Abstract
Human induced pluripotent stem cell (iPSC)-derived lung organoids, engineered to carry targeted genes, offer a robust platform for investigating mechanistic insights in lung research. Although lentiviral vectors (LVVs) are highly effective for stable expression due to their integrative properties, achieving efficient transduction in human iPSC-derived lung organoids poses a significant technical challenge, likely due to the complex structure of these organoids. In this study, we optimized a method to enhance LVV transduction efficiency by physically disrupting the organoids to increase surface area, followed by spinoculation to apply shear force during cell dissociation. This approach, combined with the use of an optimized culture medium, significantly improved transduction efficiency. The success of this method was validated at both the gene and protein levels using single-cell RNA sequencing (scRNA-seq) and various cellular and molecular assays. Our optimized transduction protocol may provide a valuable tool for investigating specific cellular and molecular mechanisms in development and disease models using human iPSCs-derived lung organoids.
Similar content being viewed by others
Introduction
Human induced pluripotent stem cells (iPSCs)-derived lung organoids are advanced three-dimensional (3D) organ-simulating systems that replicate key features of the human lung, including airway-like structures surrounded by mesenchymal and other cell types. These organoids offer a valuable in vitro model for studying cell-cell interaction and modeling lung diseases due to their complexity, reproducibility, and ability to mimic the mature human lung1,2.
These organoids can be generated from human pluripotent stem cells, including embryonic stem cells and iPSCs. They exhibit cellular polarizing and differentiation into multiple cell lineages found in lung3. The self-renewing capacity of iPSC-derived lung organoids supports sustained in vitro expansion, making them ideal for studying lung development, disease, recovery4, and for investigating respiratory viral infections such as (SARS-CoV-2)5. Additionally, iPSCs-derived lung organoids are being explored as potential sources of cells for lung repair6,7. The use of genetically modified iPSCs-derived lung organoids is particularly promising for examining the roles of specific genes and proteins in lung development and disease, especially for conditions lacking effective treatments.
Gene delivery in these systems has primarily relied on viral vectors due to their efficiency in transducing various lung cell types8,9,10. Among these, adeno-associated virus (AAV) and adenoviruses (Ads) have been widely used, though AAV’s limited packaging capacity and Ad’s safety concerns present challenges11,12,13. In contrast, lentiviral vectors (LVVs) have emerged as a compelling option14,15, offering stable, long-term gene expression through integrating into virus G protein, which enhances transduction efficiency and ensures stable gene expression with minimal immune response16. Moreover, LVVs can accommodate larger gene cassettes17, making them attractive for pulmonary applications involving gene delivery.
Previous studies have demonstrated efficient LVV transduction in iPSCs, followed by selection and differentiation into transgenic lung organoids for pathophysiological studies and gene therapy18,19. While this approach offers potential due to the self-renewal and pluripotency of stem cells19,20, it is labor-intensive, requiring 3-4 months for cell sorting or antibiotic selection, and may arise concerns of spontaneous differentiation18,21,22. This method is particularly useful for generating homogenous transgenic lung organoids to model specific conditions like surfactant protein B deficiency or cystic fibrosis transmembrane conductance regulator (CFTR) mutation23. However, for gene therapy studies requiring multiple LVVs, direct transduction of iPSC-derived lung organoids may be more practical, avoiding the need for separate iPSC differentiation processes for each vector.
The traditional methods for LVV transduction are often inadequate for iPSCs-derived lung organoids due to challenges such as limited LVV accessibility to the 3D structure, the heterogeneous cell populations within organoids, and the presence of immature cells that may be affected by gene transduction24. Some researchers have attempted to overcome these challenges by converting 3D into 2D structures and performing single-cell dissociation in intestinal and pancreatic organoids, but limited improvement25,26,27.
To address these limitations while preserving the structural integrity and cellular complexity of iPSCs-derived lung organoids, we developed a modified protocol that involves: 1) increasing the surface area of the organoids, 2) enhancing LVV diffusion, 3) improving cell survival with supplemental media, and 4) applying appropriate LVV titers. We successfully utilized this protocol to transduce a third-generation LVV encoding enhanced green fluorescent protein (eGFP) and human angiotensin converting enzyme 2 (hACE2) into iPSCs-derived lung organoids. Single-cell RNA sequencing (scRNA-seq) confirmed that the cellular composition of the organoids was retained post-transduction. Our optimized protocol may enhance the utility of iPSCs-derived lung organoids in lung research, enabling more detailed investigation into lung development, disease mechanisms, and potential therapeutic interventions.
