Abstract
Diminished bioavailability of nitric oxide (NO) contributes to the pathogenesis of cardiometabolic disorders. However, the alterations in signaling under NO deficiency remain mostly unknown. We combined genetics and proteomics to quantify changes in the heart proteome, phosphoproteome, and S-nitrosocysteine proteome in mice lacking nitric oxide synthases (NOS1, NOS2, NOS3), lacking all three enzymes (tNOS), or the alpha 1-regulatory subunit of the soluble guanylate cyclase (sGCα1). Modest changes of less than 1% in the proteome and 4% in the phosphoproteome in single NOS gene or sGCα1 null mouse hearts indicate sufficient biological compensation. In contrast, the number of S-nitrosylated proteins declined by 80%, 57%, and 35% in NOS3, NOS1, and NOS2 null mice, respectively. A 21% remodeling of the proteome and 9% of the phosphoproteome in the tNOS null mice included integral kinases that provide adaptive rewiring of signaling. The data revealed the emergence of enhanced mitogen-activated-kinases Mapk3/Mapk1 signaling, documented by increased phosphorylation of these kinases and their downstream targets. The data highlight that adaptive compensation of signaling prevents overt phenotypes during NO signaling deficits. In contrast, maladaptive signaling via Mapk3/Mapk1 may promote pathological cardiomyopathy that progressively develops in the tNOS null mice.
Similar content being viewed by others
Introduction
Reduced bioavailability of NO and downstream signaling contribute to the pathogenesis of hypertension, diminished cardiac and endothelial cell function, and cardiometabolic disorders1,2,3,4,5,6,7,8,9,10,11,12,13. Genetic predisposition or pharmacological interventions that enhance NO-signaling protect against cardiovascular diseases5,14. The canonical biochemical pathway that accomplishes NO signaling is a tiered biological process that includes three enzyme families, nitric oxide synthases (NOS), soluble guanylate cyclase (sGC; gene annotation Gucy1α1/Gucy1β1), and cGMP-dependent protein kinases (Prkg), and three small molecules NO, cyclic guanosine monophosphate (cGMP), and ATP1,2,3,4,15,16,17,18,19. The signaling begins with the enzymatic generation of NO by the three principal NOS isoforms. In the heart, NO is generated by NOS3 and NOS1, and to a lesser level by NOS2, in a highly regulated manner. NO binds to the iron in the heme center of the β subunit of the sGC, leading to the conversion of guanosine triphosphate to cGMP. Downstream of cGMP are the cGMP-dependent protein kinases, primarily the Ser/Thr kinases Prkg1 and Prkg2. Prkg1α is the predominant isoform expressed in the cardiovascular system15,16,17,18,19. The binding of cGMP to allosteric sites in the regulatory domain of the Prkg1 isozymes increases the catalytic activity, leading to the phosphorylation of client proteins15,16,17,18,19. This canonical signaling cascade is terminated in reverse order, by phosphatase-mediated dephosphorylation, removal of cGMP by phosphodiesterases, elimination of NO by competing reactions with metalloenzymes or superoxide, and termination of NO production by NOS.
The NO-mediated formation of protein S-nitrosocysteine also provides complementary signaling and regulation of protein function. This reversible post-translational modification (often designated as cysteine S-nitrosylation) is implicated in the regulation of calcium homeostasis, transcription factors, protein kinases, and metabolic pathways20,21,22,23,24,25,26,27,28,29,30. Overall, the NOS/NO signaling is exceptionally regulated and fine-tuned to provide spatial and temporal signaling that accomplishes diverse paracrine and autocrine biological functions within the cardiovascular system.
Despite considerable knowledge regarding the NOS enzymes, sGC, and Prkg1, the changes in this signaling pathway under conditions of NO deficiency are unknown. Therefore, we quantified changes in the heart proteome, phosphoproteome, and S-nitrosocysteine proteome in mice lacking all three NOS enzymes, the alpha 1-regulatory subunit of sGC, and mice null for each NOS enzyme. These mice have been well characterized, and age-dependent cardiovascular alterations that reproduce human phenotypes have been documented31,32,33,34,35,36,37,38,39,40,41,42. To acquire and analyze these proteomes, we employed established mass spectrometry-based (MS/MS) platforms and bioinformatics that we used previously in various mouse and human tissues27,30,43. Analysis of the data was centered on two questions: (1) how does the heart proteome and signaling adjust to NO/cGMP deficiencies, and (2) what aberrant changes in the proteome or signaling may explain the phenotypes described in mice with NOS and sGC genetic deletions.
Results
We acquired and analyzed the proteome, phosphoproteome, and S-nitrosocysteine proteome of hearts from male wild-type C57BL/6J mice as well as mice with known genetic deficiencies in the nitric oxide-cGMP signaling pathway. The experimental workflow for the acquisition, identification, quantification, and bioinformatic analysis of mouse heart proteomes is depicted in Fig. 1a. Simultaneous analysis of the three proteomes provides a comprehensive view of signaling landscapes under diminished NO-cGMP signaling.
a Graphical representation of the experimental workflow for the acquisition, identification, quantification, and bioinformatic analysis of mouse heart proteome, phosphoproteome, and S-nitrosocysteine proteomes. The figure was created using BioRender (license to Mohanty, I., 2024, https://biorender.com/z60z971), and the data images were generated from our proteomic analysis. b Histogram plot of the relative abundance of the 3,663 proteins quantified in all genotypes (purple) and 816 that are statistically different in the tNOS null mice, adjusted p-value < 0.05 (red). The proteome was acquired from n = 5 wild-type, n = 4 NOS1, n = 5 NOS2, n = 5 NOS3, n = 4 sGCα1 and n = 5 tNOS null mice. The gray bars represent the distribution of the manually curated and integrated reference mouse heart proteome of 7071 proteins. c Histogram of the relative abundance of phosphoproteins detected in all genotypes. 1064 proteins were identified only by the phosphorylation site (light green), whereas 1072 were detected in the proteome and phosphoproteome (blue). The phosphoproteome was acquired from n = 5 wild-type, n = 4 NOS1, n = 5 NOS2, n = 5 NOS3, n = 4 sGCα1 and n = 5 tNOS null mice. d Cumulative frequency histogram of the relative abundance of 415 S-nitrosylated proteins (red) in the wild-type mouse, n = 6.
The mouse heart proteome landscape under NO/cGMP deficiency
We detected 3973 proteins in the mouse heart across all genotypes (Supplementary Data 1 and Supplementary Fig. 1a). The detected proteins spanned at least seven orders of magnitude in relative abundance and mirrored the distribution of a curated mouse heart reference proteome (Fig. 1b). We constructed the reference mouse heart proteome by downloading the MS/MS-based Data from the PAXdb: Protein Abundance Database44, which we then mapped using UniProtKB45 to generate a list of 7071 mouse heart proteins (Supplementary Data 1).
The changes in the heart proteome of each genotypically different mouse against the wild-type mice were statistically evaluated using the Limma t-test and an adjusted p-value < 0.05 for significant differences. The mice with a single NOS gene deletion exhibited minimal changes ranging from four proteins in NOS1, three in NOS2, and none in the NOS3 null mice whereas 21 proteins were different in sGCα1 null hearts as compared to wild-type (Supplementary Fig. 1a). The MS/MS data identified expected results such as the presence of beta 1 but not of alpha 1 subunit of sGC in the sGCα1 null hearts and the absence of NOS3 from the NOS3 and tNOS null mice (Supplementary Data 1). The tNOS null mice exhibited extensive remodeling of the proteome as the relative abundance of 814 proteins was statistically different from the wild-type controls. The distribution of proteins for the pairwise comparison between the tNOS null and wild-type mice is depicted in the MA plot (Fig. 2a) and a typical volcano plot (Supplementary Fig. 1b). Principal component analysis segregated the tNOS null mice from the wild type (Supplementary Fig. 1c).
a A scatter MA plot depicting on the X-axis the relative abundance and Y-axis the log2 fold change of the 3979 proteins quantified in tNOS (n = 5) and wild type (n = 5) mice. The 816 significantly different proteins in the tNOS null mice hearts are depicted in red. b Biological processes enriched in the 366 proteins with increased relative levels in the hearts of tNOS mice compared to wild type. c Biological processes enriched in the 450 proteins, in which the relative levels were decreased in tNOS compared to wild-type mice. The X-axis indicates the enrichment ratio, the size of the circles indicates the number of proteins in the cluster, and the color heat bar indicates the false discovery rate values (FDR) of enrichment.
