mycosporine-2-glycine is the major mycosporine-like amino acid in a unicellular cyanobacterium (Euhalothece sp.) isolated from a gypsum crust in a hypersaline saltern pond. FEMS Microbiol. Lett. 208, 233–237 (2002).
Jin, H., Kim, S., Lee, D., Ledesma-Amaro, R. & Hahn, J. S. Efficient production of mycosporine-like amino acids, natural sunscreens, in Yarrowia lipolytica. Biotechnol. Biofuels Bioprod. 16, 162 (2023).
Cheewinthamrongrod, V., Kageyama, H., Palaga, T., Takabe, T. & Waditee-Sirisattha, R. DNA damage protecting and free radical scavenging properties of mycosporine-2-glycine from the Dead Sea cyanobacterium in A375 human melanoma cell lines. J. Photochem. Photobiol. B 164, 289–295 (2016).
Tarasuntisuk, S., Palaga, T., Kageyama, H. & Waditee-Sirisattha, R. Mycosporine-2-glycine exerts anti-inflammatory and antioxidant effects in lipopolysaccharide (LPS)-stimulated RAW 264.7 macrophages. Arch. Biochem. Biophys. 662, 33–39 (2019).
Holken, J. M. et al. A human 3D immune competent full-thickness skin model mimicking dermal dendritic cell activation. Front. Immunol. 14, 1276151 (2023).
Sapudom, J. et al. 3D in vitro M2 macrophage model to mimic modulation of tissue repair. NPJ Regen. Med. 6, 83 (2021).
Phuphanitcharoenkun, S., Louis, F., Sowa, Y., Matsusaki, M. & Palaga, T. Improving stability of human three dimensional skin equivalents using plasma surface treatment. Biotechnol. Bioeng. 121, 1950–1960 (2024).
Bernerd, F. & Asselineau, D. UVA exposure of human skin reconstructed in vitro induces apoptosis of dermal fibroblasts: subsequent connective tissue repair and implications in photoaging. Cell Death Differ. 5, 792–802 (1998).
Montero, P., Roger, I., Milara, J. & Cortijo, J. Damaging effects of UVA, blue light, and infrared radiation: in vitro assessment on a reconstructed full-thickness human skin. Front. Med. (Lausanne) 10, 1267409 (2023).
Mass, E., Nimmerjahn, F., Kierdorf, K. & Schlitzer, A. Tissue-specific macrophages: how they develop and choreograph tissue biology. Nat. Rev. Immunol. 23, 563–579 (2023).
Tamoutounour, S. et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013).
Baranska, A. et al. Unveiling skin macrophage dynamics explains both tattoo persistence and strenuous removal. J. Exp. Med. 215, 1115–1133 (2018).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Bosshart, H. & Heinzelmann, M. THP-1 cells as a model for human monocytes. Ann. Transl. Med. 4, 438 (2016).
Chanput, W., Mes, J. J. & Wichers, H. J. THP-1 cell line: an in vitro cell model for immune modulation approach. Int. Immunopharmacol. 23, 37–45 (2014).
Tedesco, S. et al. Convenience versus biological significance: are PMA-differentiated THP-1 cells a reliable substitute for blood-derived macrophages when studying in vitro polarization? Front. Pharm. 9, 71 (2018).
Oh, M. H., Jang, J. & Lee, J. H. Polarization of THP-1-derived macrophage by magnesium and MAGT1 inhibition in wound healing. Arch. Plast. Surg. 50, 432–442 (2023).
Horiba, S., Kami, R., Tsutsui, T. & Hosoi, J. IL-34 downregulation‒associated M1/M2 macrophage imbalance is related to inflammaging in sun-exposed human skin. JID Innov. 2, 100112 (2022).
Supino, D. et al. Negative regulation of the IL-1 system by IL-1R2 and IL-1R8: relevance in pathophysiology and disease. Front. Immunol. 13, 804641 (2022).
Etich, J. et al. Gene expression profiling of the extracellular matrix signature in macrophages of different activation status: relevance for skin wound healing. Int. J. Mol. Sci. 20, https://doi.org/10.3390/ijms20205086 (2019).