Results
Characterization of human iPSC-derived lung organoids
Human iPSCs were differentiated into lung organoids following established stepwise directed differentiation protocols24. The cellular composition of these iPSCs-derived lung organoids was validated using scRNA-seq profiling (Fig. 1) and quantitative PCR with reverse transcription (qRT-PCR) (Fig. 2a–d). Immunostaining analysis confirmed the expression of AT2-like cell marker SFTPC in the organoids (Fig. 2e). Through scRNA-seq, we identified seven distinct cell clusters within lung organoids, each characterized by specific cell type markers (Fig. 1b–h). Notably, the AT2-like cells were enriched for adult human lung AT2 cell markers, including erythroblast transformation-specific variant transcription factor 5 (ETV5), Mucin 5 (MUC5), surfactant protein B (SFTPB), surfactant protein C (SFTPC), and surfactant protein D (SFTPD) (Figs. 3–4).
a Uniform Manifold Approximation and Projection (UMAP) of visualization of the scRNA-seq transcriptome of human iPSC-derived lung organoids, highlighting distinct cell clusters: AT1-like cells, AT2-like cells, fibroblasts, stromal cells, immature cells, non-lung epithelial cells, and airway basal cells. b–h Violin plots displaying the log-normalized maximum expression levels of specific cluster markers corresponding to each cell type identified in the UMAP. Data represented in median ± IQR.
a–d Quantitative reverse transcription PCR (qRT-PCR) analysis comparing gene expression in iPSCs and iPSC-derived lung organoids across n = 3–11 biologically independent experiments. SFTPB: Surfactant Protein B; SFTPC: Surfactant Protein C; SFTPD: Surfactant Protein D. Data are presented as mean ± SEM, with statistical significance indicated (SFTPB: p = 0.0006; SFTPC: p < 0.0001; ABCA3 p = 0.0021; EPCAM p = 0.0319; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 Welch’s t test). e Confocal fluorescence microscopy images illustrating the expression of SFTPC (magenta) and nuclear staining with DAPI (blue) in human iPSC-derived lung organoids. Scale bar: 100 μm.
a–c UMAP plots depicting the expression of canonical AT2-like cell markers in different clusters: (a) SFTPB, (b) SFTPC, and (c) SFTPD. The vertical scale bar represents expression levels, with higher intensity indicating greater expression. SFTPB Surfactant Protein B, SFTPC: Surfactant Protein C, SFTPD: Surfactant Protein D.
Comparison of physical and enzymatic dissociation in human iPSCs-derived lung organoids
To enhance the surface area for LVV transduction, we compared the effects of physical dissociation (via shear force application) with (Fig. 5) with enzymatic dissociation using accutase or trypsin on human iPSCs-derived lung organoids. Five organoids of comparable sizes (1.5–2.0 mm in diameter) and morphology (dense core with smooth edges) were randomly subjected to either physical or enzymatic dissociation. Post-dissociation, the organoids were cultured in transduction media supplemented with 10 µM CHIR-99021 and 10 µM Y-27632 dihydrochloride and monitored for seven days to assess morphology changes (Fig. 6a).
a Brightfield images displaying the morphological changes in human iPSC-derived lung organoids from day 0 to day 7 following physical or enzymatic dissociation. Scale bar: 100 μm. b Brightfield and fluorescence images showing eGFP expression in human iPSC-derived lung organoids 72 h post-transduction under different spinoculation speeds. Scale bar: 300 μm. c Quantification of mean fluorescence intensity of eGFP under various transduction conditions as depicted in (b). Data are presented as mean ± SEM, with statistical significance indicated (static 1.0 h: p = 0.00248, 600g 0.5 h: p = 0.00012, 600g 1.0 h: p = 0.00001, 1200g 0.5 h: p = 0.00006, 1200g 1.0 h: p = 0.0021; *p < 0.05 vs control, Student’s t test). n = 3–5 biologically independent experiments. d Cytotoxicity in fold change of naïve and hACE2-LVV-transduced human iPSC-derived lung organoids in 72 h post-transduction under different spinoculation speeds. Data are presented as mean ± SEM. n = 4 biologically independent experiment.
On day 1 post-dissociation, all methods resulted in smaller clumps compared to the undissociated organoids (Fig. 6a). However, by days 3–7, notable morphological differences emerged between the groups. Organoids in the physical dissociation group exhibited complete recovery, with morphology similar to the original state, characterized by a dense core and smooth edges. In contrast, organoids enzymatically showed signs of stress, including rough edges (purple arrow), cellular shedding (orange arrow), and ballooning (red arrow) (Fig. 6a). We noted that polybrene exhibited toxicity to lung organoids at concentrations of 2, 4, and 8 μg/mL, with a minimum of 50% cell death observed (Supplementary Fig. 1). These finding indicate that physical dissociation is superior to enzymatic methods for reducing organoid size while maintaining structural integrity.
Optimization of LVV transduction in human iPSCs-derived lung organoids
Following physical dissociation, we optimized LVV transduction using spinoculation protocol. We assessed cell cytotoxicity, morphology, and transduction efficiency under different spinoculation conditions: 600g or 1200g for 30 min or 60 min, respectively, at room temperature. A static condition (without spinoculation) served as the control. Fluorescent intensity measurements of eGFP at day 7 post-transduction demonstrated that transduction efficiency was higher in the spinoculation groups compared to the static control group (Fig. 6b, c). Apoptosis assays revealed no significant cytotoxicity across the groups (Fig. 6d). This suggests that physical dissociation followed by spinoculation enhances LVV transduction efficiency in human iPSCs-derived lung organoids.