The analysis indicated that the relative abundance of 448 proteins decreased and 366 increased in the tNOS null compared to wild-type mice. The major biological pathways represented within the 448 proteins were cellular respiration, oxidative phosphorylation, long-chain fatty acid oxidation, and branched amino acid catabolism (Fig. 2b, and supplementary Fig. 2). Consistent with the enrichment data, 141 of the 448 proteins are localized in the mitochondria and are functionally linked to metabolic activities (Supplementary Data 1, and supplementary Fig. 2). Biological processes enriched within proteins with increased relative levels included mRNA processes essential for translation and proteins that participate in the organization of cytoskeletal structures (Fig. 2c). Collectively alterations in the tNOS null mouse heart proteome prioritize protein synthesis to maintain cellular integrity and an intact contractile apparatus while failing to secure adequate levels of proteins in metabolic pathways.
The mouse heart phosphoproteome landscape in NO/cGMP deficiency
The number of phosphosites and phosphoproteins detected in each genotype is depicted in Fig. 3a, supplementary Fig. 3a, and Supplementary Data 2. We detected 12,528 unique phosphorylation sites mapped to 2136 proteins across all six genotypes. We observed a typical distribution of phosphorylation 80.3% on serine, 17% on threonine, and 2.7% on tyrosine residues. Of the 2136 phosphoproteins, 1072 were also detected in the proteome, whereas 1064 were identified based on the detection of phosphosites. The distribution of the relative abundance of these two groups is depicted in Fig. 1c, showing a greater number of proteins with lower abundance detected only as phosphoproteins. A core of 1616 phosphoproteins representing 76% of phosphoproteins detected were shared among all genotypes (Supplementary Fig. 3b). Like the proteome, deletion of a single NOS gene resulted in small perturbations in the relative levels of phosphosites ranging from 0.84, 0.23, and 0.02% for NOS1, NOS2, and NOS3, respectively, compared to the wild type. Correspondingly 3.5, 1.1, and 0.06% of the quantified phosphoproteins differed in the NOS1, NOS2, and NOS3 null mice, respectively. Modest changes, 3.7% of phosphosites and 1.5% of phosphoproteins, were also quantified in the sGCα1 null mice. In the tNOS null mouse heart, 401 phosphorylation sites were regulated, 150 had decreased, and 251 increased phosphorylation (adjusted p-value < 0.05) compared to wild type (Supplementary Fig. 4a, Supplementary Data 2). Principal component analysis further confirms the differences between tNOS null and wild-type mice (Supplementary Fig. 4b). The 401 regulated sites were mapped to 161 proteins representing 9% of the phosphoproteins quantified in tNOS null mice. Key biological processes and pathways enriched among the 161 regulated phosphoproteins included functional clusters important in cardiomyocyte physiology such as contraction, membrane potential, cellular junction organization, and growth regulation (Fig. 3b).
a Bars represent the number of phosphosites detected in each genotype (n = 5 wild-type, n = 4 NOS1, n = 5 NOS2, n = 5 NOS3, n = 4 sGCα1, and n = 5 tNOS null mice). The number of significantly different from wild-type (adjusted p-value < 0.05) regulated sites is indicated in red and in parentheses. b Gene ontology enrichment analysis of the 161 phosphoproteins with 401 regulated phosphosites in the tNOS null mice. The data depicts four major enriched biological processes. The size of the circles indicates the number of proteins in the cluster, and the color bar signifies the fold change from the wild-type mice.
The mouse heart S-nitrosocysteine proteome under deficient production of NO/cGMP
Using our previously established chemoselective enrichment platform27,43, we precisely identified 728 S-nitrosocysteine residues in 415 proteins in the wild-type mouse hearts (Supplementary Data 3). Unlike phosphorylation, which can be derived by multiple overlapping kinase activities utilizing ATP, S-nitrosocysteine formation is restricted by nitric oxide levels. As such, single-gene deletion of NOS impacted the formation and detection of S-nitrosylated cysteine residues. We report an 85%, 68%, and 38% reduction in the S-nitrosylated cysteine residues identified in NOS3, NOS1, and NOS2 null mice, respectively. Correspondingly, an 80%, 57%, and 35% reduction in the number of S-nitrosylated proteins were quantified in the NOS3, NOS1, and NOS2 null mice respectively (Fig. 4a). The data indicated that NOS3 is a major contributor to heart protein S-nitrosylation followed by NOS1, which in part is consistent with the relative abundance of the NOS isoforms in the mouse heart under physiological conditions34,46,47. The relative abundance value from the MS/MS-based detection of NOS3 is 12.4, whereas NOS1 is 0.03, and NOS2 is non-detectable by MS/MS (Supplementary Data 1). We detected 41 sites in 41 proteins in the tNOS null heart. S-nitrosylated sites in the tNOS null may indicate the generation of nitric oxide intermediates derived from dietary nitrate/nitrite or possibly false identification of S-nitrosylated cysteine residues. Because of this uncertainty, we removed these sites and proteins from all subsequent analyses. Proteins modified by S-nitrosylation participate in the generation of ATP through actions in pyruvate metabolism, oxidative phosphorylation, long-chain fatty acid oxidation, and branch-chain amino acid metabolism (Fig. 4b), reaffirming previous observations that S-nitrosylation in the mouse heart is enriched among proteins in metabolic processes30. The sizable absence of 330 S-nitrosylated proteins in NOS3 null hearts, of which 138 proteins are assigned metabolic functions and 107 are localized in mitochondria (Fig. 4c), may explain the cardiometabolic deficits in these mice in the absence of significant deficits in the proteome and phosphoproteome. NOS3 mice but not NOS1 and NOS2 null mice have documented cardiometabolic deficits48,49,50.
a The bars indicate the number of precisely identified S-nitrosocysteine sites (red) and the corresponding proteins (blue) for each genotype (n = 6 wild-type, n = 6 NOS1, n = 6 NOS2, n = 6 NOS3, null mice). Insert indicates the relative abundance levels of the NOS isoforms in the mouse heart. b Gene Ontology analysis for biological processes enriched in the 336 proteins that are S-nitrosylated in wild-type but not in NOS3 mouse hearts. The X-axis indicates the combined score, the natural log of the p-value multiplied by the z-score, where the z-score is the deviation from the expected rank from the Enrichr95 analysis. c The same as b for the cellular component, indicating the localization of the proteins.
Proteomic and signaling changes in functional pathways
To further explore the alterations in signaling we curated major functional clusters in the cardiovascular system, specifically, the NO-cGMP pathway, cardiomyocyte contraction, type-1 angiotensin II receptor (AT1R), glucose metabolism, tricarboxylic acid and oxidative phosphorylation, lipid, and branched-chain amino acids metabolism, responses to stress and redox regulation, autophagy, cell death, and both positive and negative regulation of cardiac hypertrophy. To construct these reference sub-proteomes, we collected data from three primary databases, UniProt-KB45, the Kyoto Encyclopedia of Genes and Genomes51, and Reactome52, followed by literature-based manual curation. We then mapped changes in these clusters for the proteome, phosphoproteome, and S-nitrosocysteine proteome, for each mouse genotype (Fig. 5a, Supplementary Data 4, and Supplementary Figs. 5-7). The NOS-sGC-Prkg1/2 pathway includes 32 proteins that participate in the production of nitric oxide, the production and destruction of cGMP, the Prkg1/2 kinases, and 73 curated downstream phosphorylation clients of Prkg1/2. Of the 105 proteins, we detected 60, of which 24 were regulated in the tNOS null mice, indicating a significant disturbance in this signaling pathway under severe loss of nitric oxide (Fig. 5b). The metabolic pathways were also significantly impacted in tNOS null mice, primarily due to changes in the proteome and S-nitrosocysteine proteome. Cardiomyocyte contraction a primary function of the cardiomyocytes was also influenced (21% regulated) in tNOS null mice (Fig. 5c). Cardiac muscle contraction was also highly enriched in the regulated phosphoproteome of the NOS1 null mice, a finding consistent with the known function of this enzyme in the regulation of cardiomyocyte contraction34,46,47. Specifically, the data in NOS1 null mice showed altered phosphorylation of primary proteins participating in contraction such as troponin I, sodium channel protein type 5 subunit alpha, protein phosphatase 1 regulatory subunit 12A, myosin-6, titin, and secondary regulators such as desmin, plasma membrane calcium-transporting ATPase 1, myomesin-1, PGC-1 and ERR-induced regulator in muscle protein 1 were quantified (Supplementary Data 2).
a Manually curated sub-proteomes for major functional pathways were used to identify proteins, and phosphoproteins detected and/or regulated in the tNOS null mice. The number of regulated proteins includes S-nitrosylated proteins in the wild-type hearts, which are missing in the tNOS null mice. b Graphical depiction of the canonical signaling cascade of NO/cGMP that includes downstream targets of Prkg1 phosphorylation and a heat map depicting changes in the proteome and phosphoproteome in this pathway for all genotypes. c Heat map depicting changes in the proteome and phosphoproteome for the cardiomyocyte contraction pathway. White boxes indicate that the protein or the phosphoprotein was not detected.