Graney, P. L. et al. Macrophages of diverse phenotypes drive vascularization of engineered tissues. Sci. Adv. 6, eaay6391 (2020).
Ding, J. et al. Macrophages are necessary for skin regeneration during tissue expansion. J. Transl. Med. 17, 36 (2019).
Ledwon, J. K., Kelsey, L. J., Vaca, E. E. & Gosain, A. K. Transcriptomic analysis reveals dynamic molecular changes in skin induced by mechanical forces secondary to tissue expansion. Sci. Rep. 10, 15991 (2020).
Sukocheva, O. A. et al. Sphingosine kinase and sphingosine-1-phosphate receptor signaling pathway in inflammatory gastrointestinal disease and cancers: a novel therapeutic target. Pharm. Ther. 207, 107464 (2020).
Syed, S. N., Weigert, A. & Brune, B. Sphingosine kinases are involved in macrophage NLRP3 inflammasome transcriptional induction. Int. J. Mol. Sci. 21 https://doi.org/10.3390/ijms21134733 (2020).
Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. 2, 17023- (2017).
Hazar-Rethinam, M., Endo-Munoz, L., Gannon, O. & Saunders, N. The role of the E2F transcription factor family in UV-induced apoptosis. Int J. Mol. Sci. 12, 8947–8960 (2011).
Medina, A., Brown, E., Carr, N. & Ghahary, A. Circulating monocytes have the capacity to be transdifferentiated into keratinocyte-like cells. Wound Repair Regen. 17, 268–277 (2009).
Rosic, N., Climstein, M., Boyle, G. M., Thanh Nguyen, D. & Feng, Y. Exploring mycosporine-like amino acid UV-absorbing natural products for a new generation of environmentally friendly sunscreens. Mar. Drugs 21 https://doi.org/10.3390/md21040253 (2023).
Viola, A., Munari, F., Sanchez-Rodriguez, R., Scolaro, T. & Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019).
Yu, Q. et al. Regulations of glycolytic activities on macrophages functions in tumor and infectious inflammation. Front. Cell Infect. Microbiol. 10, 287 (2020).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, https://doi.org/10.1093/gigascience/giab008 (2021).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
S.P. gratefully acknowledges the Science Achievement Scholarship of Thailand (SAST). This research and innovation activity was funded to S.P. and T.P. by the National Research Council of Thailand (NRCT) (grant number 173851) and by the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund). This research has received funding support from the NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation [grant number B16F640117]. F.L. was supported by Kakenhi Grant-in-Aid for Early-Career Scientists (20K20199). This work was financially supported by COI-NEXT (JPMJPF2009) from JST, JPNP20004 from NEDO, Grant-in-Aid for Scientific Research (A) (20H00665), and Panasonic Holdings. We thank Jeerameth Klomsing for his help with the PCA of the data set.
Author information
Authors and Affiliations
Contributions
S.P.: conceptualization, methodology, formal analysis, investigation, visualization, writing–original draft, F.L.: conceptualization, methodology, resources, supervision, writing–reviewing and editing, Y.S., K.U., and M.K.: resources. R.W.S. and H.K.: resources, writing–reviewing and editing. M.M.: conceptualization, supervision, resources, writing–reviewing and editing, funding acquisition, T.P.: conceptualization, supervision, writing–reviewing and editing, project administration, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Joshua Morgan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Ngan Huang and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Phuphanitcharoenkun, S., Louis, F., Sowa, Y. et al. Characterization of macrophages associated with human skin models exposed to UV radiation. Commun Biol 7, 1284 (2024). https://doi.org/10.1038/s42003-024-06975-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06975-z
This article is cited by
-
Colla Corii Asini regulate collagen regeneration in UV exposure-induced skin photoaging in mice
Chinese Medicine (2025)
-
Seasonal variation in Swedish sarcoidosis patients with severe hypercalcemia
Scientific Reports (2025)
-
Propolis-loaded liposomes (ProLip) enhance macrophage-mediated immunomodulation and suppress breast cancer cell proliferation
Scientific Reports (2025)