Expression of protein of interest and transcriptomic heterogenicity in human iPSCs-derived lung organoids post-transduction
To assess the impact of LVV transduction on the transcriptomic profiles of human iPSCs-derived lung organoids, we constructed a third-generation LVV encoding the elongation factor 1 alpha 1 (EF-1α) promoter-driven eGFP reporter and the hACE2 gene tagged with 3xFLAG (Fig. 7a). scRNA-seq profiling showed no significant changes in the cell composition of organoids between the naïve and transduced groups at 7 days post-transduction (Fig. 7b, c).
a Schematic of the third-generation LVV construct encoding the full-length hACE2 gene with a 3xFLAG tag, followed by the Thosea asigna virus 2A self-cleaving peptide (T2A) and eGFP. b, c Proportional representation of cell clusters in naïve and hACE2-LVV-transduced human iPSC-derived lung organoids 7 days post-transduction. d, e Western blot analysis of eGFP and hACE2 protein levels 7 days post-transduction under different spinoculation speeds. β-actin served as a loading control. f, g Densitometric analysis of eGFP and hACE2 protein expression using Bio-Lab image lab software. Data are presented as mean ± SEM, n = 3. Statistical significance is indicated (*p < 0.05 vs. control, Student’s t test). h Confocal fluorescence microscopy images showing the expression of eGFP (green), hACE2 (red), and nuclear staining with DAPI (blue) in naïve and hACE2-LVV-transduced human iPSC-derived lung organoids 7 days post-transduction. Scale bar: 50 μm. i Quantification of mean fluorescence intensity for eGFP and hACE2, as shown in (h). Data are presented as mean ± SEM, n = 3. Statistical significance is indicated (*p < 0.05 vs. naïve, Student’s t test).
Protein expression analysis revealed significantly higher levels of both eGFP and hACE2 in the spinoculation groups compared to the static condition, with no differences observed between the two spinoculation groups at any time point (Fig. 7d, e, Supplementary Fig. 2a, b). Quantification of protein expression at expression at different spinoculation conditions and time points showed similar expression levels of eGFP and hACE2 (Fig. 7f, g). Immunostaining of 3D human iPSC-derived lung organoids 7 days post-transduction indicated that hACE2 protein expression was twice as high in the transduced group compared to the naïve group (Fig. 7h, i).
Our transduction method led to an increased expression of the hACE2 gene across various cell clusters (Fig. 8a–c), which was correlated with enhanced eGFP expression (Fig. 8d, Supplementary Fig. 3).
a Log2 normalized hACE2 expression fold change across different cell clusters in transduced lung organoids. b Log2 normalized eGFP expression across different cell clusters in transduced lung organoids. c Comparative transcriptomic analysis of hACE2 gene expression across different cell clusters between naïve and transduced human iPSC-derived lung organoids, presented as fold change in hACE2 gene expression. scRNA-seq was performed on both naïve and transduced lung organoids 7 days post-transduction. d Representative flow cytometry analysis of eGFP expression in control versus LVV-hACE2-transduced lung organoids.
Discussion
Transducing genes of interest into human iPSCs-derived lung organoids presents significant technical challenge, including suboptimal efficiency, elevated cytotoxicity, and time-consuming protocols. In this report, we present a refined and efficient method to LVV transduction into human iPSCs-derived lung organoids while preserving their 3D architecture. We utilized a third-generation LVV encoding eGFP and the human ACE2 gene as the transduction vector.
While AAV vectors are well-documented for gene delivery, their efficacy in organoid systems, particularly intestinal and lung organoids, is limited28,29. This limitation is due to low transduction efficiency, which is heavily dependent on serotype tropism and the availability of specific receptors within heterogeneous cell populations28,29,30. In contrast, LVV transduction has shown distinct advantages25,26,31,32, such as enhanced stability in gene integration and expression, as well as reduced immune responses10,16,17.
LVV-based gene delivery has been extensively optimized in 2D mammalian cell culture systems and in organoids derived from dissociated 3D structures, such as intestinal and pancreatic organoids. However, the traditional approach involves converting the 3D organoids structure into a 2D format through physical or enzymatic dissociation followed by spinoculation25,26,31,32, which can compromise the integrity of the organoids.
Our current protocol includes several refinements, as detailed in Table 120,22,33,34,35: 1) Use of non-aggressive mechanical forces through gentle pipetting to minimize damage to the organoids, 2) Enhancement of post-dissociation viability by supplementing the culture media with CHIR-99021 and Y-2763225. CHIR-99021, a WNT pathway modulator, supports cell proliferation and viability, enabling controlled expansion while minimizing premature differentiation. This selective proliferation promotes efficient LVV transduction without compromising lineage fidelity. Y-27632, a ROCK inhibitor, complements this by enhancing basal-cell proliferation, reducing apoptosis from cell dissociation, and increasing cellular resilience during manipulation, thereby improving transduction outcomes25,36. 3) Optimization of spinoculation parameters—specifically, a centrifugation rate of 600xg for 60 min to maximize LVV transduction while preserving cell viability37,38,39.