Kinases and phosphatases in NO/cGMP deficiency
Since signaling is typically a multi-tiered cascade, we inspected changes in kinases and phosphatases in the tNOS null mice. Using multiple protein database sources, we identified 293 kinases expressed in mouse hearts (Supplementary Data 5) of which 94 were detected at the proteome, 100 at the phosphoproteome, and 10 at the S-nitrosocysteine proteome (Fig. 6a). The relative levels of 20 kinases were regulated in the tNOS null mice proteome (Fig. 6b). The levels of mitochondrial pyruvate dehydrogenase kinase isozyme 4, a regulator of pyruvate metabolism, were also decreased in the sGCα1 null mice. Twenty-two phosphosites in 11 kinases were regulated in the tNOS null mice heart (Fig. 6c). These regulated phosphosites represented known sites of autophosphorylation or regulatory phosphorylation events in response to various stimuli (Supplementary Data 5). Known regulatory sites included Ser2481 in the serine/threonine-protein kinase mTOR, Thr196 in cAMP-dependent protein kinase catalytic subunit alpha (PKA C-alpha), and Thr193 in STE20-like serine/threonine-protein kinase. In wild-type mice, a total of 13 sites of S-nitrosylation were identified in 10 kinases, including previously reported S-nitrosylated kinases 5’-AMP-activated protein kinase catalytic subunit alpha-2, integrin-linked protein kinase, striated muscle-specific serine/threonine-protein kinase, and glycogen synthase kinase-3 beta25,29. Based on the known biological functions of the regulated kinases, it appears that five central nodes of signaling are regulated under severe NO signaling deficit (Supplementary Fig. 8). Kinases participating in the regulation of wingless-related integration site signaling and cell death are downregulated, suggesting potential protective adaptive signaling responses. Kinases important for signaling required for cardiomyocyte contraction are upregulated or activated. Integral multifunctional kinases AMPK, Akt, Mtor, Gsk3b, and Pdk4, which primarily coordinate and regulate metabolic processes, are also regulated, reflecting an adaptive signaling response to the proteomic changes in metabolic proteins. This coordinated kinase response likely accounts for the increased phosphorylation levels in 251 of the 401 regulated phosphosites in tNOS null mice. The increased phosphorylation is mapped to proteins that regulate cytoskeletal dynamics, contraction, calcium homeostasis, and autophagy in the heart, consistent with the known activity of the regulated kinases (Supplementary Fig. 8).
a We generated reference lists of 295 and 116 putative kinases expressed in mouse hearts by curating data from available resources. In the proteome, phosphoproteome, and S-nitrosocysteine proteome, we assess changes in 49% of the putative kinases and 35% of the phosphatases expressed in the heart. b Kinases that were significantly regulated (adjusted p-value < 0.05) at the proteome level. c Fold changes in the relative abundance of phosphosites in kinases. Phosphosites Slk_T1931, Speg_S1954, and Speg_T1956 were below the detection level in tNOS null mice. The relative levels of Stk, Camk2d, and Akt1 were also regulated at the protein level.
We also curated 116 phosphatases in cardiomyocytes of which 41 were detected in the proteome and the phosphoproteome (Fig. 6a). The relative protein levels of dual specificity protein phosphatase 23 (DUSP23), serine/threonine-protein phosphatase 2A, and serine/threonine-protein phosphatase PP1-beta catalytic subunit decreased, while the levels of serine/threonine-protein phosphatase PP1-gamma catalytic subunit, and receptor-type tyrosine-protein phosphatase mu increased in the tNOS null mice. An increase in phosphosite Ser461 in protein phosphatase Slingshot homolog 2 was the only significant change in the phosphoproteome of tNOS null mice. Four phosphatases were S-nitrosylated at a single site in wild-type mice (Supplementary Data 5).
Characterizing emergent changes in signaling during NO/cGMP deficiency
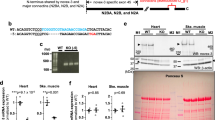
The MS/MS Data indicated increased phosphorylation of the mitogen-activated protein kinase 3 (Mapk3) and mitogen-activated protein kinase 1 (Mapk1) in both the tNOS and sGCα1 null mice (Fig. 7a,b). Mapk3/Mapk1 are activated by phosphorylation at Thr203, Tyr205, Thr208 in Mapk3 and Thr183, Tyr185, Thr188 in Mapk1 via a canonical three-tiered kinase cascade. Thr203, Tyr205, and Thr183, Tyr185, are phosphorylated by the dual specificity mitogen-activated protein kinase kinase 1 and 2 (Map2k1/2 or Mek1/2), whereas Thr208 and Thr188 represent autophosphorylation sites. The MS/MS Data was further confirmed in tNOS null mice by probing heart homogenates with antibodies that recognize phosphorylated and non-phosphorylated Mapk3 and Mapk1 (Fig. 7c). A significant increase in the ratio of phosphorylated Mapk3 (1.9-fold) and Mapk1 (2.3-fold) relative to unmodified proteins was observed in tNOS null as compared to wild-type mice. Moreover, the proteomic data indicated that the relative protein levels of Mapk3 and Mapk1 were not different among all genotypes, which is confirmed by the western blot analysis in the tNOS mice (Fig. 7c).
a MS and MS/MS extracted ion chromatograms (XIC) for the Mapk3 peptide IADPEHDHTGFLpTEpYVApTR and b for the Mapk1 peptide VADPDHDHTGFLpTEpYVApTR. In both graphs, the top tracings are the MS2, and the bottom are the MS1 XIC. These peptides correspond to the three phosphorylation sites in the Thr-Glu-Tyr-X-X-Thr motif of the activation loops, corresponding to Thr203, Tyr205, and Thr208 in Mapk3, and Thr183, Tyr185, and Thr188 in Mapk1 across all genotypes. c Western blot for phosphorylated and total Mapk3 and Mapk1 in the tNOS and wild-type mouse hearts (n = 3). Quantification of the ratio of phosphorylated to total Mapk3 and Mapk1 from three different mouse hearts (values represent mean ± standard deviation). Total Mapk3 and Mapk1 levels were normalized to GAPDH. *p value = 0.0056 for Mapk3 and p value = 0.0292 for Mapk1. d Representative western blots and quantification of the fold increase in the ratio of phosphorylated to total Mapk3 and Mapk1 in HL-1 cells after inhibition of NOS and sGC. (C) indicates no treatment control, and (I) treatment with the NOS inhibitor L-NAME and the sGC inhibitor ODQ (n = 4 independent cell cultures and treatments, values represent mean ± standard deviation).
We also utilized HL-1 cells to test if changes in nitric oxide and cGMP levels replicated the in vivo findings. HL-1 cells, an immortalized cell line derived from the AT-1 mouse atrial cardiomyocyte tumor lineage cells, retain contractile, electrophysiological, and biochemical properties of primary cardiomyocytes53. Treatment of HL-1 cells with the NOS inhibitor N(gamma)-nitro-L-arginine (L-NAME) and 1H-[1,2,4] oxadiazole [4,3-a]quinoxalin-1-one (ODQ), a selective and irreversible heme-site inhibitor of sGC, elevated the ratio of phosphorylated to total protein by 2.96 ± 0.82 fold for Mapk3, and 2.38 ± 0.90 for Mapk1 relative to untreated cells (Fig. 7d). Treatment with L-NAME increased the ratio of phosphorylated to total protein by 1.81 ± 0.41 for Mapk3 and 1.56 ± 0.42 for Mapk1, (n = 4 independent culture experiments). Treatment with ODQ increased the phosphorylated to total protein ratio by 1.49 ± 0.19 for Mapk3 and 1.60 ± 0.15 for Mapk1 (n = 3 independent culture experiments) relative to untreated cells. These data are consistent with the in vivo data and reaffirm that NO production by NOS and cGMP generation by sGC contribute to the activation of Mapk3/Mapk1 signaling.