Two pivotal factors influencing transduction efficiency in human iPSC-derived lung organoids were identified: 1) Organoid size: The multiplicity of infection (MOI) in human iPSCs-derived lung organoids cannot be accurately determined using classical cell counting methods40,41, as these organoids maintain a 3D structure. Through pilot studies, we found that gentle pipetting could reduce the size of organoids (1.5-2.0 mm in diameter) to small fragments (0.2-0.5 mm) without fully dissociating them into single cells. We further estimated the cell density per organoid by dissociating organoids into single cells using accurate, despite the associated low viability. Our findings indicated an average cell density of 1 × 106 cells per organoid of 1.5-2.0 mm in diameter24, which informed the selection of similarly sized organoids for subsequent LVV transduction studies. 2) LVV titer: Our pilot studies suggested that a minimum LVV titer of 107 transduction units per mL (TU/mL) is necessary for effective in human iPSCs-derived lung organoids.
Using this optimized LVV transduction protocol, we successfully maintained the unique transcriptomic profiles of human lung organoids post-transduction. Importantly, scRNA-seq analysis revealed that a portion of the alveolar type II cell population, previously categorized as undifferentiated cells, was clearly identified by the robust expression of hACE2 following LVV transduction. This finding suggests that the enhanced gene expression post-transduction may improve the sensitivity and specificity of scRNA-seq in identifying distinct cell phenotypes within human iPSCs-derived lung organoids.
In conclusion, we have successfully developed a high-efficiency LVV transduction protocol for human iPSCs-derived lung organoids, achieving robust expression of target genes and maintaining the structure and functional integrity of the organoids.
Methods
Cell culture
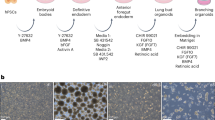
Human lung organoids were generated using a stepwise directed differentiation protocol with human induced pluripotent stem cells (iPSCs) (HDF-mRNA), provided by Drs. Snoeck and Chen24.
Human iPSC-derived lung organoids differentiation
Human iPSCs-derived lung organoids are generated from iPSCs according to a stepwise directed differentiation protocol outlined by Chen YW et al24. Briefly, undifferentiated iPSCs will be differentiated to the definitive endoderm lineage with exposure to saturating levels of Activin A. Embryoid bodies committed to the definitive endoderm will then be patterned to the anterior foregut endoderm upon sequential inhibition of transforming growth factor beta (TGFβ) and bone morphogenic protein (BMP) signaling. Following ventralization of endodermal cells, adherent cells will be adapted to three-dimensional suspension culture and promoted to the lung fate with a combination of growth and signaling factors, which includes the Wnt-agonist CHIR-99021, fibroblast growth factors and retinoic acid.
The generated human iPSCs-derived lung organoids, which contain a great propensity to give rise to AECIIs, surpass traditional immortalized and cancer AECII cell lines with regards to their expression of lung-specific markers and ability to mimic key functions (e.g., surfactant production) in vitro.
Human iPSCs-derived lung organoids are cultured in lung organoid culture media (Serum-Free Differentiation media supplemented lung maturation factors) for at least 45 days before LVV transduction procedure.
hACE2-LVV construction
A third-generation lentivirus vector containing the elongation factor 1 alpha 1 (EF-1α) promoter-driven eGFP reporter was customized and purchased from Tailored Genes (Toronto, Canada). The lentivirus vector, LVV hACE2.3xflag-T2A-eGFP, encoding the full length of the hACE2 gene (NCBI reference sequence ID, NM_021804.3) along with 3xFLAG tag (3’-GATTATAAGGATCACGATGGAGATTACAAGGACCACGACATAGACTACAAAGATGATGACGACAAG-5’), followed by Thosea asigna virus 2A self-cleaving peptide (T2A) and eGFP. qPCR and human osteosarcoma (HOS) cells are used for validation and titration. hACE2-LVV particles titer was determined by the company to be 107 TU/ml.
hACE2-LVV transduction in human iPSC-derived lung organoids
For the transduction procedure, we adapted the advice from previous protocols to freshly supplement the lung organoid culture media with 10 µM CHIR-99021 (GSK inhibitor, Cat. No 4423, Tocris, Bristol, UK), and 10 µM Y-27632 dihydrochloride (ROCKi, Cat. No 1254, Tocris, Bristol, UK) to induce proliferation and cell viability post-transduction procedure.
First, we transferred 5 high-dense large human iPSCs-derived lung organoids from low-attachment 6-well culture plate into a 1.5 ml Eppendorf tube and discarded supernatant. Human iPSCs-derived lung organoids were suspended in 100 µl of transduction culture media and were physically dissociated by gently pipetting up and down around 15 times using a 1 ml pipette. Next, we added 100 µl of high titer hACE2-LVV particles (107 TU/ml) into each Eppendorf containing the human iPSCs-derived lung organoids mixture. Then, the mixture was transferred into a separate well of a 96-well plate. For static group, we incubated the plate at room temperature for different durations (0.5 h and 1 h). For the spinoculation group we transferred the plate into a plate-centrifugation machine and centrifuged it using different speeds and duration (600g vs. 1200g and 0.5 h vs. 1 h). Thereafter, human iPSCs-derived lung organoids were collected and transferred into a well of a low-attachment 6-well culture plate and transduction culture media was completed to 2 ml of total volume. 48 h after hACE2-LVV transduction, media was collected and refreshed to lung organoid culture media.