Additional evidence for the activation of the Mapk3/Mapk1 comes from documenting increased phosphorylation of microtubule-associated protein 2 (Map2), the first known target for Mapk3, microtubule-associated protein 4 (MAP4), microtubule-associated protein tau (MAPT), and the Src substrate cortactin in the tNOS null mice (Supplementary Data 2). Moreover, previous data in the tNOS mice indicated that pharmacological inhibition of the type-1 angiotensin II receptor (AT1R) and downstream signaling reduced the development of left ventricular hypertrophy and cardiac dysfunction at 5 months of age37. Mapk3/Mapk1 activation is a well-known downstream event in the AT1R signaling or AT1R transactivation of the epidermal growth factor receptor cascade54,55,56,57,58. In addition to the increased phosphorylation of Mapk3/Mapk1, eight other proteins participating in AT1R signaling were also altered in tNOS but not in the sGCα1 null mice hearts. These include increased protein levels of the guanine nucleotide-binding protein G(q) subunit alpha, guanine nucleotide-binding protein subunit beta-5, both of which modulate signaling by guanine nucleotide-binding proteins, protein kinase C alpha type, and RAC-alpha serine/threonine-protein kinases 1 and 2 kinases. Increased phosphorylation of Kalirin, a serine/threonine-protein kinase that promotes the exchange of GDP by GTP and activates Rho GTPase proteins that are critical components of AT1R activation, was also quantified in the tNOS null mice. These data indicate that activation of the AT1R and Mapk3/Mapk1 signaling cascades may represent a prominent adaptive response during NO/cGMP deficiency in the cardiovascular system.
Discussion
Decades of research have firmly established the importance of NO/cGMP signaling in cardiovascular physiology1,2,3,4. Studies in humans and animal models have also documented that reduced NO/cGMP bioavailability resulting from debilitated production or rapid elimination of NO/cGMP signaling is central to the pathogenesis of cardiovascular disorders5,6. To gain a global appreciation of the changes during NO-signaling deficiency, we quantified for the first time changes in the proteome, phosphoproteome, and S-nitrosocysteine proteome in the hearts of wild-type, NOS1, NOS2, NOS3, all three NOS, and sGCa1 gene deletion mice. The analysis was performed in 2–4-month-old male mice. At this age, male NOS1 and tNOS null mice have increased basal contraction and increased calcium loading to the sarcoplasmic reticulum. The NOS3 and tNOS mice are hypertensive as compared with the wild-type mice31,32,33,34,35,36,37,38,39. The NOS2 null mice do not have any reported cardiovascular phenotype at the time of the analysis. Similarly, sGCα1 null mice backcrossed to the C57BL/6J background do not exhibit cardiovascular phenotypes59,60. The tNOS and, to a lesser extent, the NOS3 null mice will develop additional age-dependent progressive phenotypes, primarily left ventricular dysfunction and pathological hypertrophy37,38,39. Mice with tNOS and sGCα1 gene deletion show dysfunctional cardiac responses upon secondary challenges59,60,61,62. All the phenotypes are described in male mice, which were exclusively utilized in this study and do not apply to female mice that exhibit different responses to NO/cGMP deficiencies40,41,42.
Mice with a single NOS gene deletion exhibited minimal changes in the proteome and phosphoproteome but a substantial reduction in the S-nitrosocysteine proteome. Previous work indicated regulatory functions of protein S-nitrosylation in coordinating the flow of intermediates across different metabolic pathways to avoid metabolic gridlocks and maximize ATP production, which is critical for the heart27,28,29,30. NOS3 mice have documented cardiometabolic deficits48,49,50, which could arise from the absence of S-nitrosylated proteins since these mice have minimal perturbations in the proteome and phosphoproteome. Similarly, progressive deterioration of NO signaling via protein S-nitrosylation is likely to contribute in part to the phenotypic changes in the tNOS null mice, since these mice also exhibit deficits in the relative levels of metabolic proteins in addition to a substantial decline in protein S-nitrosylation. The metabolic deficits in the tNOS mice include reduced levels of proteins participating in triglyceride metabolism (Supplementary Data 1), consistent with the reported dyslipidemia and development of high-fat diet-induced atherosclerosis in these mice61,62. The minimal changes in the phosphoproteome in the single NOS gene deletions likely reflect the compensation provided by the expression of the other NOS isoforms. Although compensation dampens the changes in phosphorylation signaling, subtle changes such as the alteration in cardiac muscle contractility signaling are evident in NOS1 null mice. The compensation provided by other NOS is consistent with data documenting heightened age-dependent development of phenotypes in double NOS knock-out mice as compared to single NOS gene deletion33,34 and severe phenotypic changes in the triple NOS null mice37,38,39,61,62.
Modest changes in the proteome and phosphoproteome in the sGCα1 null mice indicate that, in part, signaling may proceed by cGMP generated from the sGCα2/β1 cardiac isoform63 or the membrane-associated particulate-GC. However, data have indicated compartmentalization in the production and utilization of cGMP generated by the guanylate cyclases63,64,65. Consistent with previous observations41, the phosphoproteome data (Supplementary Data 2) confirmed that known targets of Prkg1 phosphorylation, such as Ser23 and Ser24 in troponin I were not diminished in the sGCα1 null mice, indicating the redundancy in biological signaling to secure essential biological functions.
The most significant changes in the proteome and phosphoproteome were observed in the tNOS mice. The analysis revealed that 1212 of the 4782 proteins evaluated in the tNOS mice were regulated. Despite these significant perturbations, compensation afforded by increased levels and signaling by other kinases with known regulatory functions (Fig. 5d) may secure proper contraction and energy supply to overcome the NO/cGMP signaling deficiency. Moreover, the increased levels of antioxidant proteins such as the extracellular superoxide dismutase (SOD 3), glutathione peroxidase 1, and peroxiredoxin 4, may counter oxidative stress in the tNOS null mice. Although these adaptive changes may support and maintain cardiovascular function under severe NO/cGMP deficit, the tNOS null mice ultimately develop significant cardiovascular phenotypes. In part, these phenotypes are due to metabolic insufficiency resulting from remodeling of the proteome or the absence of protein S-nitrosylation, which may contribute to impaired contractile function, a hallmark of cardiomyocyte dysfunction in 5-month-old tNOS null mice37,38,39. The progressive development of phenotypes may also be due to maladaptive signaling alterations that counter adaptive responses.
One of the potential maladaptive responses is the increased phosphorylation and activation of Mapk3 and Mapk1 kinases in the tNOS and sGCa1 null mice. The amino acid sequences of Mapk3 (also commonly known as Erk1 or p44) and Mapk1 (Erk 2, or p42) are 83% identical, and since these kinases share similar upstream and downstream regulation, are commonly referred to as Mapk3/Mapk1 or Erk1/266,67,68. The canonical activation of Mapk3/Mapk1 includes the upstream receptor or stimuli-induced activation of RAF proto-oncogene serine/threonine-protein kinase (Raf1), the recruitment of scaffold proteins to form a signalosome that activates the Map2k1/2 (MEK1/2) kinases to phosphorylate Thr208, Tyr205, and Thr183, Tyr185 in Mapk3 and Mapk1, respectively. Map2k1/2 and Mapk3/Mapk1 activities are also regulated by protein-protein complex regulators such as the Lamtor 3 and 2 and Mapk organizer 169,70. The intensity and duration of Mapk signaling is also controlled by the dephosphorylation of downstream targets by the dual-specificity Mapk phosphatases (DUSP 1, 2, 14, 23). Interestingly, the levels of DUSP 23, which is shown to dephosphorylate Mapk3 in vitro71, were significantly decreased by 55% (p-adjusted value = 0.045) in tNOS mice hearts. In addition to these regulatory steps, negative feedback regulation, such as Mapk3/Mapk1 phosphorylation and inactivation of Raf1, determines the specificity and magnitude of the signaling outcome. One of the negative regulatory sites of Raf1 phosphorylation is Ser43, which is effectively phosphorylated by Prkg1, thus providing one molecular mechanism for the NO/cGMP regulation of the Mapk3/Mapk1 cascade72. The activation of the Mapk3/Mapk1 documented in the tNOS and sGCα1 null mice is consistent with previous observations in endothelial, smooth muscle cells, and blood vessels, in which inhibition of NO or cGMP synthesis increased the phosphorylation and activation of Mapk3/Mapk172,73,74,75,76,77,78. In blood vessels responding to growth factors, the activation of Mapk3/1 resulting from the inhibition of NO synthesis was associated with increased protein synthesis73, an observation consistent with the increased levels of proteins participating in protein synthesis in the tNOS mice. Collectively, these in vivo data indicate that the NO/cGMP signaling is a negative regulator of the Mapk3/1 pathway, possibly harnessing overactivation that may have pathological consequences.
Like the NO/cGMP signaling, published results document cardioprotective contributions of the Mapk3/Mapk1 signaling in heart development, the physiological responses to contraction, responses to growth factors, and productive cardiac differentiation and remodeling after injury79,80,81,82,83,84,85,86,87,88. With regards to the development of pathological cardiac remodeling and hypertrophy most but not all studies support a role for heightened Mapk3/Mapk1 signaling82,83,84,85,86,87,88. The discord may in part originate from the autonomous, non-redundant function of these two kinases as deletion of Mapk3 does not generate remarkable phenotypes, whereas Mapk1 deletion is embryonic lethal89,90. It may also relate to the relative abundance of these two kinases in the heart, as Mapk1 levels are nearly 3.5 times higher than Mapk3 (reference proteome, Supplementary Data 1). Finally, site-specific phosphorylation may dictate the outcome, as data indicated that sustained phosphorylation of Thr188 in Mapk1 (Thr205 in Mapk3) induces pathological cardiac hypertrophy in response to various stimuli86,87. Previous data have also established that Mapk3/Mapk1 signaling partially executes the hypertrophic responses in the heart after sustained activation of AT1R signaling. ATR1 signaling includes the assembly and interaction of heterotrimeric G-proteins, which then transduce signals to second messengers, including Rho GTPase family members, members of the PKC, and Akt kinases54,55,56,57. Another important signaling event includes the binding of β-arrestin 2 to the AT1 receptor, which is critical for activating the Mapk3/Mapk1 kinases91. Evidence in both the proteome and phosphoproteome supports activation of AT1R signaling in the tNOS but not in the sGCα1 null heart, consistent with the reduced pathological left ventricular hypertrophy in the tNOS mice following pharmacological attenuation of AT1R signaling37. At 5 months of age, significant left ventricular hypertrophy, increased left ventricular weight, and cardiac myocyte hypertrophy were noted in the tNOS and to a lesser extent in eNOS and null mice but not in the nNOS, or iNOS null mice, compared to wild-type mice37,38. The data suggest that regulation of AT1R and Mapk3/Mapk1 signaling by NO/cGMP opposes the development of pathological remodeling and adds another molecular mechanism for the cardioprotective functions of the NO/cGMP signaling. Although a similar increase in Mapk3/Mapk1 phosphorylation was observed in the sGCα1 null mice, these mice do not exhibit the signaling and phenotypic changes documented in the tNOS null mice. The relationship between genotype and phenotypes is confounded by the presence of NO in the sGCα1 null mice, which alleviates some of the burden placed by the reduction in cGMP levels Several lines of evidence indicate that not all biological actions of NO are mediated by cGMP, and not all actions of cGMP are derived from the NO-mediated activation of sGC including the activation of Mapk3/Mapk120,21,22,23,24,25,26,27. The data collectively revealed that adaptive responses in single NOS gene deletions restrict proteomic and signaling remodeling. However, the single NOS3 gene deletion mice exhibit a significant loss of S-nitrosylated proteins, which may account for the development of cardiometabolic phenotypes. Adaptive responses are also present in mice with deletion of sGCα1 or all three NOS isoforms. Whereas the activation of the Mapk3/Mapk1 signaling in the sGCα1 null mice may prevent the development of phenotypes, the tNOS null mice experience a profound reduction in both the NO and cGMP levels and exhibit increased activation of the AT1R and Mapk3/Mapk1 signaling pathways which may contribute to the development of pathological cardiomyocyte hypertrophy and cardiac dysfunction.
Materials and methods
Animals
All procedures were performed under the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Children’s Hospital of Philadelphia Animal Care and Use Committee. We have complied with all relevant ethical regulations for animal use. Wild-type male C57BL/6J (#000664), Nos1tm1Unc C57BL/6J (nNOS–/–: #002986), Nos2tm1Lau C57BL/6J (iNOS−/−:002609), and Nos3tm1Unc C57BL/6J (eNOS−/−: #002684) mice were obtained from Jackson Laboratories (Bar Harbor, ME). Soluble sGCα1 null and tNOS null mice were generated as described previously37,38,39,40,41,42. All the mice used in this study were male and were backcrossed to the C57BL/6J background for more than 10 generations. Mice were anesthetized using Nembutal 50 mg/kg and transcardially perfused through the left ventricle with ice-cold PBS. Mouse hearts were collected, frozen in liquid nitrogen immediately, and stored at −80 °C until use. On average, the time interval between perfusion and snap-freezing of the heart was 3 minutes.
Sample preparation for mass spectrometry analysis
Frozen heart tissue (150 mg wet weight) was cut into smaller pieces and placed into a 5 mL Eppendorf tube with two 5 mm stainless steel balls (Retsch), then frozen in liquid nitrogen. The tissue was cryopulverized using a MM400 ball mill (Retsch) for four cycles at 30 Hz for 30 s. Subsequently, 1.5 mL of lysis buffer (8 M Urea, 75 mM NaCl, 50 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM PMSF, 2 µg/mL leupeptin, 10 µM aprotinin, 10 mM sodium fluoride, 5 mM sodium butyrate, 5 mM iodoacetamide, 1:100 phosphoprotease inhibitor cocktails 2 and 3 (Sigma P5726, P0044), Benzonase (Novagen 70664-3)) was added to the frozen tissue powder and processed in the ball mill for two cycles at 30 Hz for 30 s. The samples were spun at 2500 rcf at 4 °C for 4 min to reduce foam, transferred to a 2 mL Dounce tissue grinder (Kimble), and processed with 10 strokes of pestle A and 20 strokes of pestle B. The mixture was then centrifuged at 15,000 rcf for 10 min at 4 °C. The protein concentration of the supernatant was assessed by Micro BCA (Thermo Scientific: 23235), and the remaining lysate was stored at −80 °C.
Protein hydrolysis
Lysates were spiked with chicken ovalbumin (1:1000 w/w) as a phosphoprotein internal standard, reduced with 5 mM dithiothreitol for 40 minutes, and alkylated with 20 mM iodoacetamide for 40 min in the dark. Lysates were then diluted with 50 mM Tris-HCl pH 8.0 to achieve a final urea concentration of 2 M. Samples were hydrolyzed overnight at 27 °C with a 1:1 ratio of Trypsin (Promega V5111) and LysC (FUJIFILM Wako, 129-0254) at an enzyme:substrate ratio of 1:50, followed by an additional 3 hours of digestion with the same enzyme ratio at 27 °C. After digestion, formic acid (Thermo Scientific, 28905) was added to a final concentration of 1% to acidify the samples, which were then centrifuged at 20,000 rcf for 10 min. Peptides were desalted using an Oasis HLB 96-well plate (60 mg, Waters, 186000679), which was conditioned with 400 µL 100% acetonitrile and equilibrated with 2 × 400 µL 0.1% trifluoroacetic acid (J T Baker, 9470-00). Peptides were loaded, washed with 3 × 400 µL 0.1% trifluoroacetic acid, and eluted into a new protein Lobind plate 96/2000 µL (Eppendorf, EP0030504305) with 3 × 100 µL 50% acetonitrile/0.1% trifluoroacetic acid. 5% of each sample was used for whole proteome analysis, 75% for phosphopeptide enrichment, and the remaining samples were pooled to generate a phosphoproteome spectral library. All peptides were lyophilized and stored at −80 °C. Peptides were reconstituted in 0.1% TFA with iRT peptides (Biognosys, Schlieren, Switzerland), and 2 µg was injected for whole proteome analysis.
Spectral library for phosphoproteome analysis
The pooled peptide sample was reconstituted in 20 mM ammonium formate (pH 10.0) and separated via high pH RP-HPLC using a Zorbax 300 Å Extend-C18 4.6 mm × 250 mm column (Agilent, 3.5 μm bead size) on an Acquity H-Class UPLC (Waters). Solvent A (2% acetonitrile, 5 mM ammonium formate, pH 10) and solvent B (90% acetonitrile, 5 mM ammonium formate, pH 10) were used at a flow rate of 1 ml/min. The column was monitored for separation efficiency with an in-house standard tryptic digest of BSA and bovine fetuin (Sigma). Peptide fractions were recombined into six fractions for phosphoproteome analysis, with sub-fractions created in a serpentine, concatenated pattern to match hydrophobic complexities. Fractions were lyophilized prior to IMAC enrichment.
Immobilized metal affinity chromatography (IMAC) for phosphopeptide enrichment
Peptides were dissolved in 50% acetonitrile/0.1% water and diluted (1:1) with 100% acetonitrile/0.1% TFA to a final concentration of ~0.5 µg/µL. Phos-Select iron affinity beads (Sigma, P9740-5ML) were washed twice with 30% acetonitrile/250 mM acetic acid and centrifuged at 8000 rcf for 1 min. Peptide mixtures were incubated with 58 µL of 25% IMAC beads for 1 h at 1000 rpm at room temperature, then transferred to a pre-washed 96-well glass fiber filter plate (Phenomenex, AF0-8265). Beads were washed twice with 400 µL of W/E buffer for 20 s while shaking, then buffer was removed using a vacuum manifold. Beads were washed sequentially with 400 µL of 80% acetonitrile/0.1% TFA and 400 µL of 30% acetonitrile/water. Phosphorylated peptides were eluted with 100 µL of 400 mM ammonium hydroxide/30% acetonitrile, mixed for 1 min, and collected into a protein LoBind 96-well plate (Eppendorf, 0030 504.305) using a vacuum manifold. Elution was repeated twice, and peptides were dried by vacuum centrifugation and reconstituted in 13 µL of 0.1% TFA containing iRT peptides (Biognosys, Schlieren, Switzerland).