Quantitative real-time PCR
Total RNA samples were prepared from lung organoids using the RNeasy Plus mini kit (Qiagen) according to the manufacturer’s instructions. To quantify AT2 canonical markers expression, one-step quantitative real-time PCR was performed using the Luna Universal qPCR Master Mix (New England Biolabs). Quantitative real-time PCR reactions were performed on an Applied Biosystems Quant Studio 7 Flex Real-Time PCR System. The cycling conditions were 1 cycle of denaturation at 60°C for 10 min, then 95°C for 2 min, followed by 39 amplification cycles at 95°C for 10 s and 60°C for 15 s. The delta-delta-cycle threshold (ΔΔCT) was determined relative to Gapdh and control samples. Analysis was performed using Quant Studio Real-Time PCR Software. The sequences of primers are provided in Supplementary Table 1.
Western blot
Equal number of human iPSCs-derived lung organoids were lysed with a 0.1% Triton-X buffer (Boston Bioproducts Cat. No. BP-117-250ml, MA, USA) supplied with proteinase cocktail (ThermoFisher Scientific Cat. No. PI78415, MA, USA). The total protein concentration of the cell lysates was determined using Bi-Rad protein assay (Bio-rad laboratories, Cat. No. 5000001, CA, USA) according to the manufacturer’s instructions. Equal amounts of proteins were loaded into a 10% gel, then transferred onto 0.2 µm polyvinylidene fluoride membrane (Immun-blot PVDF membrane, Bio-Rad laboratories, Cat. No 1620177, CA, USA), followed by blocking in 5% Bovine serum albumin (BSA, Cat. No 9048-46-8, Sigma-Aldrich, MO, USA) diluted in Tris buffered saline with 0.1% Tween 20 (TBST) at 25 °C for 1 h. The membranes were incubated with primary antibodies, mouse anti-GFP tag (1:1000 diluted in 1% BSA, Cat. No. MA5-15256, Invitrogen, MA, USA), Recombinant Anti-ACE2 antibody (1:1000 diluted in 1% BSA in TBST, Cat. No. ab108252, Abcam, Cambridge, UK) overnight at 4°C, and Mouse Anti β-actin (1:5000 diluted in 1% BSA in TBST, Cat. No. A5316, Sigma-Aldrich, MO, USA) for 1 h at 25 °C, then washed with TBST, and incubated with HRP-conjugated secondary goat anti-mouse IgG (1:1000 dilution in TBST, Cat. No. 115-035-003, Jackson Immunoresearch laboratories, PA, USA) or donkey anti-rabbit IgG (1:1000 dilution in TBST, Cat. No. 711-035-152, PA, USA) for 1 h at 25 °C. After 4 washes with TBST, the membranes were visualized using enhanced chemiluminescence western blotting detection reagent (Clarity Western ECL substrate, Cat. No. 1705060S, Bio-rad laboratories, CA, USA) and detected by Bio-Rad image analyzer. Densitometry was performed using ImageLab 6.0 (Bio-rad laboratories, CA, USA).
LDH cytotoxicity assay
Cytotoxicity is measured using cytotoxicity detection kit (LDH) (Cat. No. 11644793001, Roche, Mississauga, ON, Canada) which is designed to measure LDH activity and release from damaged cells once the cell membrane is disrupted. The LDH activity was performed according to the manufacturer’s suggested protocol in cell-free culture supernatant collected from human iPSCs-derived lung organoids culture 48 h after subjected to 1-h spinoculation using 600g or 1200g centrifugation speed. After addition of LDH reaction mix, a 30 min incubation was used. The optical density of the plate was read with a plate reader set at 490 nm with a 660 nm reference wavelength. The O.D results are compared to cell-free culture supernatant collected from human iPSCs-derived lung organoids and subjected to the exact similar conditions without spinoculation.
Human iPSCs-derived lung organoids live imaging
Live images of human iPSCs-derived lung organoids were visualized using ZOE cell imager (Bio-rad laboratories, CA, USA) under a ×20 objective operated with stand-alone android operating system. For each lung organoid, images were acquired from at least three different plains. Images were acquired from at least three different human iPSCs-derived lung organoids for each experiment. Images are representative of at least three separate experiments. Acquired images were exported as JPEG files, which were later processed for image analysis with the Fiji software. Fluorescent intensity is the mean fluorescent intensity of acquired fields of each human iPSCs-derived lung organoids. Fluorescent intensity was calculated by measuring integrated density per area then subtracting background mean gray value.