Mass spectrometry data acquisition
Samples were randomized and analyzed on an Exploris 480 mass spectrometer (ThermoFisher Scientific) coupled with an Ultimate 3000 nano UPLC system and EasySpray source. A 5 µL sample was loaded onto an Acclaim PepMap 100 75 µm × 2 cm trap column at 5 µL/min, then separated by reverse phase HPLC on a 75 µm id × 50 cm PepMap RSLC C18 column. Mobile phase A was 0.1% formic acid, and mobile phase B was 0.1% formic acid/acetonitrile. Peptides were eluted into the mass spectrometer at 210 nL/min using a 150-min gradient from 3% B to 45% B. DIA mass spectrometry settings included one full MS scan at 120,000 resolution with a range of 350–1200 m/z, an AGC target of 300%, and automatic maximum inject time. This was followed by variable DIA isolation windows, MS2 scans at 30,000 resolution with an AGC target of 1000%, and automatic injection time. The default charge state was 3, the first mass was fixed at 250 m/z, and the normalized collision energy for each window was set at 27.
Mass spectrometry QA/QC and system suitability
The suitability of the instrumentation was monitored using QuiC software (Biognosys; Schlieren, Switzerland) to analyze spiked-in iRT peptides. For quality control, we injected standard E. coli protein digest between samples and collected Data in Data-dependent acquisition (DDA) mode. The DDA Data were analyzed in MaxQuant92 and visualized using the PTXQC93 package to track instrumentation quality.
Mass spectrometry raw data processing for proteome, phosphoproteome, S-nitrosocysteine proteome
The DIA raw files were processed using Spectronaut 16.2. For whole proteome analysis, we used the DirectDIA mode, while for phosphoproteome analysis, we employed a merged spectral library from both DDA files from pooled samples and individual DIA files. We utilized a mouse (Mus musculus, 17,120 entries) Database with reviewed canonical Uniprot entries, supplemented with 245 common protein contaminants and iRT peptides. Enzyme specificity was set to trypsin with two allowed missed cleavages. Fixed modifications included carbamidomethylation of cysteine, and variable modifications included N-terminal acetylation, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine for phosphoproteome data. A false discovery rate limit of 1% was applied for precursors, peptides, and protein identification, with all other search parameters set to default. Proteomics data processing and statistical analysis were conducted in R. MS2 intensity values from Spectronaut were analyzed with log2 transformation and normalization by subtracting the median value for each sample. We filtered the data to include only proteins with complete values in at least one cohort.
For the phosphoproteome, the peptide quantification report was collapsed into a modification-specific peptide-like table using default settings in Spectronaut. High-confidence sites were retained with a PTM Localization Probability cutoff of 0.75. Variable modifications included phosphorylation of Ser, Thr, and Tyr, acetylation of protein N-terminal, and oxidation of Met. Phosphopeptide intensities were log2-transformed and normalized to the median for each sample, and Data were filtered to retain only phosphosites with complete values in at least one cohort. Phosphoprotein quantification involved calculating the absolute intensity of detected phosphopeptides for each protein and reporting the maximum value as the overall intensity.
A chemoselective method for direct S-nitrosocysteine conjugation utilized organomercury reagents27,43. Organic mercury selectively reacts with reduced cysteine and S-nitrosocysteine residues to form stable mercury-thiolate conjugates. After blocking reduced cysteine residues, samples are reacted with either organomercury resin or biotin-mercury conjugates which directly enriches S-nitrosylated proteins with no cross-reactivity with disulfides. The column-bound peptides are released after oxidation with performic acid, which oxidizes the cysteine residues to sulfonic acid and facilitates the precise identification of the S-nitrosylated peptide by mass spectrometry.
Data annotation for S-nitrosylated proteins (Supplementary Data 3). The precise localizations (cysteine residue modified) are indicated under cysteine location and multiple peptides from the same protein separated by commas. Most of the peptide sequences included one cysteine residue which makes the localization of the modified cysteine residue unambiguous. However, tryptic digestion occasionally produces peptides with two or more cysteine residues. Thus, these cysteine residues are included in the total count as one peptide without precise localization. The location of these cysteine residues is indicated by the / symbol. The protein lists include protein isoforms given that canonical proteins and their isoforms cannot always be differentiated. Isoforms share the same gene symbol and protein name but have a unique UniProt number. Protein isoforms were included and counted as separate proteins if the sites (peptides) were unique to the isoform. If the peptides were the same between the major protein and the isoform, then they were counted only once.
Bioinformatic analysis, construction of the mouse brain reference proteome and reference biological networks
Protein databases, PAXdb: Protein Abundance Database44, UniProt: the universal protein knowledgebase45, KEGG: Kyoto Encyclopedia of Genes and Genomes51, Reactome GSA-Efficient Multi-Omics Comparative Pathway Analysis52, and Gene Ontology94 were utilized to generate reference proteomes and functional pathways sub-proteomes. Enrichment analyses were performed using Gene Ontology94 or Enrichr: a comprehensive gene set enrichment analysis web server95.
Mouse heart processing for western blot analysis
Frozen hearts of wild-type and tNOS null mice were processed in high-speed 400 rpm homogenizer (Glass clow) using 500 µl homogenization buffer supplemented with protease inhibitor, phosphatase inhibitors 2 and 3 cocktail, and 1 mM PMSF. The homogenate was centrifuged (13,000 rpm, 4 °C) for 20 min. The supernatant was collected, and protein concentration was determined by Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA). Tissue lysates were added to Nupage sample buffer and heated for 5 min at 95 °C, followed by sample loading (40 µg of total protein), protein separation by Nupage-PAGE (10% gel), and transferred at 35 v overnight to polyvinylidene difluoride (PVDF) membranes (Merck, Rahway, NJ). The following day, the PVDF membranes were blocked with Odyssey Licor Blocking buffer (LI-COR Biotechnology, Lincoln, NE) for 1 h, followed by overnight incubation on a rocker at 4 °C with corresponding primary antibodies against p-p44/42 MAPK (T202/Y204) (D13.14.4E) XP (R)(Cell Signaling, 4370S, 1:2000), p44/42 MAPK (ERK 1/2) (137FS) (Cell Signaling, 4695S, 1:1000). Following day PVDF membranes were developed for protein band visualization using IRDye conjugated secondary Antibodies (Licor, 1:5000) and scanned using Odyssey infrared scanner. Densitometric analysis of the protein bands was performed with Image Studio (Licor, Lincoln, NE).
Cell model
HL-1 cardiomyocytes were maintained in Claycomb medium, supplemented with 2 mM glutamine, 100 U/mL penicillin/streptomycin, 100 μM norepinephrine in 30 mM L-ascorbic acid, and 10% fetal bovine serum, as described previously53. The culture dishes were precoated with 0.02% (wt/vol) gelatin containing 5 μg/mL fibronectin. Cells were grown in an atmosphere of 5% CO2 at 37 °C. Experiments were performed using cells between passages 6 and 9. Culture HL-1 cells were plated at 35,000 cells/cm2, and the following day were used for treatment at 80% confluency. In a 6-well plate, cells were incubated with 10 µM L-Arginine, and/or 100 µM L-NAME, and 30 µM ODQ for 24 h. Cells were harvested in 80 µl homogenization media per well supplemented with protease inhibitor, phosphatase inhibitors 2 and 3 cocktail, and 1 mM PMSF and were either processed for western blot analysis as described above.
Statistics and reproducibility
The number of hearts analyzed by mass-spectrometry-based proteomics is indicated in the appropriate figure legends. Differential relative abundance analysis of the proteome and phosphopeptides was conducted using a Limma t-test with p adjusted value < 0.05 as a significant cutoff. Limma uses moderated t-test, which was developed for transcriptomic data analysis and adapted to proteomic data statistical analysis. The differences in the ratio of phosphorylated to total Mapk3 and Mapk1 in Fig. 7c were evaluated using the Student’s t-test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE96 partner repository with the dataset identifier PXD060084.
Supplemental Data 1 contains the numerical source data for Fig. 1b, Fig. 2, Supplementary Fig. 1, and Supplementary Fig. 2. Supplemental Data 2 contains the numerical source data for Figs. 1c, 3, Supplementary Fig. 3, and Supplementary Fig. 4. Supplemental Data 3 contains the numerical source data for Figs. 1d, 4. Supplemental Data 4 contains the numerical source data for Fig. 5 and Supplementary Figs. 5-7. Supplemental Data 5 contains the numerical source data for Figs. 6, 7c, d. Supplementary Fig. 9 provides the full western blots for Fig. 7c, d.