Confocal imaging
Expression of eGFP fluorescent marker and hACE2 was analyzed by immunofluorescence staining of human iPSCs-derived lung organoids following hACE2-LVV transduction procedure as previously described using setting of 600g spinoculation for 1 h at 25 °C.
Briefly, whole human iPSCs-derived lung organoids were collected 7 days post-transduction procedure and washed with pre-warmed PBS and fixed in pre-warmed 4% paraformaldehyde (PFA, diluted in PBS, Cat.No 28908, Thermofisher scientific, MA, USA) for 1 h at room temperature. Following fixation, residual PFA was washed thrice with PBS and human iPSCs-derived lung organoids were thoroughly permeabilized using 0.5% Triton X-100 diluted in PBS for 1 h. Next human iPSCs-derived lung organoids were incubated with staining buffer (4% BSA + 0.05% Tween 20 dilute in PBS + 10% goat serum at pH 7.4) for 1 h. Then, human iPSCs-derived lung organoids were incubated with primary antibody, rabbit anti-ACE2 (Cat. No. ab15348, 1:100 diluted in staining buffer, Abcam, Cambridge, UK) at room temperature overnight. Following primary antibodies incubation, human iPSCs-derived lung organoids were washed trice with TBST then incubated with secondary antibodies, Alexa Fluor 594-conjugated goat anti-rabbit IgG (H + L, 1:250 in staining buffer) and nuclear stained with DAPI (4’6-diamidino-2-phenlindole; 1:500 diluted in staining buffer, Cat. No. D9542, Sigma, MO, USA) diluted in staining buffer for 4 h at room temperature, and subsequently washed trice with TBST. After the final wash human iPSCs-derived lung organoids were mounted using mounting media (Millipore Sigma, MA, USA) and an imaging chamber, then directly visualized with The Zeiss LSM700 fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany) under a 20X objective operated with the Zen Black 2.3SP1software (Carl Zeiss AG, Oberkochen, Germany). Acquired images were exported as CZI files, which were later processed (e.g., visualization of individual color channels) for figure construction with the Fiji software.
Flow cytometry
Naïve and Transduced lung organoids (5 days post-transduction) were analyzed in a CytoFLEX-LX flow cytometer (Beckman Coulter, IN, USA). This device is equipped with a 488-nm blue laser for measuring forward and side light scatter (FSC and SSC) and detecting green fluorescence. Additionally, it includes multiple lasers for multicolor analysis, such as a 640-nm red laser for far-red fluorescence detection and a 405-nm violet laser for violet fluorescence detection. The cells were acquired and gated using FSC and SSC parameters. Analysis was conducted using density plots to display eGFP fluorescence in the Fluorescence 1 channel (green) and PI fluorescence, indicating dead cells, in the Fluorescence 2 and 3 channels (red). Non-transduced cells served to set quadrant borders. All measurements were performed on a single day under identical equipment settings. Three independent experiments were performed, representative image were shown. The data were analyzed with FlowJo™ (Becton, Dickinson & Company).
Single-cell lung organoid preparation for ScRNA-seq
Human iPSCs-derived lung organoids were chemically dissociated into single cells using dispase (5 units, Cat. No. CACB354235, VWR, PA, USA) for 5 min at 37°C and cell viability percentage was estimated using Trypan Blue (Cat. No T8154, Sigma-Aldrich, MO, USA) staining and 5000 live cells were collected and resuspended with human iPSCs-derived lung organoids maturation media and collected into an Eppendorf tube and placed on ice. The live lung single-cell suspension was proceeded with the Chromium Single Cell 3′ Reagent Kit v3 (10x Genomics, product code 1000075) using a 10x Genomics Chromium Controller.
scRNA-seq data analysis for human iPSC-derived lung organoids before and after hACE2-LVV transduction
All analyses and plotting were performed using the R package Seurat version 4.0.5.
The cell ranger output (10X; https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome) for the naive and the transduced cells datasets were read in and assessed for quality. We evaluated mitochondrial RNA content, number of unique features (genes) and total counts of detected RNA molecules and selected a basic filter (nFeature_RNA > 200 & nFeature_RNA < 8000 & percent.mt < 30 & nCount_RNA > = 300) which would retain most of the cells but exclude those which would be likely to be broken or doublets. The cells available for downstream analysis were 5382 for the naive experiment, and 1036 for the transduced (from 8117 and 3019 for naive and transduced, respectively).
Each dataset was normalized in Seurat using the SCTransform function, which has been shown to improve common downstream analytical tasks. At this point, the two datasets were integrated using the standard SCT integration workflow (SelectIntegrationFeatures with nFeatures = 3000, PrepSCTIntegration, FindIntegrationAnchors, IntegrateData) to allow a more straightforward comparison.
We then performed dimensionality reduction by PCA using default parameters, and UMAP (Uniform Manifold Approximation and Projection dimensional reduction technique) embedding using the first 30 principal components. Cell neighborhoods were defined using the Seurat functions FindNeighbors, which uses a KNN (K-nearest neighbors) graph based on the Euclidean distance in PCA space and refines the edge weights between any two cells based on the shared overlap in their local neighborhoods (Jaccard similarity); clustering was performed using the FindClusters function, which iteratively groups cells together, with default algorithm (Louvain) and resolution of 0.6. This gave a total of 7 separate clusters. Clusters identification was then made by computing the overlap of differentially expressed gene markers of each cluster with human lung cell dataset (16) and using the Molecular Signatures Database (MSigDB) from https://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp.
We compared the marker genes in human iPSCs-derived lung organoids in each cluster (cluster 0. AT1-like cells (276 genes), cluster 1. AT2-like cells (136 genes), cluster 2. fibroblast cells (172 genes), cluster 3. Stromal cells (131 genes), cluster 4. Immature cells (142 genes), cluster 5. Non-lung epithelial cells (106 genes), cluster 6. Airway basal cells (157 genes) with the human lung cells (AT1 cells (1087 genes), AT2 cells (205 genes), proliferating basal cells (984 genes), alveolar fibroblasts (423 genes), myofibroblasts (290 genes) and PNECs (1585 genes). Average gene expression was calculated on the normalized RNA data using Seurat Average Expression function, which averages the normalized (feature counts for each cell are divided by the total counts for that cell and multiplied by the scale factor. This is then natural-log transformed using log1p) expression values across the cells in each dataset. Similarly, the percent of cells expressing a gene was calculated using a custom function adapted from https://github.com/satijalab/seurat/issues/371. 10X Genomics Loupe Cell Browser v.3.1.1 was used for data visualization.
Statistics and reproducibility
No statistical methods were used to predetermine sample size. n = 3 independent biological replicates were used for all experiments unless otherwise indicated. ns indicates a non-significant difference. The P values were calculated by unpaired two-tailed Student’s t-tests unless otherwise indicated. *P < 0.05; **P < 0.01; ***P < 0.001. Each experiment was repeated independently three times with similar results. Correlation of protein concentration was analyzed by linear regression with robust curve fit and Pearson coefficient test.
Data availability
scRNA-seq data for human iPSC-derived lung organoids are publicly available in the GEO repository database under the accession number GSE284689. Source data can be obtained in Supplementary data 1.
Code availability
Codes used in this study are available from https://github.com/satijalab/seurat/issues/371.
References
Dye, B. R. et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4, 1–25 (2015).
Miller, A. J. et al. In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Stem Cell Rep 10, 101–119 (2018).
McCracken, K. W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014).
Barkauskas, C. E. et al. Lung organoids: current uses and future promise. Dev. Camb. Engl. 144, 986 (2017).
Leibel, S. L. & Sun, X. Halting SARS-CoV-2: lung organoids step up to the plate. EMBO J 40, e107651 (2021).
Wong, A. P. et al. Efficient generation of functional CFTR-expressing airway epithelial cells from human pluripotent stem cells. Nat. Protoc. 10, 363–381 (2015).
Huang, S. X. et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32, 84–91 (2014).
Ostedgaard, L. S. et al. A shortened adeno-associated virus expression cassette for CFTR gene transfer to cystic fibrosis airway epithelia. Proc. Natl. Acad. Sci. USA. 102, 2952–2957 (2005).
Fischer, A. C. et al. Successful transgene expression with serial doses of aerosolized rAAV2 vectors in rhesus macaques. Mol. Ther. J. Am. Soc. Gene Ther. 8, 918–926 (2003).
Kabadi, A. M., Ousterout, D. G., Hilton, I. B. & Gersbach, C. A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res 42, e147 (2014).
Wu, Z., Yang, H. & Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. J. Am. Soc. Gene Ther. 18, 80–86 (2010).
Simon, R. H. et al. Adenovirus-mediated transfer of the CFTR gene to lung of nonhuman primates: toxicity study. Hum. Gene Ther. 4, 771–780 (1993).
Marshall, E. Gene therapy death prompts review of adenovirus vector. Science 286, 2244–2245 (1999).
Escors, D. & Breckpot, K. Lentiviral vectors in gene therapy: their current status and future potential. Arch. Immunol. Ther. Exp. (Warsz.) 58, 107–119 (2010).
Schambach, A., Zychlinski, D., Ehrnstroem, B. & Baum, C. Biosafety features of lentiviral vectors. Hum. Gene Ther. 24, 132–142 (2013).
Bañuls, L. et al. Gene therapy in rare respiratory diseases: what have we learned so far? J. Clin. Med. 9, 2577 (2020).
Vu, A. & McCray, P. B. New directions in pulmonary gene therapy. Hum. Gene Ther. 31, 921–939 (2020).
Somers, A. et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells Dayt. Ohio 28, 1728–1740 (2010).
Nethercott, H. E., Brick, D. J. & Schwartz, P. H. Derivation of induced pluripotent stem cells by lentiviral transduction. https://doi.org/10.1007/978-1-61779-201-4_6.
Leibel S. L. et al. Reversal of surfactant protein b deficiency in patient specific human induced pluripotent stem cell derived lung organoids by gene therapy. https://doi.org/10.1038/s41598-019-49696-8.