References
Furchgott, R. F. The 1996 Albert Lasker Medical Research Awards. The discovery of endothelium-derived relaxing factor and its importance in the identification of nitric oxide. JAMA 276, 1186–1188 (1996).
Murad, F. Shattuck Lecture. Nitric oxide and cyclic GMP in cell signaling and drug development. N. Engl. J. Med. 355, 2003–2011 (2006).
Ignarro, L. J. Nitric oxide: a unique endogenous signaling molecule in vascular biology. Biosci. Rep. 19, 51–71 (1999).
Moncada, S. & Higgs, E. A. The discovery of nitric oxide and its role in vascular biology. Br. J. Pharmacol. 147, S193–S201 (2006).
Lundberg, J. O., Gladwin, M. T. & Weitzberg, E. Strategies to increase nitric oxide signaling in cardiovascular disease. Nat. Rev. Drug Discov. 4, 623–641 (2015).
Huang, P. L. eNOS, metabolic syndrome, and cardiovascular disease. Trends Endocrinol. Metab. 20, 295–302 (2009).
Johnstone, M. T. et al. Impaired endothelium-dependent vasodilation in patients with insulin-dependent diabetes mellitus. Circulation 88, 2510–2516 (1993).
Petrie, J. R., Ueda, S., Webb, D. J., Elliott, H. L. & Connell, J. M. Endothelial nitric oxide production and insulin sensitivity. A physiological link with implications for pathogenesis of cardiovascular disease. Circulation 93, 1331–1333 (1996).
Oemar, B. S. et al. Reduced endothelial nitric oxide synthase expression and production in human atherosclerosis. Circulation 97, 2494–2498 (1998).
Monti, L. D. et al. Endothelial nitric oxide synthase polymorphisms are associated with type 2 diabetes and the insulin resistance syndrome. Diabetes 52, 1270–1275 (2003).
Siervo, M., Jackson, S. J. & Bluck, L. J. In-vivo nitric oxide synthesis is reduced in obese patients with metabolic syndrome: application of a novel stable isotopic method. J. Hypertens. 29, 1515–1527 (2011).
Erdmann, J. et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature 504, 432–436 (2013).
Miranda, J. A., Belo, V. A., Souza-Costa, D. C., Lanna, C. M. & Tanus-Santos, J. E. eNOS polymorphism associated with metabolic syndrome in children and adolescents. Mol. Cell Biochem. 372, 155–160 (2013).
Emdin, C. A. et al. Phenotypic consequences of a genetic predisposition to enhanced nitric oxide signaling. Circulation 137, 222–232 (2018).
Tsai, E. J. & Kass, D. A. Cyclic GMP signaling in cardiovascular pathophysiology and therapeutics. Pharmacol. Ther. 122, 216–238 (2009).
Francis, S. H., Busch, J. A. & Corbin, J. D. Cyclic GMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol. Rev. 62, 525–563 (2010).
Francis, S. H. The role of cGMP-dependent protein kinase in controlling cardiomyocyte cGMP. Circ. Res. 107, 1164–1166 (2010).
Rainer, P. P. & Kass, D. A. Old dog, new tricks: novel cardiac targets and stress regulation by protein kinase G. Cardiovasc. Res. 111, 154–162 (2016).
Hofmann, F. A concise discussion of the regulatory role of cGMP kinase I in cardiac physiology and pathology. Basic Res. Cardiol. 113, 31 (2018).
Seth, D. & Stamler, J. S. The SNO-proteome: causation and classifications. Curr. Opin. Chem. Biol. 15, 129–136 (2011).
Smith, B. C. & Marletta, M. A. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr. Opin. Chem. Biol. 16, 498–506 (2012).
Gould, N., Doulias, P. T., Tenopoulou, M., Raju, K. & Ischiropoulos, H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J. Biol. Chem. 288, 26473–26479 (2013).
Sun, J. et al. Essential role of nitric oxide in acute ischemic preconditioning: S-nitros(yl)ation versus sGC/cGMP/PKG signaling? Free Radic. Biol. Med. 54, 105–112 (2013).
Irie, T. et al. S-Nitrosylation of Calcium-Handling Proteins in Cardiac Adrenergic Signaling and Hypertrophy. Circ. Res. 117, 793–803 (2015).
Wang, S. B. et al. Protein S-nitrosylation controls glycogen synthase kinase 3β function independent of its phosphorylation state. Circ. Res. 122, 1517–1531 (2018).
Chung, H. S. et al. Transient receptor potential channel 6 regulates abnormal cardiac S-nitrosylation in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 114, E10763–E10771 (2017).
Doulias, P.-T., Tenopoulou, M., Greene, J. L., Raju, K. & Ischiropoulos, H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci. Signal. 6, rs1-7 (2013).
Bruegger, J. J., Smith, B. C., Wynia-Smith, S. L. & Marletta, M. A. Comparative and integrative metabolomics reveal that Snitrosation inhibits physiologically relevant metabolic enzymes. J. Biol. Chem. 293, 6282–6296 (2018).
Zhou, H. L. et al. Metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 565, 96–100 (2019).
Lau, B. et al. Endogenous Snitrosocysteine proteomic inventories identify a core of proteins in heart metabolic pathways. Redox. Biol. 47, 102153 (2021).
Huang, P. L. et al. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature 377, 239–242 (1995).
Shesely, E. G. et al. Elevated blood pressures in mice lacking endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. USA 93, 13176–13181 (1996).
Shankar, R. R., Wu, Y., Shen, H. Q., Zhu, J. S. & Baron, A. D. Mice with gene disruption of both endothelial and neuronal nitric oxide synthase exhibit insulin resistance. Diabetes 49, 684–687 (2000).
Barouch, L. A. et al. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature 416, 337–339 (2002).
Laubach, V. E., Shesely, E. G., Smithies, O. & Sherman, P. A. Mice lacking inducible nitric oxide synthase are not resistant to lipopolysaccharide-induced death. Proc. Natl. Acad. Sci. USA 92, 10688–10692 (1995).
Guo, Y. et al. The late phase of ischemic preconditioning is abrogated by targeted disruption of the inducible NO synthase gene. Proc. Natl. Acad. Sci. USA 96, 11507–11512 (1999).
Tsutsui, M., Shimokawa, H., Morishita, T., Nakashima, Y. & Yanagihara, N. Development of genetically engineered mice lacking all three nitric oxide synthases. J. Pharmacol. Sci. 102, 147–154 (2006).
Shibata, K. et al. Spontaneous development of left ventricular hypertrophy and diastolic dysfunction in mice lacking all nitric oxide synthases. Circ. J. 74, 2681–2692 (2010).
Tsutsui, M. et al. Significance of nitric oxide synthases: Lessons from triple nitric oxide synthases null mice. J. Pharm. Sci. 127, 42–52 (2015).
Buys, E. S. et al. Gender-specific hypertension and responsiveness to nitric oxide in sGC{alpha}1 knockout mice. Cardiovasc. Res. 79, 179–186 (2008).
Cawley, S. M. et al. sGC{alpha}1 mediates the negative inotropic effects of NO in cardiac myocytes independent of changes in calcium handling. Am. J. Physiol. Heart Circ. Physiol. 301, H157–H163 (2011).
Buys, E. S. et al. Genetic modifiers of hypertension in soluble guanylate cyclase α1-deficient mice. J. Clin. Invest. 122, 2316–2325 (2012).
Gould, N. S. et al. Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chem. Biol. 22, 965–975 (2015).
Wang, M., Herrmann, C. J., Simonovic, M., Szklarczyk, D. & von Mering, C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168 (2015).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 49, D480–D489 (2021).
Jumrussirikul, P. et al. Interaction between neuronal nitric oxide synthase and inhibitory G protein activity in heart rate regulation in conscious mice. J. Clin. Invest. 102, 1279–1285 (1998).
Sears, C. E. et al. Cardiac neuronal nitric oxide synthase isoform regulates myocardial contraction and calcium handling. Circ. Res. 92, e52–e59 (2003).
Duplain, H. et al. Insulin resistance, hyperlipidemia, and hypertension in mice lacking endothelial nitric oxide synthase. Circulation 104, 342–345 (2001).
Le Gouill, E. et al. Endothelial nitric oxide synthase (eNOS) knockout mice have defective mitochondrial beta-oxidation. Diabetes 56, 2690–2696 (2007).
Tenopoulou, M. et al. Oral nitrite restores age-dependent phenotypes in eNOS null mice. JCI Insight 3, pii: 122156 (2018).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Griss, J. et al. Reactome GSA-Efficient Multi-Omics Comparative Pathway Analysis. Mol. Cell Proteomics. 19, 2115–2125 (2020).
Claycomb, W. C. et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 95, 2979–2984 (1998).