Cao, F. et al. Comparison of gene-transfer efficiency in human embryonic stem cells. Mol. Imaging Biol. 12, 15–24 (2010).
Jacob, A. et al. Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 14, 3303 (2019).
Goldsteen, P. A., Yoseif, C., Dolga, A. M. & Gosens, R. Human pluripotent stem cells for the modelling and treatment of respiratory diseases. Eur. Respir. Rev. 30, 210042 (2021).
Chen, Y. W. et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017).
Van Lidth de Jeude J. F. et al. A protocol for lentiviral transduction and downstream analysis of intestinal organoids. J. Vis. Exp. 98, 52531 (2015).
Huang, W. et al. Pattern of invasion in human pancreatic cancer organoids is associated with loss of SMAD4 and clinical outcome. Cancer Res. 80, 2804 (2020).
Koo, B. K. et al. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods 9, 81–83 (2011).
Xian, L. et al. Genetic engineering of primary mouse intestinal organoids using magnetic nanoparticle transduction viral vectors for frozen sectioning. J. Vis. Exp. 2019, e57040 (2019).
Meyer-Berg, H. et al. Identification of AAV serotypes for lung gene therapy in human embryonic stem cell-derived lung organoids. Stem Cell Res. Ther. 11, 448 (2020).
Demayo, J. L., Wang, J., Liang, D., Zhang, R. & Demayo, F. J. Genetically engineered mice by pronuclear DNA microinjection. Curr. Protoc. Mouse Biol. 2, 245–262 (2012).
Rustanti, L. et al. Differential effects of strategies to improve the transduction efficiency of lentiviral vector that conveys an anti-hiv protein, nullbasic, in human T cells. Virol. Sin. 33, 142 (2018).
Maru, Y., Orihashi, K. & Hippo, Y. Lentivirus-based stable gene delivery into intestinal organoids. Methods Mol. Biol. Clifton NJ 1422, 13–21 (2016).
Sun, D. et al. A functional genetic toolbox for human tissue-derived organoids. eLife 10, e67886 (2021).
Sun, D. et al. SOX9 maintains human foetal lung tip progenitor state by enhancing WNT and RTK signalling. EMBO J. 41, e111338 (2022).
Liu, S., Sun, D., Butler, R. & Rawlins, E. L. RTK signalling promotes epithelial columnar cell shape and apical junction maintenance in human lung progenitor cells. Dev. Camb. Engl. 150, dev201284 (2023).
Horani, A., Nath, A., Wasserman, M. G., Huang, T. & Brody, S. L. Rho-associated protein kinase inhibition enhances airway epithelial basal-cell proliferation and lentivirus transduction. Am. J. Respir. Cell Mol. Biol. 49, 341 (2013).
Bahnson, A. B. et al. Centrifugal enhancement of retroviral mediated gene transfer. J. Virol. Methods 54, 131–143 (1995).
Wu, Y., Melton, D. W., Zhang, Y. & Hornsby, P. J. Improved coinfection with amphotropic pseudotyped retroviral vectors. J. Biomed. Biotechnol. 2009, 901079 (2009).
Guo, J., Wang, W., Yu, D. & Wu, Y. Spinoculation triggers dynamic actin and cofilin activity that facilitates HIV-1 infection of transformed and resting CD4 T cells. J. Virol. 85, 9824–9833 (2011).
Shabram, P. & Aguilar-Cordova, E. Multiplicity of infection/multiplicity of confusion. Mol. Ther. 2, 420–421 (2000).
Abedon, S. T. Multiplicity of Infection. Ref. Module Life Sci. https://doi.org/10.1016/B978-0-12-809633-8.06748-0 (2017).
Acknowledgements
Figure 6 was created with BioRender.com. The work was supported by the Canadian Institutes of Health Research grants (PJT 190130 to H.Z. and MHR-137772 to A.S.S.) H.Z. is the holder of the Robert and Dorothy Pitts Chair in Acute Care and Emergency Medicine, a joint Hospital-University Endowed Chair between the University of Toronto, Unity Health Toronto, and the St. Michael’s Foundation.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.Z. and J. Khateeb. Methodology: H.Z., J. Khateeb, J.L., Y.L., and T.T. ScRNA seq analysis: G.P. and B.T. Investigation: J. Khateeb, J.L., Y.L., J. Khang, and M.J. Resources: J. Khang and H.Z. Writing—Original Draft: J. Khateeb and J.L. Writing—Review and Editing: J.L., Y.W.C., O.R., A.S.S., and H.Z. Supervision: H.Z. Equal contribution: J. Khateeb and J.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Yuling Han, Robert Zweigerdt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Dr Ophelia Bu, Dr Pavithra Chavali.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khateeb, J., Liang, J., Li, Y. et al. Optimized gene transduction in human lung organoids: A high-efficiency method for advanced research applications. Commun Biol 8, 164 (2025). https://doi.org/10.1038/s42003-025-07461-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07461-w
This article is cited by
-
Lung-targeted RNA delivery systems: strategies and therapeutic applications
Journal of Nanobiotechnology (2026)