Duff, J. L., Berk, B. C. & Corson, M. A. Angiotensin II stimulates the pp44 and pp42 mitogen-activated protein kinases in cultured rat aortic smooth muscle cells. Biochem Biophys. Res. Commun. 188, 257–264 (1992).
Suero, J. A. & Berk, B. C. Angiotensin II prevents apoptosis by differential effects on mitogen activated protein kinases (MAPK): JNK, p38, and ERK1/2. Circulation 94, I-281 (1996).
Zou, Y. et al. Protein kinase C, but not tyrosine kinases or Ras, plays a critical role in angiotensin II-induced activation of Raf-1 kinase and extracellular signal-regulated protein kinases in cardiac myocytes. J. Biol. Chem. 271, 33592–33597 (1996).
Mehta, P. K. & Griendling, K. K. Angiotensin II cell signaling: Physiological and pathological effects in the cardiovascular system. Am. J. Physiol. Physiol. 292, C82–C97 (2007).
Gekle, M., Dubourg, V., Schwerdt, G., Benndorf, R. A. & Schreier, B. The role of EGFR in vascular AT1R signaling: From cellular mechanisms to systemic relevance. Biochem. Pharmacol. 217, 115837 (2023).
Buys, E. S. et al. sGCα1β1 attenuates cardiac dysfunction and mortality in murine inflammatory shock models. Am. J. Physiol. Heart Circ. Physiol. 297, H654–H663 (2009).
Nishida, T. et al. Protective effects of nitric oxide synthase 3 and soluble guanylate cyclase on the outcome of cardiac arrest and cardiopulmonary resuscitation in mice. Crit. Care. Med. 37, 256–262 (2009).
Nakata, S. et al. Spontaneous myocardial infarction in mice lacking all nitric oxide synthase isoforms. Circulation 117, 2211–2223 (2008).
Yatera, Y. et al. Severe dyslipidaemia, atherosclerosis, and sudden cardiac death in mice lacking all NO synthases fed a high-fat diet. Cardiovasc. Res. 87, 675–682 (2010).
Mergia, E., Friebe, A., Dangel, O., Russwurm, M. & Koesling, D. Spare guanylyl cyclase NO receptors ensure high NO sensitivity in the vascular system. J. Clin. Invest. 116, 1731–1737 (2006).
Castro, L. R., Schittl, J. & Fischmeister, R. Feedback control through cGMP-dependent protein kinase contributes to differential regulation and compartmentation of cGMP in rat cardiac myocytes. Circ. Res. 107, 1232–1240 (2010).
Götz, K. R. et al. Transgenic mice for real-time visualization of cGMP in intact adult cardiomyocytes. Circ. Res. 114, 1235–1245 (2014).
Lewis, T. S., Shapiro, P. S. & Ahn, N. G. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74, 49–139 (1998).
Johnson, G. L. & Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, andp38 protein kinases. Science 298, 1911–1912 (2002).
Rose, B. A., Force, T. & Wang, Y. Mitogen-activated protein kinase signaling in the heart: angels versus demons in a heart-breaking tale. Physiol. Rev. 90, 1507–1546 (2010).
Schaeffer, H. J. et al. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281, 1668–1671 (1998).
Vomastek, T. et al. Modular construction of a signaling scaffold: MORG1 interacts with components of the ERK cascade and links ERK signaling to specific agonists. Proc. Natl. Acad. Sci. USA 101, 6981–6986 (2004).
Takagaki, K. et al. Characterization of a novel lowmolecular-mass dual-specificity phosphatase-3 (LDP-3) that enhances activation of JNK and p38. Biochem. J. 383, 447–55 (2004).
Suhasini, M., Li, H., Lohmann, S. M., Boss, G. R. & Pilz, R. B. Cyclic-GMP-dependent protein kinase inhibits the Ras/Mitogenactivated protein kinase pathway. Mol. Cell Biol. 18, 6983–6994 (1998).
Chiu, J. J., Wung, B. S., Hsieh, H. J., Lo, L. W. & Wang, D. L. Nitric oxide regulates shear stress-induced early growth response-1. Expression via the extracellular signal-regulated kinase pathway in endothelial cells. Circ. Res. 85, 238–246 (1999).
Martens, F. M. et al. Vessel-specific stimulation of protein synthesis by nitric oxide synthase inhibition: role of extracellular signal-regulated kinases 1/2. Hypertension 39, 16–21 (2002).
Girardot, D., Demeilliers, B., deBlois, D. & Moreau, P. ERK1/2-mediated vasoconstriction normalizes wall stress in small mesenteric arteries during NOS inhibition in vivo. J. Cardiovasc. Pharmacol. 42, 339–347 (2003).
Raines, K. W. et al. Nitric oxide inhibition of ERK1/2 activity in cells expressing neuronal nitric-oxide synthase. J. Biol. Chem. 279, 3933–3940 (2004).
Bouallegue, A., Daou, G. B. & Srivastava, A. K. Nitric oxide attenuates endothelin-1-induced activation of ERK1/2, PKB, and Pyk2 in vascular smooth muscle cells by a cGMP-dependent pathway. Am. J. Physiol. Heart Circ. Physiol. 293, H2072–H2079 (2007).
Bhattacharya, I., Damjanović, M., Dominguez, A. P. & Haas, E. Inhibition of activated ERK1/2 and JNKs improves vascular function in mouse aortae in the absence of nitric oxide. Eur. J. Pharmacol. 658, 22–27 (2011).
Blanton, R. M. cGMP Signaling and Modulation in Heart Failure. J. Cardiovasc. Pharmacol. 75, 385–398 (2020).
Lee, D. I. et al. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature 519, 472–476 (2015).
Patrucco, E. et al. Roles of cGMP-dependent protein kinase I (cGKI) and PDE5 in the regulation of Ang II-induced cardiac hypertrophy and fibrosis. Proc. Natl. Acad. Sci. USA 111, 12925–12929 (2014).
Heineke, J. & Molkentin, J. D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell. Biol. 7, 589–600 (2006).
Bueno, O. F. et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 19, 6341–6350 (2000).
Purcell, N. H. et al. Genetic inhibition of cardiac ERK1/2 promotes stress-induced apoptosis and heart failure but has no effect on hypertrophy in vivo. Proc. Natl. Acad. Sci. USA 104, 14074–14079 (2007).
Ulm, S. et al. Targeted deletion of ERK2 in cardiomyocytes attenuates hypertrophic response but provokes pathological stress induced cardiac dysfunction. J. Mol. Cell Cardiol. 72, 104–116 (2014).
Lorenz, K., Schmitt, J. P., Schmitteckert, E. M. & Lohse, M. J. A new type of ERK1/2 autophosphorylation causes cardiac hypertrophy. Nat. Med. 15, 75–83 (2009).
Ruppert, C. et al. Interference with ERK(Thr188) phosphorylation impairs pathological but not physiological cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 110, 7440–7445 (2013).
Tomasovic, A. et al. Interference with ERKdimerization at the nucleocytosolic interface targets pathological ERK1/2 signaling without cardiotoxic side-effects. Nat. Commun. 11, 1733 (2020).
Pages, G. et al. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science 286, 1374–1377 (1999).
Hatano, N. et al. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells. 8, 847–856 (2003).
Shenoy, S. K. et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J. Biol. Chem. 281, 1261–1273 (2006).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Bielow, C., Mastrobuoni, G. & Kempa, S. Proteomics Quality Control: Quality Control Software for MaxQuant Results. J. Proteome. Res. 15, 777–787 (2016).
Ashburner, M. et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Kuleshov M. V., et al Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. gkw377 (2016).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2025).
Acknowledgements
This work was supported by National Institutes of Health Grants R01-HL054926 (HI); and by the project BIOMED-20 (MIS 5047236) -co-financed by Greece and the EU European Regional Development Fund (P-TD). H.I. is the Gisela and Dennis Alter Research Professor of Pediatrics. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
I.M., H.F., S.R., H.D., L.S., M.S., M. Tenopoulou, and P.-T.D., performed experiments and analyzed the data. F.I., H.Y., H.S., M. Tsutsui, contributed essential reagents and resources and edited the manuscript. H.I. conceived and designed the experiments and wrote the manuscript with important contributions from I.M., S.R., M. Tenopoulou, and P.-T.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Iraida Sharina and Patrick Sips for their contribution to the peer review of this work. Primary Handling Editors: Kaoru Ito and Dario Ummarino. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mohanty, I., Fazelinia, H., Raimo, S. et al. Proteomic surveys of the mouse heart unveil cardiovascular responses to nitric oxide/cGMP signaling deficiencies. Commun Biol 8, 817 (2025). https://doi.org/10.1038/s42003-025-08203-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08203-8
This article is cited by
-
Pannexins in the heart: cell-specific expression and contributions to disease
Cell and Tissue Research (2026)